Significance
O-GlcNAc transferase (OGT) transfers the sugar O-GlcNAc to serine and threonine residues of target proteins, a process known as O-GlcNAcylation. Using Drosophila as an animal model, we identify the protein kinase Hipk as a target protein for OGT. Further biochemical assays and genetic interaction studies reveal that OGT plays critical roles in controlling Hipk protein stability and Hipk-induced tissue growth abnormalities. This regulation is likely operative in mammals as we find that human HIPK2 is less stable when it cannot be O-GlcNAc modified. HIPK2 is implicated in several cancers and chronic fibrosis. Our work therefore opens up the possibility of exploiting the regulatory role of OGT in the treatment of HIPK2-related disorders.
Keywords: OGT, Hipk, tumorigenesis, O-GlcNAc, Drosophila
Abstract
Environmental cues such as nutrients alter cellular behaviors by acting on a wide array of molecular sensors inside cells. Of emerging interest is the link observed between effects of dietary sugars on cancer proliferation. Here, we identify the requirements of hexosamine biosynthetic pathway (HBP) and O-GlcNAc transferase (OGT) for Drosophila homeodomain-interacting protein kinase (Hipk)-induced growth abnormalities in response to a high sugar diet. On a normal diet, OGT is both necessary and sufficient for inducing Hipk-mediated tumor-like growth. We further show that OGT maintains Hipk protein stability by blocking its proteasomal degradation and that Hipk is O-GlcNAcylated by OGT. In mammalian cells, human HIPK2 proteins accumulate posttranscriptionally upon OGT overexpression. Mass spectrometry analyses reveal that HIPK2 is at least O-GlcNAc modified at S852, T1009, and S1147 residues. Mutations of these residues reduce HIPK2 O-GlcNAcylation and stability. Together, our data demonstrate a conserved role of OGT in positively regulating the protein stability of HIPKs (fly Hipk and human HIPK2), which likely permits the nutritional responsiveness of HIPKs.
Nutrients such as glucose, amino acids, and fatty acids are metabolic fuels that provide energy to cells (1). They also function as signaling molecules in various nutrient signaling pathways (1), allowing coordination between nutrient sensing and cellular behaviors like cell growth (2). One emerging nutrient signaling pathway is the hexosamine biosynthetic pathway (HBP) (3). In the HBP, glutamine:fructose-6-phosphate amidotransferases (GFATs) catalyze the first and rate-limiting step that converts glucose-derived fructose-6-phosphate into glucosamine-6-phosphate (3) (Fig. 1A). The end product of the HBP is UDP-GlcNAc, the donor nucleotide sugar substrate for various glycosyltransferases, including nucleocytoplasmic O-GlcNAc transferase (OGT) (4). OGT is the only known enzyme that catalyzes the addition of O-GlcNAc to serine and threonine residues of hundreds of target proteins in a UDP-GlcNAc–dependent manner (5). Notably, O-GlcNAcase (OGA) removes O-GlcNAc from proteins, making O-GlcNAc a dynamic and reversible posttranslational modification (PTM) (6). In this way, cellular UDP-GlcNAc levels and global O-GlcNAcylation are coordinated and are highly responsive to glucose availability, making O-GlcNAc well suited to serve as a nutrient-sensing mechanism (7, 8).
Fig. 1.
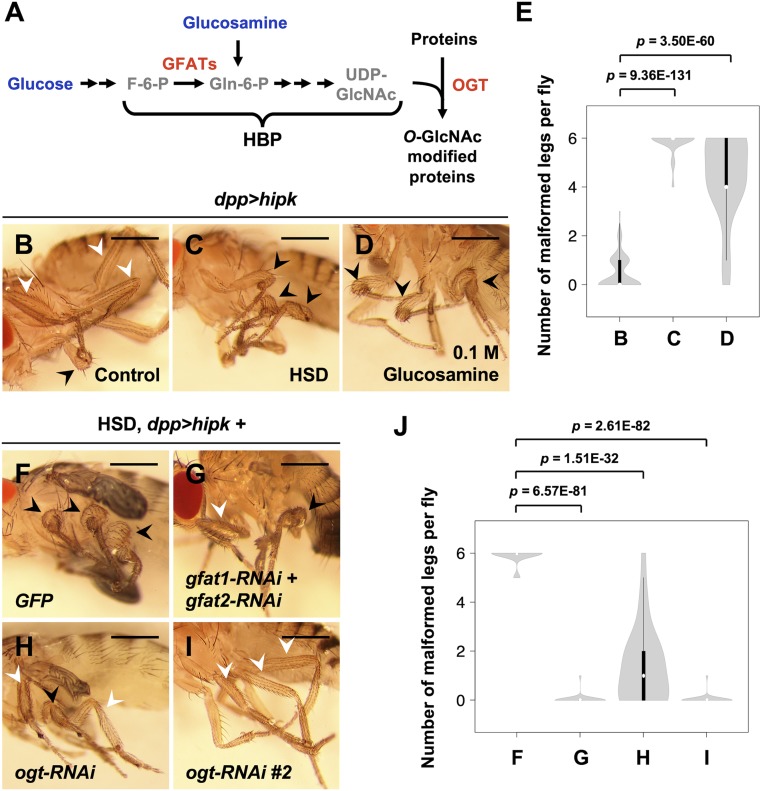
High dietary sugar potentiates Hipk-induced growth abnormalities through HBP and OGT. (A) Schematic diagram of the HBP. Dietary glucose and glucosamine are shown in blue, metabolites in gray, enzymes in red. (B–D) Adult legs of hipk-overexpressing animals (dpp > hipk) which were fed a control diet (0.15 M sucrose) (B), a HSD (1 M sucrose) (C), or a glucosamine-supplemented diet (D) from embryo hatch. White arrowheads mark the WT-like, slender leg phenotype. Black arrowheads mark the mild leg malformation phenotype. (E) A violin plot showing the malformed leg phenotype of the dpp > hipk flies raised on different diets. The letters B–D refer to the flies shown in Fig. 1 B–D. Numbers of flies counted N: (B) 173, (C) 52, and (D) 37. P values were calculated using unpaired two-tailed Student’s t test. (F–I) Adult legs of dpp > hipk + GFP (F), dpp > hipk + gfat1-RNAi + gfat2-RNAi (G), dpp > hipk + ogt-RNAi (H), and dpp > hipk + ogt-RNAi #2 (I) flies which were fed an HSD from embryo hatch. Black arrowheads mark the disfigured legs. White arrowheads mark the WT-like legs. (J) A violin plot showing the malformed leg phenotype of the indicated animals fed a HSD. The letters F–I refer to the flies shown in Fig. 1 F–I. Numbers of flies counted N: (F) 53, (G) 28, (H) 25, and (I) 29. P values were calculated using unpaired two-tailed Student’s t test. Flies were raised at 25 °C with the indicated diets. (Scale bars, 500 μm.)
Pathologically, O-GlcNAc has been implicated in cancers. For instance, hyper–O-GlcNAcylation, high levels of OGT or GFAT2, or low OGA levels are positively correlated with poor prognosis of patients with prostate or breast cancers (9, 10), aggressiveness of bladder tumors (11), or tumor recurrence of liver cancer (12). Also, many oncoproteins and tumor suppressors such as c-MYC (13), p53 (14), and YAP (15, 16) are O-GlcNAc modified. These observations link O-GlcNAc with multiple hallmarks of cancer, including sustaining proliferative signals and deregulating cellular energetics. Cancer cells are usually metabolically active. In particular they often sustain high rates of glucose uptake, a phenomenon commonly known as the Warburg effect, which is a preferential reliance on aerobic glycolysis to obtain energy (17). How elevated uptake of glucose triggers O-GlcNAc modification of cancer-related proteins and controls their activities is accordingly a topic of growing interest.
Homeodomain-interacting protein kinases (HIPKs; HIPK1–4 in mammals and Hipk in Drosophila [fruit fly]) are protein kinases involved in the regulation of signal transduction (18–21), cell proliferation and differentiation (22, 23), apoptosis (24), stress response (25), embryonic development (26), angiogenesis (27), adipogenesis (28), as well as immune homeostasis (29). The activity of HIPK2 (the most studied member of the mammalian HIPK family) is governed by multiple strategies depending on the environments. HIPK2 is normally maintained at low levels by proteasomal degradation involving various ubiquitin E3 ligases such as Siah1 and Siah2 (30, 31). During hypoxia, HIPK2 degradation is facilitated by the increased association between HIPK2 and Siah2 (31). DNA damage, on the other hand, disrupts the HIPK2–Siah1 interaction, protecting HIPK2 from degradation (30). Furthermore, high oxidative stresses modulate HIPK2 SUMOylation and acetylation states, which influence HIPK2 localization (32). Given the capacity for HIPK2 to respond to environmental cues and the growing recognition that nutrient sensing through OGT is a key regulator of cellular homeostasis, we were intrigued by the potential for O-GlcNAc modification to serve as a nutritional regulator of HIPK(s).
Our previous work has established that overexpression of Hipk in Drosophila promotes tissue growth abnormalities and several tumor-like features, including metabolic reprogramming, cell invasion-like behaviors, and cellular changes reminiscent of epithelial-to-mesenchymal transition, including up-regulation of Twist and Matrix metalloproteinase 1 (MMP1) and down-regulation of E-cadherin (19, 33, 34). Using this in vivo model, we show that OGT is not only necessary for Hipk-mediated tissue growth abnormalities, but also sufficient to synergize with mild levels of Hipk to produce tumor-like phenotypes. Furthermore, we find that HIPKs (both fly Hipk and human HIPK2) are O-GlcNAc-modified proteins. Elevated OGT results in the buildup of HIPK proteins in a posttranscriptional manner. In particular, we identify that HIPK2 O-GlcNAcylation at residues S852, T1009, and S1147 are responsible for OGT-mediated stabilization.
Results
High Dietary Sugar Potentiates Hipk-Induced Growth Abnormalities through HBP and OGT.
In Drosophila, Hipk promotes tissue growth in a dose-dependent manner (19). Using the Gal4-UAS system (35) in flies grown at different temperatures, which affect the extent of Gal4-driven transgene expression, we can generate a range of Hipk overexpression phenotypes that can be used to examine genetic and environmental interactions with Hipk (SI Appendix, Fig. S1). Using the dpp-Gal4 driver to induce UAS-hipk expression in a subset of cells in larval leg imaginal discs that give rise to adult leg appendages, we observed distinct effects at 25 °C or 29 °C. Compared with control flies (SI Appendix, Fig. S1A), flies overexpressing hipk at 25 °C (full genotype: dpp-Gal4 > UAS-hipk, abbreviated as dpp > hipk) usually displayed a wild-type (WT) slender leg phenotype (Fig. 1B, white arrowheads and SI Appendix, Fig. S1B) and rarely had one or two mildly disfigured legs (Fig. 1B, black arrowhead, Fig. 1E, and SI Appendix, Fig. S1B). We define this phenotype as mild leg malformation (SI Appendix, Fig. S1 G, Middle). In contrast, the same genotype grown at 29 °C led to a severe leg malformation phenotype (SI Appendix, Fig. S1 C and G, Right) characterized by significant distortion of pharate adult legs and loss of segmentation. Because of such severe growth abnormalities, these flies usually failed to eclose from their pupal cases. The same phenotype was seen when overexpressing two copies of Hipk at 25 °C (SI Appendix, Fig. S1D), which is associated with the previously reported cell hyperproliferation phenotypes observed in two other larval imaginal discs, wing and eye-antennal discs (33).
The Hipk phenotype seen at 25 °C indicates that when hipk-overexpressing flies were raised and fed a normal diet (0.15 M sucrose) (Fig. 1B), hipk expression levels remained close to, yet still below, the threshold required to cause significant growth abnormalities. We took advantage of the sensitized background of the mild Hipk overexpression phenotype to interrogate whether dietary sugar alters Hipk function in controlling tissue growth. Strikingly, we found that a high sucrose diet (HSD; 1 M sucrose) enhanced the abnormal leg phenotype in hipk-overexpressing flies such that nearly all legs were malformed (Fig. 1C, black arrowheads and Fig. 1E). Similarly, we observed that eye-antennal discs from HSD-fed hipk-overexpressing larvae showed an enlarged Decapentaplegic (Dpp) domain (marked by Hipk immunoreactivity) (SI Appendix, Fig. S2 C and D), indicative of augmented Hipk activities and enhanced cell proliferation.
To test if dietary sugar might modulate Hipk tissue growth through flux through the HBP, we added glucosamine to the fly media. Glucosamine bypasses the rate-limiting step governed by GFAT enzymes and directly enters the HBP (Fig. 1A) (3). We found that supplementing the normal diet (0.15 M sucrose) with glucosamine (0.1 M glucosamine) also potentiated the mild leg malformation phenotype in hipk-overexpressing flies (Fig. 1D black arrowheads, Fig. 1E) in a similar manner as feeding with a high sucrose diet. Under either feeding conditions, flies without overexpression of hipk did not show any malformed leg phenotypes (SI Appendix, Fig. S2 E–I) or increased cell proliferation (SI Appendix, Fig. S2 A and B).
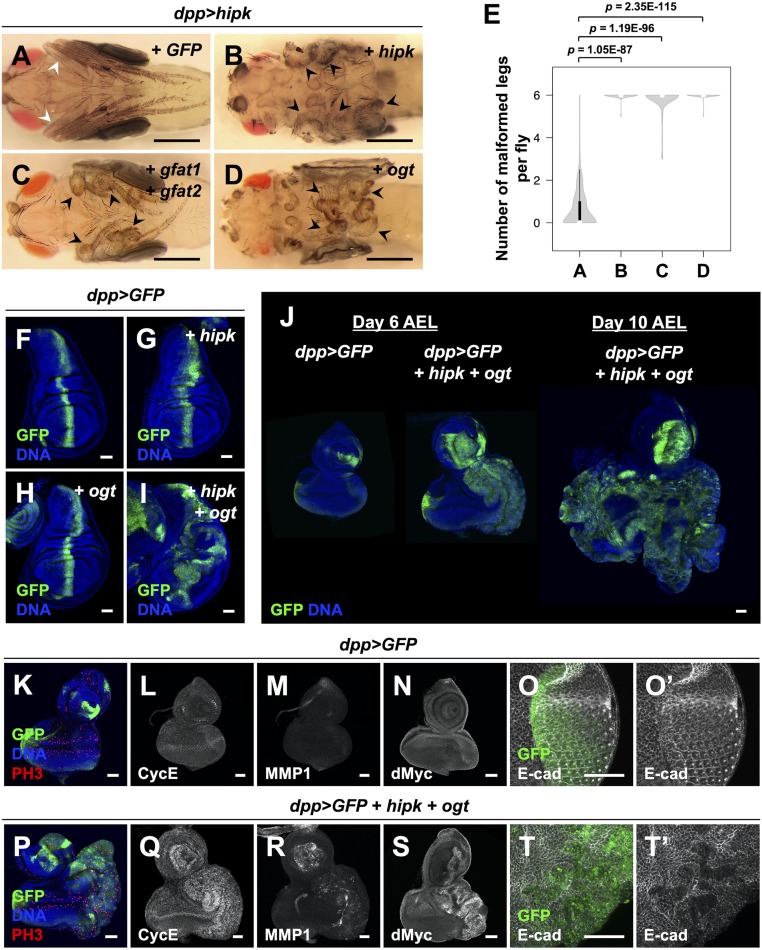
Next, we used RNA interference (RNAi) to knock down the rate-limiting enzymes in the HBP, GFATs, which are encoded in flies by gfat1 and gfat2. Notably, simultaneous knockdown of gfat1 and gfat2 suppressed the effect of high sugar on Hipk-induced malformed leg phenotypes (Fig. 1 F, G, and J). To examine if OGT downstream of the HBP is required for the stimulatory effect of high sucrose on Hipk-mediated tissue growth phenotypes, we investigated the effects of genetic inhibition of OGT. Using two independent RNAi fly lines, we found that knockdown of ogt (also known as super sex combs (sxc) in flies (36, 37)) partly rescued the leg phenotype seen after high sucrose feeding (Fig. 1 H–J). We confirmed the knockdown efficiency of the RNAi lines used in this study by qRT-PCR, Western blotting, and/or immunofluorescence (IF) analyses (SI Appendix, Fig. S3). Further supporting a link to the HBP and OGT, we also found that simultaneous overexpression of gfat1 and gfat2 (Fig. 2 C and E) or overexpression of ogt in a dpp > hipk genetic background (Fig. 2 D and E) was sufficient to produce leg malformations reminiscent of Hipk hyperactivation (Fig. 2 B and E) even under normal dietary conditions. Together, using the leg malformation phenotypes as a readout for Hipk activity, our genetic interaction studies indicate that dietary sugar promotes Hipk activity at least in part through the HBP and OGT.
Fig. 2.
OGT synergizes with Hipk to drive tumor-like tissue growth. (A–D) Adult legs of dpp > hipk + GFP (A), dpp > hipk + hipk (B), dpp > hipk + gfat1 + gfat2 (C), and dpp > hipk + ogt (D) flies. White arrowheads mark the WT-like slender leg phenotype. Black arrowheads mark the malformed legs. (Scale bars, 500 μm.) (E) A violin plot showing the malformed leg phenotype of the indicated animals. The letters A–D refer to the flies shown in Fig. 2 A–D. Numbers of flies counted N: (A) 141, (B) 47, (C) 64, and (D) 76. P values shown were calculated using unpaired two-tailed Student’s t test. (F–I) Wing discs of dpp > GFP (F), dpp > GFP + hipk (G), dpp > GFP + ogt (H), and dpp > GFP + hipk + ogt (I) larvae. GFP (green) marks the transgene-expressing cells. (J) Eye-antennal discs of dpp > GFP (Left) and dpp > GFP + hipk + ogt (Middle) larvae dissected 6 d after egg laying (AEL). dpp > GFP + hipk + ogt animals experienced an extended larval phase, and their eye disc (dissected 10 d AEL) is shown (Right). GFP (green) marks the transgene-expressing cells. (K–O) Control eye discs (dpp > GFP) stained for PH3 (red in K), CycE (gray in L), MMP1 (gray in M), dMyc (gray in N), and E-cad (gray in O and O′). (P–T) hipk- and ogt-coexpressing eye discs (dpp > GFP + hipk + ogt) stained for PH3 (red in P), CycE (gray in Q), MMP1 (gray in R), dMyc (gray in S), and E-cad (gray in T and T′). DAPI staining for DNA (blue) reveals tissue morphology. Flies were raised at 25 °C with control diet. (Scale bars, 50 μm.)
OGT Synergizes with Hipk to Drive Tumor-Like Tissue Growth.
As stated in the introduction, we previously established an in vivo fly tumor-like model with elevation of hipk (overexpression of two copies of hipk) using wing imaginal discs in larvae raised at 25 °C (33). Given the genetic interactions between Hipk and OGT observed in the adult legs (Figs. 1 H and I and 2D), we next asked whether OGTs play a role in Hipk-mediated tissue growth effects in larval wing and eye discs.
We found that in larval wing imaginal discs at 25 °C, overexpression of either hipk (single copy) (Fig. 2G) or ogt (Fig. 2H) on their own did not cause any abnormal phenotypes. Coexpression of ogt with hipk, however, resulted in profound tissue overgrowth (Fig. 2I), suggesting that they drive tumor-like growth in a synergistic manner. Coexpression of hipk and ogt also resulted in a delay in timing of pupariation (SI Appendix, Fig. S4A), a phenotype commonly seen when tumor burden is present in larvae (38). A comparable synergy was also seen in larval eye-antennal discs (Fig. 2J and SI Appendix, Fig. S4 B–E) and, in the developmentally delayed larvae, hipk and ogt coexpressing tissues grew in size continuously (Fig. 2J). Furthermore, we found that the cells coexpressing hipk and ogt were characterized by an enrichment in phosphohistone H3 (PH3, a mitotic marker), up-regulation of cyclin E (CycE), matrix metalloproteinase 1 (MMP1), and dMyc (MYC in vertebrates), as well as loss of E-cadherin (E-cad) (Fig. 2 K–T and SI Appendix, Fig. S4 B–E), which are all established markers associated with extensive cancerous growth. These data indicate that OGT is capable of augmenting the tumorigenic potential of Hipk even though hipk is mildly overexpressed in this system.
OGT Is Necessary for Hipk-Induced Tissue Growth Abnormalities.
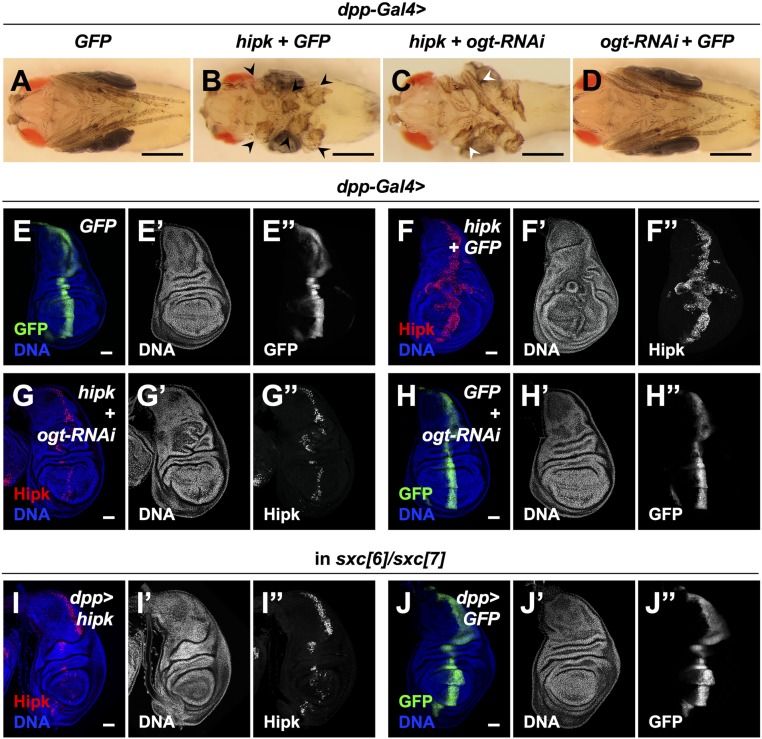
Next, we set out to evaluate the requirement of OGT for Hipk activity using the leg malformation phenotypes. We shifted the fly culture temperature from 25 °C to 29 °C such that Gal4 activity was increased to drive greater Hipk expression to reach a level sufficient to cause severe growth abnormalities (Fig. 3 A and B black arrowheads, SI Appendix, Figs. S1C and S5 A and B). Coexpression of ogt-RNAi with hipk partially rescued the severe leg malformation phenotype caused by excess Hipk as the leg segmentation was partly restored (Fig. 3C white arrowheads, SI Appendix, Fig. S5 C, E, and F). Expression of ogt-RNAi alone did not cause any abnormal leg phenotypes (Fig. 3D and SI Appendix, Fig. S5D).
Fig. 3.
OGT is necessary for Hipk-induced tissue growth abnormalities. (A–D) Adult legs of dpp > GFP (A), dpp > hipk + GFP (B), dpp > hipk + ogt-RNAi (C), and dpp > ogt-RNAi + GFP (D) flies. Black arrowheads in B mark the legs with severe malformation. White arrowheads in C mark the legs with mild malformation. See the side views and quantifications in SI Appendix, Fig. S5. (Scale bars, 500 μm.) (E–H) Wing discs of dpp > GFP (E), dpp > hipk + GFP (F), dpp > hipk + ogt-RNAi (G), and dpp > ogt-RNAi + GFP (H) larvae. The transgene-expressing domain was marked by GFP (green in E and H; gray in E″ and H″) or Hipk staining (red in F and G; gray in F″ and G″). (I and J) In an OGT mutant background (sxc[6]/sxc[7]), wing discs of dpp > hipk (I) and dpp> GFP (J) larvae are shown. The transgene-expressing domain is marked by Hipk staining (red in I; gray in I″) or GFP (green in J; gray in J″). Flies were raised at 29 °C with control diet. DAPI staining for DNA (blue; gray in E′–J′) reveals tissue morphology. (Scale bars, 50 μm.)
We then examined the requirement of OGT for Hipk-induced tumor-like tissue growth in wing discs. Compared with the control wing discs (Fig. 3E and SI Appendix, Fig. S5F), elevated Hipk (overexpression of one copy of hipk at 29 °C) induced significant abnormal overgrowth (Fig. 3F and SI Appendix, Fig. S5H). When ogt was knocked down in hipk-overexpressing cells, the overgrowth induced by elevated Hipk was largely suppressed, though mild tissue distortions remained (Fig. 3G and SI Appendix, Fig. S5I). We also used ogt mutant alleles, sxc[6] and sxc[7], which possess a point mutation at a splice acceptor site and a nonsense mutation, respectively (36, 37), to generate an ogt null genetic background. Similar to the effects of ogt knockdown, the proliferative potential of the cells with elevated Hipk was largely reduced in the ogt mutant background (Fig. 3I). Altogether, our data illustrate a requirement for OGT in Hipk-induced tissue growth abnormalities, and the requirement is general to all tissues examined.
Depletion of OGT Reduces Hipk Protein Levels.
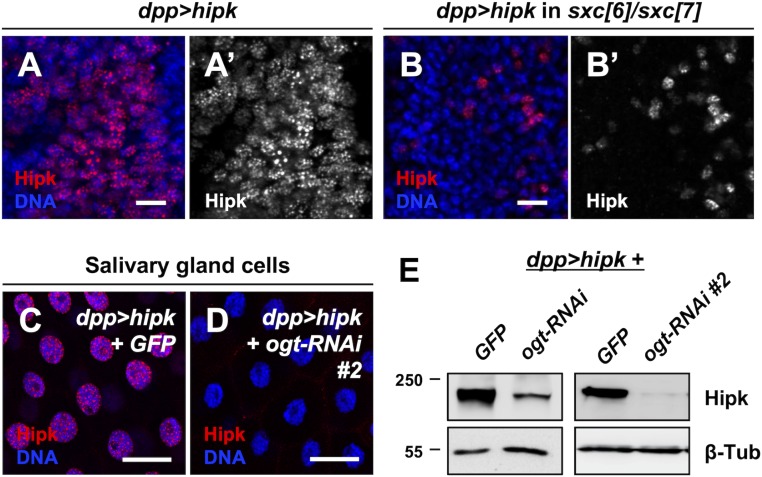
Given these results, we considered mechanisms by which OGT might influence Hipk activity and found, using immunofluorescence staining experiments, that loss of OGT caused a robust reduction of exogenous Hipk immunoreactivity (Fig. 3 F, G, and I). Compared to a control genetic background (Fig. 4A), fewer hipk-expressing cells were detected and Hipk expression levels were lower in the ogt null background (sxc[6]/sxc[7]) (Fig. 4B) or when ogt was knocked down (SI Appendix, Fig. S6A). Hipk was predominantly nuclear regardless of OGT levels (Fig. 4 A and B and SI Appendix, Fig. S6), indicating that OGT is not involved in the regulation of Hipk subcellular localization. A comparable result was obtained using additional ogt mutant alleles, sxc[4] and sxc[5] (SI Appendix, Fig. S6 B and C), which encode catalytically inactive forms of OGT (OGT-N948I and OGT-Δ1031 to 1059, respectively (36, 37)). We also examined another larval epithelial tissue, salivary glands, because their large cell size facilitates imaging. Depletion of ogt in salivary gland cells caused drastic reductions in Hipk protein levels (Fig. 4 C and D). Using immunoblotting, we confirmed that loss of OGT led to down-regulation of exogenous Hipk protein levels (Fig. 4E and SI Appendix, Fig. S6D). We noted that ogt depletion did not cause loss of dpp-Gal4 expression (Fig. 3 H and J). Thus, ogt depletion does not reduce Hipk protein levels by repressing UAS-hipk transcription. Also, reductions in OGT levels had little effects on the transcriptional levels of endogenous hipk (SI Appendix, Fig. S3B). Taken together, our data imply that the catalytic activity of OGT is required to sustain Hipk protein levels posttranscriptionally within Drosophila.
Fig. 4.
Depletion of OGT reduces Hipk protein levels. (A and B) Wing discs overexpressing hipk (dpp > hipk) in a WT background (A) or in an OGT mutant background (sxc[6]/sxc[7]) were stained for Hipk using anti-Hipk antibodies (red in A and B; gray in A′ and B′). DAPI was used to stain DNA (blue). (Scale bars, 10 μm.) (C and D) hipk-overexpressing salivary glands without (dpp > hipk + GFP) (C) or with OGT knockdown (dpp > hipk + ogt-RNAi #2) (D) were stained for Hipk using anti-Hipk antibodies (red). DAPI was used to stain DNA (blue). (Scale bars, 50 μm.) (E) Western blot analyses of the exogenous Hipk protein levels in the larval head extracts (which included imaginal discs and salivary glands) obtained from dpp > hipk + GFP, dpp > hipk + ogt-RNAi and dpp > hipk + ogt-RNAi #2 animals. β-Tubulin was used as a loading control. Flies were raised at 29 °C with control diet.
OGT Stabilizes Hipk by Preventing Hipk from Proteasomal Degradation.
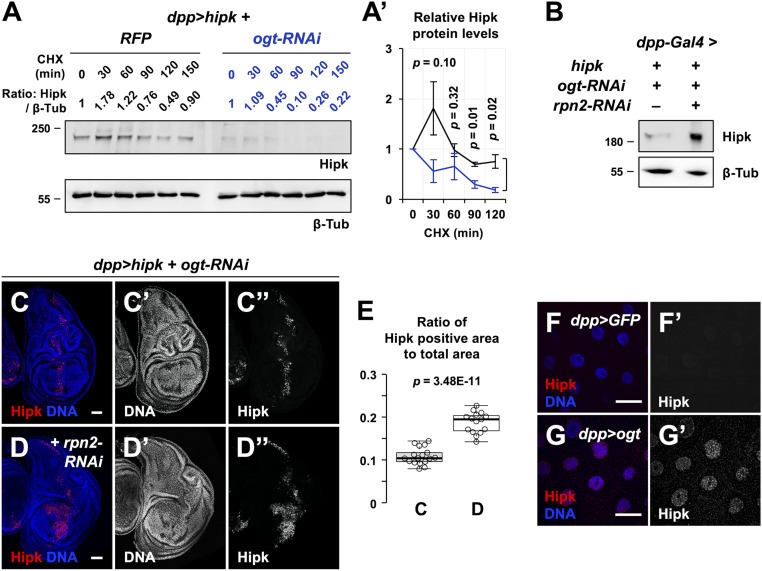
Given the dependence of Hipk protein levels on catalytically active OGT, we asked whether OGT controls Hipk protein stability. Using a cycloheximide (CHX) chase assay to observe protein stability in the absence of translation, we found that while Hipk levels dropped slightly in control cells, Hipk levels dropped precipitously when ogt was knocked down (Fig. 5 A and A′). To impair proteasomal degradation, we knocked down rpn2, a component of the 26S proteasome (39), and this restored Hipk protein levels (Fig. 5B) as well as the tumor-like growth in the ogt-depleted background (Fig. 5 C–E). Endogenous Hipk proteins were scarce and hardly detectable (Fig. 5F and SI Appendix, Fig. S7B). However, overexpression of ogt was sufficient to induce accumulation of endogenous Hipk proteins (Fig. 5G and SI Appendix, Fig. S7 C and D). Together, our results suggest that OGT activity positively regulates Hipk protein stability by reducing proteasomal degradation of Hipk.
Fig. 5.
OGT stabilizes Hipk by preventing Hipk from proteasomal degradation. (A and A′) Dissected larval heads from dpp > hipk + RFP and dpp > hipk + ogt-RNAi larvae were cultured in insect medium and exposed to 200 μg/mL CHX for the indicated times, followed by Western blot analyses of the exogenous Hipk protein levels. β-Tubulin was used as a loading control. Numeric values above the Hipk blot show the relative Hipk protein levels, which are set as 1 at 0 min of CHX treatment. (A′) Quantification of the relative Hipk protein levels of dpp > hipk + RFP (black line) and dpp > hipk + ogt-RNAi (blue line) larval head extracts in the CHX chase assay. Data are mean ± SEM and pooled from three biological replicates. At each time point, the P value shown is calculated using unpaired two-tailed Student’s t test. (B) Western blot analyses of the exogenous Hipk protein levels in the larval heads extracted from dpp > hipk + ogt-RNAi and dpp > hipk + ogt-RNAi + rpn2-RNAi animals. β-Tubulin was used as a loading control. (C and D) Wing discs of dpp > hipk + ogt-RNAi (C) and dpp > hipk + ogt-RNAi + rpn2-RNAi (D) larvae are shown. The transgene-expressing domain is marked by Hipk staining (red; gray in C″ and D″). DAPI staining for DNA (blue; gray in C′ and D′) reveals tissue morphology. (Scale bars, 50 μm.) (E) A box and whisker plot showing the ratio of Hipk-positive area to total area of the dpp > hipk + ogt-RNAi wing discs without or with rpn2 knockdown. The letters C and D refer to the fly genotypes shown in Fig. 5 C and D. P value shown is calculated using unpaired two-tailed Student’s t test. (F and G) Control (dpp > GFP) (F) and OGT-overexpressing (dpp > ogt) (G) salivary glands were stained for Hipk (red; gray in F′ and G′). DAPI was used to stain DNA (blue). (Scale bars, 50 μm.) Flies were raised at 29 °C with control diet.
OGT Binds to and O-GlcNAcylates Hipk.
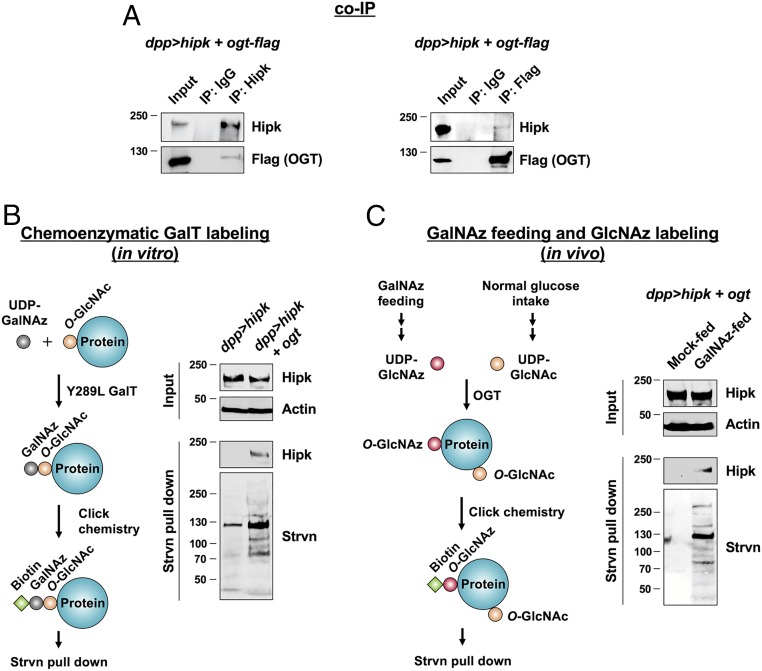
To test if Hipk and OGT physically interact with each other, we performed coimmunoprecipitation (co-IP) experiments. Using protein extracts from larval tissues expressing both proteins (dpp > hipk + ogt-flag), we found that Hipk and OGT could be reciprocally coimmunoprecipitated (Fig. 6A). A physical interaction suggested to us that Hipk may be an OGT substrate. To test whether Hipk is O-GlcNAc modified, we employed a chemoenzymatic approach (40) (Fig. 6B). Using a mutant β-1,4-galactosyltransferase (Y289L GalT), we labeled O-GlcNAc-modified proteins with N-azidoacetylgalactosamine (GalNAz). GalNAz-labeled proteins were then biotinylated using click chemistry in combination with a biotin-alkyne probe and precipitated using streptavidin resin. Subsequent immunoblot analyses of the precipitated proteins using antibodies against Hipk revealed marked Hipk O-GlcNAcylation when ogt was coexpressed (Fig. 6B). We validated the selectivity of the ligation chemistry during labeling by showing Hipk was undetectable in the precipitates when GalT was excluded from the chemoenzymatic labeling step (SI Appendix, Fig. S8).
Fig. 6.
OGT binds to and O-GlcNAcylates Hipk. Flies were raised at 25 °C with control diet (A and B) or the indicated diets (C). (A) Protein lysates extracted from dpp > hipk + ogt-flag larval heads were incubated with either IgG or anti-Hipk antibodies, followed by co-IP. Western blot analyses using anti-Hipk and anti-Flag antibodies showing that flag-tagged OGT coimmunoprecipitated with Hipk proteins (Left). Reciprocal IP showing that Hipk coimmunoprecipitated with flag-tagged OGT (Right). (B) Detection of O-GlcNAc modified Hipk by chemoenzymatic GalT labeling (schematic diagram shown on the Left). Protein lysates extracted from dpp > hipk and dpp> hipk + ogt larval heads were incubated with UDP-GalNAz and the enzyme Y289L GalT, followed by coupling with a biotin-alkyne probe by click chemistry, streptavidin (Strvn) pull-down, and Western blot analyses. Actin was used as a loading control. (C) Detection of O-GlcNAc–modified Hipk by GalNAz feeding and GlcNAz labeling (schematic diagram shown on the Left). Click chemistry was performed in protein lysates extracted from mock-fed or GalNAz-fed dpp> hipk + ogt larvae, followed by streptavidin (Strvn) pull-down and Western blot analyses. Actin was used as a loading control.
To corroborate the GalT labeling result, we adopted a metabolic feeding strategy (Fig. 6C) (41). Larvae coexpressing hipk and ogt were raised on food supplemented with either DMSO (control) or Ac4GalNAz. After feeding, the precursor Ac4GalNAz is converted in vivo through metabolism into UDP-GlcNAz (42). As UDP-GlcNAz is a close analog of UDP-GlcNAc, OGT can use this substrate to transfer GlcNAz onto proteins, to generate O-GlcNAz (42, 43). O-GlcNAz–modified proteins in the larval extracts were then chemoselectively coupled with a biotin-alkyne probe. Streptavidin pull-down followed by immunoblot analyses showed that Hipk was only present among the proteins precipitated from GalNAz-fed animals (Fig. 6C), further supporting Hipk being a substrate for OGT and being O-GlcNAc modified in vivo. Thus, our biochemical analyses confirm that fly Hipk is an O-GlcNAc–modified protein.
HIPK2 O-GlcNAcylation by OGT Promotes HIPK2 Protein Stability.
Our observations made using Drosophila indicate that Hipk is a direct target protein for OGT and that OGT is required to modulate Hipk protein stability and Hipk-induced tissue growth abnormalities. To see whether the regulation of Hipk by OGT is conserved across species, we extended our analyses to human HIPK2 using cultured mammalian cells.
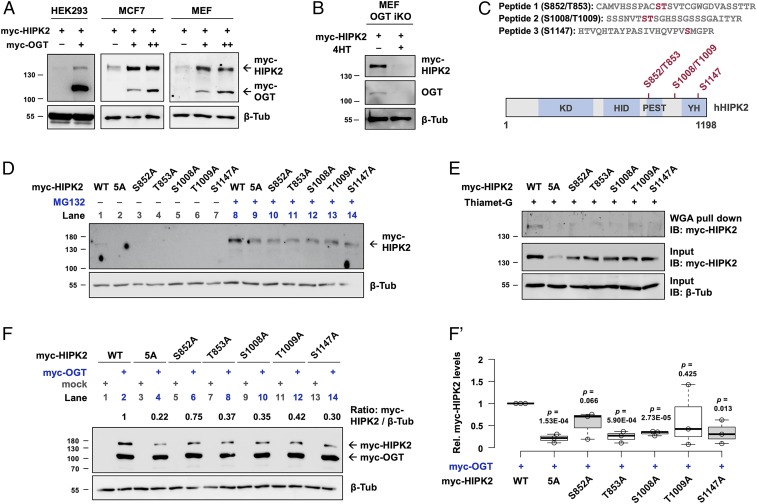
We transfected human embryonic kidney (HEK) 293 cells with OGT and found accumulation of exogenous HIPK2 proteins (Fig. 7A). Comparable results were obtained in MCF7 breast adenocarcinoma cells and mouse embryonic fibroblasts (MEFs). Next, using an MEF cell line where OGT knockout can be induced by administration of 4-hydroxytamoxifen (4HT) (44), we found that loss of OGT correlated with loss of HIPK2 (Fig. 7B). Notably, no statistically significant changes in endogenous HIPK2 mRNA levels were found in HEK293 and MEF cells upon genetic manipulation of OGT levels (SI Appendix, Fig. S9 A and C). In MCF7 cells, however, OGT induced HIPK2 transcriptional up-regulation (SI Appendix, Fig. S9B). Together, our data suggest that, whereas OGT generally promotes HIPK2 protein buildup posttranscriptionally, the transcriptional response of HIPK2 to OGT seems to be cell-type specific.
Fig. 7.
HIPK2 O-GlcNAcylation by OGT promotes HIPK2 protein stability. (A) Increasing amounts of myc-tagged OGT were cotransfected with myc-tagged HIPK2 in HEK293, MCF7, and MEF cell lines. Forty-eight hours after transfection, protein levels of myc-tagged HIPK2 and OGT were analyzed by Western blotting using anti-myc antibodies. β-Tubulin was used as a loading control. (B) Myc-tagged HIPK2 was transfected in mock (methanol)-treated or 4-hydroxytamoxifen (4HT)-treated MEF cells (an OGT inducible knockout [iKO] strain). Forty-eight hours after transfection, protein levels of myc-tagged HIPK2 were analyzed by Western blotting using anti-myc antibodies. OGT knockout efficiency was evaluated by examining the endogenous OGT levels. β-Tubulin was used as a loading control. (C) Mass spectrometry analyses revealed three O-GlcNAc–modified peptides of HIPK2. Schematic diagram of the human HIPK2 protein (amino acids 1 to 1,198) with its domains. KD, kinase domain; HID, homeobox-interacting domain; PEST, proline, glutamic acid, serine, and threonine-rich domain; YH, tyrosine, histidine-rich domain (also known as auto-inhibitory domain [AID]). (D) WT or alanine mutant variants of myc-tagged HIPK2 were transfected in HEK293 cells. Twenty-four hours after transfection, indicated cells were treated with MG132. Forty-eight hours after transfection, protein levels of myc-tagged HIPK2 were analyzed by Western blotting using anti-myc antibodies. β-Tubulin was used as a loading control. (E) Detection of O-GlcNAc levels of HIPK2. HEK293 cells were transfected with WT or alanine mutant variants of myc-tagged HIPK2 and treated with Thiamet-G. O-GlcNAc–modified proteins were pulled down by WGA-conjugated beads. Myc-tagged HIPK2 in the precipitates and in the input were analyzed by Western blotting. β-Tubulin was used as a loading control. (F and F′) WT or alanine mutant variants of myc-tagged HIPK2 were cotransfected with mock or myc-tagged OGT as indicated. Twenty-eight hours after transfection, protein levels of myc-tagged HIPK2 and OGT were analyzed by Western blotting using anti-myc antibodies. β-Tubulin was used as a loading control. (F′) A box and whisker plot showing the relative protein levels of the WT or alanine mutant variants of myc-tagged HIPK2 in the presence of myc-OGT cotransfection. Data are pooled from three biological replicates. P values showing the significant differences between the WT and the mutant protein levels were calculated using unpaired two-tailed Student’s t test.
We next mapped the positions of O-GlcNAc modification on HIPK2 purified from transfected HEK293 cells, which were treated with Thiamet-G, an OGA selective inhibitor (45), to stabilize O-GlcNAc modifications and facilitate mapping sites of modification. By using higher energy collision dissociation (HCD) product-ion triggered electron transfer/higher energy collision dissociation (EThcD) tandem mass spectrometry (MS/MS) analyses, we identified three O-GlcNAc-modified peptides containing five putative OGT target sites (S852, T853, S1008, T1009, and S1147) (Fig. 7C and SI Appendix, Fig. S10). Subsequent analyses confirmed that S852, T1009, and S1147 are O-GlcNAc modified (SI Appendix, Fig. S10 B–D). All of the sites identified are located outside of the kinase domain. S852 and T853 are within a conserved PEST region implicated in protein stability and S1147 within the YH protein interaction domain (sometimes referred to as an autoinhibitory region) (46). Notably, these sites are conserved among mammals (SI Appendix, Fig. S11).
To assess the functional relevance of HIPK2 O-GlcNAcylation, we replaced these serine and threonine residues with alanine by site-directed mutagenesis. Since we could not definitively rule out that all five residues are targets of OGT, we mutated the three confirmed (S852, T1009, and S1147) and two adjacent sites (T853 and S1008) to alanine to address the possibility of these sites being modified. Individual single alanine mutants and a quintuplet alanine (5A) mutant were generated (SI Appendix, Fig. S12). We found that while WT HIPK2 was weakly expressed (Fig. 7D, lane 1), the mutants were expressed below detectable limits (Fig. 7D, lanes 2 to 7). Even in the presence of MG132 (used to impair proteasomal degradation), the mutants were expressed at reduced levels when compared with WT HIPK2 (Fig. 7D, lanes 8 to 14).
To determine the O-GlcNAc states of WT HIPK2 and the mutants, we used higher concentrations of plasmids for transfection, combined with Thiamet-G treatment to prevent the removal of O-GlcNAc modifications (Fig. 7E). Using wheat germ agglutinin (WGA) to selectively pull down O-GlcNAc–modified proteins, we found that WT HIPK2, unlike the alanine mutants, was readily detectable in the precipitates (Fig. 7E), indicating that the serine and threonine residues identified by MS can be O-GlcNAc modified and losses of these residues rendered HIPK2 more resistant to O-GlcNAcylation.
If the O-GlcNAc sites identified are responsible for maintaining HIPK2 protein stability through OGT, we would expect the mutants to be less responsive to OGT-mediated stabilization. Strikingly, OGT overexpression had much reduced effects on the stabilization of the mutants when compared with WT HIPK2 (Fig. 7F, even-numbered lanes, Fig. 7F′). Nevertheless, all mutant variants, like WT HIPK2, could be stabilized to some extent by OGT (Fig. 7F, compare even- with odd-numbered lanes), suggesting the presence of additional, not yet identified, OGT-mediated mechanisms contributing to HIPK2 stability. These data point to the importance of direct O-GlcNAcylation of HIPK2 in the positive regulation of HIPK2 protein stability by OGT.
Discussion
Cancer cells often vigorously consume glucose, aided by metabolic reprogramming involving multiple processes, for example, elevated expression of glucose transporters (17). Notably, an ample supply of glucose not only facilitates aerobic glycolysis within tumors, but also drives increased flux through parallel pathways, including the HBP (47). As a consequence, increased levels of UDP-GlcNAc and subsequent hyper–O-GlcNAcylation are found in nearly all cancers examined, including breast, prostate, colon, lung, liver, pancreatic, and leukemias (48). Indeed, global O-GlcNAcylation is emerging as a node linking glucose availability and cancer progression (48).
The link between dietary sugar and increased tumorigenesis is found to be conserved in Drosophila (49–51). Using various fly cancer models, researchers have shown that excess sugar intake affects cancer progression through multiple pathways. For example, high dietary sugar has been shown to drive the malignancy of Ras/Src tumors and neoplasia of EGFR-driven tumors through evasion of insulin resistance and lactate dehydrogenase-dependent aerobic glycolysis, respectively (49–51). The involvement of hexosamine signaling in fly tumorigenesis, however, has not been explored. Here we report that the activity of Hipk, a proliferation-promoting protein in flies, is controlled by the HBP–OGT axis in response to metabolic nutrients (Fig. 1). Using the fly Hipk tumor-like model, we demonstrate that OGT is both necessary and sufficient for Hipk-induced growth abnormalities (Figs. 2 and 3). Thus, our work defines the HBP–OGT axis as a glucose-dependent mechanism regulating Hipk-mediated growth control. This is consistent with several studies of O-GlcNAc–dependent regulation of growth-promoting proteins such as YAP in mammals (15, 16).
In addition, we observed a mild up-regulation of OGT in the Hipk-overexpressing cells (SI Appendix, Fig. S5H), inferring that OGT may be a downstream target of Hipk and that the up-regulation of OGT may sustain ectopic Hipk expression and form a positive feedback loop. Recently, we reported that fly Hipk tumor-like cells display elevated aerobic glycolysis driven by up-regulation of dMyc (MYC in vertebrates) (34). It is therefore tempting to speculate that the increased glucose metabolism could stimulate HBP flux and generate more UDP-GlcNAc for O-GlcNAcylation, and together with the mild OGT up-regulation, the combined effects may ultimately feedforward to sustain high Hipk protein levels and reinforce tumor-like proliferation. Future studies may shed light on this mechanism.
Although Hipk proteins are proposed to be maintained at low levels through degradation by the proteasomal machinery, minimal amounts of Hipk are indispensable for normal growth and development as flies homozygous mutant for hipk are not viable (21). This hints at the existence of cellular mechanisms that oppose Hipk protein degradation. Our work demonstrates that OGT stabilizes both endogenous and exogenous Hipk and protects Hipk from proteasomal degradation (Figs. 4 and 5). We confirm that fly Hipk is an O-GlcNAc–modified protein (Fig. 6). While the fly Hipk O-GlcNAc motifs and their roles remain unknown, we show that OGT-mediated stabilization of human HIPK2 relies on direct O-GlcNAcylation of HIPK2 at residues S852/T853, S1008/T1009, and S1147 (Fig. 7). This report describes HIPK2 O-GlcNAcylation as a molecular mechanism that controls HIPK2 protein stability. Given the nutrient-sensing nature of O-GlcNAc, it is conceivable that HIPK2 O-GlcNAcylation might contribute to the nutrient sensitivity of HIPK2 abundance. Intriguingly, a study shows that HIPK2 protein levels increased in a mouse model of diabetes, and the authors attributed the effect to down-regulation of the E3 ubiquitin ligase Siah-1 (52). Hence, the nutritional regulation of HIPK2 might be achieved by multiple strategies like direct O-GlcNAcylation and impairment of proteasome system, or their cooperative effects.
In summary, our work illustrates the conserved regulation of HIPK (fly Hipk and human HIPK2) protein stability by OGT and demonstrates the functional consequences of this regulation in tumor-like events using an in vivo fly model. The roles of human HIPK2 in cancers are context dependent and yet to be fully understood. On the one hand, HIPK2 activates the tumor suppressor p53 after UV irradiation (53, 24). Also, HIPK2 acts as a tumor suppressor upon induction of two-stage skin carcinogenesis (54) or γ-radiation–induced tumorigenesis (55). On the other hand, HIPK2 promotes cytoprotection in cancer cells when challenged with chemotherapeutic drugs (25). Elevation of HIPK2 is also associated with malignancy of pilocytic astrocytomas (56) and cervical carcinogenesis (57). These results raise concerns that targeting HIPK2 may have paradoxical effects. Elevated HIPK2 is also implicated in fibrosis in kidney (58), lung (59), and liver (60). Thus, depending on the context, we believe that the dynamic control of HIPK2 abundance by O-GlcNAc modification reported in our study can be of great interest and exploited in the treatment of HIPK2-related disorders with the use of dietary control or metabolic drugs targeting the HBP–OGT axis.
Materials and Methods
Fly Culture.
Crosses were kept at 25 °C or 29 °C as indicated on the food. Food recipes and fly strains are provided in SI Appendix, SI Materials and Methods.
Immunofluorescence Microscopy.
Larval imaginal discs or salivary glands were dissected in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde (PFA) for 15 min at room temperature, followed by standard immunofluorescence staining protocol. Details are provided in SI Appendix, SI Materials and Methods.
Quantitative Real Time-PCR.
Total RNA isolation was performed using the RNeasy Mini Kit (Qiagen 74104), followed by first-strand cDNA synthesis using the PrimeScript Kit (Clontech 6110A). qRT-PCR was performed using the SensiFAST SYBR Kit (Bioline 92005) on the StepOne Real-Time PCR System (Applied Biosystems). See SI Appendix, Table S1 for primers used in this study.
Western Blotting.
Cells or tissues were lysed with 1× Cell Lysis Buffer (Cell Signaling Technology), supplemented with 1× Protease Inhibitors (Roche) and 1 mM phenylmethylsulfonyl fluoride. Protein lysates with 1× SDS sample buffer were resolved on SDS/PAGE and then transferred to nitrocellulose membranes. Membranes were blocked with 5% skimmed milk or 5% bovine serum albumin before primary and secondary antibody incubation. Images were acquired by a FujiFilm LAS-4000 Chemiluminescent Scanner or the Li-Cor Odyssey Imaging System. A list of antibodies is provided in SI Appendix, SI Materials and Methods.
Cycloheximide (CHX) Chase Assay.
Wandering third-instar larvae were rinsed in PBS and dissected. The anterior half was cultured in insect medium (HyClone) supplemented with CHX (Sigma, 200 µg/mL) for 0 to 120 (or 150) min at room temperature. Tissues were then homogenized in lysis buffer by brief sonication and used for Western blot analyses.
Coimmunoprecipitation and Detection of Hipk and HIPK2 O-GlcNAcylation.
Protein extracts were incubated with indicated antibodies at 4 °C overnight, followed by incubation with a mixture of protein A (Invitrogen 10002D) and protein G (NEB S1430S) magnetic beads for 3 h at 4 °C. The beads were washed with lysis buffer and boiled in 3× SDS sample buffer. The supernatants were then analyzed by Western blotting. Details of WGA pull down, GalNAz feeding, and GalT labeling for detection of Hipk or HIPK2 O-GlcNAcylation are provided in SI Appendix, SI Materials and Methods.
Cell Lines and Cell Culture.
HEK293 and MCF7 were cultured in Dulbecco’s modified eagle medium (DMEM) high glucose (25 mM glucose) (Gibco 11965-092) supplemented with 10% fetal bovine serum (FBS). All cells were cultured in a humidified, 5% CO2 incubator at 37 °C. Cells were transiently transfected with mock (empty vector) or indicated plasmids using Lipofectamine 3000 (Invitrogen) or TransIT-2020 (Mirus Bio) according to the manufacturers’ instructions. Cells were harvested 48 h after transfection unless otherwise indicated. pcDNA3.1-Myc-HIPK2 and pcDNA3.1-Myc-HIPK2-S1008A were purchased from GenScript. pCMV-Myc-OGT-WT has been described (61).
MEF cells containing lentivirus encoding mutated estrogen receptor (mER)-Cre-2A-GFP construct were described previously (44). Cells were cultured in DMEM (high glucose, Gibco) with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin at 37 °C in a water-jacketed, humidified CO2 (5%) incubator. Cells were plated at 30 to 50% confluency. Unless otherwise noted, Cre-recombinase was activated to excise OGT through incubation with 2 µM 4-hydroxytamoxifen (4HT in methanol, Bioshop) for 72 h after plating. Fresh medium and 4HT were changed every 24 h.
Site-Directed Mutagenesis.
For single-site directed mutagenesis, PCRs were run for 18 cycles of 15 s at 95 °C, 30 s at 60 °C, and 18 min at 72 °C using Q5 High-Fidelity DNA Polymerase (NEB M0491). In addition to GC enhancer, 3.5% DMSO was included in the PCR mix because of the GC-rich regions in the HIPK2 insert. The PCR products were treated with FastDigest DpnI (Thermo Scientific FD1704) to remove the template DNA, followed by transformation into One-Shot MAX Efficiency DH5α-T1 Competent Cells (Invitrogen 12297016). Multisite-directed mutagenesis was performed using the GeneArt Site-Directed Mutagenesis PLUS System (Invitrogen A14604) with slight modifications. DNA methylation was not included prior to PCR. Instead, template DNA was removed using DpnI digestion, followed by in vitro recombination reaction. See SI Appendix, Table S2 for the mutagenic primers used. Mutations were then verified by DNA sequencing.
Mapping of O-GlcNAc Sites.
HEK293 cells were transfected with pcDNA3.1-Myc-HIPK2, followed by treatment with Thiamet-G to block the removal of physiological relevant O-GlcNAc attachments. HIPK2 harvested from the cells was purified through immunoprecipitation using anti-Myc tag antibodies and succinylated wheat germ agglutinin (sWGA) enrichment. The resulting purified HIPK2 was run on gels, followed by in-gel tryptic digestion and then LC-MS/MS. Data were obtained from two biological samples. Details for in-gel digestion and MS to map the O-GlcNAcylation sites of HIPK2 are provided in SI Appendix, SI Materials and Methods.
Statistical Analysis.
The data were represented by mean ± SEM. The P values were determined by two-tailed Student’s t tests. P < 0.05 was considered statistically significant. Bar graphs were generated by Microsoft Excel. Violin plots and box and whisker plots to represent the distribution of data were generated by BoxPlotR (62).
Supplementary Material
Acknowledgments
We are grateful to the Developmental Studies Hybridoma Bank, the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, and Jackson ImmunoResearch Laboratories for providing fly strains and antibodies. We thank M. Alteen for production of chemical materials. We appreciate all the discussion and input from our laboratory’s current and past members, including Dr. G. Tettweiler, Dr. E. Hall, S. Kinsey, L. Madonsela, R. Tse, T. Wiens, L. Zhe, J. Blaquiere, N. Wray, and E. Hoesing. We also thank J. Blaquiere and R. Chung for conducting a genetic screen to search for Hipk modifiers. This work was funded by operating grants from the Canadian Institutes of Health Research, PJT-156204 and PJT-148732. Mass spectrometry data were acquired at the Academia Sinica Common Mass Spectrometry Facilities for Proteomics and Protein Modification Analysis located at the Institute of Biological Chemistry, Academia Sinica, supported by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-107).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.A.H. is a guest editor invited by the Editorial Board.
Data deposition: Data and materials are available on the Open Science Framework at https://osf.io/epkmj/.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912894117/-/DCSupplemental.
References
- 1.Marshall S., Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: A nutritional perspective of diabetes, obesity, and cancer. Sci. STKE 2006, re7 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Yuan H.-X., Xiong Y., Guan K.-L., Nutrient sensing, metabolism, and cell growth control. Mol. Cell 49, 379–387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Love D. C., Hanover J. A., The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci. STKE 2005, re13 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Bond M. R., Hanover J. A., A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardivillé S., Hart G. W., Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20, 208–213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X., Qian K., Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu K., Paterson A. J., Chin E., Kudlow J. E., Glucose stimulates protein modification by O-linked GlcNAc in pancreatic beta cells: Linkage of O-linked GlcNAc to beta cell death. Proc. Natl. Acad. Sci. U.S.A. 97, 2820–2825 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walgren J. L. E., Vincent T. S., Schey K. L., Buse M. G., High glucose and insulin promote O-GlcNAc modification of proteins, including α-tubulin. Am. J. Physiol. Endocrinol. Metab. 284, E424–E434 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Kamigaito T., et al. , Overexpression of O-GlcNAc by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis. 17, 18–22 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Onodera Y., Nam J.-M., Bissell M. J., Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-GlcNAc pathways. J. Clin. Invest. 124, 367–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozanski W., et al. , Prediction of bladder cancer based on urinary content of MGEA5 and OGT mRNA level. Clin. Lab. 58, 579–583 (2012). [PubMed] [Google Scholar]
- 12.Zhu Q., et al. , O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med. Oncol. 29, 985–993 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Chou T. Y., Dang C. V., Hart G. W., Glycosylation of the c-Myc transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 92, 4417–4421 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W. H., et al. , Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Peng C., et al. , Regulation of the Hippo-YAP pathway by glucose sensor O-GlcNAcylation. Mol. Cell 68, 591–604.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., et al. , The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat. Commun. 8, 15280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu P. P., Sabatini D. M., Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Swarup S., Verheyen E. M., Drosophila homeodomain-interacting protein kinase inhibits the Skp1-Cul1-F-box E3 ligase complex to dually promote Wingless and Hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 9887–9892 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Verheyen E. M., Homeodomain-interacting protein kinase regulates Yorkie activity to promote tissue growth. Curr. Biol. 22, 1582–1586 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Lee W., Swarup S., Chen J., Ishitani T., Verheyen E. M., Homeodomain-interacting protein kinases (Hipks) promote Wnt/Wg signaling through stabilization of beta-catenin/Arm and stimulation of target gene expression. Development 136, 241–251 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Lee W., Andrews B. C., Faust M., Walldorf U., Verheyen E. M., Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev. Biol. 325, 263–272 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Hattangadi S. M., Burke K. A., Lodish H. F., Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood 115, 4853–4861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinaldo C., et al. , HIPK2 controls cytokinesis and prevents tetraploidization by phosphorylating histone H2B at the midbody. Mol. Cell 47, 87–98 (2012). [DOI] [PubMed] [Google Scholar]
- 24.D’Orazi G., et al. , Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 4, 11–19 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Torrente L., et al. , Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene 36, 6204–6212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isono K., et al. , Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Mol. Cell. Biol. 26, 2758–2771 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang Y., et al. , Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-β-TAK1-dependent mechanism. PLoS Biol. 11, e1001527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sjölund J., Pelorosso F. G., Quigley D. A., DelRosario R., Balmain A., Identification of Hipk2 as an essential regulator of white fat development. Proc. Natl. Acad. Sci. U.S.A. 111, 7373–7378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao L., et al. , HIPK2 is necessary for type I interferon–mediated antiviral immunity. Sci. Signal. 12, eaau4604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter M., et al. , Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat. Cell Biol. 10, 812–824 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Calzado M. A., de la Vega L., Möller A., Bowtell D. D. L., Schmitz M. L., An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat. Cell Biol. 11, 85–91 (2009). [DOI] [PubMed] [Google Scholar]
- 32.de la Vega L., et al. , A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol. Cell 46, 472–483 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Blaquiere J. A., Wong K. K. L., Kinsey S. D., Wu J., Verheyen E. M., Homeodomain-interacting protein kinase promotes tumorigenesis and metastatic cell behavior. Dis. Model. Mech. 11, dmm031146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong K. K. L., Liao J. Z., Verheyen E. M., A positive feedback loop between Myc and aerobic glycolysis sustains tumor growth in a Drosophila tumor model. eLife 8, e46315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Sinclair D. A. R., et al. , Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gambetta M. C., Oktaba K., Müller J., Essential role of the Glycosyltransferase Sxc/Ogt in Polycomb repression. Science 325, 93–96 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Menut L., et al. , A mosaic genetic screen for Drosophila neoplastic tumor suppressor genes based on defective pupation. Genetics 177, 1667–1677 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenzweig R., Bronner V., Zhang D., Fushman D., Glickman M. H., Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 287, 14659–14671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khidekel N., et al. , A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J. Am. Chem. Soc. 125, 16162–16163 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Liu T.-W., et al. , Genome-wide chemical mapping of O-GlcNAcylated proteins in Drosophila melanogaster. Nat. Chem. Biol. 13, 161–167 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Boyce M., et al. , Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 3141–3146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vocadlo D. J., Hang H. C., Kim E.-J., Hanover J. A., Bertozzi C. R., A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U.S.A. 100, 9116–9121 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kazemi Z., Chang H., Haserodt S., McKen C., Zachara N. E., O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3beta-dependent manner. J. Biol. Chem. 285, 39096–39107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuzwa S. A., et al. , A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Blaquiere J. A., Verheyen E. M., Homeodomain-interacting protein kinases: Diverse and complex roles in development and disease. Curr. Top. Dev. Biol. 123, 73–103 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Ma Z., Vosseller K., Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem. 289, 34457–34465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrer C. M., Sodi V. L., Reginato M. J., O-GlcNAcylation in cancer biology: Linking metabolism and signaling. J. Mol. Biol. 428, 3282–3294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirabayashi S., Baranski T. J., Cagan R. L., Transformed Drosophila cells evade diet-mediated insulin resistance through wingless signaling. Cell 154, 664–675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirabayashi S., Cagan R. L., Salt-inducible kinases mediate nutrient-sensing to link dietary sugar and tumorigenesis in Drosophila. eLife 4, e08501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eichenlaub T., et al. , Warburg effect metabolism drives neoplasia in a Drosophila genetic model of epithelial cancer. Curr. Biol. 28, 3220–3228.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Oh H. J., et al. , Inhibition of the processing of miR-25 by HIPK2-Phosphorylated-MeCP2 induces NOX4 in early diabetic nephropathy. Sci. Rep. 6, 38789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofmann T. G., et al. , Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4, 1–10 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Wei G., et al. , HIPK2 represses beta-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 13040–13045 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao J.-H., et al. , Hipk2 cooperates with p53 to suppress γ-ray radiation-induced mouse thymic lymphoma. Oncogene 31, 1176–1180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deshmukh H., et al. , High-resolution, dual-platform aCGH analysis reveals frequent HIPK2 amplification and increased expression in pilocytic astrocytomas. Oncogene 27, 4745–4751 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Al-Beiti M. A., Lu X., Expression of HIPK2 in cervical cancer: Correlation with clinicopathology and prognosis. Aust. N. Z. J. Obstet. Gynaecol. 48, 329–336 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Jin Y., et al. , A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat. Med. 18, 580–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ricci A., et al. , Homeodomain-interacting protein kinase2 in human idiopathic pulmonary fibrosis. J. Cell. Physiol. 228, 235–241 (2013). [DOI] [PubMed] [Google Scholar]
- 60.He P., Yu Z. J., Sun C. Y., Jiao S. J., Jiang H. Q., Knockdown of HIPK2 attenuates the pro-fibrogenic response of hepatic stellate cells induced by TGF-β1. Biomed. Pharmacother. 85, 575–581 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Martinez-Fleites C., et al. , Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat. Struct. Mol. Biol. 15, 764–765 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Spitzer M., Wildenhain J., Rappsilber J., Tyers M., BoxPlotR: A web tool for generation of box plots. Nat. Methods 11, 121–122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.