Significance
The bactericidal activity of macrolide antibiotics correlates with the presence of an extended alkyl-aryl side chain, which accounts for their slow departure rate from the ribosome. Here, we found that the base pair between 23S ribosomal RNA (rRNA) nucleotides 752 and 2609 located in the macrolide binding site is important for the ribosome functionality and for establishing the unique interactions with the extended side chain of macrolide antibiotics. Disruption of the 752-2609 base pair accelerates the departure of extended macrolides from the ribosome and reduces their cidality. Our results demonstrate that not only the chemical features of the antibiotic, but also the structure of the target site contribute to the ability of the inhibitor to kill bacteria.
Keywords: ribosome, antibiotics, translation, protein synthesis, macrolide
Abstract
While most of the ribosome-targeting antibiotics are bacteriostatic, some members of the macrolide class demonstrate considerable bactericidal activity. We previously showed that an extended alkyl-aryl side chain is the key structural element determining the macrolides’ slow dissociation from the ribosome and likely accounts for the antibiotics’ cidality. In the nontranslating Escherichia coli ribosome, the extended side chain of macrolides interacts with 23S ribosomal RNA (rRNA) nucleotides A752 and U2609, that were proposed to form a base pair. However, the existence of this base pair in the translating ribosome, its possible functional role, and its impact on the binding and cidality of the antibiotic remain unknown. By engineering E. coli cells carrying individual and compensatory mutations at the 752 and 2609 rRNA positions, we show that integrity of the base pair helps to modulate the ribosomal response to regulatory nascent peptides, determines the slow dissociation rate of the extended macrolides from the ribosome, and increases their bactericidal effect. Our findings demonstrate that the ability of antibiotics to kill bacterial cells relies not only on the chemical nature of the inhibitor, but also on structural features of the target.
One of the parameters that define the therapeutic potential of an antibiotic is its ability to kill the pathogen (1–3). Bacteriostatic drugs stop bacteria from growing but do not prevent them from resuming proliferation once the inhibitor is removed. In contrast, only few cells, if any, can resume growth upon treatment with bactericidal antibiotics. While cidality is related to the mode of antibiotic action, even inhibitors of the same class can differ significantly in their ability to kill bacteria (4), a concept particularly applicable to macrolide antibiotics (5).
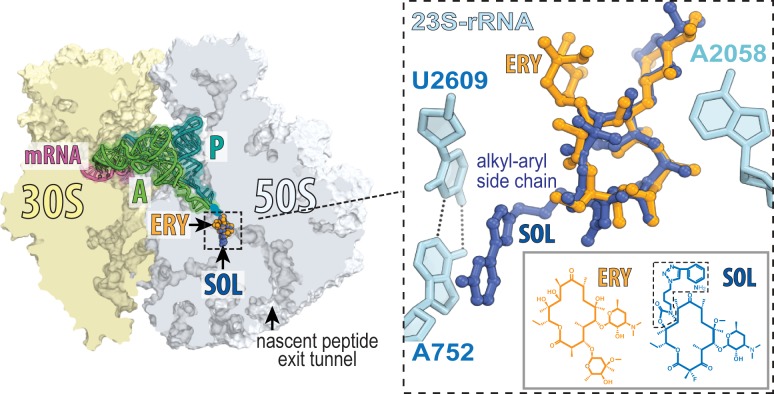
Macrolides inhibit protein synthesis and bacterial growth by binding in the nascent peptide exit tunnel (NPET) of the ribosome (6) (Fig. 1). While many macrolides, e.g., erythromycin (ERY), are largely bacteriostatic (7, 8), drugs of later generations, e.g., solithromycin (SOL), exhibit a more pronounced bactericidal activity (5, 9). We recently showed that the cidal capacity of macrolides depends on their dissociation kinetics from the ribosome (10). The faster dissociating macrolides tend to be bacteriostatic, whereas the slower dissociating ones are considerably more bactericidal. Importantly, the rate of dissociation from the ribosome and the cidality of the macrolides critically depend on the presence of an extended alkyl-aryl side chain in the antibiotic structure (10) (Fig. 1).
Fig. 1.
The binding site of macrolides in the ribosome. A cross-section of the E. coli ribosome showing ERY (PDB ID code 4V7U) (11) and SOL (PDB ID code 4WWW) (12) bound in the nascent peptide exit tunnel. The zoomed-in image shows the interaction of the alky-aryl side chain (boxed) of SOL with the A752-U2609 base pair.
Crystallographic studies showed that in the nontranslating Escherichia coli ribosome the macrolide side chain may interact with a putative 23S ribosomal RNA (rRNA) base pair formed by residues A752 and U2609 belonging to the distant domains II and V, respectively (11, 12) (Fig. 1). However, both of these residues are partially accessible for modifications by chemical reagents that target single-stranded RNA (13–15), and it remains unknown whether this base pair forms in a ribosome that is engaged in protein synthesis. Residues A752 and U2609 were rendered unpaired in the initial crystallographic structures of the vacant E. coli ribosome (16) but were presented in base-paired configuration in later reconstructions of the ribosome complexed with macrolides (11, 12). In the structures of ribosomes from other bacteria, these residues have been rendered unpaired, partially paired, or fully paired (17–20). Irrespective of the interaction status assigned on the basis of static crystallographic structures, the existence of the A752-U2609 base pair in the translating ribosome, its importance for macrolide binding, and its contribution to the mode of antibiotic action have not been established with any certainty.
Here, we demonstrate that in the translating ribosome the distant residues A752 and U2609 likely form a base pair, which contributes to nascent peptide sensing. We show that this long-range base pair is critical for the dissociation kinetics and cidality of macrolides with extended side chains, likely due to specific interactions between this base pair and the drug. Our finding may guide a strategy for developing new macrolides with improved bactericidal properties through optimization of the idiosyncratic interactions with the ribosome.
Results and Discussion
Disruption of the A752-U2609 Base Pair Affects the Ribosomal Response to Regulatory Nascent Peptides.
We engineered two E. coli strains to carry the single mutations A752G or U2609C that would partially or completely disrupt the putative base pair, and a third strain where mutations A752G and U2609C were introduced simultaneously to restore the base-pairing potential. Because in the engineered strains the rRNA is exclusively expressed from a plasmid (21), the cells contain pure populations of mutant ribosomes.
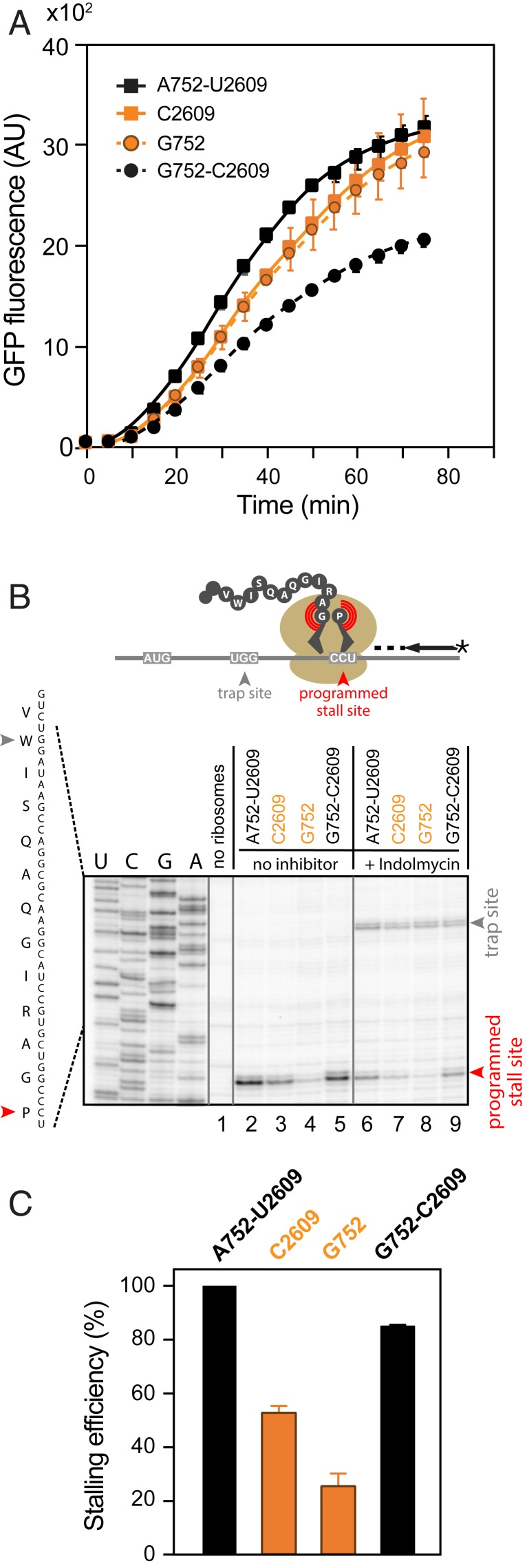
The doubling time of all three mutants in rich media was practically indistinguishable from that of the WT cells (τ ∼ 32 min), and the in vitro activity of the isolated wt and mutant ribosomes was comparable (Fig. 2A). Thus, neither the identity of the 23S rRNA residues 752 or 2609, nor their potential to form a base pair, is critical for cell growth or for general ribosomal functions.
Fig. 2.
Role of the 752-2609 base pair in ribosomal activities. (A) In vitro translation of the sf-GFP protein by WT and mutant ribosomes monitored by fluorescence. Error bars represent SD of three independent experiments. (B) Toeprinting analysis of the SecM-mediated programmed ribosome stalling during translation of the secM gene by WT and mutant ribosomes. Bands representing SecM-arrested ribosomes (Pro codon in the A site) are marked by a red arrowhead. Reactions in lanes 6–9 contained indolmycin, an inhibitor of tryptophanyl-tRNA synthetase, which leads to trapping ribosomes at the Trp codon (gray arrowhead) prior to the secM programmed arrest site. The comparable intensity of the trap-site bands shows that the general translation of secM is not affected by the ribosomal mutations. (C) The bar graph showing relative intensity of the secM arrest bands in samples 2–5. Error bars represent SEM of two independent experiments.
Since nucleotides A752 and U2609 are located in the NPET, where they likely interact with growing proteins, we wondered whether they could play a role in the ribosomal response to regulatory nascent peptides (22). Indeed, disruption of the A752-U2609 base pair decreases the efficiency of translational arrest mediated by SecM (Fig. 2B, lanes 3 and 4), a nascent peptide involved in sensing secretion stress (23). Remarkably, SecM-mediated stalling was reduced in single mutants but was restored in the A752G/U2609C double mutant (Fig. 2B, lane 5). These findings parallel the previous observation that the integrity of the 752-2609 base pair is important for ribosome stalling during translation of the regulatory leader peptide TnaC of the tryptophanase operon (24). Altogether, our data argue that the A752-U2609 base pair does form in the translating ribosome, facilitating the response to nascent peptides.
The Ribosomes with an Intact or Disrupted 752-2609 Base Pair Bind Macrolides with Comparable Affinities.
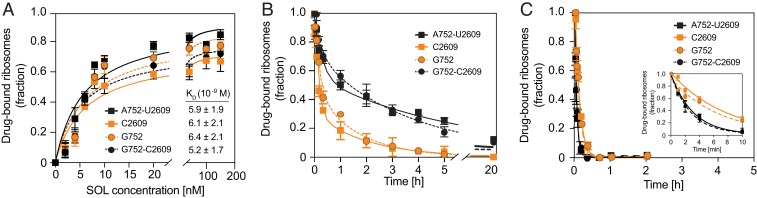
Because crystallographic studies have indicated that the alkyl-aryl side chain of macrolides can interact with the paired A752/U2609 nucleotides (11, 17) (Fig. 1), it was suggested that disruption of this base pair would decrease the affinity of the drug for the ribosome (11). However, equilibrium binding of SOL to the ribosome was minimally affected by the mutations (Fig. 3A). Hence, varying the identities of the 23S rRNA residues 752 and 2609 or disrupting their base-pairing capacity does not impact the macrolides overall affinity for the ribosome.
Fig. 3.
Effect of the 752-2609 base pair on the interactions of macrolides with the ribosome. (A) Equilibrium binding of SOL. Ribosomes were equilibrated for 2 h with varying [14C]-SOL concentrations and then the amount of bound antibiotic was measured (10). KD values determined from the binding curves are indicated. SOL (B) and ERY (C) dissociation kinetics. Following preequilibration of ribosomes with [14C]-SOL (B) or [14C]-ERY (C), an excess of the corresponding unlabeled antibiotic was added, and ribosome-associated radioactivity was monitored over time. Error bars represent SD of three independent replicates.
The Integrity of the A752-U2609 Base Pair Slows the Departure of the Extended Macrolides from the Ribosome.
To further characterize the interactions between macrolides and ribosomes with or without the A752G/U2609C base pair, we measured the rate of antibiotic dissociation. Consistent with our previous data (10), SOL slowly dissociates from WT ribosomes with biphasic kinetics, likely reflecting the existence of fast and slowly dissociating ribosome–drug complexes (Fig. 3B and Table 1). Similar kinetic was observed for the mutant ribosomes with the restored A752G/U2609C base pair (Fig. 3B and Table 1). In contrast, the single A752G or U2609C mutations accelerated the SOL off rate in both the fast-dissociating and slow-dissociating populations. In addition, the fast-dissociating population became predominant, leading to an overall significantly expedited departure of SOL from the ribosome (Fig. 3B and Table 1). Unlike their effect on dissociation kinetics of SOL, these mutations had only a minor influence on the monophasic dissociation kinetics of ERY (Fig. 3C and Table 1), a drug that lacks the alkyl-aryl side chain (Fig. 1). These data suggest that the effect of the 752-2609 base pair on the rate of drug dissociation from the ribosome is specifically mediated by the interaction established with the macrolide side chain.
Table 1.
Kinetic parameters of dissociation of ERY and SOL from WT or mutant E. coli ribosomes
| Mutation(s) | ERY* | SOL† | ||
| k, min−1 | kfast, min−1 | kslow, min−1 | Fraction of fast population, %) | |
| WT(A752/U2609) | 0.32 ± 0.020 | 0.063 ± 0.018 | 0.0036 ± 0.0008 | 42.7 ± 6.3 |
| U2609C | 0.15 ± 0.015 | 0.120 ± 0.019 | 0.0085 ± 0.0002 | 69.1 ± 5.4 |
| A752G | 0.13 ± 0.010 | 0.089 ± 0.020 | 0.0101 ± 0.0002 | 53.1 ± 8.2 |
| A752G/U2609C | 0.36 ± 0.023 | 0.063 ± 0.045 | 0.0050 ± 0.0001 | 26.9 ± 11.2 |
Disruption of the 752-2609 Base Pair Alleviates Macrolide Cidality.
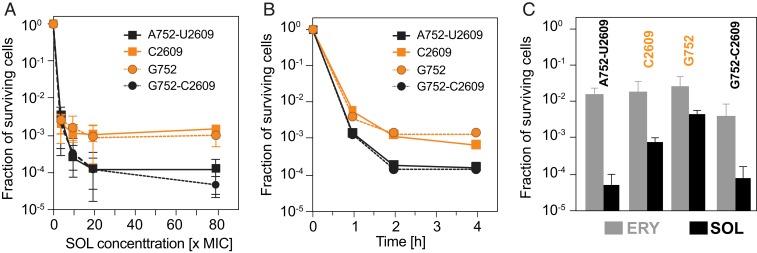
We asked whether disrupting the ability of 752-2609 nucleotides to base pair would also interfere with the cidality of the extended macrolides. First, we determined the minimal inhibitory concentration (MIC) of SOL and found that neither the individual A752G or U2609C, nor the compensatory A752G/U2609C mutations had any pronounced effect on the sensitivity to the drug (MICSOL = 0.25–0.5 µg/mL). [Of note, the previously reported 4- to 8-fold resistance of the U2609C mutant to the extended macrolides (15) was observed only in strains with the intact tolC gene, whereas it was not manifested in the ΔtolC cells used in the present experiments.]
Having established that the rRNA mutations do not affect MICSOL in our strains, we examined the survival of the WT and mutant cells after their exposure to SOL. When exposed to high concentrations of SOL, only ∼0.01% of the cells with the 752-2609 base pair could resume growth whereas, in contrast, ∼10 times more cells survived the equivalent antibiotic treatment when the 752-2609 base pair was weakened or disrupted by the single mutations (Fig. 4A). The same trend was observed when cells were exposed for varying time intervals to a fourfold MIC of SOL (Fig. 4B). This result shows that the integrity of the 752-2609 base pair defines not only the dynamics of the interaction between the drug and the ribosome but also the bactericidal activity of the drug against E. coli. The mutations had only minimal effect on cidality of the side chain-lacking ERY (Fig. 4C).
Fig. 4.
The base pairing ability of the 23S rRNA residues 752 and 2609 impacts the bactericidal properties of SOL. (A) Concentration dependence of SOL cidality after 4 h drug exposure. (B) Time dependence of the cidal effect of 4× MIC of SOL. (C) Cell survival after 4 h treatment with 7× MIC of ERY (gray bars) or SOL (black bars). Error bars represent SD of three independent replicates.
Although in ∼95% of analyzed bacterial genomes the identities of the 23S rRNA residues 752 and 2609 (E. coli numbering) support the base pair formation, the pairing ability is not universal (25). Nevertheless, macrolides with extended side chain can exhibit cidality even against some bacteria lacking the base pair (26). This agrees well with our data showing that although the disruption of the 752-2609 base pair reduced SOL cidality, this antibiotic still killed the mutant cells more readily than ERY, which lacks the alkyl-aryl side chain (Fig. 4C). Conceivably, when the 752-2609 base pair cannot be formed, the macrolide's extended side chain may maintain stacking interaction with one of the unpaired bases or, alternatively, may reorient and interact with other nearby rRNA residues (20, 27). It is also possible that even in the species where pairing of the 23S rRNA residues 752 and 2609 can occur, the base pair may form only transiently in response to additional cues, e.g., the presence of a nascent protein with a specific amino acid sequence in the NPET or the binding of specific small molecules, including the extended-chain macrolides (6). The dynamic nature of this base pair could account for the biphasic kinetics of SOL dissociation from the E. coli ribosomes observed in our experiments (Fig. 3B), as well as biphasic binding mode of extended macrolides reported previously (28), where the off and on rates of the antibiotic could depend on the formation or disruption of the pairing of the 752 and 2609 residues.
Although our experiments have been carried out only with SOL, we expect that similar effects would be observed with other extended macrolide antibiotics, e.g., telithromycin, whose alkyl-aryl side chain establishes equivalent interactions with the A752-U2609 base pair (11, 12). Altogether, our findings suggest that optimizing the interactions with the A752-U2609 base pair by modifying the side chain of macrolides or other drugs binding in a similar ribosomal location could be a strategy to improve the antibiotics cidal activity.
Materials and Methods
Construction of Mutant Strains.
Single A752G or U2609C or double A752G/U2609C mutations were introduced by site-directed mutagenesis into 23S rRNA gene of the pAM552 plasmid (29) using the QuikChange Lightning Multi Site-directed mutagenesis kit (Agilent Technologies). Plasmids were transformed into the ΔtolC E. coli SQ171 strain lacking chromosomal rRNA alleles (21, 30), and transformants were cured off the resident pCSacB plasmid encoding WT rRNA (31). The presence of the mutation and the purity of the ribosome population carrying the mutant rRNA was verified by sequencing and by primer extension on the total cellular rRNA.
Ribosome Preparation and Binding Studies.
WT and mutant ribosomes were purified according to ref. 32. Equilibrium and kinetic binding studies were performed as described in ref. 10.
In Vitro Translation and Toeprinting.
Translation and toeprinting reactions were carried out in the Δribosome PURExpress system (New England Biolabs) supplemented with isolated WT or mutant ribosomes as described in ref. 29. Plasmid pY71-sfGFP (33) was used as a template for translation of sfGFP reporter protein. Expression of sfGFP was continuously monitored by fluorescence (488ex/520em nm) in a microplate reader (Tecan). Toeprinting analysis was carried out as described in ref. 34 using a DNA template prepared by PCR encoding the last 29 codons of the secM gene.
MIC Determination and Cidality Testing.
MIC was determined in 96-well plates by serial dilution of antibiotic and incubating plates overnight at 37 °C without shaking. The optical density of the starting bacterial cultures was A600 = 0.001. For the analyses of bactericidal action, overnight cultures were diluted 1:500 and grown at 37 °C to A600 ∼ 0.2. Various concentrations of SOL or ERY were added and culture dilutions were plated after 4 h incubation. For the time-kill measurements, cells were incubated with 4× MICSOL (2 µg/mL for WT, A752G, and A752G/U2609C mutants, 1 µg/mL for U2609C mutant). Aliquots were withdrawn, and culture dilutions were plated. Colonies were counted following 48- to 72-h incubation at 37 °C. For comparison of SOL and ERY cidality, cells were incubated for 4 h with 7× MIC concentrations of the drugs: SOL (3.5 µg/mL for WT, A752G, and A752G/U2609C mutants, 1.75 µg/mL for U2609C mutant); ERY (7 µg/mL for WT, A752G, and A752G/U2609C mutants, 3.5 µg/mL for U2609C mutant).
Data Availability.
The engineered strains are available upon request.
Acknowledgments
We thank Dorota Klepacki, Tanja Florin, and Prabha Fernandes for help and advice; and Yury Polikanov for helpful discussions and help with figure preparation. This work was supported by Grant R21 AI137584 from the National Institutes of Health.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
References
- 1.Osmon D. R., Antimicrobial prophylaxis in adults. Mayo Clin. Proc. 75, 98–109 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Sinner S. W., Tunkel A. R., Antimicrobial agents in the treatment of bacterial meningitis. Infect. Dis. Clin. North Am. 18, 581–602 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Hook E. W. 3rd, Roberts R. B., Sande M. A., Antimicrobial therapy of experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 8, 564–570 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohanski M. A., Dwyer D. J., Wierzbowski J., Cottarel G., Collins J. J., Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandes P., Martens E., Bertrand D., Pereira D., The solithromycin journey-It is all in the chemistry. Bioorg. Med. Chem. 24, 6420–6428 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Vázquez-Laslop N., Mankin A. S., How macrolide antibiotics work. Trends Biochem. Sci. 43, 668–684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawks G. H., Antibiotic therapy of staphylococcal infections. Can. Med. Assoc. J. 93, 848–853 (1965). [PMC free article] [PubMed] [Google Scholar]
- 8.Norcia L. J., Silvia A. M., Hayashi S. F., Studies on time-kill kinetics of different classes of antibiotics against veterinary pathogenic bacteria including Pasteurella, Actinobacillus and Escherichia coli. J. Antibiot. (Tokyo) 52, 52–60 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Woosley L. N., Castanheira M., Jones R. N., CEM-101 activity against Gram-positive organisms. Antimicrob. Agents Chemother. 54, 2182–2187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svetlov M. S., Vázquez-Laslop N., Mankin A. S., Kinetics of drug-ribosome interactions defines the cidality of macrolide antibiotics. Proc. Natl. Acad. Sci. U.S.A. 114, 13673–13678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunkle J. A., Xiong L., Mankin A. S., Cate J. H., Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 107, 17152–17157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llano-Sotelo B., et al. , Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis. Antimicrob. Agents Chemother. 54, 4961–4970 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong L., Shah S., Mauvais P., Mankin A. S., A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31, 633–639 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Hansen L. H., Mauvais P., Douthwaite S., The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31, 623–631 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Garza-Ramos G., Xiong L., Zhong P., Mankin A., Binding site of macrolide antibiotics on the ribosome: New resistance mutation identifies a specific interaction of ketolides with rRNA. J. Bacteriol. 183, 6898–6907 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., Dunkle J. A., Cate J. H., Structures of the ribosome in intermediate states of ratcheting. Science 325, 1014–1017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulkley D., Innis C. A., Blaha G., Steitz T. A., Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 107, 17158–17163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eyal Z., et al. , Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 112, E5805–E5814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms J., et al. , High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Berisio R., et al. , Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 185, 4276–4279 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quan S., Skovgaard O., McLaughlin R. E., Buurman E. T., Squires C. L., Markerless Escherichia coli rrn deletion strains for genetic determination of ribosomal binding sites. G3 (Bethesda) 5, 2555–2557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito K., Chiba S., Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 82, 171–202 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Nakatogawa H., Ito K., The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Martínez A. K., et al. , Crucial elements that maintain the interactions between the regulatory TnaC peptide and the ribosome exit tunnel responsible for Trp inhibition of ribosome function. Nucleic Acids Res. 40, 2247–2257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quast C., et al. , The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gustafsson I., Engstrand L., Cars O., In vitro pharmacodynamic studies of activities of ketolides HMR 3647 (Telithromycin) and HMR 3004 against extracellular or intracellular Helicobacter pylori. Antimicrob. Agents Chemother. 45, 353–355 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu D., Blaha G., Moore P. B., Steitz T. A., Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121, 257–270 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Kostopoulou O. N., Petropoulos A. D., Dinos G. P., Choli-Papadopoulou T., Kalpaxis D. L., Investigating the entire course of telithromycin binding to Escherichia coli ribosomes. Nucleic Acids Res. 40, 5078–5087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orelle C., et al. , Protein synthesis by ribosomes with tethered subunits. Nature 524, 119–124 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Kannan K., Vázquez-Laslop N., Mankin A. S., Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell 151, 508–520 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Zaporojets D., French S., Squires C. L., Products transcribed from rearranged rrn genes of Escherichia coli can assemble to form functional ribosomes. J. Bacteriol. 185, 6921–6927 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohashi H., Shimizu Y., Ying B. W., Ueda T., Efficient protein selection based on ribosome display system with purified components. Biochem. Biophys. Res. Commun. 352, 270–276 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Bundy B. C., Swartz J. R., Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjug. Chem. 21, 255–263 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Orelle C., et al. , Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 41, e144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The engineered strains are available upon request.