Significance
Voltage-gated ion channels (VGICs) initiate and transmit action potentials in nerve and muscle cells. Because they are generally activated by depolarization of the membrane potential, their structures usually represent activated channel states since they are necessarily determined in the absence of a voltage gradient. We determined the structure of a resting intact VGIC at 3.2 Å resolution by cryogenic electron microscopy. This structure provides the first resting state of a voltage sensor to be observed without the need for any inhibitory ligand, independent of any chemical, or mutagenic alteration.
Keywords: ion channel, cryoEM, voltage sensors, structure, electrophysiology
Abstract
Voltage-gated ion channels endow membranes with excitability and the means to propagate action potentials that form the basis of all neuronal signaling. We determined the structure of a voltage-gated sodium channel, two-pore channel 3 (TPC3), which generates ultralong action potentials. TPC3 is distinguished by activation only at extreme membrane depolarization (V50 ∼ +75 mV), in contrast to other TPCs and NaV channels that activate between −20 and 0 mV. We present electrophysiological evidence that TPC3 voltage activation depends only on voltage sensing domain 2 (VSD2) and that each of the three gating arginines in VSD2 reduces the activation threshold. The structure presents a chemical basis for sodium selectivity, and a constricted gate suggests a closed pore consistent with extreme voltage dependence. The structure, confirmed by our electrophysiology, illustrates the configuration of a bona fide resting state voltage sensor, observed without the need for any inhibitory ligand, and independent of any chemical or mutagenic alteration.
Voltage-gated ion channels (VGICs) use changes in electrical potential across biological membranes to modulate their open probability (1, 2). These channels use a voltage sensing domain (VSD) containing basic residues to couple changes in transmembrane voltage to dilation of a central pore domain through which ions diffuse (3, 4). Excitable membranes contain VGICs which usually activate upon membrane depolarization and most exhibit maximal open probability at ∼0 mV (5). While many groups have studied voltage activation using a variety of biophysical techniques (1, 6–10), resting conformations have eluded structural characterization (1, 2). VSD activation is typically described as charge translocation across a charge transfer center (CTC) whereby positively charged arginine side chains in the resting state move away from an intracellular negative cluster, across an insulating hydrophobic constriction site (HCS) to form interactions with an extracellular negative cluster (ENC). Recent structures from the two-pore channel family (11, 12) and of toxin-bound NaV1.7 channels (13, 14) provided the first glimpses of resting state VSDs by virtue of VSD-binding ions or ligands that inhibit voltage activation until above 0 mV, the condition in a crystal lattice or single particle electron microscopy (EM) experiment.
The two-pore channel family comprises three cation channels (TPC1–3), each dimers of tandem Shaker-like domains, that dimerize to form a functional channel. TPC1/3 are voltage-gated by virtue of positively charged arginine-containing voltage sensing domains, whereas TPC2 is lipid gated (15, 16). TPCs were first identified in Arabidopsis thaliana where a slow-activating depolarization-elicited current in the vacuole was detected (17). The channel responsible was TPC1, which, in plants, forms a nonselective voltage-gated cation channel in the vacuolar membrane that undergoes diverse regulation by Ca2+, lipids, and phosphorylation (12, 16). Vertebrate and mammalian TPCs are Na+ selective channels primarily localized to endolysosomal membranes (18, 19), and their activity is tuned by PIP2 and association with, or phosphorylation by, mTORC1 (20). Despite encompassing only three members, the TPC family exhibits remarkable functional diversity, and the physiological roles of these channels are still largely unknown (16, 21, 22).
Cang and colleagues (23) provided the first characterization of the molecular properties of TPC3, which they determined is plasma membrane localized, Na+ selective, and activated only at very large depolarization (e.g., V50 ∼ +75 mV). Owing to functional similarity, they established that TPC3 forms the endogenous voltage-gated Na+ channel on the plasma membrane of Xenopus laevis oocytes. Mammalian TPC3, however, is primarily localized to endolysosomal membranes and contributes to the regulation of membrane sorting, suggesting that the role of TPC3 may vary significantly across species (24). TPC3 does not inactivate and is probably responsible for generating or maintaining ultralong action potentials, the phenomenon of prolonged membrane depolarization (23). Such processes appear to be widespread in the eggs of many species, including during fertilization-induced excitation in Xenopus (25) and Ca2+-activated currents that last multiple minutes in worms (26, 27). Similar phenomena are also observed in TPC1 which generates ultralong action potentials in plant vacuoles (28).
By virtue of the unique hyperdepolarized activation of TPC3, we hypothesized that we could structurally capture TPC3 in a VSD resting conformation (Fig. 1E). Unlike AtTPC1, whose resting state was crystallographically accessed by occupation of an external Ca2+-binding site (11, 12) that positively shifts the activation potential, TPC3 does not require ions or small molecules to attenuate its voltage sensitivity, suggesting that the channel contains an intrinsic mechanism to suppress activation. Therefore, we turned to TPC3 as a first example of a stable resting state VGIC channel. We purified glycodiosgenin-solubilized TPC3 from HEK293F cells and used electron cryomicroscopy to determine a structure to 3.2 Å resolution (Fig. 1A), that enabled complete and unambiguous placement of residues and side chains in all transmembrane helices. In conjunction with voltage clamp electrophysiology, we define the role of the VSDs in voltage activation and provide a snapshot of a resting state VSD that does not depend on any ion, toxin, or ligand.
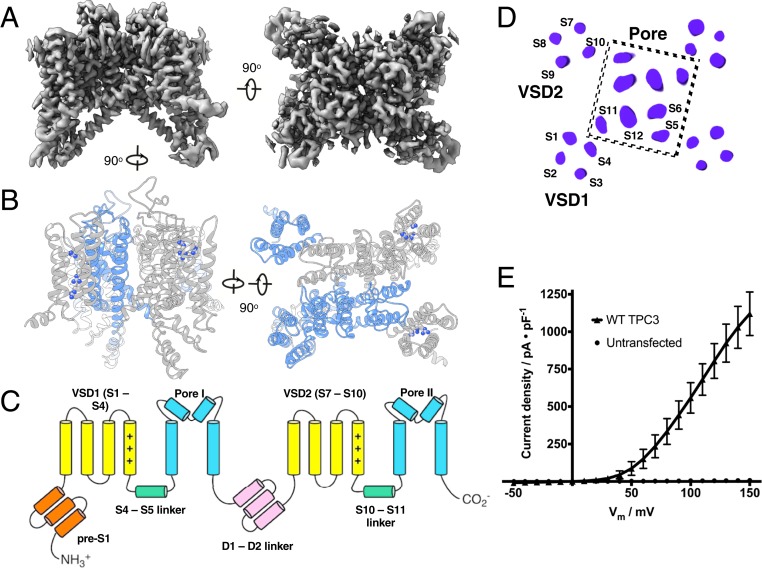
Fig. 1.
Cryo-EM structure of two-pore channel 3. (A) Orthogonal views of the Coulomb potential map of TPC3 sharpened with a B factor of −90 Å2. (B) Molecular model of TPC3 colored by a subunit to illustrate domain swapping of pore and voltage sensor domains (Right) and oriented to bring VSDs of the gray subunit to the front (Left). VSD2 is on the left. Arginine side chains of S4 and S10 are shown. (C) Annotated schematic of domain arrangement. (D) A 2D arrangement of helices, viewed normal to a central slice through the channel. (E) Current density of wild-type (WT) TPC3 showing its characteristic right-shifted response, determined by a whole cell voltage clamp on HEK293T cells. All recordings use symmetric [Na+] (n = 6 cells).
TPC3 is a C2 symmetric channel with “domain-swapped” voltage sensing and pore domains (Fig. 1 B–D). Domains I and II of TPC3 contain homologous voltage sensing domains, each composed of three arginine residues in the fourth TM helix: R166, R169, and R172 in S4 (corresponding to R2–R4 of the Shaker K+ channel) and R522, R525, and R528 in S10 (corresponding to R3–R5 of the Shaker K+ channel).
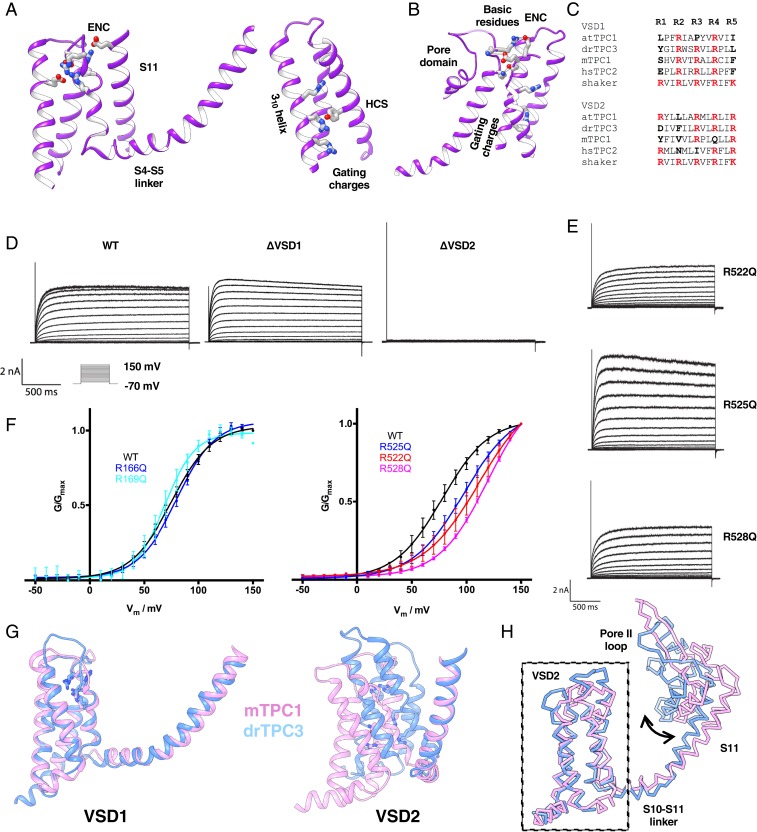
By electrophysiology, we show that only one voltage sensor, VSD2, is responsible for voltage activation and that R3, R4, and R5 within S10 each sensitize the channel to voltage. To interrogate the role of each VSD arginine in voltage sensing, we neutralized each one, in turn, and recorded currents from cells expressing mutants. VSD1 has no effect: R166Q and R169Q (R3 and R4 of VSD1) have no effect on voltage dependence relative to WT (Fig. 2E). We had difficulty recording from R172Q (R5 of VSD1) because of intracellular retention, probably due to misfolding of the protein, however, in rare cases, we did observe WT-like currents. R522Q, R525Q, and R528Q in VSD2 each shift the voltage dependence further positive (Fig. 2 D and E), indicating that all three S10 arginines cooperate in activation. Triple mutants (R to Q) on either of the VSDs (denoted ΔVSD1/2) confirmed that VSD2 alone controls voltage gating. These data contrast with mammalian TPC1 where analogous mutations in VSD2 either inhibit or constitutively activate the channel, whereas in TPC3 they mimic the plant TPC1 phenotype where all R to Q mutations inhibit voltage activation (11, 21, 29).
Fig. 2.
Structural and functional asymmetries in the voltage sensing domains. (A) Structure of VSD1 showing the interactions of the arginines with the extracellular negative cluster (Left). Structure of VSD2 showing interactions of R525 with the hydrophobic constriction site Y455 and R528 with the intracellular solvent (Right). (B) Implied interactions between basic residues on the S11 helix and the extracellular negative cluster in VSD2. (C) Sequence alignment of the S4 (Top) and S10 helices (Bottom) of zebrafish TPC3, Arabidopsis TPC1, mouse TPC1, and human TPC2 with respect to the Shaker KV VSD. R1–R5 of the Shaker convention are in bold, and basic residues are colored red. (D) Currents obtained from a voltage step protocol of WT and arginine to glutamine triple mutants in VSD1 and VSD2. The VSD1 triple mutant resembles WT while the VSD2 triple mutant is silent. (E) Currents from the VSD2 R-Q point mutants. (F) Conductance curves of the channel with R-Q mutants in VSD1 (Left) and VSD2 (Right), calculated from their whole cell steady state currents using a voltage step protocol. The mean conductance of six to eight cells each with SD is shown, and curves are fitted using a single Boltzmann sigmoid. (G) Conformational comparisons of VSD1 (Left) and VSD2 (Middle) of TPC3 (blue) and mTPC1 (PDB: 6C96) (pink), based on alignment of the S4-S5 linker and S5 or the S10-S11 linker and S11. (H) Comparison of the TPC3 versus mTPC1 pore domains based on alignment of their VSD2s to emphasize the differences between the two-domain conjugates in resting versus putatively activated/preactivated states.
In the inactive VSD1, S4 adopts an α-helical fold near the S4-S5 linker, but lacks secondary structure at the S3-S4 loop. The arginines in S4 are clearly accessible to extracellular solvent and make strong Coulombic interactions with the ENC. R169 forms a strong interaction with E566 of S11, noncovalently stapling S4 to the pore domain. The arrangement of both the basic and the acidic residues aligns very well with that of VSD1 in mTPC1 and both lack the common features of a voltage sensing helix, congruent with voltage independence (Fig. 2A).
In VSD2, S10 of TPC3 contains three arginines, unlike mammalian TPC1 that has only two. Y455 forms the HCS of the CTC. R4 and R5 are situated below Y455 with R4 forming a cation-π interaction with the phenol ring (Fig. 2A). The most extracellular arginine, R3, points upwards but is slightly displaced from interaction with the extracellular negative cluster. This is in contrast to the active state VSDs of NaVAb (30) and NaVRh (31) in which R3 faces outward and is in contact with the ENC, while R1 and R2 even more toward the extracellular side, point toward the outside surface (an active state VSD conformation). S10 adopts a 310-helical fold as is observed in other S4 helices such that the arginines stack into the interior of the four-helix bundle (Fig. 2A). Three basic residues, K219, K223, and R224, on the extracellular extreme of the pore helix S11, are in proximity with the ENC, although the weak density for such residues precludes accurate placement of the side chains. Such basic residues are not present in TPC1, and the hypothetical interaction is located where the inhibitory Ca2+ acts in plant TPC1 (32). Hence, such interactions may either exclude the S10 arginines from forming their active state interactions with the ENC or potentially couple the electrical state of VSD2 to the pore domain.
While VSD2 of mTPC1 and TPC3 align well with each other as rigid bodies in isolation, alignment of the two channels based on the pore domain shows that the two domains are out of register (Fig. 2E). Alignment of the two channels based on one VSD2 bundle shows high similarity in VSD2 (∼0.9 Å rmsd) and large differences in the rest of the channel (∼5.5 Å rmsd), including in the other VSD2 (Fig. 2F). If mTPC1 and TPC3 share common activation processes, this may represent an early conformation during activation, prior to the electromechanical coupling step of pore opening. Activation of VSD2 may, hence, involve a combination of rigid body motion of the entire four-helix bundle relative to the pore and the upwards translocation of R3 toward the ENC and R4 across the HCS. The potential aforementioned interactions between the VSD2 ENC and the S11 basic residues may hinder such a conformation rearrangement, helping to explain the large energetic barrier to voltage activation.
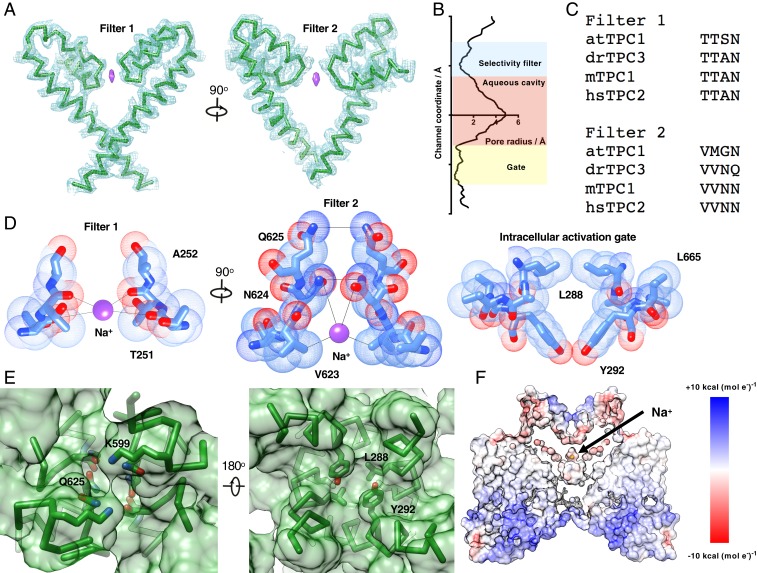
The channel pore is tightly closed, consistent with a nonconducting state (Fig. 3 A and B). The selectivity filter, formed by filter 1 and filter 2 of domains 1 and 2, respectively, is defined by a coin slot entry suitable for partially hydrated Na+ ions, similar to that observed in recent sodium-selective TPC structures. Filter 2 is formed by orthogonal Q625 and N624 pairs with distances of 5.6 and 4.3 Å. Below the asparagine pair is filter 1, consisting of the amide carbonyl of A252 and the amide carbonyl of T251 with pairwise distances of 7.1 and 6.2 Å, respectively. A putative sodium ion (the only monoatomic cation in solution) is situated between the carbonyl oxygen and the side chain hydroxyl of T251. Below the filter is an aqueous cavity lined by small hydrophobic side chains. Beneath the cavity is the gate, formed by constriction points at L288 (3.79 Å), L665 (4.02 Å), and Y292 (5.58 Å), too narrow to allow the passage of hydrated ions (Fig. 3 B–D).
Fig. 3.
Structural features of the ion conduction pathway. (A) Structure of the pore 1 (Left) and pore 2 (Right) with sharpened potential map. (B) Pore radius as a function of channel coordinate, roughly aligned to the molecule shown in A calculated using HOLE (45). (C) Sequence alignment of TPC selectivity filters. (D) Chemical features of the selectivity filter and gate with Van der Waals surface shown. (E) Surface representation illustrates a solvent-excluding selectivity filter (Left) and intracellular activation gate (Right). (F) Surface representation colored by the Coulombic potential calculated in UCSF Chimera, illustrating the ion conduction pathway.
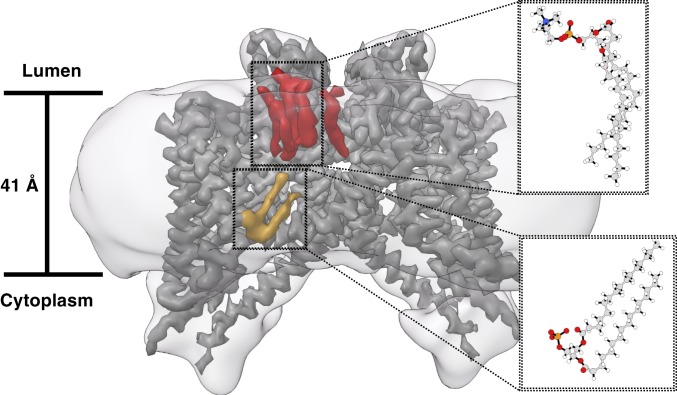
We observe multiple well-ordered lipids tightly associated with the channel (Fig. 4). One upper leaflet lipid is located in the same site that inhibitor NED-19 occupies in the Arabidopsis TPC1 crystal structure. We observe a lipidlike density at the S4-S5 linker (i.e., lower leaflet), similar in location to the PIP2 regulatory site in mTPC1. The lipid, with a small head group poised toward the membrane, looks distinct from the phosphatidylcholinelike upper leaflet lipids and resembles a phosphatidic acid. This would be consistent with the observed lower leaflet phosphatidic acid at the S10-S11 linker in AtTPC1 and the proposed functional role of phosphatidic acids in TPC physiology (12, 16). No lipids were supplemented during purification so all observed density belongs to copurified species. We confirmed the observation of Cang et al. that TPC3 is insensitive to PI(3,5)P2 and PI(4,5)P2 administered through the pipette solution during whole cell voltage clamp recordings (33), and we determined that PI(3,4)P2 has no apparent effect at concentrations up to 100 μM.
Fig. 4.
Lipids interacting with the channel. Sharpened potential map of TPC3 at high contour overlaid with lipidic densities at low contour colored orange (lower leaflet) and red (upper leaflet). The orange density resembles a phosphatidic acid with the generic chemical structure shown. The upper leaflet lipids have large head groups, similar to those in phosphatidylcholinelike lipids with the generic chemical structure shown. The upper leaflet species on the right resembles a glycodiosgenin molecule given its single tail. The detergent micelle (low contour map) is shown to approximate the boundaries of the bilayer.
The structure of TPC3 should represent a profoundly resting state VSD configuration as demonstrated by electrophysiology. Since most structures of VGICs represent a VSD-activated state, this and the structural rearrangements observed during AtTPC1 voltage activation (34) suggest that the mechanisms of voltage activation in TPCs may involve more complex conformational changes than the classical models of voltage sensors would predict. Substantial motions in the VSD, driven by a positive membrane potential, may be necessary to open the activation gate. These may include in-plane motions of the VSD which can be sensed either through the S10-S11 linker or the extracellular salt bridges or the noncovalent interactions between S10 and the pore domain. Understanding the exact nature of these conformational changes and how they couple to gating charge translocation will provide fundamental insights into how “modular” voltage sensing domains can exhibit exquisite functional diversity.
Methods
Protein Expression and Purification.
Orthologs of TPC3 were screened for expression and biochemical behavior using fluorescence size exclusion chromatography (35). HEK 293T cells were transfected with eGFP fusions of orthologs in a pEG_Bacmam vector using a JetPrime transfection reagent. Some 48 h posttransfection, cells were washed with ice cold PBS and solubilized in 20 mM Tris pH 7.4, 150 mM NaCl, 1 mM PMSF, and 1% β-dodecyl maltoside, overnight. The suspension was centrifuged at 40,000 rpm (TLA-55 rotor) for 1 h before injection over a Superose 6 gel filtration column in line with a fluorescence detector (ex/em = 488 nm/510 nm). Additive and detergent screens were performed in a similar manner to determine optimal buffer conditions for purification. TPC3 from Danio rerio (DrTPC3) was expressed as a C-terminal eGFP fusion in a pEG plasmid using the Bacmam system in HEK GnTi− cells. Cells were harvested 48 h postinduction with 10 mM sodium butyrate. The cell pellets were resuspended in buffer A (20 mM Tris pH 7.4 and 150 mM NaCl) and stored at −80 °C until use. All subsequent steps were performed at 4 °C. Cells were thawed, phenylmethylsulfonyl fluoride (PMSF) was added to 1 mM, and Roche cOmplete protease inhibitor mixture was added at one tablet per 100 mL. Cells were lysed by sonication on ice at 1 s−1 for 5 min. Glycodiosgenin was added to the lysate to a final concentration of 1%. After 8 h of solubilization, the lysate was centrifuged at 30,000 rpm for 40 min (Ti45 rotor) to remove insoluble material. The clarified lysate was filtered through a 0.22 μm filter and batch bound to anti-GFP nanobody-conjugated Sepharose (made in house) for 2 h with gentle nutation. The flow through was collected, and the resin was washed with 20 column volumes of buffer B (20 mM Tris pH 7.4, 150 mM NaCl, and 0.06% glycodiosgenin). The washed resin was resuspended in buffer B with 2 mM CaCl2 and 200 U thrombin for on-column cleavage overnight. The eluate was collected and concentrated before injection over a Superose 6 gel filtration column. Peak fractions were pooled and concentrated to ∼3 mg mL−1 for vitrification. Grids for electron cryomicroscopy were prepared using a Mark IV Vitrobot in which 4.5 μL of sample was applied to freshly glow discharged Quantifoil Cu R1.2/1.3 holey carbon grids, blotted for 4 s at 100% humidity and plunge frozen in liquid N2-cooled ethane. Grids were screened for ice quality on a Talos Arctica microscope before extensive data collection.
Microscopy.
Movies were acquired on a 300 keV Titan Krios equipped with a Gatan Imaging Filter (20 eV slit width) and K2 Summit camera (operating in superresolution mode). The pixel size on the specimen was 0.407 Å pix−1. Movies were collected with 100 frames over 10 s (0.1 s dose fractionation) using a per frame dose of 0.92 e− Å−2 (92 e− Å−2 accumulated dose) over an underfocus range of 0.8–2.0 μm. Movies were drift corrected and dose weighted with MotionCor2 (36) using 5×5 patches and twice Fourier binned to a pixel size of 0.814 Å pix−1. SerialEM (Mastronarde) and Focus (37) were used for data acquisition and on-the-fly drift correction, respectively.
Image Processing.
CTF estimation was performed using CTFFIND4, and 1,201,080 particles were picked ab initio from 7118 micrographs using cisTEM (38). The particles were subjected to 2D classification in cisTEM from which 706,801 particles were selected. These particles were used to calculate a 3D ab initio template in cisTEM. The coordinates were exported to Relion 3.0 (39), and particles were extracted twice binned, resulting in a box size of 216 × 216 pixels and a pixel size of 1.628 Å pixel−1. Unmasked 3D classification into six classes was performed in Relion 3.0 using the twice down-sampled ab initio map as a reference, low-pass filtered to 60 Å. Two classes constituting 213,328 particles were selected and subjected to 3D autorefine in Relion 3.0 resulting in a 3.75 Å reconstruction. This map was used to calculate a micelle-excluding mask for use in a “skip-align” 3D classification into eight classes in Relion 3.0. Two classes representing 170,446 particles were selected. The particles were reextracted in Relion unbinned and exported to cisTEM using “relion stack create” and refined while applying the aforementioned mask (down-weighting and filtering the exterior to 0.1 and 30 Å, respectively), excluding frequencies past 7 Å during alignment. Multiple rounds of masked refinement in cisTEM resulted in a 3.2 Å reconstruction according to Fourier shell correlation using a 0.143 cutoff. Simultaneously, the Relion particle stack was subjected to nonuniform refinement in cryoSPARC v2 (40), resulting in a 3.1 Å reconstruction. The cisTEM and cryoSPARC maps are of similar quality, nominal global resolution, and both were used for model building.
Model Building.
The TPC3 atomic model was built de novo using a MODELER-generated homology model (41) as a guide. All model building was performed in Coot (42). The final model was refined using phenix_real_space_refine (43). Map and model figures were prepared using University of California, San Francisco (UCSF) Chimera (44). Residues 320–407, corresponding to the cytosolic linker between domains 1 and 2, were not modeled due to poor resolution.
Electrophysiology.
Voltage clamp recording of DrTPC3 and mutants was performed in a whole cell configuration using an Axoclamp 200B amplifier and a Digidata 1440A controlled by pClamp (Molecular Device). HEK293T cells were transiently transfected with DrTPC3-eGFP variants using the JetPrime transfection reagent. Some 48 h posttransfection, the cells were trypsinized and plated at appropriate density on poly-l-lysine-coated coverslips. Cells with plasma membrane-localized GFP expression were selected for recording. The pipette and bath solutions were adapted from Cang et al. (23) with symmetric [Na+]. The bath solution consisted of 145 mM Na+ methanesulphonate, 5 mM NaCl, 10 mM Hepes, and 10 mM glucose pH 7.4. The pipette solution consisted of 130 mM Na-methanesulphonate, 2.5 mM MgCl2, 10 mM Hepes, 10 mM EGTA, and 20 mM NaOH pH 7.2. Patch pipettes were pulled from borosilicate glass and flame polished to a resistance of 2–4 MΩ, and recordings were made from cells with an access resistance below 10 MΩ. Currents were acquired using a step protocol with 2 s steps from −50 mV to +150 mV in 10 mV increments from a holding potential of −70 mV. Data were analyzed using Clampfit (Molecular device). Conductance was calculated using Ohm’s law from steady state currents and fitted using a single Boltzmann sigmoid.
Data and Materials Availability.
The sharpened and unsharpened maps are available in the Electron Microscopy Data Bank (EMDB), https://www.ebi.ac.uk/pdbe/emdb (accession no. 21015); and the model has been deposited into the Protein Data Bank (PDB), https://www.rcsb.org/ (accession no. 6V1Q). The DrTPC3, wild type, and mutant, pEG_BacMam plasmids are available upon request.
Acknowledgments
We thank Alexander Kintzer for help with construct design and screening; Phuong Nguyen and Alisa Bowen for assistance with mammalian cell culture; Axel Brilot for data processing advice; Michael Braunfeld and David Bulkley for maintenance of the UCSF EM facility; and the NIH for their support of our facility. We also thank Janet Finer-Moore, Sergei Pourmal, and Evan Green for critical reading of the paper. The research was funded by NIH Grant R01 GM24485 (to R.M.S.). M.S.D. acknowledges a NSF graduate research fellowship.
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Protein Data Bank (PDB), https://www.rcsb.org/ (accession no. 6V1Q), and the Electron Microscopy Data Bank (EMDB), https://www.ebi.ac.uk/pdbe/emdb (accession no. 21015).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915144117/-/DCSupplemental.
References
- 1.Hille B., Ion Channels of Excitable Membranes (Sinauer Associates, Sunderland, Mass, ed. 3, 2001). [Google Scholar]
- 2.Catterall W. A., Wisedchaisri G., Zheng N., The chemical basis for electrical signaling. Nat. Chem. Biol. 13, 455–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezanilla F., The voltage sensor in voltage-dependent ion channels. Physiol. Rev. 80, 555–592 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Bezanilla F., How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9, 323–332 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Ahern C. A., Payandeh J., Bosmans F., Chanda B., The hitchhiker’s guide to the voltage-gated sodium channel galaxy. J. Gen. Physiol. 147, 1–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stühmer W., et al. , Structural parts involved in activation and inactivation of the sodium channel. Nature 339, 597–603 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Seoh S. A., Sigg D., Papazian D. M., Bezanilla F., Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron 16, 1159–1167 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Gandhi C. S., Isacoff E. Y., Molecular models of voltage sensing. J. Gen. Physiol. 120, 455–463 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villalba-Galea C. A., Sandtner W., Starace D. M., Bezanilla F., S4-based voltage sensors have three major conformations. Proc. Natl. Acad. Sci. U.S.A. 105, 17600–17607 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao X., Lee A., Limapichat W., Dougherty D. A., MacKinnon R., A gating charge transfer center in voltage sensors. Science 328, 67–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo J., et al. , Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature 531, 196–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kintzer A. F., Stroud R. M., Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana. Nature 531, 258–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clairfeuille T., et al. , Structural basis of α-scorpion toxin action on Nav channels. Science 363, eaav8573 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Xu H., et al. , Structural basis of Nav1.7 inhibition by a gating-modifier spider toxin. Cell 176, 702–715 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Calcraft P. J., et al. , NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 459, 596–600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kintzer A. F., Stroud R. M., On the structure and mechanism of two-pore channels. FEBS J. 285, 233–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedrich R., Neher E., Cytoplasmic calcium regulates voltage-dependent ion channels in plant vacuoles. Nature 329, 833–836 (1987). [Google Scholar]
- 18.Wang X., et al. , TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H., Ren D., Lysosomal physiology. Annu. Rev. Physiol. 77, 57–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cang C., et al. , mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell 152, 778–790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.She J., et al. , Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature 556, 130–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.She J., et al. , Structural mechanisms of phospholipid activation of the human TPC2 channel. eLife 8, e45222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cang C., Aranda K., Ren D., A non-inactivating high-voltage-activated two-pore Na+ channel that supports ultra-long action potentials and membrane bistability. Nat. Commun. 5, 5015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogunbayo O. A., et al. , Organelle-specific subunit interactions of the vertebrate two-pore channel family. J. Biol. Chem. 290, 1086–1095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baud C., Kado R. T., Marcher K., Sodium channels induced by depolarization of the Xenopus laevis oocyte. Proc. Natl. Acad. Sci. U.S.A. 79, 3188–3192 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyazaki S., Takahashi K., Tsuda K., Calcium and sodium contributions to regenerative responses in the embryonic excitable cell membrane. Science 176, 1441–1443 (1972). [DOI] [PubMed] [Google Scholar]
- 27.Jaffe L. A., Kado R. T., Kline D., A calcium-activated sodium conductance produces a long-duration action potential in the egg of a nemertean worm. J. Physiol. 381, 263–278 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaślan D., et al. , Voltage-dependent gating of SV channel TPC1 confers vacuole excitability. Nat. Commun. 10, 2659 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaślan D., et al. , Gating of the two-pore cation channel AtTPC1 in the plant vacuole is based on a single voltage-sensing domain. Plant Biol (Stuttg) 18, 750–760 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Payandeh J., Scheuer T., Zheng N., Catterall W. A., The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., et al. , Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486, 130–134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dadacz-Narloch B., et al. , A novel calcium binding site in the slow vacuolar cation channel TPC1 senses luminal calcium levels. Plant Cell 23, 2696–2707 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cang C., Bekele B., Ren D., The voltage-gated sodium channel TPC1 confers endolysosomal excitability. Nat. Chem. Biol. 10, 463–469 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Kintzer A. F., et al. , Structural basis for activation of voltage sensor domains in an ion channel TPC1. Proc. Natl. Acad. Sci. U.S.A. 115, E9095–E9104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawate T., Gouaux E., Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biyani N., et al. , Focus: The interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Sali A., Blundell T. L., Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360(1996). [DOI] [PubMed] [Google Scholar]