Significance
Rabies virus (RABV) and other viruses with single-segment, negative-sense, RNA genomes have a multi-functional polymerase protein (L) that carries out the various reactions required for transcription and replication. Many of these viruses are serious human pathogens, and L is a potential target for antiviral therapeutics. Drugs that inhibit polymerases of HCV and HIV-1 provide successful precedents. The structure described here of the RABV L protein in complex with its P-protein cofactor shows a conformation poised for initiation of transcription or replication. Channels in the molecule and the relative positions of catalytic sites suggest that L couples a distinctive capping reaction with priming and initiation of transcription, and that replication and transcription have different priming configurations and different product exit sites.
Keywords: rabies lyssavirus, NNS RNA viruses, vesicular stomatitis virus, transcription, replication
Abstract
Nonsegmented negative-stranded (NNS) RNA viruses, among them the virus that causes rabies (RABV), include many deadly human pathogens. The large polymerase (L) proteins of NNS RNA viruses carry all of the enzymatic functions required for viral messenger RNA (mRNA) transcription and replication: RNA polymerization, mRNA capping, and cap methylation. We describe here a complete structure of RABV L bound with its phosphoprotein cofactor (P), determined by electron cryo-microscopy at 3.3 Å resolution. The complex closely resembles the vesicular stomatitis virus (VSV) L-P, the one other known full-length NNS-RNA L-protein structure, with key local differences (e.g., in L-P interactions). Like the VSV L-P structure, the RABV complex analyzed here represents a preinitiation conformation. Comparison with the likely elongation state, seen in two structures of pneumovirus L-P complexes, suggests differences between priming/initiation and elongation complexes. Analysis of internal cavities within RABV L suggests distinct template and product entry and exit pathways during transcription and replication.
Rabies virus (RABV) and other viruses with nonsegmented, negative-strand (NNS) RNA genomes have, as the catalytic core of their replication machinery, a large, multifunctional RNA polymerase (L). Many of these viruses are serious human pathogens, including Ebola virus, respiratory syncytial virus, and measles. Vaccines are available for some, including descendants of the storied work on rabies by Louis Pasteur and Pierre Paul Émile Roux, but specific, small-molecule therapeutics are still under development. By analogy with inhibitors of viral polymerases of many other types, L would be a suitable candidate for inhibitor development, for which structural and mechanistic studies are essential precursors.
For all mononegaviruses, including the rhabdoviruses RABV and vesicular stomatitis virus (VSV), L associates with a cofactor known as P (for phosphoprotein). P bridges L with the viral nucleocapsid, an antisense RNA genome fully encapsidated by the viral nucleoprotein (N). The complete structure of L from VSV (1), the sole example published so far of an NNS viral polymerase in which all five domains are clearly resolved, shows the global features that sequence comparisons suggest it has in common with most other NNS RNA viral L proteins. Its multidomain organization includes three enzymatic modules: an RNA-dependent RNA polymerase (RdRp), a capping domain (CAP), and a dual-specificity methyltransferase (MT) domain. CAP is a GDP:polyribonucleotidyltransferase (PRNTase) that transfers a 5′ monophosphate of the nascent RNA transcript onto a GDP acceptor (2); the single MT domain methylates the ribose 2′-O position on the first nucleotide of the transcript and then the N-7 position of the capping guanylate (3). Transcription initiation and cap addition require several residues within or proximal to a priming loop from CAP, which extends into the RdRp core (4). Residues at related positions in the RABV L amino acid sequence are similarly essential for its activities. A connector domain (CD) and a C-terminal domain (CTD), both with nonenzymatic functions, flank the MT domain and, in conjunction with P, likely facilitate the large structural rearrangements that appear to coordinate the three enzymatic activities during transcription and replication.
We report here an atomic model of L from RABV SAD-B19 in complex with a 91-residue, N-terminal fragment of P (P1–91) from electron cryo-microscopy (cryo-EM) at an average resolution of 3.3 Å. The atomic structure resembles that of VSV L-P in many respects, consistent with the 34.1% amino acid sequence conservation between the two L proteins. As in the VSV L-P complex, binding of P1–91 locks the CD, MT domain, and CTD into a fixed, “closed” arrangement with respect to the large RdRp-CAP module. This closed conformation appears to represent the L protein poised for initiation at the 3′ end of the genome or antigenome. Comparison with structures of two recently published pneumovirus L-P complexes (5, 6) as well as with that of a VSV L-P reconstruction determined at 3.0 Å resolution (7) suggests that replication and transcription have alternative priming configurations and alternative product exit sites.
Results and Discussion
Identification of a Stable L-P Complex.
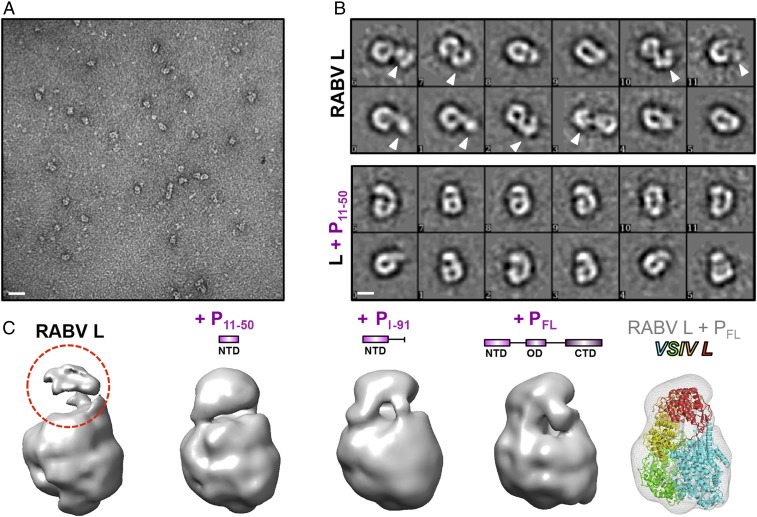
We have determined previously that a minimal fragment of the N-terminal domain of RABV P, P11–50, is sufficient to stimulate processivity of RABV L on a nonencapsidated RNA template derived from the 3′ leader sequence of the RABV genome (8). In our prior studies of VSV, we found a minimal fragment of VSV P that stimulated VSV L activity in vitro and also stably anchored its C-terminal globular domains (CD, MT, and CTD) against the N-terminal RdRp and CAP domains (9, 10). We visualized RABV L alone or in complex with various fragments of P by negative-stain electron microscopy (Fig. 1) and found that, in the absence of P, the C-terminal domains of RABV L, like those of VSV L, have a range of positions with respect to the ring-like RdRp-CAP core (Fig. 1B). While P11–50 stabilized the C-terminal domains of RABV L, a longer fragment (P1–91) shifted their positions further, to a degree equivalent to that achieved by adding full-length P (PFL) (Fig. 1C). To fit the structure of VSV L (obtained in complex with VSV P35–106) into a low-resolution map for RABV L-P11–50, we needed to fit the C-terminal domains at an ∼80° offset (data not shown), but we could readily fit the low-resolution maps of RABV L-P1–91 and RABV L-PFL with the structure of VSV L as a single rigid body (Fig. 1 C, Right). We inferred that the RABV L-P1–91 and VSV L-P35–106 complexes reflected analogous states and therefore used P1–91 to determine the RABV L structure by cryo-EM.
Fig. 1.
Effect of P on conformation of L. (A) Electron micrograph of negatively stained RABV L. (Scale bar, 25 nm.) (B) Representative 2D class averages of L alone or in complex with P11–50. (Scale bar, 5 nm.) Arrowheads indicate flexible conformations of the three C-terminal globular domains of L. (C) Three-dimensional reconstructions of negatively stained L alone or in complex with the indicated fragment of P. Red dashed circle indicates poor density for the disordered C-terminal globular domains in the absence of any P. (Right) The VSV L atomic model fit into the RABV L-PFL reconstruction.
Cryo-EM Structure Determination.
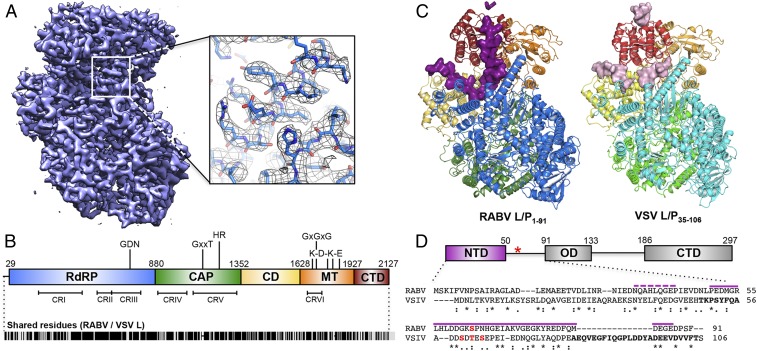
We immobilized purified complexes of RABV L-P1–91 in vitreous ice and visualized them by cryo-EM (SI Appendix, Fig. S1 A and B) as described in Materials and Methods. We created an initial three-dimensional (3D) reference de novo using the ab-initio module in cisTEM (11) from dose-fractionated movies obtained at 200 kV acceleration and calculated a 6.7 Å resolution map from 22,000 particles after classification and refinement using RELION (12) (SI Appendix, Fig. S1C). In extending the resolution from movies recorded at 300 kV acceleration, preferred orientation of the particles in the ice complicated particle selection and 3D reconstruction. By using an angular binning procedure to decrease representation of common views (SI Appendix, Figs. S1D and S2), we obtained a reasonably isotropic reconstruction using cisTEM. After per-particle, projection-based contrast transfer function (CTF) and motion correction (Bayesian polishing) in RELION 3.0 (13, 14), refinement in cisTEM yielded a map with an average resolution of 3.3 Å (Fourier shell correlation = 0.143 criterion; Fig. 2A and SI Appendix, Figs. S1E and S2).
Fig. 2.
Structure of the RABV L-P complex. (A) A 3.3-Å cryo-EM map of RABV L-P1–91 and model fit into the density with clear side-chains even on exterior residues. (B) Domain organization of RABV L, with key catalytic residues listed. Numbers give amino acid positions at domain boundaries. (Below) Locations of conserved regions (CRI-VI) throughout NNS RNA virus L proteins and amino acid similarity score with VSV L (Indiana strain); black lines indicate identical residues (∼35%). (C) Structure of RABV L-P1–91 (Left) and the model of VSV L-P35–106 from a 3.0-Å cryo-EM map (Right) (7). Domains are colored as in B. P for both structures is shown in surface representation, colored in plum (RABV) and light pink (VSV). The five unassigned residues of RABV P are represented in plum ribbon. (D) Domain organization of RABV P (Top) amino acid positions (red asterisk, position of Ser63P, typically phosphorylated). (Below) Alignment with VSV P (purple lines, modeled RABV P residues; boldface letters, modeled VSV P residues; asterisks, shared residues; colons, chemically similar residues; periods, weakly similar residues; red, phosphorylation sites).
To generate an atomic model, we aligned and threaded the sequence of RABV L onto the atomic coordinates of VSV L and crudely fit the model to the map using Rosetta (15–17). After manual adjustment, we could fit nearly all of the 2,127-residue polypeptide chain of L from residue 29 to the C terminus (Fig. 2). We assigned residues for regions of continuous, but disordered, density in the CAP priming loop (1,177 to 1,184) and the long linker between the CD and MT domains (1,624 to 1,635) because there was little ambiguity about the general course of the polypeptide chain, although the Cα positions and the side-chain orientations are imprecise. Several interdomain linkers that were ambiguous in the VSV L map have continuous and ordered density for the analogous regions in the RABV L map, permitting assignment of their residues with high confidence. The clarity of side-chains throughout all five domains of L indicates that regions of the map with poor density reflect local flexibility rather than wholesale domain movements (SI Appendix, Fig. S3). Domain-specific refinement from a much larger portion of the dataset using RELION Multi-Body showed significant interdomain flexibility (Movie S1) and suggested that the comparatively small fraction of the dataset used for the 3.3-Å reconstruction reflects a particular configuration within a somewhat broader ensemble.
Rigorous selection of this small set of homogeneous particles for the 3.3-Å map yielded clear and nearly continuous density for the C-terminal half of P1–91. Visible density for some side-chains on a stretch of P bound to the CTD permitted assignment of the sequence register in this region, from which we have inferred the approximate residue positions for the remainder of visible P density. We have therefore assigned 37 of the 91 residues of P included in the complex (Fig. 2 C and D), not including a stretch of 5 residues, which we could not assign, bound to the top of the CTD. The cryo-EM data collection and model validation statistics for the map and complete RABV L-P structure are summarized in SI Appendix, Table S1.
The structures of RABV and VSV L in complex with their respective P fragments are very similar (Fig. 2C). Rigid-body alignment of these structures yields a root-mean-square deviation (rmsd) of 1.905 Å based on 1,539 Cα residues; individual domain rmsds are 0.91 Å (595 Cα), 1.07 Å (352 Cα), 2.09 Å (186 Cα), 1.50 Å (233 Cα), and 3.68 Å (120 Cα), respectively, for the RdRp, CAP, CD, MT, and CTD domains superposed independently (SI Appendix, Fig. S4). There is nearly complete conservation of the secondary structure elements from end to end, owing to strong sequence conservation between each of the five domains for both proteins: shared amino acid identities are 36.1, 41.9, 25.4, 31.2, and 20.1% for the RdRp, CAP, CD, MT, and CTD domains, respectively. The overall course of P as it transits through L is also similar for both RABV and VSV, despite broad differences in sequence among the residues that could be assigned for the respective P fragments and the mere 11.8% shared amino acid identities between them overall.
Phosphoprotein Fragment P1–91.
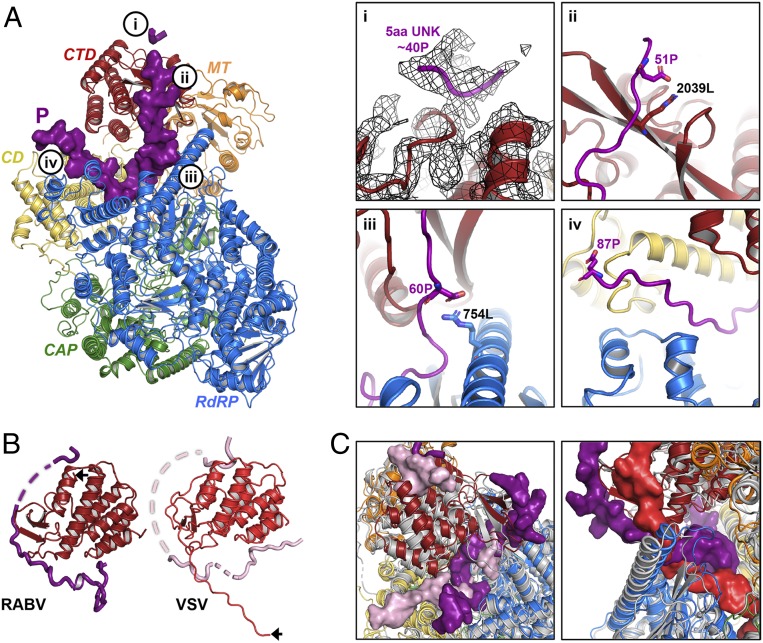
We detect two segments of density for the P1–91 polypeptide chain (Fig. 3). One segment binds in a groove on the outward-facing surface of the CTD above the C-terminal residues of L (Fig. 3 A, i). This segment is too short to assign its sequence with confidence, but biochemical experiments that measured RABV L processivity on naked RNA templates in the presence of N-terminal fragments of P suggest that this interaction involves at least some of the residues immediately following Gln40 of P (8). The second segment is much longer, with a continuous stretch of 37 residues. It contains about 11 residues bound on the proximal face of the CTD, probably in part through a salt bridge between Glu51P and Arg2039L (Fig. 3 A, ii); about 3 residues in contact with a long RdRp helix (residues 746 to 773), probably including a salt bridge between Asp60P and Arg754L (Fig. 3 A, iii); and about 13 additional residues in the groove between the RdRp and CD (Fig. 3 A, iv). The C terminus of the ordered segment that we could build is Glu87P. The closer association of P with the proximal face of the CTD of L for RABV relative to VSV suggests that RABV P partially compensates for the C-terminal tail that is “missing” in RABV L (Fig. 3B).
Fig. 3.
Association of RABV P1–91 with L. (A, Left) RABV L-P1–91 as in Fig. 2C. Bubbled letters indicate the regions shown in greater detail in i–iv on the Right. (A, Right) (i) Unassigned residues of P at the top of the CTD with the 3.3-Å map in black mesh. (ii–iv) Indicated residues of L (black) and P (purple) in stick representation. (B) CTDs of RABV and VSV L; P is in plum for RABV and light pink for VSV. Black arrows indicate the C-terminal residues of each L. (C) Superposition of RABV L (colored cartoon, as in Fig. 2) and VSV L (gray cartoon). (Left) Different paths taken by RABV P (plum, surface) and VSV P (light pink, surface) through L. (Right) RABV P (plum, surface) is partially coincident with the VSV C-tail (red, surface) in the interdomain cavity of L, while VSV P occupies far less of the same cavity.
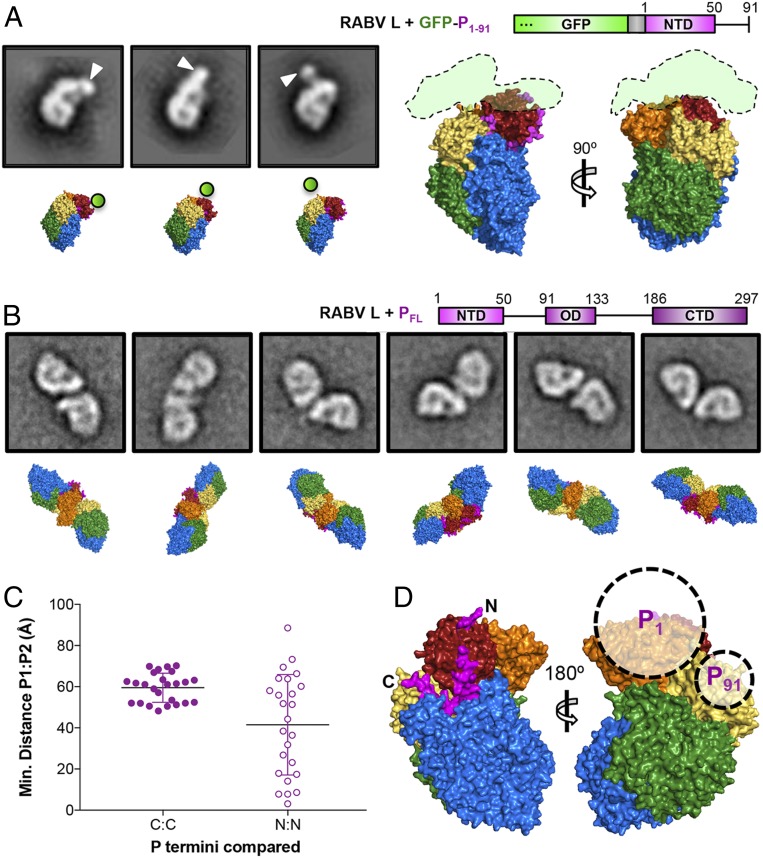
As a first check on the polarity that we had assigned to P, we fused GFP to the N terminus of P1–91 and visualized its complex with L by negative-stain EM (Fig. 4A). A low-resolution 3D reconstruction showed diffuse density for the globular GFP at the top of L, radiating from a position near the top of the long CD:MT linker (Fig. 4A and SI Appendix, Figs. S5 and S6). This result is consistent with our assignment of the direction of P residues in the density and with our assignment of residues near Gln40P to the segment bound on the CTD outward-facing surface. The map for the L-GFP-P1–91 complex otherwise resembled that of the L-P1–91 complex without GFP (SI Appendix, Fig. S6), indicating that N-terminal fusion of GFP to P1–91 had not disrupted its binding to L.
Fig. 4.
Analysis of negatively stained L-GFP-P1–91 and L-P dimers. (A, Left) Two-dimensional class averages of negatively stained L-GFP-P1–91 and matched projections of the RABV L atomic model with GFP position approximated (SI Appendix, Figs. S5 and S6). (A, Right) GFP trace from a 3D reconstruction of negatively stained RABV L-GFP-P1–91 (SI Appendix, Fig. S6) superimposed on the RABV L atomic model. (B) Two-dimensional classes of negatively stained RABV L-PFL dimers and matched projections of the RABV L atomic model (SI Appendix, Figs. S7–S9), as described in legend to SI Appendix, Fig. S7. Square boxes in A and B are 336 Å on a side. (C) Minimal distances between Cα atoms of either P87 (C:C) or the N terminus of the P segment bound at the CTD apex (N:N) for each projection-matched dimer. (D) Approximate locations of P1 and P91 with respect to L determined from the projection-matching analysis.
We confirmed the location of the C terminus of P1–91 by visualizing RABV L in complex with PFL by negative-stain EM, which showed dimers of RABV L that closely resembled those of VSV L complexed with full-length VSV P (10). The central, oligomerization domain of RABV P (POD) includes residues 91 to 133, which form a homodimer (18). We matched projections of the high-resolution RABV L-P1–91 map to each L monomer in the negative-stain EM two-dimensional (2D) class averages (Fig. 4B and SI Appendix, Figs. S7–S9). The distance between P91 residues in the crystal structure of the homodimeric RABV POD is 25 Å, and the last ordered residue in our model is 87, giving a spacing (assuming a flexible but extended chain for 87 to 91) of about 55 Å for the exit points of P on two L molecules in the L-PFL dimer. Projection matching of the dimer partners in negative-stain EM images yielded spacings of ∼60 ± 10 Å, while the distances between the N termini of the short, bound segment of P varied widely from 0 to 90 Å (Fig. 4C). We also found several different dimer arrangements (SI Appendix, Fig. S9), all compatible with the polarity of the segments of P that we have modeled, with our assignment of the C-terminal residues of P1–91 and with flexibility of P residues 87 to 91.
RNA-Dependent RNA Polymerase.
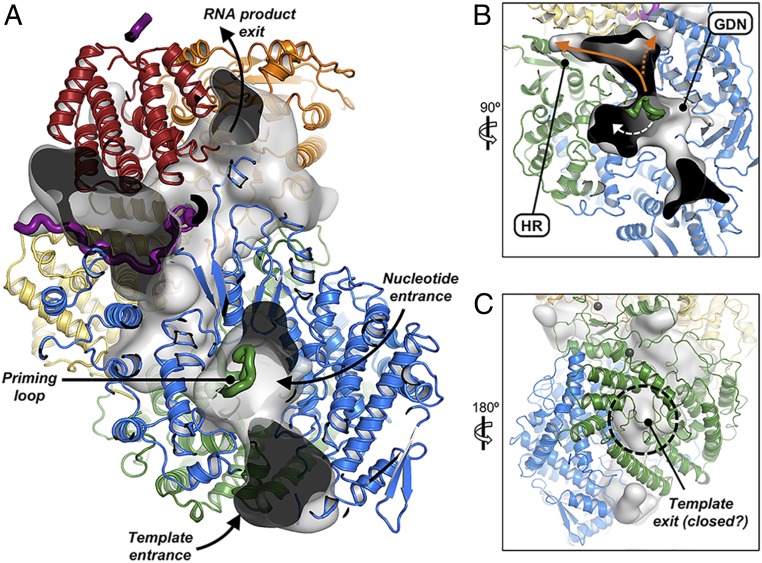
The RdRp domains of RABV and VSV L have nearly coincident 3D structures (SI Appendix, Fig. S4), as expected from their amino acid sequences, which are identical at 36.1% of the aligned positions. They have the familiar finger-palm-thumb structure at their core, augmented by a substantial N-terminal subdomain and by a C-terminal transition into the capping domain (SI Appendix, Fig. S10). The catalytic site (GDN motif) is within a central hollow, connected to the exterior of the molecule by four channels. Comparison with double-stranded RNA virus polymerases, for which structures of initiating and elongating complexes are known (19, 20), has allowed assignment of these channels to the four molecular-passage functions required for RdRp activity: template entry, template exit, nucleotide substrate entry, and product exit (Fig. 5) (21). In the initiation-competent configuration of RABV L that we describe here, the product exit channel is occluded by a retractable priming loop from CAP (Fig. 5B, discussed below), while the putative template exit channel appears “closed” by a grouping of small loops. The exiting template RNA could push these loops aside during elongation (Fig. 5C), as shown for template exiting from influenza virus polymerase (22). A substantially more open channel is present in VSV L (7).
Fig. 5.
Proposed RNA transit through L. (A–C) Cutaways showing continuous internal cavities (gray, translucent surface) through L (colored, as in Fig. 2C) when the priming loop residues (green ribbon) are deleted from the model. Black arrows indicate nucleotide and template entrance channels and the presumed RNA product exit channel. (B) Cavity into which the priming loop may retract during elongation (white dashed arrow). Nascent RNA may continue toward the CAP active site (HR motif) following internal initiation for mRNA transcription (solid orange arrow) or directly upward toward the MT domain following terminal de novo initiation for genomic RNA replication (dashed orange arrow). (C) The presumed template exit channel at the back of the CAP domain (black dashed circle) is occupied by small loops, which we propose move aside as polymerization progresses.
The region with weakest conservation, both of sequence and of structure, surrounds the template entrance channel, the position at which the template must separate from N and thread through the catalytic site. Evidence that virus-specific features of L may facilitate this process comes from the observation that RABV L cannot transcribe RNAs from the VSV nucleocapsid, even when the RNP-binding region of P is replaced by that of VSV (8). The N-terminal 28 residues of RABV L are disordered, as are the initial 34 residues of VSV L. Their adjacency to the template entrance channel suggests a role in nucleocapsid engagement and template insertion; the low isoelectric point of the disordered peptide further suggests that it might facilitate separation of the template RNA from N.
Capping Domain.
The RABV L capping enzyme catalyzes the formation of GTP-capped pre-mRNA by a mechanism that differs from eukaryotic capping. The nascent RNA transcript with a 5′ triphosphate is first covalently linked to a catalytic histidine residue (H1241) in a reaction that leads to a monophosphate RNA-L intermediate. This linkage is subsequently thought to be attacked by a GTP molecule, resulting in addition of GDP to the monophosphate-RNA to form a GTP-capped pre-mRNA (23). Several residues in both RABV and VSV CAP that affect cap addition have been identified, including the GxxT and HR motifs broadly conserved throughout Mononegavirales (1, 24). The histidine in the latter motif is the site of covalent attachment. While we initially supposed that the GxxT motif would support the substrate GTP, the precise role of the motif in capping is unclear, as mutation of the threonine, essential for VSV capping (25), is less critical for capping by RSV L (26). The respective CAP domains are extremely similar (SI Appendix, Figs. S4 and S11), although there are small differences in the positioning of certain loops, including those in or near the active site (SI Appendix, Fig. S11A). The two sites that coordinate structural zinc ions in VSV L are also present in RABV L (SI Appendix, Fig. S11B). Each site includes two pairs of residues separated by approximately 200 residues in the polypeptide chain, thereby joining separate regions within the CAP domain.
The GxxT motif leads into the ∼20-residue-long priming loop, which extends from CAP toward the RdRp active site (SI Appendix, Fig. S11C), as we also found for VSV L (1). Relatively poor density in both maps indicates conformational variability. The state captured in this structure is evidently a preinitiation or initiation conformation, as elongation after priming requires that the priming loop withdraw from the RdRp catalytic cavity to make room for nascent product to reach the CAP active site and extend upward into the MT domain (Fig. 5). The catalytic cavity of CAP appears to be large enough to accommodate a retracted priming loop (Fig. 5B), together with a short segment of nascent transcript, even without displacing CAP from RdRp.
Connector Domain.
The RABV CD has no known enzymatic function and probably has a largely organizational role. The CD of RABV L consists of eight helices; long linkers at either end connect it to the CAP and MT domains (SI Appendix, Fig. S12). Whereas the CAP-CD linker is sufficiently ordered to assign rotamers to several larger side-chains, much of the longer CD-MT linker is disordered, and even main-chain coordinates can be assigned only approximately. Nevertheless, the general course of the latter linker is clear, and we have therefore included the poorly ordered residues in the model (SI Appendix, Fig. S12A).
Opposite the HR motif at the CAP active site are three tightly grouped basic resides in the final helix of the CD (SI Appendix, Fig. S12B). The first two, Arg1610 and His1611, conserved among all Lyssavirus L, are also present in VSV L. The third, R1614, is either arginine or lysine in all lyssaviruses; it is a lysine in VSV L. The proximity of these basic residues in the CD to the catalytic HR motif in CAP, as well as their chemical composition, suggest that they may help guide the nascent transcript toward His1241, the position of covalent attachment. Alternatively, they could direct GTP-capped mRNAs into the MT active site.
Methyltransferase Domain.
The RABV MT is likely to be a dual-function enzyme that methylates the GTP cap of viral mRNAs, first at the 2′-O and then at the N-7 position, as shown for the VSV MT (3). The structurally conserved RABV and VSV MT domains closely resemble other dual-function viral MTases, including those of West Nile Virus (WNV), Dengue Virus (DENV) (SI Appendix, Fig. S13), and human metapneumovirus (27–29). A shared GxGxG motif forms the binding site for the methyl donor, S-adenosylmethionine (SAM), and a conserved set of charged residues (K-D-K-E; RABV residues Lys1685, Asp1797, Lys1829, and Glu1867) forms the catalytic tetrad for methyl group addition (30–33). The K-D-K-E tetrads in RABV and VSV MTs are slightly further from the SAM-binding site than in WNV and DENV MTs, possibly due to the presence of bound S-adenosylhomocysteine (modeled) in the WNV and DENV MT crystals; there is no such density in the RABV and VSV L cryo-EM maps.
In addition to the K-D-K-E and GxGxG motifs, we identified neighboring residues in RABV L that are interspersed with the K-D-K-E residues and are close to the presumptive cap-binding site [indicated by the location of ribavirin in the DENV MTase structure (27)]: R1674, Y1831, T1860, and Y1869 (SI Appendix, Fig. S13B). These residues balance charges of the catalytic residues and may support cap positioning for methylation and/or reaction order. They are conserved throughout Rhabdoviridae and match similarly positioned residues in WNV and DENV.
C-Terminal Domain.
The CTDs of RABV and VSV L are the least conserved of the five domains; only 20.1% of the amino acid residues at corresponding positions are the same. Nonetheless, direct alignment of the CTDs shows they are close structural homologs (SI Appendix, Fig. S3) with an rmsd of 3.7 Å.
Density for the CTD was less well defined than it was for most of the rest of the molecule. Multibody refinement of an expanded particle dataset showed that both the CTD and the MT explore a range of conformations, flexing with respect to the other domains (Movie S1). While the MT flexes as a rigid domain, the CTD appears to have “open” and “closed” conformations that would involve remodeling the α helix spanning residues 2,092 to 2,105 (see the upper left in the middle and right panels, Movie S1). This hinge-like behavior may account for the lack of well-ordered density for P at the apex of the CTD in the high-resolution map. Elements of the CTD closest to the MT domain, such as the helix spanning residues 2,019 to 2,033, follow the MT domain away from the rest of the CTD when open, consistent with the close contact between the two sequential domains. The open state in our particle set could represent a fluctuation toward the fully open conformational ensemble seen in the absence of P, in which the CD, MT, and CTD all move away from the RdRp-CAP module. We suggest below that capping and methylation may require opening of L in this way.
The C-terminal tail that anchors the CTD in VSV L is absent in RABV L, which has 24 fewer residues. Whereas the C-terminal tail in VSV L-P projects into a cavity in the core of the complex bounded by each of the five domains (Fig. 3 B and C), the C terminus of RABV L terminates at the surface of the globular CTD (Fig. 3B). A segment of P that penetrates the gap between CTD and CD, and somewhat closer packing of the adjacent domains, together appear to compensate for the “missing” C-terminal arm (Fig. 3C).
Priming, Initiation, Elongation, and Capping.
The structure described here, together with those of VSV L-P at 3.0 Å resolution (7) and of two recently published pneumovirus L-P complexes (5, 6), suggests a mechanism for switching between replication and transcription, with alternative priming configurations and alternative sites for product exit.
The “priming loop” (residues 1,170 to 1,186 in the CAP domain) projects into the RdRp catalytic cavity, closing off a channel that connects it with the catalytic cavity of the CAP domain. Model building in the homologous VSV L-P complex shows that an initiating nucleotide would stack on a tryptophan (W1167), corresponding to residue W1180 in RABV L, at the tip of the projecting loop (7). Mutation of this residue compromises end initiation, but not internal initiation or capping (4). Elongation beyond formation of the initial dinucleotide requires that the priming loop retract into the CAP catalytic cavity. The recent pneumovirus L-P structures show just such a retracted configuration (5, 6). A nascent transcript can then pass across the retracted loop. Moreover, analysis of cavities in the L-P complex (Fig. 5) shows that, after priming loop retraction, a continuous tunnel leads from the connected catalytic cavities of RdRp and CAP to a likely exit site for the full-length replication products (antigenomic and genomic RNA). Proximity of this putative exit site to the N-terminal end of P would then allow prompt delivery of N protein, bound near the N terminus of P, coating and protecting the emerging replication product (or uncapped leader transcript).
The N-protein delivery pathway suggested above depends on the site at the apex of the CTD that anchors the N-terminal segment of P. That site is a shallow pocket, which accommodates just four or five residues. In the VSV L-P complex, a tyrosine side-chain packs against the base of the pocket; mutation of that tyrosine to small residues severely impairs viral growth, but does not affect RdRp activity (7). We have suggested that failure to anchor P in that site may impede replication (but not transcription) by interfering with efficient delivery of N to the product. That function might explain the conservation of the position of the site, but not its chemistry.
Priming for internal initiation and capping and priming for end initiation may depend on different configurations and different mechanisms for supporting the priming GTP. Internal initiation is insensitive to mutation of Trp1180 (4), suggesting that the priming loop remains retracted after termination of the leader transcript and that support for the priming GTP comes from some other source, such as the 3′ end of the preceding transcript, still base-paired with the template (see a related, early proposal in ref. 34), or from some aspect of the posttermination conformation of the template-engaged RdRp domain. Each of the products of internal initiation has a 5′-AACA sequence and forms a covalent attachment with His1241. A noteworthy characteristic of the “closed” initiation competent structure seen here and for VSV L is the absence of an obvious GTP-binding site in the capping domain. We suggest that the retracted priming loop and elongated nascent transcript might create such a site, perhaps linked to the covalent attachment of the 5′ end of the transcript to His1241. As long as the 5′ attachment to His1241 is present, elongation will produce a loop that will fill the CAP catalytic cavity and force the smaller domains (CD, MT, CTD) to swing outward (SI Appendix, Fig. S14)—as they do in the absence of P (Fig. 1). This transition to a more open complex will allow GTP to diffuse into the CAP active-site cavity (if it is not already there), and it will also expose the methyltransferase catalytic site more completely than in the closed structure. These steps may account for the observation that capping of VSV mRNA occurs only after 31 nucleotides have been transcribed (35).
The scheme proposed here provides a simple mechanism for switching between replication mode, in which full-length product acquires an N-protein coat, and transcription mode, in which an mRNA product acquires a 5′ cap. It also provides an evolutionary rationale for the distinctive capping mechanism (polyribonucleotide transferase rather than guanylyl transferase) found in NNS viruses. Retraction of the priming loop creates a continuous cavity shared by the CAP and RdRp catalytic sites. Covalent attachment of the 5′ end of the transcript then allows ongoing RNA polymerization to fill this cavity with product strand, generating the force needed to release the CD-MT-CTD module. Tests of this model will require structures of transcribing intermediates.
Materials and Methods
Protein Expression and Purification.
We expressed 6xHis-tagged recombinant RABV SAD-B19 L alone or with variants of RABV SAD-B19 P in SF9 or SF21 insect cells from baculovirus vectors constructed using the pFastBac-Dual recombination system (Thermo Fisher) as described previously (8). The N-terminal fragments of RABV SAD-B19 P, P11–50, and P1–91 were expressed in Escherichia coli and purified as previously described (8). The P variants GFP-P1–91, PFL, and PΔOD with StrepII affinity tags were coexpressed with L in insect cells and copurified with L. For GFP-P1–91, with an N-terminal StrepII tag, L-GFP-P1–91 complexes were purified first on nickel and then on Strep-Tactin resin (GE Healthcare). For PFL, with a C-terminal StrepII tag, L-PFL complexes were purified first on Strep-Tactin and then on nickel resin. For PΔOD, with a hemagglutinin (HA) epitope tag followed by an internal HRV 3C protease cleavage site in place of the deleted oligomerization domain of P (P residues 92 to 131), as well as a C-terminal StrepII tag, L-PΔOD complexes were immobilized on Strep-Tactin resin and then cleaved from the resin in the presence of HRV 3C protease to yield purified complexes of L-P1–91. The nine-residue HA tag, not used for purification, remained attached to P1–91 following proteolytic cleavage. For cryo-EM specimens, these L-PΔOD–derived L-P1–91 complexes were further purified on a Superdex 200 Increase column, and the peak fractions were concentrated to 0.3 mg/mL immediately before grid preparation. For each grid, 3 μL of sample were applied to glow-discharged, C-Flat 400-mesh copper grids coated with 40-nm-thick holey carbon (1.2/1.3 spacing) and plunge-frozen in liquid ethane in a CP3 cryo-dipper.
Electron Microscopy.
For the medium-resolution initial reconstruction, images of RABV L-P1–91 were recorded on a Tecnai F20 electron microscope (FEI) operated at 200 kV, using UCSF Image4 (Yuemin Li, University of California, San Francisco) to collect movies on a K2 Summit direct detector (Gatan) in superresolution mode with dose fractionation. For each 8-s exposure, we collected 32 frames at 250 ms each with a total electron dose of 72 e/Å2. Collection was performed at a nominal magnification of 29,000×, with a calibrated pixel size of 1.28 Å/pixel. Movie frames were gain-subtracted, Fourier-binned 2×, and motion-corrected using MotionCor2 with 5 × 5 patch correction.
For the high-resolution dataset, images were recorded on a Tecnai F30 Polara electron microscope (FEI) operated at 300 kV, using SerialEM to collect movies on a K2 Summit direct detector (Gatan) in superresolution mode with dose fractionation. For each 8-s exposure, we collected 32 frames at 250 ms each with a total electron dose of 72 e/Å2. Collection was performed at a nominal magnification of 31,000× with a calibrated physical pixel size of 1.234 Å/pixel.
Image Processing.
Micrograph movie frames were gain-subtracted, Fourier-binned to physical pixel size, and motion-corrected using MotionCor2 with 5 × 5 patch correction. Micrograph CTF coefficients were calculated using CTFFIND4 (36), and particle images were CTF-corrected as needed by the processing software used.
For the medium-resolution reconstruction, a single dataset was collected from one of four identical grids prepared in parallel from a single protein sample. We picked 43,000 particles by hand from 316 motion-corrected movies using EMAN2.1 (37). A de novo 3D reference was obtained using the ab-initio procedure in cisTEM (11) following preliminary 2D classification, and the hand was subsequently flipped to yield a reference bearing strong resemblance to VSV L-P. Final 2D and 3D classifications using the flipped reference were then carried out in RELION 2.1 (38), yielding a 6.7 Å (medium-resolution) map from 23,000 particles.
For the high-resolution reconstruction, we collected a total of 10,949 movies from three datasets using the remaining three grids from the initial set of four. Selected 20-Å low-pass-filtered projections of the 6.7-Å map were used as particle-picking templates for autopicking in RELION (38). Autopicked particles on each micrograph were manually cleaned of junk, contamination, noise, drift, and otherwise bad or damaged particles using SamViewer, an interactive image analysis program written in wxPython by Maofu Liao, Harvard Medical School. From 5.3 M remaining particles, coarse 2D classification at a high-resolution cutoff of 25 Å was performed in RELION (38) to remove additional poor particles, and classes representing dominant views were separated from the remaining good classes, as described in SI Appendix, Fig. S2. Three-dimensional classification was performed on 1.47 M nondominant view particles, and the best class of six was selected, containing 649 k particles. These particles were added to the separated 408 k dominant view particles; a final round of 3D classification then yielded a total of 963 k particles from the best class of three. This particle set was then subjected to 3D refinement, per-particle CTF refinement, and Bayesian polishing in RELION 3.0 (13), and the resulting stack was imported into cisTEM (11) along with corresponding Euler angles and offsets for further refinement, as described in SI Appendix, Figs. S1 and S2). The anisotropic 3D reconstruction from refinement in RELION 3.0 (13) was used as a starting reference for manual refinement in cisTEM (11), and particles were removed from the reconstruction at each refinement iteration using the SCORE parameter. We quickly removed the worst-scoring 50% of particles. Because dominant view particles had higher scores (due to anisotropy of the map caused by a preferred orientation), we applied an angular binning script to cap the number of particles per 4° × 4° bin, keeping those with the highest scores. Further manual refinement using these remaining 177 k particles with flattened angular distribution led to reduced anisotropy and improved map quality, with the best 25% of particles by score giving the highest-resolution map. A final refinement using this set of 44,500 particles at an alignment resolution cutoff of 4.5 Å yielded a reasonably isotropic map (despite a majority of particles having the preferred orientation) with an average resolution of 3.3 Å. The Electron Microscopy Data Bank (EMDB) accession number for the deposited map is EMD-20753.
Model Building.
To generate the atomic model of RABV L, we first replaced the sequence of VSV L with RABV L in the model of VSV L (PDB ID 6U1X) (7), adding simple loops where discontinuities in the alignment occurred. We fit these coordinates as a single rigid body into the 3.3-Å RABV L-P1–91 density map and then divided the coordinates into the five structural domains (RdRp, CAP, CD, MT, and CTD) and refit each domain as a rigid body. We then performed iterative real-space refinement using PHENIX (39, 40) at increasing resolution, from 40 to 5 Å. The resulting model was then used as an input model for ROSETTA (17). To improve loop fitting, we also input libraries of 3- and 9-mer peptide structures corresponding to all 3- and 9-amino acid sequences contained in the 2,127-residue RABV L sequence (41). From 5,000 simulations, we selected a model with very good map-to-model agreement and with visibly strong fitting of secondary structure elements throughout. We then performed several rounds of manual adjustment in Coot, alternating with real-space refinement in PHENIX, using secondary structure restraints throughout and Ramachandran restraints in the final rounds. The final model was validated using PHENIX and MolProbity (SI Appendix, Table S1) and deposited in the Protein Data Bank under PDB ID 6UEB.
Projection Matching.
Molecules of RABV L in complex with GFP-P1–91 were visualized by negative-stain EM at ∼0.01 mg/mL total protein in the presence of 0.7% uranyl formate. Twenty-eight 2D class averages of picked particles showed clear density for the GFP moiety at the N terminus of P (Fig. 4A and SI Appendix, Figs. S5 and S6). We aligned the 2D class averages with obvious GFP density to a single reference to standardize their orientation in the image plane and applied a circular noise mask to obscure the GFP signal as much as possible without obstructing signal from L. To determine the viewing angle of the RABV L atomic model corresponding to each 2D class, we projected the high-resolution RABV L-P1–91 cryo-EM map over all viewing angles, incremented every ∼7.2° for a total of 800 projections. Projections were low-pass-filtered to 20 Å and then scaled and clipped to match the pixel and box size of the 2D classes, and a circular noise mask was applied to obscure the clipping edges. Cross-correlation coefficients between projections and 2D classes were obtained using e2simmx.py from EMAN2 (37), allowing for rotation and translation only. The top projection match for each 2D class is shown in Fig. 4 and SI Appendix, Fig. S5, with GFP modeled as a best approximation.
L-PFL dimers, representing about 25% of L species on micrographs of negatively stained, purified RABV L/PFL, were picked by hand and subjected to 2D class averaging using EMAN2.1 (37). From 29 high-quality 2D class averages of dimers, we extracted each of the 58 dimeric monomer averages as individual particles (hereafter, DMAPs). We applied a circular noise mask to each DMAP to obscure excess signal from the paired DMAPs present in each particle image. We then projected the high-resolution RABV L-P1–91 cryo-EM map over all viewing angles, incremented every ∼7.2° for a total of 800 projections. Projections were low-pass-filtered to 20 Å and then clipped and scaled to match the box and pixel size of the negative-stain EM DMAPs, and a circular noise mask was applied to obscure the clipping edges. Cross-correlation coefficients between projections and DMAPs were obtained using e2simmx.py, allowing for rotation and translation only. The resulting matrix was then normalized within each DMAP over all projections, from which we constructed spherical heat maps in the coordinate system of the RABV L-P1–91 cryo-EM map to facilitate visualization of peaks reflecting high cross-correlation scores. For about half of the DMAPs, a single peak predominated, and application of the angular coordinates from the peak to the atomic model of RABV L-P1–91 returned an obvious match to the corresponding DMAP. When more than one high-scoring peak was observed, the best apparent visual match was selected. Of the 58 DMAP:projection comparisons, only 6 were matched to a secondary peak, and 5 could not be reliably matched. In total, we successfully matched 24 of the 29 DMAP pairs. Using PyMOL (Schrödinger, LLC), we then applied the angular coordinates from each match to an atomic model of RABV L-P1–91 for each DMAP in a pair and approximated the relationship between the two modeled DMAPs using only two dimensions (X and Y), as suggested by the original 2D class average. We then measured the minimum distance between C-alpha atoms of either P87 or the mapped, but unassigned, N-terminal-most residue of P in each modeled pair by translating one of the models along the z axis. The results of these projection-matching analyses are summarized in Fig. 4B and illustrated in detail in SI Appendix, Fig. S8.
Visualization of Internal Cavities.
We used the program VOIDOO (42) to probe internal cavities, substrate entry, and product exit channels of the RABV L-P1–91 complex. We used a probe radius of 1.8 Å to calculate a probe-occupied volume.
Data Availability.
Data for EM 3D reconstruction have been deposited in the Electron Microscopy Data Bank (accession no. EMD-20753), and data for coordinates fit to 3D reconstruction have been deposited in the Protein Data Bank (PDB ID 6UEB).
Supplementary Material
Acknowledgments
We thank Louis-Marie Bloyet for discussions of Mononegavirus biology and advice on L-protein preparation. J.A.H. is an Amgen Fellow of the Life Sciences Research Foundation. The work was supported by NIH Grant R37 AI059371 (to S.P.J.W.) and Grant R01 CA13202 (to S.C.H.). S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
Data deposition: Data for the cryo-EM 3D reconstruction reported in this paper have been deposited in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (accession no. EMD-20753), and data for coordinates fit to 3D reconstruction have been deposited in the Protein Data Bank, https://www.rcsb.org/ (PDB ID 6UEB).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918809117/-/DCSupplemental.
References
- 1.Liang B., et al. , Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogino T., Banerjee A. K., Formation of guanosine(5′)tetraphospho(5′)adenosine cap structure by an unconventional mRNA capping enzyme of vesicular stomatitis virus. J. Virol. 82, 7729–7734 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahmeh A. A., Li J., Kranzusch P. J., Whelan S. P., Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 83, 11043–11050 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogino M., Gupta N., Green T. J., Ogino T., A dual-functional priming-capping loop of rhabdoviral RNA polymerases directs terminal de novo initiation and capping intermediate formation. Nucleic Acids Res. 47, 299–309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilman M. S. A., et al. , Structure of the respiratory syncytial virus polymerase complex. Cell 179, 193–204.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan J., et al. , Structure of the human metapneumovirus polymerase phosphoprotein complex. Nature, 10.1038/s41586-019-1759-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenni S., et al. , Structure of the vesicular stomatitis virus L protein in complex with its phosphoprotein cofactor. Cell Rep., 10.1016/j.celrep.2019.12.024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin B., Liang B., Gardner E., Ross R. A., Whelan S. P. J., An in vitro RNA synthesis assay for rabies virus defines ribonucleoprotein interactions critical for polymerase activity. J. Virol. 91, e01508-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahmeh A. A., et al. , Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 20075–20080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahmeh A. A., et al. , Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 109, 14628–14633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakane T., Kimanius D., Lindahl E., Scheres S. H., Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohl C. A., Strauss C. E., Chivian D., Baker D., Modeling structurally variable regions in homologous proteins with Rosetta. Proteins 55, 656–677 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Rohl C. A., Strauss C. E., Misura K. M., Baker D., Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Song Y., et al. , High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov I., Crépin T., Jamin M., Ruigrok R. W., Structure of the dimerization domain of the rabies virus phosphoprotein. J. Virol. 84, 3707–3710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X., et al. , Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 16, 1678–1688 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao Y., Farsetta D. L., Nibert M. L., Harrison S. C., RNA synthesis in a cage: Structural studies of reovirus polymerase lambda3. Cell 111, 733–745 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Reguera J., Gerlach P., Cusack S., Towards a structural understanding of RNA synthesis by negative strand RNA viral polymerases. Curr. Opin. Struct. Biol. 36, 75–84 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Kouba T., Drncová P., Cusack S., Structural snapshots of actively transcribing influenza polymerase. Nat. Struct. Mol. Biol. 26, 460–470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogino T., Green T. J., Transcriptional control and mRNA capping by the GDP polyribonucleotidyltransferase domain of the rabies virus large protein. Viruses 11, E504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neubauer J., Ogino M., Green T. J., Ogino T., Signature motifs of GDP polyribonucleotidyltransferase, a non-segmented negative strand RNA viral mRNA capping enzyme, domain in the L protein are required for covalent enzyme-pRNA intermediate formation. Nucleic Acids Res. 44, 330–341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Rahmeh A., Morelli M., Whelan S. P., A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 82, 775–784 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun M. R., et al. , RNA elongation by respiratory syncytial virus polymerase is calibrated by conserved region V. PLoS Pathog. 13, e1006803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benarroch D., et al. , A structural basis for the inhibition of the NS5 dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphate. J. Biol. Chem. 279, 35638–35643 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y., et al. , Structure and function of flavivirus NS5 methyltransferase. J. Virol. 81, 3891–3903 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paesen G. C., et al. , X-ray structure and activities of an essential mononegavirales L-protein domain. Nat. Commun. 6, 8749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Rahmeh A., Brusic V., Whelan S. P., Opposing effects of inhibiting cap addition and cap methylation on polyadenylation during vesicular stomatitis virus mRNA synthesis. J. Virol. 83, 1930–1940 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Chorba J. S., Whelan S. P., Vesicular stomatitis viruses resistant to the methylase inhibitor sinefungin upregulate RNA synthesis and reveal mutations that affect mRNA cap methylation. J. Virol. 81, 4104–4115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., Wang J. T., Whelan S. P., A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 103, 8493–8498 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Fontaine-Rodriguez E. C., Whelan S. P., Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 79, 13373–13384 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shuman S., A proposed mechanism of mRNA synthesis and capping by vesicular stomatitis virus. Virology 227, 1–6 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Tekes G., Rahmeh A. A., Whelan S. P., A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5′ processing. PLoS Pathog. 7, e1002073 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludtke S. J., Single-particle refinement and variability analysis in EMAN2.1. Methods Enzymol. 579, 159–189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimanius D., Forsberg B. O., Scheres S. H., Lindahl E., Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D. E., Chivian D., Baker D., Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleywegt G. J., Jones T. A., Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for EM 3D reconstruction have been deposited in the Electron Microscopy Data Bank (accession no. EMD-20753), and data for coordinates fit to 3D reconstruction have been deposited in the Protein Data Bank (PDB ID 6UEB).