Often described as nature’s most beautiful cofactor (1), vitamin B12 (cobalamin) is a complex and fascinating organometallic molecule that, although made only by some prokaryotes, has key functional roles in microbes, animals, and humans (2). Its two major biological forms, methylcobalamin (MeCbl) and adenosylcobalamin (AdoCbl), have a central cobalt atom in a corrin ring that is coordinated via a cobalt–carbon bond to an upper axial methyl or 5′-deoxyadenosyl group, respectively; in both forms the lower axial ligand is the 5,6-dimethylbenzimidazole (DMB) base of a nucleotide tail attached to the corrin ring (2–5). Finely controlled enzymatic or photolytic cleavage of the upper axial cobalt–carbon bond underlies the use of B12 as an enzyme cofactor or, in a new twist, as the light-sensing molecule in a photoreceptor protein (2–5). As an enzyme cofactor, MeCbl is used for methyl transfer reactions by methyltransferases and AdoCbl for radical-based transformations by mutases, dehydratases, deaminases, and ribonucleotide reductases (2–4). In a broad range of organisms and biological processes, B12-dependent methyltransferases catalyze the transfer of a methyl group from a donor to a final acceptor, utilizing MeCbl or its methylcobamide analogs (with a base other than DMB) as methyl carriers. In these enzymes the nucleotide base in B12 is always displaced, often by a histidine ligand of a conserved DxHxxG protein motif (6–8). Methionine synthase, a well-studied MeCbl-dependent methyltransferase present in many bacteria and mammals, is a single polypeptide with four modules that uses methyltetrahydrofolate as donor and homocysteine as acceptor to catalyze the terminal step in methionine biosynthesis (6, 7). In many microbes that thrive in anaerobic habitats, such as methanogens and acetogens, growth and energy production rely on B12-dependent methyltransferase systems that employ a variety of methyl donors (e.g., methanol, methylamines, and methyltetrahydromethanopterin) and acceptors (coenzyme M and tetrahydrofolate) (6, 7). Rather than a single polypeptide, these B12-dependent methyltransferases are often multiprotein enzyme complexes with one subunit each for binding the methyl donor, the B12 cofactor, and the methyl acceptor (6, 7). In PNAS, Wang et al. (9) report on a B12-dependent methyltransferase system that targets estrogen, a previously unsuspected methyl acceptor, in an anaerobic denitrifying bacterium. As a result, the estrogen gets transformed into androgen for subsequent degradation and use, thus linking B12 with anaerobic bacterial steroid catabolism.
The sex steroids, androgens and estrogens (Fig. 1), are cholesterol-derived hormones with critical roles in mammalian physiology, development, reproduction, and behavior (10). Steroid biosynthesis typically occurs in eukaryotes but is rare in bacteria, where it is observed in Gemmata obscuriglobus and in some methanotrophs and myxobacteria (11–13). Because steroids are generally recalcitrant to degradation and can disrupt endocrine functions even at low concentrations in a variety of organisms including humans, the constant release of natural and synthetic steroids into the environment due to agriculture, industry, and sewage is of ever-increasing concern. Hence, bacteria capable of degrading steroids are valuable bioremediation agents (14, 15). How bacteria deal with steroids is also important in the context of host–microbe metabolic interdependencies, including in pathogenesis (16). Thus, the findings by Wang et al. (9) not only add estrogen methylation to the repertoire of B12-dependent methyltransferase activities but are also relevant in bioremediation/wastewater treatment and in host–microbe metabolic interactions.
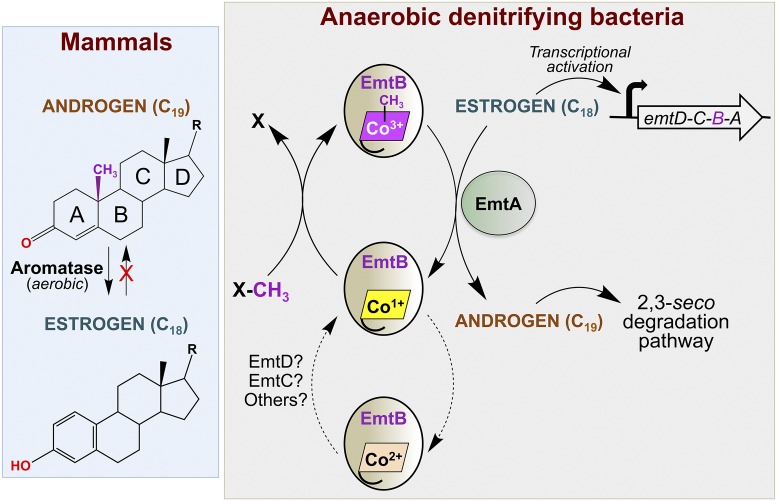
Fig. 1.
Proposed B12-dependent methylation of estrogen to androgen in anaerobic denitrifying bacteria. The putative catalytic and B12-binding units are EmtA and EmtB, respectively, which are encoded by the estrogen–up-regulated emtABCD operon. As in well-characterized B12-dependent methyltransferases, the cobalt in B12 is expected to cycle between the +3 and +1 oxidation states. The latter is a potent nucleophile that suffers occasional oxidative inactivation to the +2 state and requires reductive activation to reenter the catalytic cycle (dotted arrows). This may be mediated by EmtD (and EmtC) and/or other candidate proteins, whose genes are up-regulated in estrogen-fed cells. “X” is a methyl donor, whose identity is as-yet unknown. For comparison, a simplified schematic of the aerobic, irreversible, aromatase-catalyzed conversion of androgen to estrogen in mammals is shown on the left.
Bacterial steroid degradation mechanisms, pathways, and intermediates are well characterized for androgens but less so for estrogens, and aerobic pathways are better charted than anaerobic ones, with most insights emerging from studies of some Actinobacteria and a few α-, β-, and γ-Proteobacteria (14, 15). Thus, androgens are degraded aerobically via the 9,10-seco pathway (bond cleavage between ring B carbon atoms C9 and C10 to generate the corresponding secosteroid) or anaerobically in denitrifying bacteria via the 2,3-seco pathway (bond cleavage between ring A atoms C2 and C3) (14, 15). Estrogens are more refractory to degradation due to their stable aromatic A-ring and are degraded in some aerobic bacteria by oxygenase-catalyzed cleavage of the bond between ring A atoms C4 and C5 (the 4,5-seco pathway) (14, 15). Anaerobic estrogen degradation had been reported only in two bacterial species, and the genes and mechanisms involved have remained unknown. The study in PNAS (9) contributes toward filling this gap by identifying a degradation strategy that is based on “retroconversion” of estrogens to androgens for subsequent degradation via the anaerobic 2,3-seco pathway (Fig. 1). Such an estrogen-to-androgen conversion represents a challenging reaction that has not been reported previously. Indeed, in eukaryotes an oxygen/NADPH-dependent cytochrome P450 aromatase (or estrogen synthase) specifically targets the androgen “A” ring for the biosynthesis of estrogens with one less carbon, in a complex three-step hydroxylation–demethylation–aromatization reaction that is considered irreversible (refs. 17 and 18 and Fig. 1).
In the study in PNAS (9), an anaerobic β-proteobacterium, Denitratisoma sp. strain DHT3, which exhibits efficient B12-dependent estrogen degradation under denitrifying conditions, was isolated from a municipal wastewater treatment plant. Its genome revealed a complete set of genes for the 2,3-seco androgen degradation pathway and for B12 transport and utilization but a lack of most genes for B12 biosynthesis. Transcriptomic analysis indicated that while 2,3-seco pathway genes were similarly expressed under both estrogen- and androgen-fed conditions, genes for B12 transport and utilization, and those in a cluster denoted emtABCD (emt for estradiol methylation), were up-regulated under estrogen-fed conditions. Consistent with a role in estrogen metabolism, emtABCD, which is transcribed as a polycistronic RNA, also occurs in the other two known denitrifying, estrogen-degrading microbes: the β-proteobacterium Denitratisoma oestradiolicum and the γ-proteobacterium Steroidobacter denitrificans. Protein sequence analysis suggested that EmtA may be the catalytic subunit of a putative B12-dependent methyltransferase complex, whose B12-binding subunit would be EmtB, based on the presence of the DxHxxGx41/42SxLx24–28GG motif found in MeCbl-dependent methione synthase and many B12-binding proteins (3–8). EmtC is a hypothetical protein, and EmtD resembles the F420/FMN-dependent oxidoreductase involved in the reductive activation of B12-dependent methyltransferases, required to regenerate the active cofactor +1 state upon its sporadic oxidation to the +2 state (Fig. 1 and refs. 6 and 7).
In PNAS, Wang et al. report on a B12-dependent methyltransferase system that targets estrogen, a previously unsuspected methyl acceptor, in an anaerobic denitrifying bacterium.
Evidence that EmtA mediates B12-dependent methylation of estrogen to androgen, which is then channeled into its established anaerobic degradation pathway, came from data obtained using genetic, biochemical, and ultraperformance liquid chromatography–high resolution mass spectrometry methods (9). Thus, disrupting the emtA gene abrogated the ability to utilize estrogen, and metabolite profiling for 13C-labeled estrogen-fed bacteria revealed characteristic intermediates of the 2,3-seco androgen degradation pathway. Moreover, a decrease in estrogen levels with a concomitant appearance of androgens was observed when estrogens were incubated anaerobically with cell extracts from the wild-type strain (but not from the emtA-disrupted one) in the presence of MeCbl and of cofactors for the reductive activation of B12-dependent methyltransferases. The effect of propyl iodide, an inhibitor of B12-dependent enzymes in the dark but not in the light, provided further support for the requirement of B12 in the estrogen-to-androgen conversion reaction.
Although the evidence presented for B12-dependent methylation of estrogen and its conversion to androgen is persuasive, the study also raises a slew of exciting new questions that remain to be addressed. Functions of the Emt proteins and the proposed catalytic mechanisms were hypothesized based on sequence comparisons and phylogenetic analysis and need to be demonstrated. Whether the proteins are subunits of larger complexes, as with various B12-dependent methyltransferase systems, must be assessed. The molecular basis for the specific recognition and methylation of estrogens, ascribed to EmtA, has to be determined, as does also the identity of the initial methyl donor involved in the catalytic cycle. These will require purification, biochemical characterization, and in vitro reconstitution studies, as well as structure elucidation of these proteins, besides genetic analyses. From the standpoint of gene regulation, it will be interesting to decipher how estrogens up-regulate expression of the emt cluster and of the genes for B12 transport, salvage, and reductive activation. Nonetheless, the discovery of estrogen as the methyl acceptor of a putative B12-dependent methyltransferase and its conversion to androgen, a reaction thought to be formidably difficult, is noteworthy and fills an important niche in bacterial steroid metabolism.
The B12–steroid degradation link identified in the study has other ramifications. Because the anaerobic bacteria that degrade estrogen require B12 but cannot synthesize it de novo they must necessarily depend on exogenous B12 to “digest” estrogen. Thus, they may have to rely on other microbes in their natural milieu that do produce B12, a frequently shared (and precious) resource in microbial consortia (19). Such intermicrobe nutritional interactions will be relevant not only in optimizing any bioremediation strategy against environmental estrogen but also in the host–microbe metabolic interactions that are subject to modulation by sex steroids (16) and by B12 (19).
Acknowledgments
I thank S. Padmanabhan for comments on the manuscript. This work is supported by grants PGC2018-094635-B-C21 and PGC2018-094635-B-C22 from the Agencia Estatal de Investigación of Spain and the European Regional Development Fund and grant 20992/PI/18 from Fundación Séneca of Murcia, Spain.
Footnotes
The author declares no competing interest.
See companion article on page 1395 in issue 3 of volume 117.
References
- 1.Stubbe J., Binding site revealed of nature’s most beautiful cofactor. Science 266, 1663–1664 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Smith A. D., Warren M. J., Refsum H., Vitamin B12. Adv. Food Nutr. Res. 83, 215–279 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Bridwell-Rabb J., Drennan C. L., Vitamin B12 in the spotlight again. Curr. Opin. Chem. Biol. 37, 63–70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruber K., Puffer B., Kräutler B., Vitamin B12-derivatives-enzyme cofactors and ligands of proteins and nucleic acids. Chem. Soc. Rev. 40, 4346–4363 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Padmanabhan S., Jost M., Drennan C. L., Elías-Arnanz M., A new facet of vitamin B12: Gene regulation by cobalamin-based photoreceptors. Annu. Rev. Biochem. 86, 485–514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthews R. G., Koutmos M., Datta S., Cobalamin-dependent and cobamide-dependent methyltransferases. Curr. Opin. Struct. Biol. 18, 658–666 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ragsdale S. W., Catalysis of methyl group transfers involving tetrahydrofolate and B12. Vitam. Horm. 79, 293–324 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drennan C. L., Huang S., Drummond J. T., Matthews R. G., Ludwig M. L., How a protein binds B12: A 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science 266, 1669–1674 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Wang P.-H., et al. , Retroconversion of estrogens into androgens by bacteria via a cobalamin-mediated methylation. Proc. Natl. Acad. Sci. U.S.A. 117, 1395–1403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauvais-Jarvis F., Role of sex steroids in β cell function, growth, and survival. Trends Endocrinol. Metab. 27, 844–855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bode H. B., et al. , Steroid biosynthesis in prokaryotes: Identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca. Mol. Microbiol. 47, 471–481 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Lee A. K., et al. , C-4 sterol demethylation enzymes distinguish bacterial and eukaryotic sterol synthesis. Proc. Natl. Acad. Sci. U.S.A. 115, 5884–5889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson A., Budin M., Brocks J. J., Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U.S.A. 100, 15352–15357 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang Y. R., Wei S. T., Wang P. H., Wu P. H., Yu C. P., Microbial degradation of steroid sex hormones: Implications for environmental and ecological studies. Microb. Biotechnol., 10.1111/1751-7915.13504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivera E. R., Luengo J. M., Steroids as environmental compounds recalcitrant to degradation: Genetic mechanisms of bacterial biodegradation pathways. Genes (Basel) 10, 512 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.vom Steeg L. G., Klein S. L., Sex steroids mediate bidirectional interactions between hosts and microbes. Horm. Behav. 88, 45–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh D., Egbuta C., Lo J., Testosterone complex and non-steroidal ligands of human aromatase. J. Steroid Biochem. Mol. Biol. 181, 11–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas M. P., Potter B. V., The structural biology of oestrogen metabolism. J. Steroid Biochem. Mol. Biol. 137, 27–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gude S., Taga M. E., Multi-faceted approaches to discovering and predicting microbial nutritional interactions. Curr. Opin. Biotechnol. 62, 58–64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]