Abstract
Objective:
To study the prevalence of inducible clindamycin along with vancomycin and methicillin resistance and assessment of hyper variable region (HVR) of mecA gene among different clinical isolates of Staphylococcus spp.
Methods:
A total of 176 clinical isolates of Staphylococci were collected from Pakistan Institute of Medical Sciences (PIMS), Islamabad during 2014-2015. The sample sources were pus, blood, urine, sputum, tracheal secretions and tissue fluids. Bacterial identification was done by colony morphology and biochemical tests. Kirby-Bauer disc-diffusion method was carried out to assess the susceptibility against different antibiotics. Minimal inhibitory concentrations (MICs) were done for vancomycin resistance. Double Disk Diffusion test (D-test) was used to detect the clindamycin inducible resistance. PCR was performed to detect erm(C), mecA and HVR genes.
Results:
Clindamycin inducible resistance among Staphylococcal isolates was found to be 7%, whereas in S. aureus it was 4%, and in coagulase negative Staphylococci (CoNS) it was 11%. The highest resistance was observed against fosfomycin, fusidic acid and cefoxitin. Vancomycin resistance was observed in 23 isolates (13%) of Staphylococci. erm(C), mecA and HVR genes were found in 18%, 50% and 42% respectively.
Conclusions:
D-test must be performed routinely to avoid clindamycin failure. A high level of resistance against vancomycin in Staphylococcal isolates is a concern for public health.
Keywords: Clindamycin inducible resistance, D-test, HVR region, mecA, Staphylococci
INTRODUCTION
S. aureus and Coagulase-negative Staphylococci (CoNS) are considered to be one of the leading causes of hospital and community acquired infections. Resistance to methicillin and vancomycin in Staphylococci is well-known and it is important to find alternatives that can be used in case of methicillin resistant S. aureus (MRSA) and vancomycin resistant S. aureus (VRSA). MRSA strains carry mobile genetic elements, recognized as Staphylococcal cassette chromosome mec (SCCmec) that has a number of genes where the most important is mecA and genes that regulate the expression of mecA.1 mecA gene is responsible for resistance to methicillin and other β-lactam drugs in S. aureus and encodes a 78-kDa penicillin-binding protein 2a (PBP2a).2
In 1991, a new region between IS431-mec and the mecA gene in SCCmec was described;3 which was named as the hyper variable region (HVR) due to DNA length polymorphism. One MRSA strain’s HVR region was sequenced and ten repeat units of 40bp each were identified.3 Most of the studies either have analyzed the hybridization patterns of these direct repeat units PCR products or compared the gel band sizes4 while direct repeat unit (dru) region was sequenced by one study.5 Since the number of these repeat units may be different among isolates, the amplification of HVR region can be used to type and classify MRSA strains.
The macrolide-lincosamide-streptogramin B (MLSB) is a group of antibiotics used to treat different infections caused by Staphylococci.6 Clinicians avoid clindamycin administration, when erythromycin resistance is present, as erythromycin can induce clindamycin resistance in clindamycin sensitive strains by activating erm(C) gene. erm(C) is usually located on small plasmids and is responsible for inducible or constitutive resistance to erythromycin.7 This study was designed to analyze the prevalence of clindamycin inducible resistance along with vancomycin and methicillin resistance and assessment of hyper variable region in MRSA, among different clinical isolates of Staphylococci spp.
METHODS
Bacterial isolates
A total of 176 clinical isolates of Staphylococci spp. were collected from Pakistan Institute of Medical Sciences (PIMS), Islamabad during 2014-2015. Out of these isolates, 138 (78%) were from out-patients and 38 (22%) were from in-patients. Main sample sources were pus followed by blood, urine, tracheal secretions, sputum and tissue fluids.Ethical Approval (No. F.1-1/2015/ERB/SZABMU/ dated May 20, 2016) was obtained from Ethics Review Board of Shaheed Zulfiqar Ali Bhutto Medical University, PIMS, Islamabad.
Bacterial identification
Identification was done by colony morphology on mannitol salt agar and biochemical tests that included catalase, slide and tube coagulase, and DNase tests.
Antibiotic Susceptibility Testing
Antibiotic susceptibility was done by Kirby-Bauer disk diffusion method8 against a panel of antibiotics which included cefoxitin (30µg), linezolid (30µg), rifampin (5µg), fusidic acid (10µg), fosfomycin (50µg), tigecycline (15µg), tetracycline (30µg), chloramphenicol (30µg), ciprofloxacin (5µg), gentamicin (10µg), clindamycin (2µg), erythromycin (15µg), and sulfamethoxazole/trimethoprim (25µg) (Oxoid, UK). S. aureus ATCC 25923 was used as quality control strain. Results were interpreted according to the Clinical and Laboratory Standard Institute (CLSI) guidelines 2015. Minimum inhibitory concentrations (MICs) were performed for vancomycin (Sigma-Aldrich) using agar dilution method.9
Double Disk Diffusion test (D-test)
D-test was performed to detect the inducible clindamycin resistance. Bacterial lawn was prepared on Mueller Hinton agar and antibiotic discs of clindamycin (2µg), and erythromycin (15µg) were placed 15-20 mm apart. Plates were incubated at 37°C overnight. Three phenotypes were interpreted according to CLSI guidelines 2014. Macrolide Type-B streptogramin (MSB) phenotype: resistance to erythromycin (zone size ≤ 13mm) and sensitivity to clindamycin (zone size ≥ 23mm) with a circular zone of inhibition around clindamycin (D-test negative). Inducible macrolides-lincosamides-streptogramin B (iMLSB) phenotype: resistance to erythromycin (zone size ≤ 13mm) and sensitive to clindamycin (zone size ≥ 23mm) with D-shaped zone of inhibition around clindamycin (D-test positive). Constitutive macrolides-lincosamides-streptogramin B (cMLSB) phenotype: resistance to both erythromycin (zone size ≤ 13mm) and clindamycin (zone size ≤ 14mm) with circular zone of inhibition following CLSI guidelines 2015.
Molecular detection of clindamycin inducible resistance erm(C), mecA and HVR genes
Genomic DNA was isolated using lysis method previously described.10 Phenotypically D-test positive isolates were screened for the presence of erm(C) gene while mecA and HVR genes were screened in MRSA isolates by PCR using already published primers.11-13HYPERLINK \l “_ENREF_12” \o “Senna, 2002 #21” PCR conditions were as following; initial denaturation at 95°C for 5 minutes, 35 cycles of 95°C for one minute, annealing at 47°C for one minute for erm(C), 59°C for mecA and 58°C for HVR, extension at 72°C for 1 minute followed by the final extension at 72°C for 10 minutes. PCR products were run on 2% agarose gel and visualized in UV transilluminator (UVItec, EEC).
Statistical Analysis
Statistical analysis was carried out to find association between resistance profiles of organisms using GraphPad Prism software version 7.04. A p-value less than 0.05 was considered statistically significant.
RESULTS
Out of a total of 176 Staphylococcal isolates 51% (n=90) were identified as S. aureus of which 69% (n=62) were MRSA and 31% (n=28) were methicillin sensitive S. aureus (MSSA). A total of 49% (n=86) isolates were identified as CoNS, in which 93% (n=80) were methicillin resistant coagulase negative Staphylococci (MRCoNS) and 7% (n=6) were methicillin sensitive coagulase negative Staphylococci (MSCoNS). Major sample sources were pus (48%), followed by blood (29%), tracheal secretions (8%), urine (4%), sputum (3%), tip of drain (3%), tissue fluids (2%), catheter tip (1.3%), nasal swabs (1%) and semen (0.7%).
Antibiotic Resistance Profile and Minimal inhibitory concentrations (MICs)
Among all S. aureus strains, maximum resistance was observed against fosfomycin followed by fusidic acid, cefoxitin, tetracycline and ciprofloxacin. Least resistance was observed against chloramphenicol, gentamicin and rifampicin. Among CoNS, the maximum resistance was observed against fosfomycin and cefoxitin followed by linezolid, gentamicin, fusidic acid and tetracycline. The least resistance was observed against chloramphenicol and tigecycline. We found a significant correlation of antibiotic resistance with CoNS as compared to S. aureus (p=0.001). The detailed resistance profile of all isolates and statistical comparison are shown in Table-I. From a total of 176, 23 isolates of Staphylococci showed resistance to vancomycin using Kirby Bauer method so MICs were performed on these 23 resistant isolates using agar dilution method, which showed that 6 (7%) isolates were VRSA having MICs more than 16 µg/mL and 17 (20%) were vancomycin resistant coagulase negative Staphylococci (VRCoNS). Out of 17 isolates of VRCoNS, one isolate showed intermediate resistance with MIC of 8 µg/mL and 16 isolates had MICs greater than 32 µg/mL.
Table-I.
Resistance profile of all Staphylococcal isolates.
| Antibiotics | Resistant profile of S. aureus (%) | Resistant profile of CoNS (%) | p-value |
|---|---|---|---|
| Linezolid (LZD) | 37 | 90 | 0.0001 |
| Tigecycline (TGC) | 39 | 33 | 0.372 |
| Tetracycline (TE) | 51 | 66 | 0.0301 |
| Ciprofloxacin (CIP) | 50 | 59 | 0.1739 |
| Rifampin (RD) | 28 | 43 | 0.0174 |
| Fosfomycin (FOS) | 90 | 95 | 0.1795 |
| Fusidic Acid (FD) | 87 | 81 | 0.3408 |
| Clindamycin (DA) | 37 | 52 | 0.0227 |
| Erythromycin (ER) | 42 | 66 | 0.0007 |
| Cefoxitin (FOX) | 71 | 94 | 0.0001 |
| Gentamicin (CN) | 28 | 83 | 0.0001 |
| Chloramphenicol (C) | 4 | 30 | 0.0001 |
| Sulfamethoxazole (SXT) | 49 | 58 | 0.1566 |
Phenotypes of Staphylococci on the basis of D-test
Out of a total of 176 staphylococcal isolates, 152 isolates were tested for inducible clindamycin resistance using D-test (Fig.1). Out of 152 isolates, MSB phenotype was observed in 27 (18%) isolates, cMLSB phenotype was observed in 114 isolates (75%) while 11 (7%) isolates showed iMLSB phenotype. Among these 11 isolates, three isolates were MRSA, one was MSSA and seven were MRCoNS. Overall, inducible clindamycin resistance in S. aureus was 4% and among CoNS it was 11%.
Fig.1.
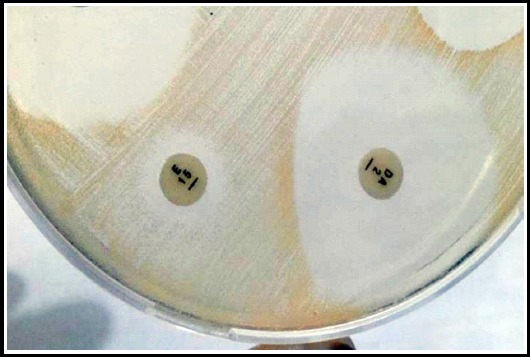
A representative Mueller Hinton agar plate showing positive D-test which is a D type zone around clindamycin (DA) disc.
Molecular detection of HVR, erm(C) and mecA gene
Out of 11 isolates that showed iMLSB phenotype, only two (18%) isolates (D-test positive) showed the presence of erm(C) gene (Fig.2C) and these were MRSA. Out of 62 MRSA, 50% were positive for mecA gene (Fig.2A) and 48% isolates were found negative for mecA gene. We also screened MSSA for mecA gene and all were found to be negative. Out of 31 mecA positive isolates, 68% were from male patients while 32% were from female patients. Out of MRSA isolates 26 (42%) were found to be positive for HVR (Fig.2B) whereas 36 (58%) isolates did not reveal the presence of HVR.
Fig.2.
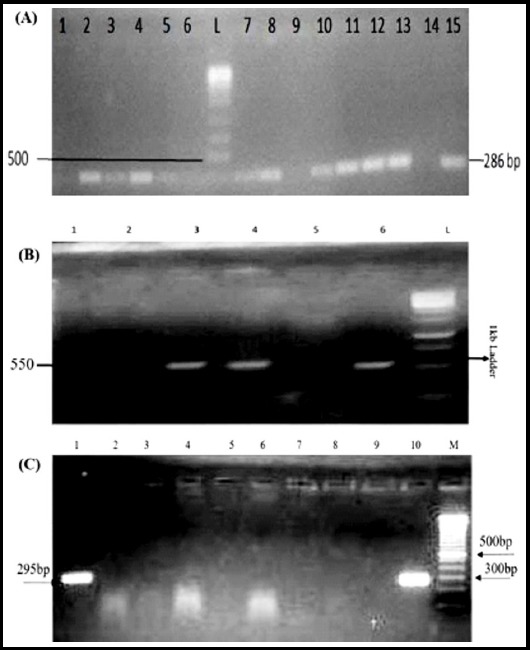
Representative gel images showing PCR products of mecA, HVR and erm(C) genes. (A) Lane L shows ladder of 1kb, Lanes 2-8, 10-13, and 15 show shows amplification of mecA gene (286bp) and Lane-1 shows negative control. (B) Lane L shows ladder of 1 kb, Lanes 3, 4 and 6 show amplification of HVR gene (550 bp) Lanes 1, 2 and 5 show negative results. (C) Lane M shows 100 bp ladder, Lanes 1 and 10 show amplification of erm(C) gene (295bp), Lanes 2-9 show negative results.
DISCUSSION
The prevalence of MRSA in this study was reported as 69% which is higher than previous reports from Pakistan where a range of 5% in 1989 and up to 52% in 2017 have been reported.14,15 This indicates a continuous increase in the circulation of this organism in clinical settings. Prevalence of methicillin resistance among CoNS in this study was 93% which is higher than previous reports of 70% oxacillin resistance in CoNS.16 The prevalence of inducible clindamycin resistance among all Staphylococci was observed to be 7% while among S. aureus isolates it was 4%. All of the CoNS were methicillin resistant. Clindamycin inducible resistance is difficult to be noticed in routine antibiotic susceptibility testing if clindamycin and erythromycin discs are not placed adjacent to each other. Clinicians end up prescribing clindamycin without knowledge of whether the particular strains of Staphylococci are positive for clindamycin inducible resistance or not. D-test is used to detect inducible resistance in which both antibiotic discs are placed adjacent to each other with the distance of 15-20mm. Inducible resistance is one of the factors of clindamycin therapeutic failure. So D-test must be performed in routine diagnostic laboratories for analysis of clinical Staphylococci.
One study from Peshawar has reported 16% inducible clindamycin resistance in MRSA isolates17 while a study from Karachi reported 72% inducible clindamycin resistance phenotype in S. aureus isolates.18 Prevalence of clindamycin inducible resistance in CoNS in this study was 11%. There is no data available on prevalence of clindamycin inducible resistance among CoNS from Pakistan while a study from India reported 7.56% inducible clindamycin resistance in CoNS and 16.4% in S. aureus.19 This shows that prevalence of clindamycin inducible resistance varies from region to region. In the present study, erm(C) gene was detected in only two isolates showing phenotypic inducible clindamycin resistance. In one study from Iran, three S. aureus isolates with positive D-test were negative for erm genes by PCR.20 Similar findings have been reported where 17%21 and 33%22 of S. aureus strains were negative for erm(A) and erm(C) genes. A positive D-test suggests the presence of an erm gene that could result in inducible clindamycin resistance and clinical failure. Absence of erm genes in positive D-test isolates suggests the role of other factors or genes.
In the current study, prevalence of mecA gene was reported as 50% which is slightly higher from the report of 48.1% from Peshawar.17 MRSA isolates subjected to HVR-PCR generated amplification products that ranged from 290bp to 650bp in size with 3 HVR types: HVR 2 (400bp), HVR 5 (550bp), HVR 6 (600bp). One study reported eight types of HVR which were HVR types 3, 5, 7, 8, 9, 10, 11, and 12 direct repeat units (DRUs).12 Another study from Iran reported HVR types 1-11.23 The differences in the HVR region could be due to the geographical variation, different samples size, and host genetic factor(s).
In the current study, vancomycin resistance in MRSA was recorded as 7% while in CoNS it was 20%. One study from Pakistan recorded high levels of vancomycin resistance in MRSA as 13% using E-test method.24 Reason for increase in rate of resistance to vancomycin could be inadequate use of vancomycin and transfer of vancomycin resistance genes (vanA-C) from other vancomycin resistant organisms such as Enterococcus. Particularly our clinicians should be concerned about these reports on vancomycin resistance so that they can prescribe vancomycin accordingly to avoid therapeutic failure. Resistance to tigecycline against S. aureus in the present study was observed to be 39% which is a concern as previous reports from Pakistan have reported 100% susceptibility to tigecycline.15,25 In the current study, resistance to linezolid was reported as 37% in S. aureus isolates while in CoNS it was 90%. Previous study conducted in Pakistan reported cfr gene in 78% of linezolid resistant S. aureus isolates24 which is higher than our results, although we observed high resistance of linezolid in CoNS. The increase in rate of resistance against linezolid is very alarming, particularly among CoNS which are developing resistance to other antibiotics as well.
CONCLUSION
This is the first report of clindamycin inducible resistance in coagulase negative Staphylococci from local strains in Islamabad. Increasing levels of resistance in coagulase negative Staphylococci is a point of concern for public health and the prescription of antibiotics in these cases should be given with caution. We also suggest the use of D-test in routine antibiotic susceptibility analysis in order to cater the inducible clindamycin resistance.
Author`s Contribution:
AAK: Performed the experiments and prepared draft of paper.
JF: Performed the experiments, analyzed data and helped in writing the manuscript draft.
MA: Performed experiments, analyzed data.
RZ: Conception and design of research, analysis interpretation of the data, approval of final version, is responsible for integrity of research.
Footnotes
Conflicts of interest: None.
Source of funding: Higher Education Commission, Pakistan.
REFERENCES
- 1.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45(5):1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. doi:10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger-Bachi B, Rohrer S. Factors influencing methicillin resistance in staphylococci. Arch Microbial. 2002;178(3):165–167. doi: 10.1007/s00203-002-0436-0. doi:10.1007/s00203-002-0436-0. [DOI] [PubMed] [Google Scholar]
- 3.Ryffel C, Bucher R, Kayser F, Berger-Bachi B. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol. 1991;173(23):7416–7422. doi: 10.1128/jb.173.23.7416-7422.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishi J, Miyanohara H, Nakajima T, Kitajima I, Yoshinaga M, Maruyama I, et al. Molecular typing of the methicillin resistance determinant (mec) of clinical strains of Staphylococcus based on mec hypervariable region length polymorphisms. J Lab Clin Med. 1995;126(1):29–35. doi:10.1016/j.lab.2006.01.003. [PubMed] [Google Scholar]
- 5.Nahvi MD, Fitzgibbon JE, John JF, Dubin DT. Sequence analysis of dru regions from methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococcal isolates. Microbial Drug Resistance. 2001;7(1):1–12. doi: 10.1089/107662901750152684. doi:10.1089/107662901750152684. [DOI] [PubMed] [Google Scholar]
- 6.Fiebelkorn K, Crawford S, McElmeel M, Jorgensen J. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41(10):4740–4744. doi: 10.1128/JCM.41.10.4740-4744.2003. doi:10.1128/JCM.41.10.4740-4744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westh H, Hougaard D, Vuust J, Rosdahl V. Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob Agents Chemother. 1995;39(2):369–373. doi: 10.1128/aac.39.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer A, Kirby W, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. doi:10.1093/ajcp/45.4_ts.493. [PubMed] [Google Scholar]
- 9.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. doi:10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 10.Lu JJ, Perng CL, Lee SY, Wan CC. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol. 2000;38(6):2076–2080. doi: 10.1128/jcm.38.6.2076-2080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA) FEBS Letters. 1992;298(2-3):133–136. doi: 10.1016/0014-5793(92)80039-j. doi:10.1016/0014-5793(92)80039-J. [DOI] [PubMed] [Google Scholar]
- 12.Senna J, Pinto C, Carvalho L, Santos D. Comparison of pulsed-field gel electrophoresis and PCR analysis of polymorphisms on the mec hypervariable region for typing methicillin-resistant Staphylococcus aureus. J Clin Microbio. 2002;40(6):2254–2256. doi: 10.1128/JCM.40.6.2254-2256.2002. doi:10.1128/JCM.40.6.2254-2256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen LB, Frimodt-Møller N, Aarestrup FM. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol Letters. 1999;170(1):151–158. doi: 10.1111/j.1574-6968.1999.tb13368.x. doi:10.1111/j.1574-6968.1999.tb13368.x. [DOI] [PubMed] [Google Scholar]
- 14.Siddiqui T, Muhammad IN, Khan MN, Naz S, Bashir L, Sarosh N, et al. MRSA:Prevalence and susceptibility pattern in health care setups of Karachi. Pak J Pharm Sci. 2017;30(Suppl 6):2417–2421. [PubMed] [Google Scholar]
- 15.Khan S, Rasheed F, Zahra R. Genetic Polymorphism of agr Locus and Antibiotic Resistance of Staphylococcus aureus at two hospitals in Pakistan. Pak J Med Sci. 2014;30(1):172–176. doi: 10.12669/pjms.301.4124. doi:10.12669/pjms.301.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehsan MM, Memon Z, Ismail MO, Fatima G. Identification and antibiotic susceptibility pattern of coagulase-negative staphylococciin various clinical specimens. Pak J Med Sci. 2013;29(6):1420–1424. doi: 10.12669/pjms.296.4064. doi:10.12669/pjms.296.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ullah A, Qasim M, Rahman H, Khan J, Haroon M, Muhammad N, Khan A, Muhammad N. High frequency of methicillin-resistant Staphylococcus aureus in Peshawar Region of Pakistan. Springerplus. 2016;5(1):600. doi: 10.1186/s40064-016-2277-3. doi:10.1186/s40064-016-2277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasih N, Irfan S, Zafar A, Khan E, Hasan R. Inducible clindamycin resistance due to expression of erm genes in Staphylococcus aureus:report from a tertiary care hospital Karachi, Pakistan. J Pak Med Assoc. 2010;60(9):750–753. [PubMed] [Google Scholar]
- 19.Baragundi Mahesh C, Kulkarni Ramakant B, Sataraddi Jagadeesh V. The prevalence of inducible and constitutive clindamycin resistance among the nasal isolates of staphylococci. J Clin Diagn Res. 2013;7(8):1620–1622. doi: 10.7860/JCDR/2013/6378.3223. doi:10.7860/JCDR/2013/6378.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moosavian M, Shoja S, Rostami S, Torabipour M, Farshadzadeh Z. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus due to erm genes, Iran. Iran J Microbiol. 2014;6(6):421–427. [PMC free article] [PubMed] [Google Scholar]
- 21.Aktas Z, Aridogan A, Kayacan CB, Aydin D. Resistance to macrolide, lincosamide and streptogramin antibiotics in staphylococciisolated in Istanbul, Turkey. J Microbiol. 2007;45(4):286–290. [PubMed] [Google Scholar]
- 22.Saderi H, Emadi B, Owlia P. Phenotypic and genotypic study of macrolide, lincosamide and streptogramin B (MLSB) resistance in clinical isolates of Staphylococcus aureus in Tehran, Iran. Med Sci Monit. 2011;17(2):BR48–BR53. doi: 10.12659/MSM.881386. doi:10.12659/MSM.881386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nia NZ, Pourmand MR, Afrough P. Comparison of hypervariable region (hvr) of meca gene in staphylococcus aureus isolated from nasal carriers and clinical samples. Jundishapur J Microbiol. 2013;6(9) doi:10.5812/jjm.7686. [Google Scholar]
- 24.Azhar A, Rasool S, Haque A, Shan S, Saeed M, Ehsan B, Haque A. Detection of high levels of resistance to linezolid and vancomycin in Staphylococcus aureus. J Med Microbiol. 2017;66(9):1328–1331. doi: 10.1099/jmm.0.000566. doi:10.1099/jmm.0.000566. [DOI] [PubMed] [Google Scholar]
- 25.Saleem F, Fasih N, Zafar A. Susceptibility pattern of methicillin resistant Staphylococcus aureus to vancomycin and other alternate agents:Report from a private sector hospital laboratory. J Pak Med Assoc. 2017;67(11):1743–1746. [PubMed] [Google Scholar]


