Key Points
SP induces the HMGB1 gene in BM stroma.
NK-A, via HMGB1, negatively regulates SP effects on hematopoiesis.
Humanized mice confirmed HMGB1 exerting negative hematopoietic effects.
Abstract
Hematopoiesis is tightly regulated by the bone marrow (BM) niche. The niche is robust, allowing for the return of hematopoietic homeostasis after insults such as infection. Hematopoiesis is partly regulated by soluble factors, such as neuropeptides, substance P (SP), and neurokinin A (NK-A), which mediate hematopoietic stimulation and inhibition, respectively. SP and NK-A are derived from the Tac1 gene that is alternately spliced into four variants. The hematopoietic effects of SP and NK-A are mostly mediated via BM stroma. Array analyses with 2400 genes indicated distinct changes in SP-stimulated BM stroma. Computational analyses indicated networks of genes with hematopoietic regulation. Included among these networks is the high-mobility group box 1 gene (HMGB1), a nonhistone chromatin-associated protein. Validation studies indicated that NK-A could reverse SP-mediated HMGB1 decrease. Long-term culture-initiating cell assay, with or without NK-A receptor antagonist (NK2), showed a suppressive effect of HMGB1 on hematopoietic progenitors and increase in long-term culture-initiating cell assay cells (primitive hematopoietic cells). These effects occurred partly through NK-A. NSG mice with human hematopoietic system injected with the HMGB1 antagonist glycyrrhizin verified the in vitro effects of HMGB1. Although the effects on myeloid lineage were suppressed, the results suggested a more complex effect on the lymphoid lineage. Clonogenic assay for CFU– granulocyte-monocyte suggested that HMGB1 may be required to prevent hematopoietic stem cell exhaustion to ensure immune homeostasis. In summary, this study showed how HMGB1 is linked to SP and NK-A to protect the most primitive hematopoietic cell and also to maintain immune/hematopoietic homeostasis.
Introduction
Bone marrow (BM) is the major site of hematopoiesis, with hematopoietic cells organized in a hierarchical clustering of cells beginning with hematopoietic stem cells (HSCs) (1, 2). Despite new technologies revising the hematopoietic hierarchy, the basic principle of HSC differentiation to immune and blood cells remain undisputed (2). Hematopoietic activity occurs in the endosteal and perivascular region of the central sinus (3, 4). Hematopoiesis is supported by the BM niche that includes cells, collectively referred as stroma, such as fibroblasts, macrophages, adipocytes, endothelial cells, and mesenchymal stem cells (4, 5). Stroma support hematopoiesis via secretome such as cytokines, extracellular matrices, and microvesicles (6). Innervated fibers also regulate hematopoiesis by producing neuropeptides belonging to the tachykinin family as well as others from the adrenergic system (7–9).
The tachykinins are small peptides that are derived from peptidergic fibers and other nonneural cells such as BM stroma (10–14). The tachykinins can modulate immune and hematopoietic responses, mostly through substance P (SP) and neurokinin (NK)-A, with each acting antagonistically to the other (7, 15–18). The Tac1 gene is alternately spliced into α-, β-, λ-, and δ-mRNAs (19). SP is encoded by Exon 3 of each transcript and NK-A from Exon 6 of β- and λ-transcripts (19). SP and NK-A show binding preference for the seven transmembrane G-protein–coupled NK1 and NK2 receptors, respectively (20).
NK1 and NK2 mediate hematopoietic regulation directly or indirectly via the production of cytokines in stroma (18, 21–24). In stroma, there is a yin–yang relationship between NK1 and NK2 (24). NK1 is induced by cytokines, including those linked to hematopoietic stimulation (17). The increase in NK1 correlates with decreased NK2, and the latter required hematopoietic suppressors, such as TGF-β and MIP1, for its expression (18). Additionally, NK1 and NK2 mediate intracellular cross-talk such that one receptor regulates the expression of the other, consequently impacting hematopoietic regulation (24).
The high-mobility group box 1 protein (HMGB1) is a member of nonhistone, chromatin-associated HMGB1 family that is evolutionally conserved (25). Nuclear HMGB1 binds to the minor groove of DNA to facilitate the assembly of other transcriptional complexes such as p53 and NF-κB (25). Membrane HMGB1 can control cell movement (26, 27). In malignant cells, HMGB1 can exert both suppressor and oncogenic functions (26, 28, 29). HMGB1 is also linked to inflammation with its release in necrotic cells as alarmins (30–35). HMGB1 can mediate intracellular signaling via TLR4 and RAGE receptor (36, 37). HMGB1 can be released from nonnecrotic monocytes by exocytosis of microvesicles (38).
Array studies indicated decreased HMGB1 in BM stroma, leading us to test the link between conserved HMGB1 and Tac1 peptides. We proposed that HMGB1 negatively regulates hematopoietic stimulation because SP, which is a hematopoietic stimulator, decreased HMGB1. Because NK-A can negatively regulate SP-mediated hematopoietic stimulation, we determined the relationship between NK-A and HMGB1 (18).
BM stromal support of hematopoiesis was selected to investigate the SP–HMGB1–NK-A axis because of the key role in regulating SP/NK-A hematopoietic regulation (17). Furthermore, monocytes, which can differentiate into macrophages to comprise stroma, can express HMGB1 (38). Indeed, the results showed NK-A blunting SP effects to decrease HMGB1. Long- and short-term hematopoietic assays with stroma transfected with an inducible HMGB1 lentivirus, as well as studies with immune-deficient mice with a human hematopoietic system, confirmed a mediating role for NK-A in the ability of HMGB1 to suppress hematopoietic progenitors and protect the more primitive hematopoietic cells.
Materials and Methods
Reagents
DMEM, α-MEM, penicillin–streptomycin, hydrocortisone, Ficoll-Hypaque, glutamine, FBS, accutase, 0.05% trypsin–EDTA, and mammalian protein extraction reagent (M-PER) were purchased from Thermo Fisher Scientific (Waltham, MA); horse sera, geneticin, and G418 were purchased from HyClone Laboratories (Logan, UT); PBS, glycyrrhizin (Gly), vasoactive intestinal peptide (VIP), SP, and NK-A were purchased from Sigma-Aldrich (St Louis, MO); hydrocortisone and polybrene were purchased from MilliporeSigma; SYBR Green PCR Master Mix was purchased from Applied Biosystems and Thermo Fisher Scientific; tetracycline-free FCS, 7-aminoactinomycin D was purchased from BioLegend (San Diego, CA); and doxycycline (Dox) was purchased from Takara Bio (Fitchburg, WI). NK2-specific antagonist (SR 48968) was obtained from Sanofi Recherche & Developpement (Montpellier Cedex, France) and NK1 receptor enantiomer (CP-100,263-1) was provided by Pfizer (Groton, CT). The method to dilute SP, NK-A, VIP, CP-100,263-1 and SR48968 was previously described (17, 24, 39).
Abs
Human anti-rabbit HMGB1 Ab and anti-rabbit β-actin, from Cell Signaling Technology, were used as primary Abs for Western blots. HMGB1 Ab was used at a final dilution of 1:1000, and β-actin was used at 1:3000 final dilution. Anti-rabbit IgG, HRP-linked Ab from Cell Signaling Technology, was used as secondary Ab. Mouse anti-human-CD45-allophycocyanin, -CD3-allophycocyanin, -CD56-PE, -HLA-DR mAb, -CD3-PerCP-Cy5.5, -CD38-PE, -CD4-PE, CD33–allophycocyanin, and -CD19-PE were purchased from BD Biosciences (San Jose, CA).
Flow cytometry
Cells were directly labeled with fluorochrome-tagged anti-CD45, -CD34, -CD38, pan-T cell (-CD3), -CD4, -CD19, -CD33 and NK cell (-CD56). Cells were washed in cold PBS, resuspended in 500 μl of 1% formaldehyde, and then immediately analyzed on the FACSCalibur (BD Biosciences).
Cell line
HEK 293T cells were purchased from the American Type Culture Collection and propagated as per their instruction.
Western blot
Whole cell extracts were isolated with M-PER (Thermo Fisher Scientific). The protein samples were quantified using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Extracts (10 μg) were electrophoresed along with a prestained protein ladder and Thermofluor protein ladder in Mini-PROTEAN precast gels (Bio-Rad Laboratories, Hercules, CA) for 1.5 h at 100 V using a PowerPac HC power supply (Bio-Rad Laboratories). After this, the proteins were electrically transferred to membrane (Immun-Blot polyvinylidene fluoride membrane) (Bio-Rad Laboratories).
To reduce the noise by nonspecific binding of the Abs, the membranes were blocked by incubating in 5% milk in PBS for 20 min at room temperature. Membranes were incubated overnight with primary Ab diluted to 1:1000 for HMGB1 and 1:3000 for β-actin at 4°C. On the next day, membrane was washed three to four times with PBS plus 0.1% Tween for 5 min. The HRP-conjugated secondary Ab (anti-rabbit IgG) was used at 1:3000 final dilution for 2 h at room temperature. To remove the excess Ab PBS plus 0.1% Tween, washing was done twice. Proteins were detected by chemiluminescence using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). Images were taken and stored using the molecular imager ChemiDoc XRS system (Bio-Rad Laboratories).
Real-time PCR
Total RNA was isolated with RNeasy Mini Kit (QIAGEN) and then reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Applied Biosystems). Real-time PCR was performed with 200 ng of cDNA and SYBR Green PCR Master Mix on a 7300 Real-Time PCR System (Applied Biosystems). The cycle conditions were 50°C for 2 min as initial activation and initial denaturation at 95°C for 10 min followed by 35 cycles of denaturation at 95°C for 15 s, primer annealing/extension at 60°C for 60 s. The following primers were obtained from Sigma-Aldrich: HMGB1, 5′-GCC AGC TTT TCA AAC AAA-3′ (forward) and 5′-TGC CAA ATT GTT CCC TAA-3′ (reverse); β-Actin, 5′-GCC GAG GAC TTT GAT TGC AC-3′ (forward) and 5′-TGC TAT CAC CTC CCC TGT GT-3′ (reverse); ATF-6, 5′-AGT TGC CAT GCC AGA TTA-3′ (forward) and 5′-TTA TTG TCA GCC CCA AGA-3′ (reverse); APP, 5′-AGC GAC AGT GAT CGT CAT-3′ (forward) and 5′-TGG TTT TGC TGT CCA ACT-3′ (reverse). The relative expression was calculated using 2ΔΔCT . This was calculated as follows: β-actin was selected as the gene of reference (gor) and HMGB1 as the gene of interest (goi). The relative expression was determined as 2(gor–goi).
Vectors
The HMGB1 expressing pCMV6-AC-GFP was purchased from Origene (Rockville, MD), and pLVX-TetOne-Puro, from was purchased from Takara Bio (Mountainview, CA). The coding sequence of HMGB1 was amplified by RT-PCR using the pCMV6 template: 5′-CCC TCG TAA AGA ATT CAT GGG CAA AGG AGA TCC TAA GAA G-3′ (forward) and 5′-GCA GAG ATC TGG ATC CCT ATT CAT CAT CAT CAT CTT CTT C-3′ (reverse). The primers contained 15 bp for BamH1 and EcoR1. The amplified region was inserted into pLVX with the In-Fusion HD Cloning Kit (Takara Bio). The insert was confirmed by DNA sequencing at GENEWIZ (South Plainfield, NJ). The final vector was designated pLVX–HMGB1/GS.
Viral propagation and quantitation
pLVX–HMGB1/GS was produced with the Lenti-X Packing Single Shots (VSV-G) (Takara Bio). Lentiviral DNA (7 μg) was incubated with the VSV-G packaging plasmids in 600 μl MilliQ water for 10 min at room temperature. The DNA complex was added to 80% confluent HEK 293T cells in 100-mm3 dishes containing 8 ml DMEM with 10% tetracycline-free FCS (propagation media). The plates were incubated at 37°C. After 8 h, 6 ml of fresh propagation media was added to the dish. After 48 h, the supernatant was collected and filtered through 0.45-μm followed by concentration using the Lenti-X Concentrator (Takara Bio). pLVX-TetOne-HMGB1/GS was quantified with the Lenti-X p24 Rapid Titer Kit, as described in manufacturer’s protocol (Takara Bio).
Transduction and induction of HMGB1
Stromal cells at 70% confluence in six-well tissue culture plates were transduced with pLVX-TetOne-HMGB1/GS at a multiplicity of infection of 10:1 in the presence of 4 μg/ml polybrene. The transduction was performed in 2 ml of stromal media (see below). The plates were immediately centrifuged at 1200 × g for 60 min at 32°C. After 24 h, HMGB1 was induced with fresh media containing 10 ng/ml Dox. The dose was selected with the results of dose-response curves for 24 h and Dox concentrations of 1, 10, 50, 100, and 1000 ng/ml.
Array and data analyses
The microarray data and analyses were partly reported (40). The data from the hybridized slides were acquired with a software by NEN Life Sciences (Boston, MA). The data are available in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131410). The differentially expressed genes in BM stroma between SP-stimulated and vehicle groups were analyzed using several methods. The accession number for the 2400 genes along with their C5 and C3 values were uploaded as the input data set into the respective software. Normalization was based on the average of the replicates and log2 transform of the data were performed as described (41). Hierarchical clustering using the average linkage method with a Pearson-correlated distance metric was performed using TM4 MultiExperiment Viewer to produce a dendrogram of closely related genes (42). Genes with log >1 and < −1 were considered as upregulated or downregulated, respectively. After this, we used a ±2-fold cutoff of gene expression.
Pathway analyses with Ingenuity Pathway Analysis (IPA) software (QIAGEN, Redwood City, CA) used genes with the accession number mapped to the corresponding gene in the Ingenuity Pathways Knowledge Base. Networks were assigned a score according to the relevance with the genes used as input. The output networks or pathways show each gene or gene product as a node for connecting to other functional nodes, which are represented by different shapes. The genes were filtered based on hematopoietic function with the networks set to a display 35 molecules. The input of genes in IPA generated canonical pathways based on the following: 1) the ratio of the number of genes from the input data to the total number of genes in the pathway; 2) calculated p value as per Fischer exact test.
Human subjects
The use of human BM aspirate was approved by Rutgers Human Subjects Protection Program and Institutional Review Boards. The aspirates were obtained from healthy donors ages 20–35 y. All donors signed the informed consent forms.
Stromal culture
Stroma was prepared from BM aspirates of healthy donors 18–25 y of age, as described previously (18). Unfractionated cells from BM aspirates were cultured at 37°C in α-MEM with 12.5% FCS, 12.5% horse serum, 0.1 μM hydrocortisone, 0.1 mM 2-ME, and 1.6 mM glutamine. At day 3, RBCs and granulocytes were eliminated by Ficoll-Hypaque density gradient, and the mononuclear fraction was replaced into the tissue culture flask. Cultures were reincubated with weekly replacement of 50% medium until confluence.
Culture of mesenchymal stem cell
Mesenchymal stem cells (MSCs) were cultured from BM aspirates, as previously described (43). Aliquots of BM aspirates added to DMEM with 10% FCS in vacuum gas plasma-treated plates (BD Falcon; Franklin Lakes, NJ). After 3 d, the RBCs and granulocytes were removed by Ficoll-Hypaque density gradient centrifugation, and the mononuclear fractions were readded to the respective dishes. At 80% confluence, the adherent cells were serially passaged. At the fifth passage, the cells were used in the assay. Prior to this, MSCs were characterized by phenotype and multilineage differentiation (adipogenic and osteogenic lineages), as described previously (44). MSCs were negative for CD34 and CD45 and positive for CD29, CD73, CD90, CD105, and CD44.
Autophagy detection
BM stroma, transduced with the HMGB1–pLVX/GS and then induced with Dox was assessed for autophagy, using the CYTO-ID Autophagy Detection Kit (Enzo, Farmingdale, NY). Briefly, after 24 h of induction with Dox or vehicle, the cells were detached with acutase and washed twice with PBS. Positive control cells were treated with 500 nM rapamycin and 20 μM chloroquine for 16 h. The use of the reagent in the kit followed manufacturer’s recommendation and then was immediately analyzed by flow cytometry.
ELISA
Samples were studied for SP and NK-A by competitive ELISA. NK-A quantitation was previously described (45). SP quantitation used a kit from R&D Systems (Minneapolis, MN). Briefly, samples and standards were added to wells and SP with immobilized Ab. The captured peptides were detected with a specific enzyme-linked mAb. The substrate was analyzed using absorption at 450 nm. The quantity of the peptide was calculated using a standard curve established with known standards.
Modified long-term culture-initiating cells
Confluent stromal cells were untransfected or ectopically expressed for HMGB1 in six-well plates. The stromal cells were subjected to 150 Gy delivered by a cesium source. After 16 h, the floating cells were washed, and 1 × 107 BM mononuclear cells were added to the gamma-irradiated stroma. At weeks 6, 10, and 16, aliquots of mononuclear cells were assayed for CFU–granulocyte-monocyte (CFU-GM). Clonogenic assays for CFU-GM were performed as described in methylcellulose matrices (13). The cultures were supplemented with 3 U/ml GM-CSF (R&D Systems). Colonies were counted by a blinded observer. Each colony contained >15 cells.
In vivo studies
The reporting of in vivo studies followed the ARRIVE guidelines (46). The use of mice was approved by the Institutional and Animal Care Committee of Rutgers University (Newark, NJ). Female NOD/SCID BALB/c (NSG) mice (4 wk old) were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited facility. The mice were housed for 1 wk and then humanized as follows: mice (n = 10) were exposed to 150 cGy with a cesium source. After 16 h, the mice were injected i.v. via the tail vein with 6 × 105 human CD45+CD34+ cells (isolated from BM aspirate) in 200 μl of sterile PBS. Human chimerism was examined at weeks 8 and 12 by flow cytometry with blood for human CD45. At week 12, chimerism was confirmed. The mice (five per group) were injected i.p. with two doses of vehicle or Gly, 3 mg/kg in 200 μl volume (47, 48). After 3 d, the mice were euthanized, and the femurs and spleen were harvested. Cells were flushed from the femurs and analyzed by flow cytometry and clonogenic assay for CFU-GM.
Statistical analyses
Data were analyzed using a two tailed Student t test to determine the significance (p value) between experimental values. A p value ≤0.05 was considered significant.
Results
Gene validation
SP and other tachykinin members regulate hematopoiesis, directly or indirectly through BM stroma (18, 49, 50). Indirect effects occurred through cytokine production, activators, such as HIF1α, in addition to maintaining bone homeostasis (17, 40, 51). To understand SP-mediated hematopoietic regulation, we analyzed 16-h SP-stimulated BM stroma with arrays of 2400 genes. The analyses used stroma from three different human donors.
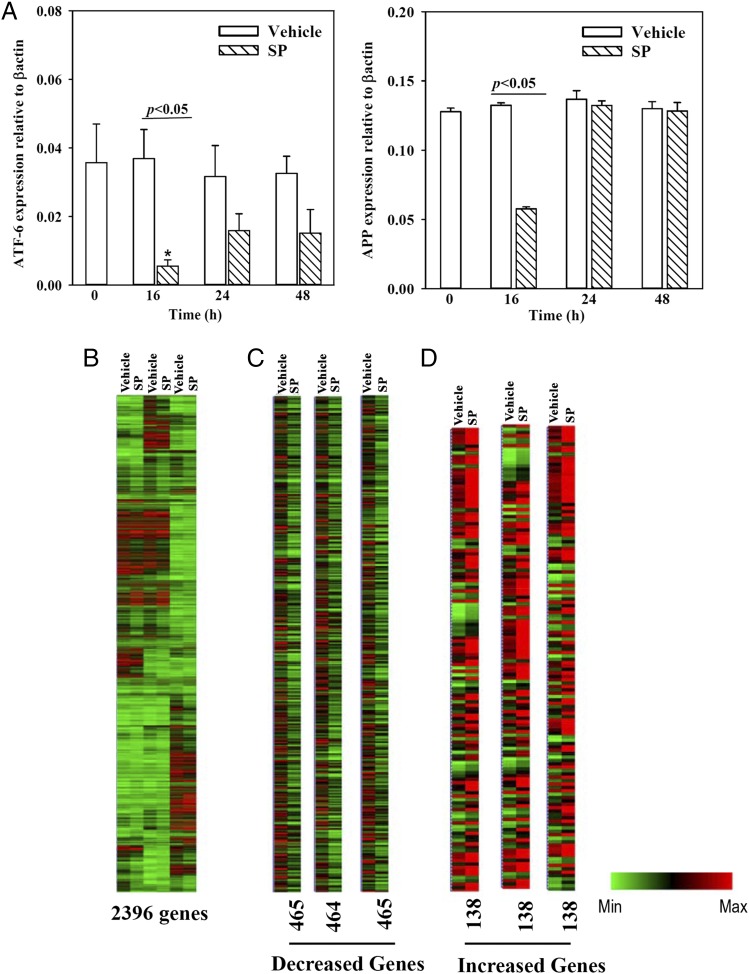
The array was validated by randomly selecting ATF-6 and APP for quantitation by real-time PCR in time-course studies. SP-stimulated BM stroma showed significant (p < 0.05) decreases for both genes after 16 h, consistent with the array data (Fig. 1A). ATF-6 was increased at 48 h, suggesting SP-mediated changes could be reversed in BM stroma. Similarly, the effects on APP were reversed to control level (unstimulated and vehicle) by 24 h (Fig. 1A). The results allowed us to proceed with further analyses and functional studies.
FIGURE 1.
Array validation and heatmap of the gene arrays. (A) Time-course studies were done with BM stromal cells stimulated with 10 nM SP. Total RNA was extracted and then studied by real-time PCR for APP and ATF-6. The results are presented as the mean ± SD (n = 5); each experiment was performed with stromal cells from a different donor. Each donor was tested in triplicate, and the mean of each was incorporated as one experimental point. The control (vehicle) values are shown for each time point. *p < 0.05 versus 24 and 48 h time points. (B) Heat map of microarray gene expression profile of all genes based on SP stimulation or vehicle treated in three biological replicates (each with a different BM donor). (C) Downregulated genes from the data in (B), and (D) upregulated genes based on the data in (B).
Hierarchical clustering of genes
A heat map with the total gene set showed variations among the three donors who were healthy and within the same age (22–25 y). Although tight clustering, the results showed differences between baseline and SP stimulation (Fig. 1B). Heat maps of genes with 2-fold increases and decreases indicated distinct changes between BM stroma stimulated with vehicle and SP (Fig. 1C, 1D).
Hematopoietic-associated genes in SP-stimulated stroma
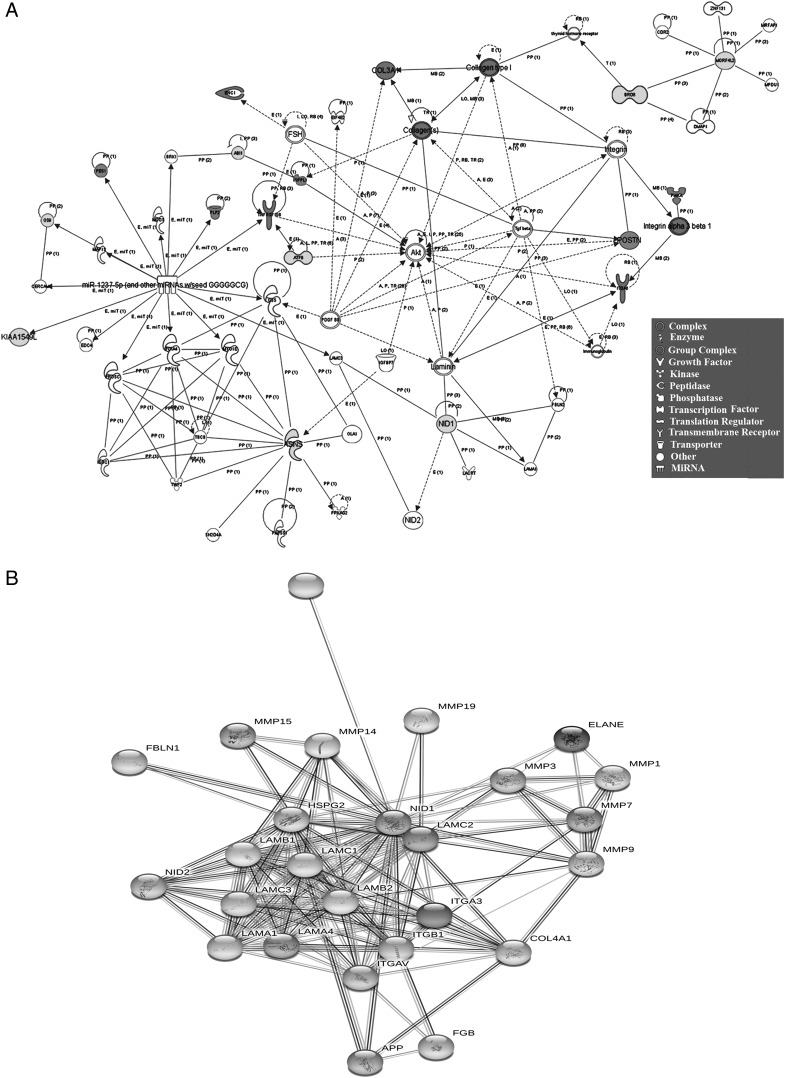
This section determined how the gene expression in SP-stimulated stroma predicted hematopoietic regulation. We subjected the lowest and highest expressing 20 genes (vehicle/SP) to IPA (Supplemental Table I). The predicted networks were consistent with hematopoietic regulation: intercellular communication, assembly and organization, tissue development, adhesion, migration, and proliferation (Fig. 2A, Supplemental Table II). The genes linked to apoptosis were decreased, consistent with a survival role for SP (52). The growth factors within the network underscored the complex mechanism by which SP stimulates hematopoiesis (17). The decrease in nidogen-1 (NID-1) by SP was significant because of the functional role of NID-1 in differentiation and its link to the integrins (Fig. 2B, Supplemental Table I). As a hematopoietic stimulator, SP would require dissociation of HSCs from stromal extracellular matrices. Hence, by lowering of NID-1, SP would be able to initiate hematopoiesis through disassociation of HSCs from the stromal compartment.
FIGURE 2.
Functional network with the highest and lowest gene expression. (A) IPA established a network of upregulated and downregulated genes (Supplemental Table I). (B) String analysis established the identity of proteins linked to NID-1.
We expanded the analyses with genes showing 1.5 fold changes (increased and decreased). IPA output provided >50 clusters with >300 overlapping genes. Narrowing the analyses for hematopoietic regulation resulted in groups with significant p values, 10−19–10−13 (Supplemental Table II). The networks were associated with blood cell development (175 genes), granulocytes (165 genes) and mononuclear cells (147 genes). The latter were further divided into total lymphocytes (145 genes) and T cells (128 genes). Overall, the analyses showed a significant number of genes linked to hematopoiesis.
Predicted hematopoietic effects of HMGB1
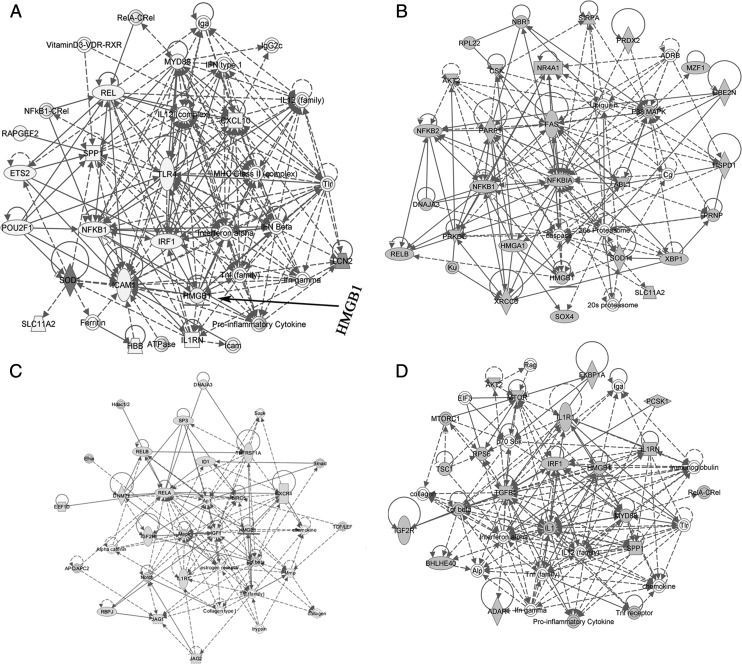
HMGB1 was identified in the networks that link hematopoiesis (Supplemental Fig. 1, Supplemental Table II). Because HMGB1 was decreased in SP-stimulated stroma, we performed further analyses by subjecting the overlapping 278 genes to IPA. The output indicated genes linked to inflammation and hematopoiesis, such as IFN-γ, NF-κB, and HMGB1 (Fig. 3A). We next narrowed the analyses with 35 genes based on top scoring networks in IPA associated with lymphocyte development. This indicated HMGB1’s involvement with lymphocyte differentiation (Fig. 3B, 3C). The high scores for HMGB1 in lymphocyte development was reduced in a network for all leukocytes (Fig. 3D). Because lymphocytes are a subset of leukocytes, the data suggested that the effects of HMGB1 might be specific to particular hematopoietic subsets. Furthermore, an examination of pathways formed by differentially expressed genes indicated a p value of 4.09 × 10−12 in networks with HMGB1 in several functions linked to hematopoiesis (Supplemental Fig. 1). Overall, the predicted networks suggested that HMGB1 could be key to tachykinin-mediated hematopoietic regulation.
FIGURE 3.
Genes selected by IPA with functional link between HMGB1 and hematopoiesis. The genes linked to hematopoiesis were established as a network using IPA. (B) The genes in (A) with a cut off score of 53 were used to establish a network of lymphocyte development. (C) Similar network for lymphocyte development using a score of 31. (D) Network established with the genes shown in (A) for leukocyte development.
SP-mediated effects on HMGB1 expression
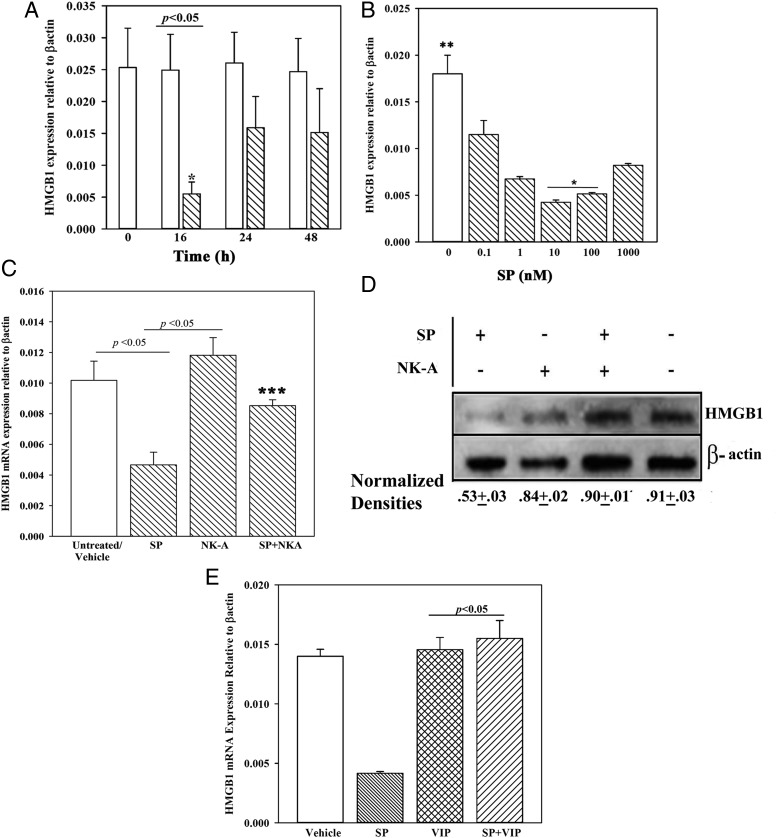
Because HMGB1 is predicted to be involved in a network with SP-mediated hematopoietic effects, we validated the array data. Time-course studies were performed by stimulating BM stroma with optimum SP (10 nM) or vehicle (PBS). HMGB1 mRNA was significantly (p < 0.05) decreased at 16 h relative to baseline (unstimulated/vehicle). These findings were consistent with the array data (Fig. 3). At each time point, the controls (unstimulated/vehicle) were similar, and the data were therefore plotted on the same bar. This decrease was reversible because significant (p < 0.05) increases were noted at the 24- and 48-h time point relative to 16 h (Fig. 4A).
FIGURE 4.
Time-course and dose-response effects of SP on HMGB1 mRNA and effects of NK-A and/or SP on HMGB1 expression in BM stroma. (A) Real-time PCR using total RNA in time-course studies with stromal cells stimulated with SP (10 nM) in media with 10% FCS. The values of controls at each time point in which stroma was unstimulated and stimulated with vehicle (PBS) were similar and are there plotted together. The data were normalized with β-actin. Each experimental point is presented as the mean ± SD (n = 4), each with a different BM donor. Each donor was tested in triplicates, and the data point was entered as the mean value. (B) The studies in (A) were repeated, except with BM stroma stimulated for 16 h with different concentrations of SP in media containing 10% FCS. The results represent four independent experiments (mean ± SD), each with a different BM donor. (C) BM stromal cells were stimulated with 10 nM SP and/or 10 nM NK-A. Controls were stimulated with vehicle. At 16 h, total RNA was isolated and then analyzed by real-time PCR for HMGB1 mRNA. The values of unstimulated and vehicle were similar and were therefore plotted together (n = 8). The results are shown for four independent experiments (mean ± SD), each with stroma from a different donor. Each donor was tested in triplicates, and the mean was used for each experimental point. (D) Western blots for HMGB1 were performed with whole cell extracts from BM stroma stimulated for 16 h with 10 nM SP and/or 10 nM NK-A. Controls were unstimulated or treated with vehicle. The membranes were stripped and reprobed for β-actin. Densitometric analyses of normalized bands for three independent studies are shown below the graph. Each study was performed with a different BM donor. (E) The studies in (C) were repeated with 10 nM VIP replacing NK-A. The normalized values are presented as mean ± SD (n = 4); each experiment was performed with a different donor. *p < 0.05 versus other concentrations of SP, **p < 0.05 versus SP stimulation, ***p < 0.05 versus SP alone.
Because SP can be degraded with proteases in sera, we repeated the 16-h time-point studies in dose-response analyses with 2 and 10% FCS. SP stimulation in 10% FCS showed a significant (p < 0.05) decrease at all SP levels as compared with vehicle but significantly (p < 0.05) less at 10 and 100 nM SP (Fig. 4B). All experiments were therefore performed with 10% FCS. The increase in HMGB1 at high SP levels could be explained as follows: increased HMGB1 at the higher SP levels may be explained by the need for HMGB1 to exert negative feedback on hematopoiesis. A more plausible explanation is that a high concentration of SP could lead to receptor desensitization. If so, this would allow for the return to baseline HMGB1. The decrease of HMGB1 in SP-stimulated stroma at hour 16 will allow for hematopoietic stimulation, but this was transient because of an increase in HMGB1 by hour 24.
Antagonizing effect of NK-A on HMGB1 induction
SP and NK-A can exert opposite hematopoietic effects, partly by antagonizing the effects of each other (23, 45, 53). Because SP, a hematopoietic stimulator, lowered HMGB1 expression in stroma, we asked whether NK-A can blunt SP’s effect on HMGB1. Stromal cells were stimulated for 16 h with 10 nM SP, 10 nM NK-A, or 10 nM SP plus 10 nM NK-A. NK-A significantly (p < 0.05) reversed the suppressive effects of SP on HMGB1 mRNA (Fig. 4C). HMGB1 mRNA was similar (p > 0.05) between NK-A treatment and baseline/vehicle change. These changes at the mRNA were verified at the protein level by Western blots with normalized band densities (Fig. 4D). Because NK-A and SP are peptides from the same gene, we selected another peptide (VIP) with hematopoietic function similar to NK-A (39). The results were similar to those of NK-A, suggesting that HMGB1 might be a key factor in negative hematopoiesis (Fig. 4E).
Effects of HMGB1 on long-term culture-initiating cell assay
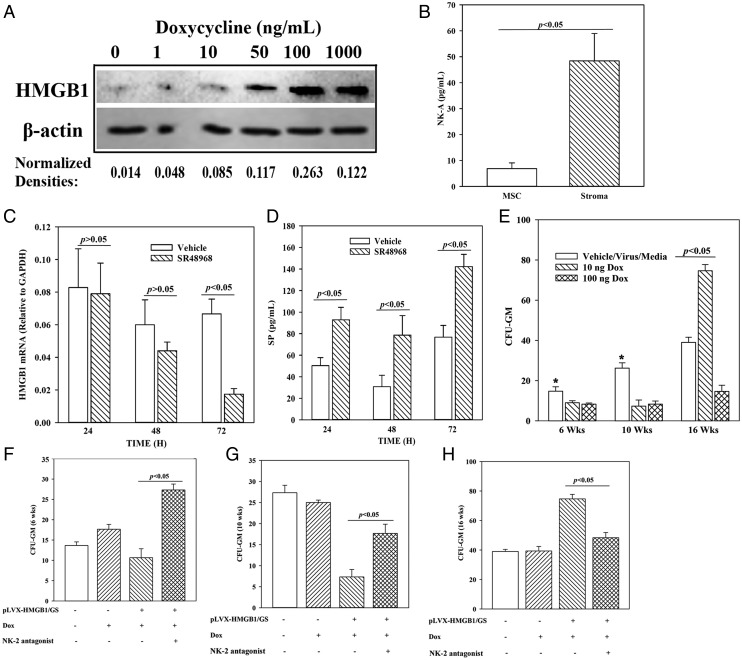
We investigated a role for HMGB1 in hematopoiesis and asked whether NK-A can mediate such function. This question was addressed with the long-term culture-initiating cell (LTC-IC) assay, which recapitulates in vivo HSC function (54). Because stromal cells support the function of LTC-ICs, we induced HMGB1 in stromal cells and observed necrosis with constitutive HMGB1 expression. We therefore modified the approach with an inducible system that regulated HMGB1 level (pLVX–HMGB1/GS). Dose-response studies evaluated HMGB1 by Western blot using stromal extracts and selected 10 and 100 ng/ml Dox (Fig. 5A, Supplemental Fig. 2). We omitted the concentration with the highest HMGB1 levels because of its necrotic effects on stroma (data not shown).
FIGURE 5.
Effects of HMGB1 and NK-A on LTC-ICs. (A) Dose-response analyses for HMGB1 levels in pLVX–HMGB1/GS–transduced stroma, which were cultured with different concentrations of Dox. After 24 h of Dox treatment, Western blot for HMGB1 was performed with whole cell extracts. The normalized band densities for two independent experiments are shown below the blot. (B) BM stroma and MSCs were analyzed for baseline NK-A by ELISA. The results are presented as mean ± SD pg/ml (n = 9). Cells were obtained from three different BM donors, with each donor studied in triplicate. (C) BM stroma was cultured in with 100 nM SR48968 or vehicle. At different times, media were collected and then studied for HMGB1 mRNA by real-time PCR. The results are presented as the mean ± SD (n = 3). Each experiment was performed with stroma from a different donor. (D) The studies in (C) were repeated, and instead, the culture media were analyzed for SP level by ELISA. The results are presented as the mean ± SD picogram per milliliter (n = 3). (E) LTC-IC cultures were established with stromal cells and transduced with an inducible HMGB1 (pLVX–HMGB1/GS) or vector alone. The transduced stroma was studied with or without 10 nM NK-2 antagonist (SR48968). HMGB1 was induced with 10 ng and 100 ng of Dox at initiation of culture. At 6, 10, and 16 wk, mononuclear cells were analyzed for CFU-GM in methylcellulose cultures. The results for cultures with vehicle, media alone, and virus without the HMGB1 insert (virus) were similar, and the data were combined (controls). The data are the mean ± SD (n = 4), each experiment performed with a different donor. Each experiment was performed with stroma from a different donor and tested in sextuplets, and the mean was added as one experimental point. *p < 0.05 versus Dox. (E–H) The cultures in (C) were repeated, except for HMGB1 induction with 10 ng Dox, in the presence or absence of 100 nM SR48968 (NK-2 antagonist). The total number of experiments and the results were similarly presented for cultures without antagonists (E), and those cultures with antagonists at weeks 6 (F), 10 (G), and 16 (H).
As NK-A is proposed to be a negative feedback to SP via HMGB1, we first assessed baseline NK-A in stroma. We compared it with MSCs because of their role as additional support of hematopoiesis (55). ELISA for NK-A indicated ∼5-fold more in stroma as compared with MSCs, indicating a major role for the former in the NK-A–HMGB1 axis (Fig. 5B). Next, we asked whether stroma-derived NK-A induced HMGB1 by an autocrine mechanism. This was addressed by assessing HMGB1 mRNA levels in the presence or absence of NK-A receptor (NK2) antagonist. Indeed, at 72 h, there was a significant decrease in HMGB1 mRNA (Fig. 5C). We previously reported on the regulation of SP and NK-A in stroma (24, 45). We therefore asked whether blocking autocrine release of NK-A with the antagonist would decrease SP. ELISA for SP showed a significant increase in SP in the presence of NK2 antagonist, indicating a role for autocrine NK-A in the decrease of SP (Fig. 5D).
Prior to assessing the role of NK-A in the LTC-IC function, we first studied the role of HMGB1 in regulating HSC function. LTC-IC assays were performed with pLVX–HMGB1/GS–transduced gamma-irradiated BM stroma. HMGB1 was induced with 10 and 100 ng/ml Dox at day 0 in the cultures with BM mononuclear cells (HSCs, progenitors, and immune cells) on top of the stromal cells. Radiation prevented stromal cell proliferation without compromising metabolic activity (56). At weeks 6, 10, and 16, aliquots of mononuclear cells were analyzed for CFU-GM. Because progenitors were present in the seeding mononuclear cells, the colonies at week 6 mostly represented the seeding progenitors. Those from week 16 cultures were from the seeded HSCs because the original progenitors would not survive by week 16. The values for vehicle, backbone lentivirus, and media were similar and were plotted together (control). HMGB1 induction significantly (p < 0.05) decreased CFU-GM at weeks 6 and 10 as compared with controls (Fig. 5E). Week 16 CFU-GM colonies were increased with 10 ng/ml Dox but decreased with 100 ng/ml Dox (Fig. 5E). The decrease in CFU-GM by 100 ng/ml Dox was probably because of increased cell death (Supplemental Fig. 2). Based on this, subsequent studies used 10 ng/ml Dox.
Next, we investigated a role for NK2 in HMGB1-mediated increase in LTC-ICs (Fig. 5E). The LTC-IC assay was performed in the absence or presence of NK2 antagonist (10 nM SR48968). NK2 antagonist significantly (p < 0.05) increased CFU-GM at 6- and 10-wk cultures, regardless of HMGB-1 induction, indicating a mediator role for NK-A in HMGB1-mediated hematopoietic suppression (Fig. 5F, 5G). At week 16, there was a significant (p < 0.05) increase in CFU-GM with induced HMGB1 as compared with baseline, and this increase was returned to baseline with NK2 antagonist (Fig. 5H). The increase at week 16 indicated that, unlike the suppressive effects of HMGB1 on progenitors (weeks 6 and 10), HMGB1 enhanced HSC/LTC-IC function. The increased activity of HSC/LTC-IC at week 16 was partly because of NK2 activation because its inhibitor significantly (p < 0.05) reduced the colonies to baseline. In summary, HMGB1 via NK-A maintained LTC-ICs but decreased hematopoietic progenitors.
In vivo hematopoietic effects of HMGB1
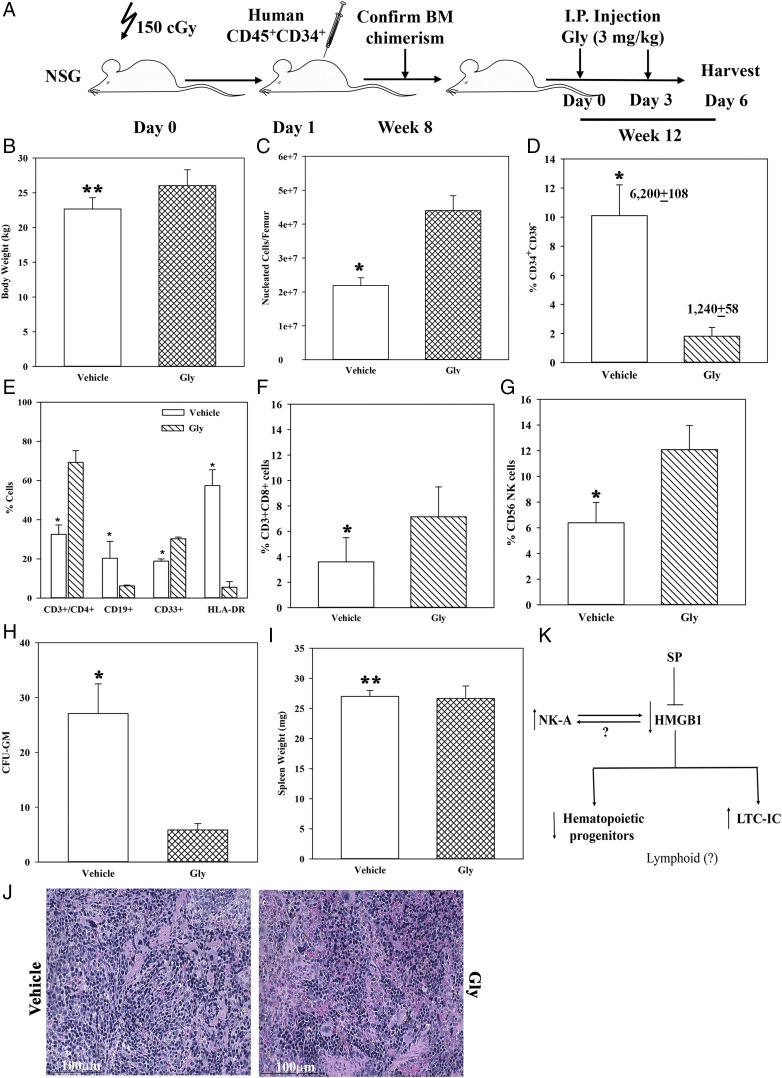
The in vitro studies indicated that HMGB1 can protect the most primitive HSCs but suppressed granulocytic-monocytic progenitors (Fig. 5). These findings were tested in vivo using NSG mice with human hematopoietic system. The in vivo method recapitulates physiological sources of tachykinins, which could be neural and nonneural sources within the femurs of NSG mice (40). Thus, hematopoietic activity by human cells could be induced by murine tachykinins because of the evolutionary conserved nature of these peptides. Because Gly inhibited HMGB1, the humanized NSG model examined the effects of HMGB1 at baseline/homeostasis hematopoietic activity.
Mice were treated with a HMGB1 antagonist (Gly) as per Fig. 6A. Gly did not cause lethargy or weight change (Fig. 6B). At 3 d after the last Gly injection, total nucleated cells were significantly (p < 0.05) increased when HMGB1 was blocked with Gly as compared with vehicle (Fig. 6C). This increase was in line with the removal of suppressor HMGB1 to increase the proliferation of hematopoietic progenitors (Fig. 5).
FIGURE 6.
Effects of HMGB1 on hematopoiesis in humanized NSG female BALB/c. (A) The outline of humanizing five NSG mice with CD34+ cells from BM aspirates and injection with HMGB1 inhibitor (Gly) is shown in the diagram. (B) Mean ± SD (n = 5) body weight of mice. (C) Total nucleated cells/femur (mean ± SD; n = 5). (D) Percentage of CD34+CD38− cells/femur is shown in addition to the absolute numbers of CD45+ cells on top of each bar (mean ± SD; n = 5). (E) Percentage of CD3+CD4+ T cells, CD19 B cells, CD33+ myeloid cells, and HLA-DR (mean ± SD; n = 5). (F) Percentage of CD3+CD8+ T cells (mean ± SD; n = 5); (G) Percentage of CD56+ NK cells (mean ± SD; n = 5). (H) CFU-GM/1 × 105 nucleated cells from femurs. The results are presented as the mean ± SD (n = 5). Each clonogenic assay was performed in duplicate, and the mean value was included as one experimental point. (I) Spleen weight, mean ± SD (n = 5). (J) H&E staining of spleen sections from mice treated with Gly or vehicle. The images (original magnification ×40) were taken with an Evos FL2 Auto Imager and represented five mice. (K) Summary of key findings. *p < 0.05 versus Gly treatment, **p > 0.05 versus Gly treatment
Phenotypic analyses for human CD45+ hematopoietic cells in femurs were performed by flow cytometry. CD34+CD38− cells were significantly (p < 0.05) reduced by Gly as compared with vehicle, suggesting loss of the most primitive cells when HMGB1 was absent (Fig. 6D versus Fig. 5E). There was a significant (p < 0.05) increase in CD3+CD4+ T cells with Gly, consistent with a suppressive effect of HMGB1, although in this group, T cell lymphoid lineage appeared to be also affected by the absence of HMGB1 (Fig. 6E). This effect contrasted CD19+ B cells, suggesting that HMGB1 could enhance B cell lymphoid lineage (Fig. 6E). CD33+ cells (myeloid) were significantly (p < 0.05) increased with Gly, consistent with a suppressive effect by HMGB1 on in vitro myeloid progenitors (Fig. 6E versus Fig. 5C). Because HLA-DR could be associated with cell activation, we assessed the total population and showed marked reduction with Gly (Fig. 6E). Because of the enhanced effects on T lymphoid cells, we analyzed the femurs for CD3+CD8+ T cells and CD56+ NK cells. The results indicated a significant (p < 0.05) increase for both subsets in mice treated with Gly (Fig. 6F, 6G).
In the next set of studies, we performed clonogenic assays using human GM-CSF. The results showed a significant (p < 0.05) decrease in CFU-GM with Gly as compared with vehicle (Fig. 6H). This decrease could not be explained by extramedullary hematopoiesis because there was no difference in the weight of the spleens or gross pathology between the two group (Fig. 6I). In total, the results showed a role for HMGB1 on in vivo hematopoiesis in the experimental model of humanized NSG mice.
Discussion
We report on a central role for HMGB1 in Tac1 peptide-mediated hematopoietic regulation. The model used an inducible system to express HMGB1, and the level selected did not cause necrosis of stromal cells. In fact, stromal cells underwent necrosis with a vector that constitutively expressed HMGB1 (data not shown). The criterion used to select the concentration of Dox was based on the highest level that showed no evidence of necrosis. There was no change in autophagy despite the link between HMGB1 and autophagy (57) (Supplemental Fig. 2). HMGB1 regulation in stroma involves SP and NK-A, evolutionary peptides derived from the Tac1 gene. Although NK-A, which is encoded by the same gene as SP, was able to reverse the suppression of HMGB1 in stromal cells, this feedback mechanism might not be limited to NK-A. The use of another peptide (VIP), shown to negatively regulate hematopoiesis also blunted SP-mediated decrease of HMGB1 (Fig. 4E) (39). This suggested that HMGB1 might be central to other negative hematopoietic regulators and opens avenues for further research. Baseline NK-A in BM stroma appeared to regulate HMGB1 by autocrine mechanisms and also ensured low levels of SP (Fig. 5B–D). This indicated a method by which the stromal compartment prevented hematopoietic exhaustion. In total, unlike VIP, which requires additional studies, we concluded that NK-A links SP–HMGB1–mediated hematopoietic homeostasis. The links are based on the findings in this study as well as reports on SP and NK-A regulating hematopoietic balance (17, 18). Increased HMGB1 in NK-A–stimulated stroma at 24 and 48 h is in line with NK-A as a negative regulator of SP-mediated hematopoietic stimulation (Fig. 4A). The delayed decrease of HMGB1 in the presence of NK2 antagonist was not a surprise because NK2 activation is required to maintain its expression via cytokine production (24). Thus, it would take some time for NK2 to be decreased with the antagonist.
SP as a hematopoietic stimulator was negatively regulated by two other peptides, NK-A and VIP, and this occurred partly via HMGB1. The effects were specific because an inactive enantiomer of NK1 showed no effect (data not shown). SP and CGRP are generally colocalized nerve fibers. Our ongoing studies have identified CGRP as a hematopoietic stimulator. However, we did address the hematopoietic effects of SP and CGRP and also whether the negative regulator peptides can blunt the stimulatory effects of SP and CGRP. Additionally, we did not determine whether CGRP mediates hematopoietic stimulation by decreasing HMGB1. These are ongoing studies of high significance because the noted peptides can also be released from the innervated nerve fibers in BM (7–9).
The tachykinins regulate hematopoiesis partly through the production of cytokines from BM stroma (17, 18). This study expanded on these mechanisms by selecting genes with >2-fold changes from an array of 2400 genes (Fig. 1). The selected genes, when subjected to computational studies, predicted hematopoietic functions (Fig. 2, Supplemental Table II). This gene set, when grouped into networks with low p values, indicated a low probability that the genes were randomly linked within the predicted pathways (Figs. 2, 3). The unfiltered gene set, when presented as a hierarchical clustering, showed distinct differences among the three stromal donors despite similarities in age and health (Fig. 1B). Despite these differences, HMGB1 showed a p value <0.05. Additionally, there were common genes among the top decrease and increase.
The networks led us to examine HMGB1, which was decreased in the array with SP-stimulated stroma. Computational analyses indicated HMGB1 as a key molecule in hematopoietic regulation (Fig. 3, Supplemental Table II). The networks, combined with a ubiquitous role for HMGB1 in hematopoietic cell development, underscored the importance of understanding the SP–NK-A–HMGB1 hematopoietic axis. An understanding of this axis in the model using healthy cells is highly significant because the information could lead to insights on hematological disorders and development. As an example, the finding of NID-1 in SP-mediated hematopoietic stimulation was important because of its role in fetal liver hematopoiesis (58). Also, because the NK-A–HMGB1 axis acts as a negative regulator of SP, it will be interesting going forward to examine how NID-1 is linked to the positive and negative regulating effects of the tachykinins.
The decrease in HMGB1 by SP in BM stroma was reversible because the level returned to baseline by 24 h (Fig. 4A). However, addition of exogenous NK-A to SP-stimulated stroma blunted the ability of SP to decrease HMGB1. This suggested the opposing effects of SP and NK-A in HMGB1 expression (Fig. 4D, 4E).
The regulation between SP and NK-A in HMGB1 expression was explained in LTC-IC cultures to assess HMGB1’s effects on progenitors and LTC-ICs, which are an in vitro surrogate of HSCs. HMGB1 decreased hematopoietic progenitors (6- and 10-wk cultures) but increased the LTC-IC at week 16 cultures (Fig. 5). These results indicated a tight control of HMGB1 in hematopoiesis, maintaining the HSCs and acting as a negative regulator of progenitor proliferation. The role of HMGB1 appeared to be mediated by NK-A because its receptor antagonist reversed the effects of HMGB1 on hematopoietic progenitors and LTC-ICs (Fig. 5).
The in vitro findings were tested with NSG mice, humanized with CD34+ cells (Fig. 6A). The mice were treated with an inhibitor of HMGB1 (Gly) and then examined for hematopoietic activity. The use of the chemical did not result in evidence of lethargy or change in body weight in mice. The total number of nucleated cells was increased, and this was consistent with loss of hematopoietic suppression when HMGB1 was inhibited (Fig. 6C). The in vitro studies on the role for HMGB1 in hematopoiesis was validated in vivo because the number of CD34+CD38− cells was significantly decreased with HMGB1 inhibition (Figs. 5E, 6D). The increase in cellularity was not explained by increased progenitors, which were shown to be decreased when HMGB1 was inhibited (Fig. 6H). The increased cellularity might be due to increased differentiation to mature cells and suppression of HSC activity. However, the effects of HMGB1 appeared to depend on the lineage because blocking of HMGB1 increased T cell subsets and NK cells (CD4, CD8, and CD56), and decreased B cells (CD19) (Fig. 6E–G).
The question is whether interrupting HMGB1 could cause extramedullary hematopoiesis, which would be an effect of BM inhibition. The answer to this was briefly addressed with spleen weight that showed no difference between Gly and vehicle. The issue of extramedullary hematopoiesis will require long-term observation of the mice. However, the results of this manuscript would be important in the study of aging as indicated by the SP–HMGB1 link to development. Indeed, our unpublished studies indicated a decrease in HMGB1 in the aged hematopoietic cells as compared with the young human cohort. This observation is consistent with the suppressive role of HMGB1 in hematopoietic progenitors and its role in maintaining the LTC-ICs, which are the in vitro equivalent of HSCs. Decreased HMGB1 in the aged could lead to increased proliferation of HSCs, as noted in the BM of older individuals (59). Also of significance, there was increase in CD33 myeloid cells, similar to aged BM (Fig. 6E) (59).
The investigation in this study adds to our previous validation of HIF-1α, which showed SP through stromal cells influencing hematopoiesis (40). In our previous report, the selection of a HIF-1 alone lacked robust analyses to determine how SP through stroma regulated hematopoiesis. This gap has now been fulfilled in the more robust studies linking HMBG1. The heat map developed from the array data indicated variations among the three donors despite similarities in age and health (Fig. 1). Regardless of these variations, there was consistency in the SP–HMGB1–NK-A axis with respect to hematopoietic regulation.
Figure 6J summarizes the key findings in the paper. During hematopoietic stimulation. SP, which has been shown to induce the production of cytokines and hematopoietic stimulation, blocked HMGB1. By doing so, SP removed the hematopoietic suppressor HMGB1, explaining previous studies reporting on SP’s role as a hematopoietic stimulator (17). The ability of NK-A receptor (NK2) antagonist to block the hematopoietic effects of HMGB1 suggested there is a positive correlation between HMGB1 and NK-A levels. These two molecules can similarly affect hematopoiesis to attain homeostasis, leading to decrease in progenitor proliferation and maintenance of LTC-IC. The dual effects of HMGB1 on the humanized NSG mice on T and B cell types indicate that the effects on lymphoid lineage at this time is unclear. This will require further studies, including thymic cultures and CFU–B as well as CFU–pre-B assays.
This paragraph discusses the findings in the context of NK1 and NK2 receptors (Fig. 6J). We have reported on NK1 being induced by growth factors with hematopoietic stimulation and NK2 by hematopoietic suppressors (17, 18, 24). The findings in this report add to the already reported network among SP, NK-A, cytokines, and NK receptors in hematopoietic homeostasis. We showed cytokines inducing NK2 receptor as well as activating the ligand (40). It appears that once NK-A is increased, it activates NK2 to induce HMGB1 (Fig. 5). Indeed, the tachykinins can interact with NK1 and NK2. However, we showed specificity of these receptors for SP and NK-A (13, 24, 45, 60).
The in vitro studies dissected the tachykinins and determined how they interact with HMGB1. The tachykinins can be released from nerve fibers. The findings, therefore, have implications for processes in which the nerve fibers are activated, such as infection or peripheral injury. Indeed, surgical insults have been shown to affect hematopoiesis, and such findings can benefit from this report. We have begun to determine how physiological SP and NK-A can interact with HMGB1 to affect hematopoiesis. This began with the humanized NSG mice, in which HMGB1 inhibitor (Gly) was injected into mice. In this model, regardless of the tachykinin source, its effects on HMGB1 were determined and confirmed the negative hematopoietic effects. Physiologically, the tachykinins, as well as the other peptides, could be the source of hematopoietic regulation with a network involving the nerve fibers, hematopoietic niche cells, and hematopoietic stem/progenitors. The findings will be the basis to further investigate the HMGB1–tachykinin axis in hematopoiesis.
Supplementary Material
Acknowledgments
This work is in partial fulfillment for a Ph.D. thesis for M.G.
The online version of this article contains supplemental material.
- BM
- bone marrow
- CFU-GM;
- CFU–granulocyte-monocyte
- Dox
- doxycycline
- Gly
- glycyrrhizin
- HMGB1
- high-mobility group box 1
- HSC
- hematopoietic stem cell
- IPA
- Ingenuity Pathway Analysis
- LTC-IC
- long-term culture-initiating cell
- NID-1
- nidogen-1
- NK-A
- neurokinin A
- SP
- substance P
- VIP
- vasoactive intestinal peptide.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Eaves C. J. 2015. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood 125: 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y., Gao S., Xia J., Liu F. 2018. Hematopoietic hierarchy - an updated roadmap. Trends Cell Biol. 28: 976–986. [DOI] [PubMed] [Google Scholar]
- 3.Winkler I. G., Barbier V., Wadley R., Zannettino A. C., Williams S., Lévesque J. P. 2010. Positioning of bone marrow hematopoietic and stromal cells relative to blood flow in vivo: serially reconstituting hematopoietic stem cells reside in distinct nonperfused niches. Blood 116: 375–385. [DOI] [PubMed] [Google Scholar]
- 4.Morrison S. J., Scadden D. T. 2014. The bone marrow niche for haematopoietic stem cells. Nature 505: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendelson A., Frenette P. S. 2014. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 20: 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eltoukhy H. S., Sinha G., Moore C. A., Gergues M., Rameshwar P. 2018. Secretome within the bone marrow microenvironment: a basis for mesenchymal stem cell treatment and role in cancer dormancy. Biochimie 155: 92–103. [DOI] [PubMed] [Google Scholar]
- 7.Berger A., Benveniste P., Corfe S. A., Tran A. H., Barbara M., Wakeham A., Mak T. W., Iscove N. N., Paige C. J. 2010. Targeted deletion of the tachykinin 4 gene (TAC4-/-) influences the early stages of B lymphocyte development. Blood 116: 3792–3801. [DOI] [PubMed] [Google Scholar]
- 8.Liu K., Castillo M. D., Murthy R. G., Patel N., Rameshwar P. 2007. Tachykinins and hematopoiesis. Clin. Chim. Acta 385: 28–34. [DOI] [PubMed] [Google Scholar]
- 9.Cosentino M., Marino F., Maestroni G. J. 2015. Sympathoadrenergic modulation of hematopoiesis: a review of available evidence and of therapeutic perspectives. Front. Cell. Neurosci. 9: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabarowski Z., Gibson-Berry K., Felten S. Y. 1996. Noradrenergic and peptidergic innervation of the mouse femur bone marrow. Acta Histochem. 98: 453–457. [DOI] [PubMed] [Google Scholar]
- 11.Felten D. L., Felten S. Y., Carlson S. L., Olschowka J. A., Livnat S. 1985. Noradrenergic and peptidergic innervation of lymphoid tissue. J. Immunol. 135(2 Suppl.): 755s–765s. [PubMed] [Google Scholar]
- 12.Qian J., Yehia G., Molina C., Fernandes A., Donnelly R., Anjaria D., Gascon P., Rameshwar P. 2001. Cloning of human preprotachykinin-I promoter and the role of cyclic adenosine 5′-monophosphate response elements in its expression by IL-1 and stem cell factor. J. Immunol. 166: 2553–2561. [DOI] [PubMed] [Google Scholar]
- 13.Joshi D. D., Dang A., Yadav P., Qian J., Bandari P. S., Chen K., Donnelly R., Castro T., Gascon P., Haider A., Rameshwar P. 2001. Negative feedback on the effects of stem cell factor on hematopoiesis is partly mediated through neutral endopeptidase activity on substance P: a combined functional and proteomic study. Blood 98: 2697–2706. [DOI] [PubMed] [Google Scholar]
- 14.Leeman S. E., Ferguson S. L. 2000. Substance P: an historical perspective. Neuropeptides 34: 249–254. [DOI] [PubMed] [Google Scholar]
- 15.Ohtake J., Kaneumi S., Tanino M., Kishikawa T., Terada S., Sumida K., Masuko K., Ohno Y., Kita T., Iwabuchi S., et al. 2015. Neuropeptide signaling through neurokinin-1 and neurokinin-2 receptors augments antigen presentation by human dendritic cells. J. Allergy Clin. Immunol. 136: 1690–1694. [DOI] [PubMed] [Google Scholar]
- 16.Onaga T. 2014. Tachykinin: recent developments and novel roles in health and disease. Biomol. Concepts 5: 225–243. [DOI] [PubMed] [Google Scholar]
- 17.Rameshwar P., Ganea D., Gascón P. 1993. In vitro stimulatory effect of substance P on hematopoiesis. Blood 81: 391–398. [PubMed] [Google Scholar]
- 18.Rameshwar P., Gascón P. 1996. Induction of negative hematopoietic regulators by neurokinin-A in bone marrow stroma. Blood 88: 98–106. [PubMed] [Google Scholar]
- 19.Rameshwar P. 2012. The tachykinergic system as avenues for drug intervention. Recent Patents CNS Drug Discov. 7: 173–180. [DOI] [PubMed] [Google Scholar]
- 20.Patacchini R., Maggi C. A. 1995. Tachykinin receptors and receptor subtypes. Arch. Int. Pharmacodyn. Ther. 329: 161–184. [PubMed] [Google Scholar]
- 21.Hiramoto M., Aizawa S., Iwase O., Nakano M., Toyama K., Hoque M., Nabeshima R., Kaidow A., Imai T., Hoshi H., Handa H. 1998. Stimulatory effects of substance P on CD34 positive cell proliferation and differentiation in vitro are mediated by the modulation of stromal cell function. Int. J. Mol. Med. 1: 347–354. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Lu L., Furlonger C., Wu G. E., Paige C. J. 2000. Hemokinin is a hematopoietic-specific tachykinin that regulates B lymphopoiesis. Nat. Immunol. 1: 392–397. [DOI] [PubMed] [Google Scholar]
- 23.Greco S. J., Corcoran K. E., Cho K. J., Rameshwar P. 2004. Tachykinins in the emerging immune system: relevance to bone marrow homeostasis and maintenance of hematopoietic stem cells. Front. Biosci. 9: 1782–1793. [DOI] [PubMed] [Google Scholar]
- 24.Bandari P. S., Qian J., Oh H. S., Potian J. A., Yehia G., Harrison J. S., Rameshwar P. 2003. Crosstalk between neurokinin receptors is relevant to hematopoietic regulation: cloning and characterization of neurokinin-2 promoter. J. Neuroimmunol. 138: 65–75. [DOI] [PubMed] [Google Scholar]
- 25.Thomas J. O. 2001. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 29: 395–401. [DOI] [PubMed] [Google Scholar]
- 26.Campana L., Bosurgi L., Rovere-Querini P. 2008. HMGB1: a two-headed signal regulating tumor progression and immunity. Curr. Opin. Immunol. 20: 518–523. [DOI] [PubMed] [Google Scholar]
- 27.Rauvala H., Rouhiainen A. 2007. RAGE as a receptor of HMGB1 (Amphoterin): roles in health and disease. Curr. Mol. Med. 7: 725–734. [DOI] [PubMed] [Google Scholar]
- 28.Kang R., Zhang Q., Zeh H. J., III, Lotze M. T., Tang D. 2013. HMGB1 in cancer: good, bad, or both? Clin. Cancer Res. 19: 4046–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang D., Kang R., Zeh H. J., III, Lotze M. T. 2010. High-mobility group box 1 and cancer. Biochim. Biophys. Acta 1799: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lotze M. T., Tracey K. J. 2005. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 5: 331–342. [DOI] [PubMed] [Google Scholar]
- 31.Sims G. P., Rowe D. C., Rietdijk S. T., Herbst R., Coyle A. J. 2010. HMGB1 and RAGE in inflammation and cancer. Annu. Rev. Immunol. 28: 367–388. [DOI] [PubMed] [Google Scholar]
- 32.Lotze M. T., Zeh H. J., Rubartelli A., Sparvero L. J., Amoscato A. A., Washburn N. R., Devera M. E., Liang X., Tör M., Billiar T. 2007. The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol. Rev. 220: 60–81. [DOI] [PubMed] [Google Scholar]
- 33.Ivanov S., Dragoi A. M., Wang X., Dallacosta C., Louten J., Musco G., Sitia G., Yap G. S., Wan Y., Biron C. A., et al. 2007. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood 110: 1970–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiraldi M., Raucci A., Muñoz L. M., Livoti E., Celona B., Venereau E., Apuzzo T., De Marchis F., Pedotti M., Bachi A., et al. 2012. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J. Exp. Med. 209: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kokkola R., Andersson A., Mullins G., Ostberg T., Treutiger C. J., Arnold B., Nawroth P., Andersson U., Harris R. A., Harris H. E. 2005. RAGE is the major receptor for the proinflammatory activity of HMGB1 in rodent macrophages. Scand. J. Immunol. 61: 1–9. [DOI] [PubMed] [Google Scholar]
- 36.Apetoh L., Ghiringhelli F., Tesniere A., Criollo A., Ortiz C., Lidereau R., Mariette C., Chaput N., Mira J. P., Delaloge S., et al. 2007. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol. Rev. 220: 47–59. [DOI] [PubMed] [Google Scholar]
- 37.Chavakis E., Hain A., Vinci M., Carmona G., Bianchi M. E., Vajkoczy P., Zeiher A. M., Chavakis T., Dimmeler S. 2007. High-mobility group box 1 activates integrin-dependent homing of endothelial progenitor cells. Circ. Res. 100: 204–212. [DOI] [PubMed] [Google Scholar]
- 38.Gardella S., Andrei C., Ferrera D., Lotti L. V., Torrisi M. R., Bianchi M. E., Rubartelli A. 2002. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 3: 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rameshwar P., Gascon P., Oh H. S., Denny T. N., Zhu G., Ganea D. 2002. Vasoactive intestinal peptide (VIP) inhibits the proliferation of bone marrow progenitors through the VPAC1 receptor. Exp. Hematol. 30: 1001–1009. [DOI] [PubMed] [Google Scholar]
- 40.Qian J., Ramroop K., McLeod A., Bandari P., Livingston D. H., Harrison J. S., Rameshwar P. 2001. Induction of hypoxia-inducible factor-1alpha and activation of caspase-3 in hypoxia-reoxygenated bone marrow stroma is negatively regulated by the delayed production of substance P. J. Immunol. 167: 4600–4608. [DOI] [PubMed] [Google Scholar]
- 41.Quackenbush J. 2002. Microarray data normalization and transformation. Nat. Genet. 32(S4 Suppl.): 496–501. [DOI] [PubMed] [Google Scholar]
- 42.Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- 43.Bliss S. A., Sinha G., Sandiford O. A., Williams L. M., Engelberth D. J., Guiro K., Isenalumhe L. L., Greco S. J., Ayer S., Bryan M., et al. 2016. Mesenchymal stem cell-derived exosomes stimulate cycling quiescence and early breast cancer dormancy in bone marrow. Cancer Res. 76: 5832–5844. [DOI] [PubMed] [Google Scholar]
- 44.Potian J. A., Aviv H., Ponzio N. M., Harrison J. S., Rameshwar P. 2003. Veto-like activity of mesenchymal stem cells: functional discrimination between cellular responses to alloantigens and recall antigens. J. Immunol. 171: 3426–3434. [DOI] [PubMed] [Google Scholar]
- 45.Murthy R. G., Greco S. J., Taborga M., Patel N., Rameshwar P. 2008. Tac1 regulation by RNA-binding protein and miRNA in bone marrow stroma: implication for hematopoietic activity. Brain Behav. Immun. 22: 442–450. [DOI] [PubMed] [Google Scholar]
- 46.Kilkenny C., Browne W. J., Cuthill I. C., Emerson M., Altman D. G. 2010. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8: e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vitali R., Palone F., Cucchiara S., Negroni A., Cavone L., Costanzo M., Aloi M., Dilillo A., Stronati L. 2013. Dipotassium glycyrrhizate inhibits HMGB1-dependent inflammation and ameliorates colitis in mice. PLoS One 8: e66527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida S., Lee J. O., Nakamura K., Suzuki S., Hendon D. N., Kobayashi M., Suzuki F. 2014. Effect of glycyrrhizin on pseudomonal skin infections in human-mouse chimeras. PLoS One 9: e83747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger A., Frelin C., Shah D. K., Benveniste P., Herrington R., Gerard N. P., Zúñiga-Pflücker J. C., Iscove N. N., Paige C. J. 2013. Neurokinin-1 receptor signalling impacts bone marrow repopulation efficiency. PLoS One 8: e58787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mei G., Xia L., Zhou J., Zhang Y., Tuo Y., Fu S., Zou Z., Wang Z., Jin D. 2013. Neuropeptide SP activates the WNT signal transduction pathway and enhances the proliferation of bone marrow stromal stem cells. Cell Biol. Int. 37: 1225–1232. [DOI] [PubMed] [Google Scholar]
- 51.Niedermair T., Schirner S., Seebröker R., Straub R. H., Grässel S. 2018. Substance P modulates bone remodeling properties of murine osteoblasts and osteoclasts. Sci. Rep. 8: 9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J., Nie J., Fu S., Liu S., Wu J., Cui L., Zhang Y., Yu B. 2017. The fast track to canonical Wnt signaling in MC3T3-E1 cells protected by substance P against serum deprivation-induced apoptosis. Cell Biol. Int. 41: 71–78. [DOI] [PubMed] [Google Scholar]
- 53.Patel N., Castillo M., Rameshwar P. 2007. An in vitro method to study the effects of hematopoietic regulators during immune and blood cell development. Biol. Proced. Online 9: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ploemacher R. E. 1997. Stem cells: characterization and measurement. Baillieres Clin. Haematol. 10: 429–444. [DOI] [PubMed] [Google Scholar]
- 55.Wu J., Zhang W., Ran Q., Xiang Y., Zhong J. F., Li S. C., Li Z. 2018. The differentiation balance of bone marrow mesenchymal stem cells is crucial to hematopoiesis. Stem Cells Int. 2018: 1540148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Os R. P., Dethmers-Ausema B., de Haan G. 2008. In vitro assays for cobblestone area-forming cells, LTC-IC, and CFU-C. Methods Mol. Biol. 430: 143–157. [DOI] [PubMed] [Google Scholar]
- 57.Xu T., Jiang L., Wang Z. 2018. The progression of HMGB1-induced autophagy in cancer biology. OncoTargets Ther. 12: 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomte L. T., Annatshah Y., Schlüter N. K., Miosge N., Herken R., Quondamatteo F. 2006. Hematopoietic cells are a source of nidogen-1 and nidogen-2 during mouse liver development. J. Histochem. Cytochem. 54: 593–604. [DOI] [PubMed] [Google Scholar]
- 59.Morrison S. J., Wandycz A. M., Akashi K., Globerson A., Weissman I. L. 1996. The aging of hematopoietic stem cells. Nat. Med. 2: 1011–1016. [DOI] [PubMed] [Google Scholar]
- 60.Bandari P. S., Qian J., Yehia G., Seegopaul H. P., Harrison J. S., Gascon P., Fernandes H., Rameshwar P. 2002. Differences in the expression of neurokinin receptor in neural and bone marrow mesenchymal cells: implications for neuronal expansion from bone marrow cells. Neuropeptides 36: 13–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.