Significance
The current progress of genome editing techniques accelerates the knock-out and knock-in studies in animal models and production of genetic modifications in livestock. Increased resistance to viral pathogens is a particular goal because many monogenic host cell factors are necessary for productive infection and pathogenesis. For example, virus receptors with their specific virus binding sites are direct targets for the CRISPR/Cas9 gene editing. We introduced a single amino acid deletion into the gene encoding the receptor that is required for avian leukosis virus subgroup J to infect chicken cells. Here, we demonstrate that this mutation confers the resistance of chickens to avian leukosis virus subgroup J, an important pathogen in poultry. In addition, we present highly efficient genome-editing technology in chicken.
Keywords: avian leukosis virus subgroup J, Na+/H+ exchanger type 1, CRISPR/Cas9 genome editing in chicken, primordial germ cells, disease resilience in poultry
Abstract
Avian leukosis virus subgroup J (ALV-J) is an important concern for the poultry industry. Replication of ALV-J depends on a functional cellular receptor, the chicken Na+/H+ exchanger type 1 (chNHE1). Tryptophan residue number 38 of chNHE1 (W38) in the extracellular portion of this molecule is a critical amino acid for virus entry. We describe a CRISPR/Cas9-mediated deletion of W38 in chicken primordial germ cells and the successful production of the gene-edited birds. The resistance to ALV-J was examined both in vitro and in vivo, and the ΔW38 homozygous chickens tested ALV-J–resistant, in contrast to ΔW38 heterozygotes and wild-type birds, which were ALV-J–susceptible. Deletion of W38 did not manifest any visible side effect. Our data clearly demonstrate the antiviral resistance conferred by precise CRISPR/Cas9 gene editing in the chicken. Furthermore, our highly efficient CRISPR/Cas9 gene editing in primordial germ cells represents a substantial addition to genotechnology in the chicken, an important food source and research model.
Background
Since its identification during the first outbreak in the United Kingdom (1), avian leukosis virus subgroup J (ALV-J) has been an important concern for the poultry industry. European and American ALV-J strains, such as the prototypic HPRS103, mostly induced myelocytomatosis in broiler chickens. In China and Southeast Asia, however, ALV-J evolved into various strains with diversified pathologies in both meat- and egg-type chickens (2). The last big ALV-J outbreak in China, with surprisingly high mortality of infected chickens, occurred in 2018, despite an eradication program geared toward the virus (3).
Chicken Na+/H+ exchanger type 1 (chNHE1) has been recognized as a receptor for ALV-J, and its prominent and glycosylated extracellular loop 1 is necessary for virus entry (4). Comparison of its amino acid sequence in ALV-J–susceptible species (domestic chicken, jungle fowls, turkey) and resistant species (most galliform birds) pointed to W38 being a residue critical for the receptor function (5). The single amino acid deletion of W38 introduced by CRISPR/Cas9 in cultured chicken cells rendered the cells resistant to ALV-J and confirmed the importance of W38 for virus entry (6). Unfortunately, chNHE1 is well conserved in a wide range of chicken breeds, including indigenous breeds of Asian origin (7). As a result, there is no known source of genetic variability to be used to selectively breed an ALV-J–resistant chicken. This obstacle might be overcome by precise gene editing using CRISPR/Cas9.
The current approach to chicken genome manipulation relies on primordial germ cell (PGC) technology (8). Male PGCs are derived from chicken embryos infected with a retroviral vector or DNA-transfected during in vitro culture and finally returned to the embryos, where they differentiate and mature into functional sperms in the cockerels after hatching. This is now a well-established technology providing transgenic chickens (9) and even genetic knockouts (10). However, the knock-in technology and CRISPR/Cas9 editing of endogenous loci in the chicken remained to be expanded, with the low efficiency of embryonal PGC application being the main obstacle. This has now changed with our method of orthotopic PGC transplantation into sterilized adult cockerels (11). This technique improves the efficiency of transgenesis in the chicken and makes such gene editing feasible. In this report, we describe the preparation of a chicken line with CRISPR/Cas9-introduced ΔW38 into chNHE1. This chicken line is fully resistant to ALV-J infection.
Results
Generation of chNHE1-Edited Chickens.
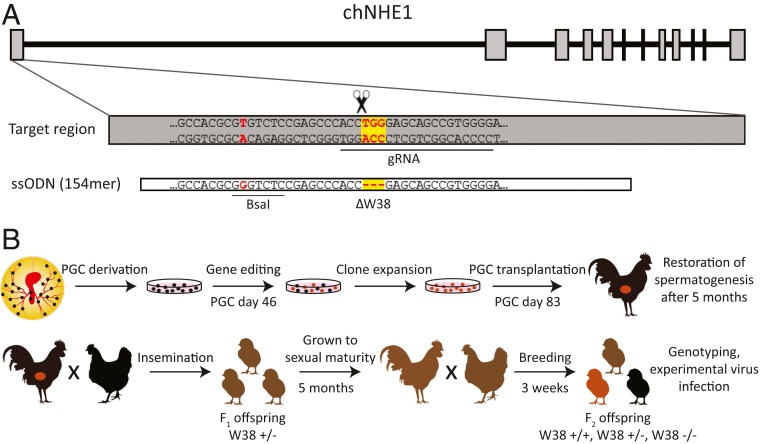
We derived PGCs from blood aspirations of 14- to 17-stage (H & H) male embryos of inbred line CB (SI Appendix, Fig. S1), which is homozygous for the wt chNHE1 allele and susceptible to ALV-J infection. For the introduction of ΔW38 into the endogenous chNHE1 allele, we constructed a suitable GFP-transducing CRISPR/Cas9 vector, and, together with the single-strand oligonucleotide (ssODN) as the recombination template, we transfected it into the PGCs. The design of the guide RNA sequence and ssODN and their correspondence to the chNHE1 are shown in Fig. 1A. We observed a very high efficiency of homologous recombination and found that 88% (30/34) of the clones had obtained the precise three-nucleotide deletion in both chNHE1 alleles. Only four clones displayed incorrect deletion of two, five, eight, or 24 nucleotides present in both (three clones) or just one (one clone) chNHE1 allele (SI Appendix, Table S1).
Fig. 1.
Design of guide RNA and homologous recombination ssODN for CRISPR/Cas9 gene editing of chicken ALV-J receptor chNHE1 in primordial germ cells. (A) The structure of coding exons and introns of chNHE1 (Top), the CRISPR/Cas9 target sequence of exon 1 with the guide RNA (gRNA) complementary sequence (underlined) and the TGG triplet encoding W38 (red on the yellow background; Middle), and the central part of the ssODN template for homologous recombination with deleted TGG triplet and a single nucleotide substitution (in red) creating the BsaI restriction site (Bottom). (B) Preparation of ΔW38 chickens: schematic representation of the workflow and timeline.
Two of the clones that tested homozygous for ΔW38 (W38−/−) were expanded and orthotopically transplanted each into one rooster previously sterilized by gamma irradiation. Both roosters showed restored spermatogenesis, one after 8 wk and the second after 24 wk, and the ΔW38 allele was detected in DNA isolated from their ejaculates. The rooster that showed restored spermatogenesis earlier (no. 32) was used for further breeding (Table 1); the second rooster (no. 31) was not used because of constantly low semen concentration. After artificial insemination of 18 Barred Leghorn hens, we obtained 21 heterozygous W38+/− chickens in the offspring of the F1 generation. One founder male and one founder female, both W38+/−, were mated to produce F2 generation offspring. In F2, the observed ratio of W38 +/+, +/−, and −/− genotypes (29:61:28, respectively) matched the expected Mendelian ratio (Pearson’s χ2 test, P < 0.05). We did not observe either phenotypic side effects or increased mortality in W38−/− chickens, three of which now reached the age of 5 mo. Up to now, only one W38−/− male had reached sexual maturity. According to the semen analysis parameters, he was normospermic (5.2 × 109 of morphologically normal and motile spermatozoa per milliliter of ejaculate) and fully fertile, with 92% egg fertilization and 81% hatchability after intravaginal insemination of laid hens. The whole procedure resulting in ΔW38 chickens is summarized in Fig. 1B. All experiments were performed in accordance with the Czech legislation for animal handling and welfare.
Table 1.
Summarized data on the production of ΔW38 chickens
| Rooster no./PGC clone | Restoration of spermatogenesis after transplantation, wk | Hens inseminated* | Fertilized eggs* | Hatched chickens* |
| 31/47 | 24 | Not done | Not done | Not done |
| 32/13 | 8 | 13/5 | 18/6 | 17/4 |
Intramagnal/intravaginal insemination.
In Vitro Cultured ΔW38 Embryo Fibroblasts Resistant to ALV-J.
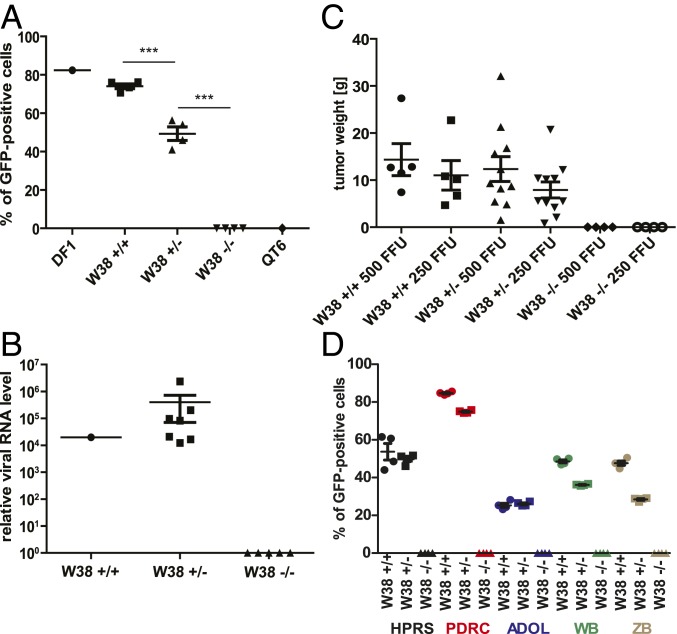
In vitro knockout of chNHE1 or W38 deletion by CRISPR/Cas9 in chicken DF-1 cells confers resistance to ALV-J (6, 12). We tested the resistance of W38−/− chickens to ALV-J in a similar way by in vitro infection of chicken embryo fibroblasts derived from 10-d embryos, four of each genotype. The J subgroup retroviral vector RCASBP(J)GFP (5) was used to challenge the cultured fibroblasts, and the susceptibility to ALV-J was measured as the percentage of GFP-transduced cells. The W38−/− cells were fully resistant to RCASBP(J)GFP, whereas W38+/+ cells displayed normal spread of the virus (Fig. 2A and SI Appendix, Fig. S2). Embryo fibroblasts derived from W38+/− chickens were susceptible to ALV-J, but significantly (P < 0.0006) less than W38+/+ cells.
Fig. 2.
Resistance of W38−/− chickens to ALV-J infection. (A) In vitro infection of fibroblasts derived from W38+/+, W38+/−, and W38−/− embryos with GFP-transducing J subgroup-specific retrovirus RCASBP(J)GFP. Chicken DF1 and Japanese quail QT6 cells were used as susceptible and resistant controls, respectively. Each value represents the mean of two technical replicates. Each group is characterized as mean ± SD (***P < 0.001, analysis of variance followed by unpaired t test). (B) RT-PCR quantification of ALV-J env gene in the serum of RCASBP(J)GFP-infected chickens. Each value represents relative units as the mean of triplicate quantifications of individual chickens. For the group of W38+/− chickens, the means ± SD are shown. (C) In vivo tumor induction in W38+/+, W38+/−, and W38−/− chickens. Weights of tumors induced with either 500 or 250 FFU are given as individual values and means ± SD. (D) In vitro infection of fibroblasts derived from W38+/+, W38+/−, and W38−/− embryos with GFP-transducing J subgroup-specific retroviruses RCASBP(J)GFP (HPRS; black), RCASBP(JPDRC)GFP (PDRC; red), RCASBP(JADOL)GFP (ADOL; blue), RCASBP(JWB)GFP (WB; green), and RCASBP(JZB)GFP (ZB; brown). Each value represents the mean of four technical replicates. Each group is characterized as mean ± SD.
In Vivo Resistance of ΔW38 Chickens to ALV-J.
Next, we tested whether the J subgroup ALV replicates in vivo and establishes viremia in W38−/− chickens. We challenged four W38−/− chickens, seven W38+/− chickens, and one W38+/+ chicken aged 14 to 28 d by i.v. injection of 106 IU of RCASBP(J)GFP and detected the virus quantitatively in blood sera collected 6 and 13 d post infection (p.i.). The viral genomes were detected by quantitative reverse transcriptase PCR (qRT-PCR) with J subgroup env-specific primers, and the replication-competent virus was detected by a complementation assay. The qRT-PCR demonstrated viremia in six W38+/− chickens and the one W38+/+ chicken 6 d p.i. Thirteen days p.i., all W38+/− chickens and the one W38+/+ chicken were positive, whereas all W38−/− chickens remained aviremic (Fig. 2B). In accordance with the qRT-PCR experiment, virus complementation demonstrated low titers (101 to 102 IU/mL) of infectious virus in blood samples from two W38+/− chickens and the one W38+/+ chicken 6 d p.i. The virus titers and the incidence of viremia increased 13 d p.i., when all W38+/− chickens and the one W38+/+ chicken were positive (102 to 103 IU per mL). The W38−/− chickens did not develop viremia, confirming their resistance to ALV-J (Table 2).
Table 2.
ALV-J viremia in chickens inoculated with RCASBP(J)GFP
| chNHE1 genotype | Chicken no. | Age of chicken, d | Virus titer, IU/mL | |
| 6 d p.i. | 13 d p.i. | |||
| W38−/− | 604 | 28 | 0 | 0 |
| W38−/− | 606 | 28 | 0 | 0 |
| W38−/− | 611 | 20 | 0 | 0 |
| W38−/− | 612 | 20 | 0 | 0 |
| W38−/− | 615 | 14 | 0 | 0 |
| W38+/− | 601 | 28 | 0 | 101 |
| W38+/− | 603 | 28 | 0 | 102 |
| W38+/− | 605 | 28 | 0 | 101 |
| W38+/− | 610 | 20 | 102 | 103 |
| W38+/− | 616 | 20 | 0 | 103 |
| W38+/− | 618 | 14 | 0 | 102 |
| W38+/− | 619 | 14 | 101 | 103 |
| W38+/+ | 617 | 14 | 102 | 103 |
In Vivo Resistance to ALV-J–Pseudotyped Transforming Virus.
In addition to the in vivo induction of viremia by ALV-J, we also tested virus resistance by inducing tumors with a v-src–transducing virus pseudotyped with the J subgroup envelope (13). The titer of the transforming pseudotyped virus was determined by in vitro focus assay as focus-forming units (FFUs), and each chicken in this experiment was inoculated with 250 FFU into one wing web and with 500 FFU into the other wing web. Four W38−/−, eleven W38+/−, and five W38+/+ chickens aged 13 to 56 d were included in the experiment. All W38+/− and W38+/+ chickens developed bilateral rapidly growing tumors (SI Appendix, Fig. S3) and had to be euthanized 16 d p.i. The weight of the tumors induced by 500 FFU was greater than that of the tumors induced by 250 FFU in every chicken, but the mean differences were insignificant, as were the mean differences between the tumors induced in W38+/+ and W38+/− chickens. On the contrary, none of the four W38−/− chickens, including the youngest ones, developed tumors at the site of inoculation, and all remained healthy without tumors for the next 4 mo, after which they were euthanized and autopsied (Fig. 2C and SI Appendix, Table S2).
W38−/− Cells Are Resistant to a Wide Range of ALV-J Strains.
Because of the rapid diversification of the ALV-J subgroup, some virus strains might expand their host range. We therefore tested the resistance of W38−/− embryo fibroblasts to further ALV-J strains in the form of RCAS vectors. In addition to RCASBP(J)GFP, which bears the env gene from the prototypic ALV-J strain HPRS103, we challenged the cultured fibroblasts with vectors RCASBP(JPDRC)GFP, RCASBP(JADOL)GFP, RCASBP(JWB)GFP, and RCASBP(JZB)GFP bearing the env genes from ALV-J strains PDRC59831, ADOL7501, WB11016j, and ZB110604-5, respectively (7). As in the case of RCASBP(J)GFP, the W38−/− cells were fully resistant to all J subgroup RCAS vectors, whereas W38+/+ and W38+/− cells displayed normal spread of the virus (Fig. 2D).
Assessment of Off-Target Mutations and Vector Integration.
To explore the level of non-NHE1 mutations introduced into the genome of W38−/− chickens by the CRISPR/Cas9 system, we selected potential off-target sites and analyzed the nucleotide sequence at these sites in the DNA of W38−/− PGC clone 13, which was used for the successful orthotopic transplantation. In total, eight most probable off-target sites were selected based on the cutting frequency determination (CFD) scores (SI Appendix, Table S4). All off-target sites shared 16 to 17 matched nucleotides and 10 to 16 consecutive matched nucleotides with the 20-bp chNHE1 gRNA sequence. We performed PCR amplification of potential off-target sites, sequenced the PCR products, and compared the resulting sequence with the NCBI database sequence and our sequence of the CB inbred chicken line. We did not identify any mutation in any potential off-target site of the W38−/− PGC line. To exclude genome integration of the CRISPR/Cas9 construct, we tested the absence of GFP sequence in cultured PGC clone 13 and in chNHE1-edited chickens by PCR. As expected, we did not amplify any GFP-specific PCR product in DNAs from the PGC clone 13 sampled at the time of transplantation, W38+/− chickens of F1 generation, and W38−/− chicken of F2 (SI Appendix, Fig. S4). This indicates either no integration of the GFP-transducing CRISPR/Cas9 plasmid or its loss during the PGC clone expansion in vitro.
Discussion
In conclusion, we prepared chickens bearing the homozygous deletion of W38 in the first extracellular loop of chNHE1 and demonstrated by three independent assays that these chickens are fully resistant to ALV-J infection. At the same time, we did not observe any visible side effect of this single amino acid deletion that could affect the breeding and utility of W38−/− chickens. This phenotypic assessment is preliminary, with only one ΔW38 chicken sexually mature and fully fertile at the age of 5 mo, and further analysis will be necessary to exclude that some side effects appear at the adult age. We, however, do not expect that the physiological function of chNHE1 could be affected by the W38 deletion, because this polymorphism normally exists in nonchicken galliform birds (5). We suggest that the use of these genome-edited chickens in the poultry industry could help reduce ALV-J–related economic losses and might even contribute to elimination of the virus in the domestic chicken.
What still remains to be examined is the resistance of W38−/− chickens under standard breeding conditions and with natural virus transmission, which probably introduces repeated but much lower doses of infectious virus than those used in our experiments. Second, the chNHE1 editing approach should also be applied on broiler chickens with respect to the possible differences in ALV-J infection of meat- and egg-type breeds. Third, of cardinal importance is also the possible adaptation of ALV-J to using ΔW38 chNHE1 as an entry receptor. We tested the resistance of W38−/− cells to the RCAS vector with the envelope gene of the prototypic HPRS103 strain and four additional ALV-J isolates. The observed resistance to all virus variants suggested that the existing diversification of the ALV-J subgroup did not lead to such an adaptation. Long-term in vitro and in vivo experiments will be required to answer the question whether ALV-J could evolve and overcome the resistance in W38−/− chickens by structural alterations of its envelope glycoproteins.
Potential off-target mutations are of less concern in domestic animals because they are expected to disappear by random drift during the breeding. Nevertheless, we analyzed at least the most probable off-target sites by PCR amplification and sequencing, and we found our CRISPR/Cas9 editing as very specific with no off-target mutations. Similarly, we did not detect any integration of the CRISPR/Cas9 construct in the genome of transplanted PGCs and F1/F2 animals. Even more important, the GC-rich ssODN used for homologous recombination could produce microhomology-based off-target insertions into the host genome. We will test the efficiency of CRISPR/Cas9 editing with lower doses of ssODN in our next experiments.
In addition to W38 deletion, amino acid substitutions could lead to the same level of antiviral resistance (5). Furthermore, other amino acid residues close to the W38 are also critical for ALV-J entry and might be used for creation of resistant chickens (14, 15). The turkey (unlike the Guinea fowl, the domestic duck, or the goose) is susceptible to ALV-J similarly as the chicken, which correlates with the presence of W38 in the turkey NHE1. Thus, the same strategy of W38 deletion could be applied in the turkey to confer the ALV-J resistance.
Our study is a proof of the principle that editing the gene for a virus receptor could confer resistance to the virus and to the virus-associated diseases. It has been demonstrated previously that modifications of CD163, receptor for porcine reproductive and respiratory syndrome virus, protected genome-edited pigs from the virus infection (16).
Last but not least, chicken is the most important source of food among domestic animals, but birds have been lagging behind mammals in terms of genome-editing technology. The CRISPR/Cas9 technology combined with PGC was used for loxP site introduction into the chicken IgH locus (17) and for targeting the ovomucoid gene (18). Quite recently, the CRISPR/Cas9 technology has been employed to introduce a Z chromosome marker (19). We would like to point out the enormously high efficiency of CRISPR/Cas9-mediated homologous recombination in chicken PGCs (SI Appendix, Table S1), which, combined with orthotopic transplantation of manipulated PGCs, opens the way toward precisely manipulated receptors or restriction factors for many other poultry pathogens, such as avian leukosis virus subgroup A, Marek’s disease virus, or avian influenza virus.
Materials and Methods
Experimental Animals and Breeding.
White Leghorn embryos of the CB inbred line were used as a source of PGCs. As recipient roosters of PGCs, we used two hybrids of White Leghorn inbred lines (CC × L15) at the age of 7 mo. The hens used for insemination were outbred Barred Leghorns (population SH) at the age of 7 to 9 mo. All inbred lines (CB, CC, and L15) and the SH population (20) were maintained at the Institute of Molecular Genetics, Czech Academy of Sciences, Prague, Czech Republic. We maintained the experimental cockerels in deep litter individual cages, whereas inseminated hens were housed individually in battery cages. Standard husbandry conditions (16 h light/8 h dark cycle and food/water provided ad libitum) were applied. Eggs were incubated in a forced air incubator (BIOS MIDI). We conducted all experiments and procedures in accordance with the Czech legislation for animal handling and welfare. The Animal Commodities Department of the Ministry of Agriculture of the Czech Republic approved all animal experiments described in this study (approval no. 6413/2018-MZE-17214).
PGC Derivation.
PGCs were derived from the blood of CB chicken embryos incubated for 2.5 d (Hamburger & Hamilton stage 16) using the procedures described previously (21). We aspirated 5 μL of blood from the dorsal aorta and transferred it to 150 μL of Avian KO-DMEM (Thermo Fisher Scientific) supplemented with growth factors as described elsewhere (22) in 48-well plates. After that, we cultured the blood samples for 2 to 3 wk. At the end of this period, the successfully derived PGC lines expanded up to 105 cells, and aliquots of PGCs were used for DNA isolation (PureGene kit; Qiagen) and sex determination. The W chromosome was detected as a 664-bp fragment amplified using the primers W4-forward and W4-reverse (SI Appendix, Table S3), and cycling conditions were as follows: 98 °C for 45 s, 38 cycles of 15 s at 98 °C, 30 s annealing at 54 °C, and 30 s amplification at 72 °C. One established and characterized line of PGCs was selected for further work and, at the age of 46 d in culture, subjected to homologous recombination procedure.
CRISPR/Cas9-Mediated Homologous Recombination.
We prepared the CRISPR/Cas9 constructs by cloning the chNHE1-specific guide RNA sequences into the sg scaffold of PX458 vector (pSpCas9BB-2A-GFP; AddGene no. 48138; ref. 23). The oligonucleotides used for annealing the guide RNA sequences, gRNA1 and gRNA2 (SI Appendix, Table S3), were designed into the first exon of chNHE1 using the CRISPR Design Tool (https://zlab.bio/guide-design-resources) (24) and CRISPOR (http://crispor.tefor.net/) (25). Both oligonucleotides were phosphorylated, hybridized, and ligated into pX458 cleaved by BbsI to form pX458-NHE1-4. To prepare PGCs carrying the precise deletion of W38 in NHE1, we used single-stranded oligodeoxynucleotides (ssODN) containing the W38 deletion and a silent mutation (T96G) introducing a BsaI restriction site as the template for homologous recombination (ref. 6 and SI Appendix, Table S3).
We resuspended 2.5 × 106 PGCs in Nucleofector Solution V (Lonza) and mixed them with 5 μg of pX458-NHE1-4 and 0.5 nmol of ssODNΔW38 (SI Appendix, Table S4) in a total volume of 100 μL. Electroporation was carried out using the AMAXA device program A-27. The PGCs with highest GFP fluorescence intensity were single-cell sorted 3 d post transfection. Within 2 to 3 wk, we expanded and analyzed 34 clones. Genomic DNAs from individual clones were extracted, and chNHE1 regions targeted with CRISPR/Cas9 were amplified by PCR using oligonucleotides ECL1 forward and ECL1 reverse (SI Appendix, Table S4). PCR products were purified, digested by BsaI, and sequenced. The efficiency of homologous recombination turned out to be so high that the preselection of PGC clones by BsaI digestion was not necessary (SI Appendix, Table S1).
Irradiation of Roosters.
To suppress endogenous spermatogenesis of the recipient roosters, we used local gamma-ray irradiation as described previously (11, 21). The Teragram K-01 radiation unit (Škoda) with 60Co as a source of gamma rays was employed for the irradiation procedure. We irradiated each rooster with five doses of 8 Gy over 14 d. Two weeks after the last irradiation, we started monitoring the decline of spermatogenesis. Semen samples were collected using the dorsoabdominal massage. When these samples were assessed as azoospermic in four consecutive observations, roosters were considered ready for transplantation. None of the sterilized roosters died during the irradiation procedure, and none of them showed any side effects related to the irradiation.
PGC Transplantation.
We applied the total dose of 0.9 to 2.5 × 106 of PGCs in 250 μL of cell culture medium into each testis of anesthetized recipient roosters according to the previously published procedures (11, 21). Briefly, the cell suspension was injected through tunica albuginea randomly at four to five different sites of each testis. No side effects or mortality associated with PGC transplantation or anesthesia were observed.
Breeding of ΔW38 Chickens.
Three weeks after the PGC transplantation, we started collecting semen samples using dorsoabdominal massage. As soon as the semen concentration reached 104 to 105 sperms per milliliter, we performed intramagnal insemination to increase the probability of fertilization. The insemination dose ranged from 0.1 to 0.4 mL of fresh undiluted semen per hen. Later, after the recovery of spermatogenesis, we also applied intravaginal insemination.
Virus Propagation.
The stocks of RCASBP(J)GFP, RCASBP(JPDRC)GFP, RCASBP(JADOL)GFP, RCASBP(JWB)GFP, and RCASBP(JZB)GFP viruses used in this study were propagated by transfection of the vector plasmid DNA into DF-1 cells (26) using the XtremeGENE transfection reagent (Roche), harvested on day 9 or 10 post transfection, cleared of debris by centrifugation at 2,000 × g for 10 min at 10 °C, and stored as aliquots at −80 °C. The virus titer was determined by terminal dilution and subsequent infection of DF-1 cells as 106 IU/mL. The transforming virus of subgroup J for in vivo sarcoma induction was produced by rescuing replication-defective BH-RSV present in the 16Q cell line (27). DF-1 cells were infected with RCASBP(J)GFP, and virus spread was monitored by GFP fluorescence. After 4 d, the infected GFP-positive DF-1 cells were mixed with 16Q cells and cultivated for a further 5 d. The viral stock containing both GFP-reporter viruses and transforming viruses of the same subgroup was centrifuged at 2,000 × g for 10 min at 10 °C and stored at −80 °C. The titer of the transforming virus was quantitated by an src focus assay in Brown Leghorn CEFs as 104 FFU/mL.
In Vitro Infection of Chicken Embryo Fibroblasts.
Chicken embryo fibroblasts (CEFs) were individually prepared from ten 10-d-old embryos as described previously (28), and their NHE1 genotypes were determined. CEFs, chicken permanent cell line DF-1, and Japanese quail QT6 cells were grown in a mixture of two parts of DMEM and one part of F-12 medium supplemented with 5% calf serum (CS), 5% FCS, 1% chicken serum, and penicillin/streptomycin (100 μg/mL each) in a 5% CO2 atmosphere at 37 °C. The RCASBP(J)GFP virus transducing GFP was used for infection in 100 μL of media. Virus infection and spread were observed as an increasing proportion of GFP-positive cells and quantified by flow cytometry on day 3 p.i.
In Vivo Infection of Chickens.
W38 +/+, +/−, and −/− chickens aged 14 to 28 d were challenged by i.v. injection of 106 IU of RCASBP(J)GFP. Six- and 13-d p.i., blood samples were collected, and sera were prepared. Viral RNA was isolated from the chicken serum by a QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer’s protocol. Five microliters of isolated RNA was reversely transcribed using Protoscript II Reverse Transcriptase (NEB) and 3′cDNA synthesis primer (SI Appendix, Table S4). The amount of 1.5 μL of cDNA was then used for quantitative PCR with MESA GREEN qPCR MasterMix Plus (Eurogentec) and primers for the J subgroup env (SI Appendix, Table S3). The PCR was run in a Bio-Rad CFX96TM real-time cycler as follows: 8 min at 95 °C, then 45 cycles of 15 s at 95 °C, 25 s at 51 °C, and 35 s at 72 °C and final polymerization at 72 °C for 10 min. Cycles of quantification (Cq) values were analyzed by the CFX Manager software. The specificity of the PCR product was confirmed by melting curve analysis.
Because of transient viremia and low titer of circulating virus in ALV-J–infected chickens, we employed indicator DF-1 cells harboring integrated env-less RCAS vector for amplification and complementation of infectious virus in serum samples. The env-less vector was produced from the RCASBP(C)GFP by elimination of the XhoI-BstBI part of the env-C gene. The indicator DF-1 cells do not produce infectious virus unless complemented for the env deficiency by the virus present in serum. The chicken sera were serially diluted, and 0.1-mL aliquots were applied for infection. The indicator cells were subcultured 3 d p.i., and the cell supernatants were harvested 10 d p.i. Intact DF-1 cells were then infected with 1 mL of collected media filtered through Millipore 0.45-μm filter. Three to four days later, the GFP positivity was assessed microscopically, and the titer of virus was calculated as infectious units (IUs) per milliliter of serum.
In Vivo Tumor Induction in Chickens.
W38 +/+, +/−, and −/− chickens at the age of 13 to 56 d were inoculated with 250 or 500 FFU of transforming virus rescued from 16Q cells in 0.1 mL of Iscove’s DMEM, s.c. into the left and right wing web, respectively. The growth of sarcomas at the site of inoculation was monitored and quantitated by weighting the dissected sarcomas from euthanized birds.
Selection of Off-Target Sites and Mutation Analysis.
We screened the NCBI chicken genome using the CRISPOR tool (http://crispor.tefor.net/) and identified the most probable off-target sites. Eight candidates with the highest CFD scores containing at least 10 consecutive nucleotides matched with the 20-bp chNHE1 gRNA sequence and the NGG protospacer adjacent motif were selected (SI Appendix, Table S4A). We designed PCR amplification primers for each of the potential off-target sites (SI Appendix, Table S4B) and amplified the respective off-target regions using 100 ng of genomic DNA of the W38−/− PGC clone 13 and CB embryo fibroblasts as a control. The PCR amplification was done under the following conditions: 3 min at 98 °C, 40 cycles of 10 s at 98 °C, 30 s at 62 °C, 30 s at 72 °C, and final amplification for 5 min at 72 °C. Sequence analysis of the resulting PCR products was performed, and the sequences were aligned to the respective NCBI NHE1 sequence (SI Appendix, Table S4C). The GFP sequence was amplified as follows: 3 min at 95 °C, 30 cycles of 15 s at 95 °C, 30 s at 58 °C, 45 s at 72 °C, and final amplification for 7 min at 72 °C. The sequences of primers GFP-F and GFP-R are given in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank Filip Šenigl and Zdeněk Cimburek (Institute of Molecular Genetics, Prague) for their kind help with FACS and cell sorting experiments; Björn Schuster and Petr Kašpárek (Institute of Molecular Genetics, Prague) and Benjamin Schusser (Technische Universität München, Freising) for valuable consultation in regard to the technical details of CRISPR/Cas9 technology and PGC manipulations; and Daniel Elleder (Institute of Molecular Genetics, Prague) for critical reading and comments on the manuscript. This study was supported by the Czech Science Foundation (grant no. 15-23993S); Ministry of Education, Youth and Sports of the Czech Republic (project NPU I no. LO1419); Ministry of Agriculture of the Czech Republic (grant no. QK1810344); and Czech Academy of Sciences (Premium Academiae Award to J.H.).
Footnotes
Competing interest statement: A.K., P.T., J.M., M.R., J.P., J.K., D.K., and J.H. are inventors in a patent application related to this work (CZ patent application no. PV 2019-392). The other authors declare no competing interests.
This article is a PNAS Direct Submission. S.H.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913827117/-/DCSupplemental.
References
- 1.Payne L. N., et al. , A novel subgroup of exogenous avian leukosis virus in chickens. J. Gen. Virol. 72, 801–807 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Payne L. N., Nair V., The long view: 40 years of avian leukosis research. Avian Pathol. 41, 11–19 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Zhou D., et al. , Outbreak of myelocytomatosis caused by mutational avian leukosis virus subgroup J in China, 2018. Transbound. Emerg. Dis. 66, 622–626 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Chai N., Bates P., Na+/H+ exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc. Natl. Acad. Sci. U.S.A. 103, 5531–5536 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kučerová D., et al. , Nonconserved tryptophan 38 of the cell surface receptor for subgroup J avian leukosis virus discriminates sensitive from resistant avian species. J. Virol. 87, 8399–8407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H. J., et al. , Precise gene editing of chicken Na+/H+ exchange type 1 (chNHE1) confers resistance to avian leukosis virus subgroup J (ALV-J). Dev. Comp. Immunol. 77, 340–349 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Reinišová M., et al. , Genetic diversity of NHE1, receptor for subgroup J avian leukosis virus, in domestic chicken and wild anseriform species. PLoS One 11, e0150589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Lavoir M. C., et al. , Germline transmission of genetically modified primordial germ cells. Nature 441, 766–769 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Collarini E. J., Leighton P. A., Van de Lavoir M. C., Production of transgenic chickens using cultured primordial germ cells and gonocytes. Methods Mol. Biol. 1874, 403–430 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Schusser B., et al. , Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc. Natl. Acad. Sci. U.S.A. 110, 20170–20175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trefil P., et al. , Male fertility restored by transplanting primordial germ cells into testes: A new way towards efficient transgenesis in chicken. Sci. Rep. 7, 14246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koslová A., et al. , Genetic resistance to avian leukosis viruses induced by CRISPR/Cas9 editing of specific receptor genes in chicken cells. Viruses 10, 605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinišová M., et al. , Intronic deletions that disrupt mRNA splicing of the tva receptor gene result in decreased susceptibility to infection by avian sarcoma and leukosis virus subgroup A. J. Virol. 86, 2021–2030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plachý J., et al. , Identification of New World quails susceptible to infection with avian leukosis virus subgroup J. J. Virol. 91, 02002–02016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan X., et al. , Residues 28 to 39 of the extracellular loop 1 of chicken Na+/H+ exchanger type I mediate cell binding and entry of subgroup J avian leukosis virus. J. Virol. 92, e01627-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitworth K. M., et al. , Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Dimitrov L., et al. , Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. PLoS One 11, e0154303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oishi I., Yoshii K., Miyahara D., Kagami H., Tagami T., Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 6, 23980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H. J., et al. , Targeted gene insertion into Z chromosome of chicken primordial germ cells for avian sexing model development. FASEB J. 33, 8519–8529 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Plachý J., The chicken–A laboratory animal of the class Aves. Folia Biol. (Praha) 46, 17–23 (2000). [PubMed] [Google Scholar]
- 21.Mucksová J., et al. , Conservation of chicken male germline by orthotopic transplantation of primordial germ cells from genetically distant donors. Biol. Reprod. 101, 200–207 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Whyte J., et al. , FGF, insulin, and SMAD signaling cooperate for avian primordial germ cell self-renewal. Stem Cell Reports 5, 1171–1182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu P. D., et al. , DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeussler M., et al. , Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Himly M., Foster D. N., Bottoli I., Iacovoni J. S., Vogt P. K., The DF-1 chicken fibroblast cell line: Transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248, 295–304 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Moscovici C., et al. , Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11, 95–103 (1977). [DOI] [PubMed] [Google Scholar]
- 28.Federspiel M. J., Hughes S. H., Retroviral gene delivery. Methods Cell Biol. 52, 179–214 (1997). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.