Significance
Fibrinogen hexamers are major components of blood clots. After release of fibrinopeptides resulting in fibrin monomers, clot formation occurs through fibrin oligomerization followed by lateral aggregation, packing into fibrin fibers, and consequent branching. Shedding light on fibrin clots by in situ cross-linking mass spectrometry and structural modeling extends our current knowledge of the structure of fibrin with regard to receptor-binding hotspots. Further restraint-driven molecular docking reveals how fibrin oligomers laterally aggregate into clots and uncovers the molecular architecture of the clot to albumin interaction. We hypothesize this interaction is involved in the prevention of clot degradation. Mapping known mutations validates the generated structural model and, for a subset, brings their molecular mechanisms into view.
Keywords: cross-linking mass spectrometry, XlinkX, DSSO, fibrinogen, fibrin clot
Abstract
Upon activation, fibrinogen forms large fibrin biopolymers that coalesce into clots which assist in wound healing. Limited insights into their molecular architecture, due to the sheer size and the insoluble character of fibrin clots, have restricted our ability to develop novel treatments for clotting diseases. The, so far resolved, disparate structural details have provided insights into linear elongation; however, molecular details like the C-terminal domain of the α-chain, the heparin-binding domain on the β-chain, and other functional domains remain elusive. To illuminate these dark areas, we applied cross-linking mass spectrometry (XL-MS) to obtain biochemical evidence in the form of over 300 distance constraints and combined this with structural modeling. These restraints additionally define the interaction network of the clots and provide molecular details for the interaction with human serum albumin (HSA). We were able to construct the structural models of the fibrinogen α-chain (excluding two highly flexible regions) and the N termini of the β-chain, confirm these models with known structural arrangements, and map how the structure laterally aggregates to form intricate lattices together with the γ-chain. We validate the final model by mapping mutations leading to impaired clot formation. From a list of 22 mutations, we uncovered structural features for all, including a crucial role for βArg’169 (UniProt: 196) in lateral aggregation. The resulting model can potentially serve for research on dysfibrinogenemia and amyloidosis as it provides insights into the molecular mechanisms of thrombosis and bleeding disorders related to fibrinogen variants. The structure is provided in the PDB-DEV repository (PDBDEV_00000030).
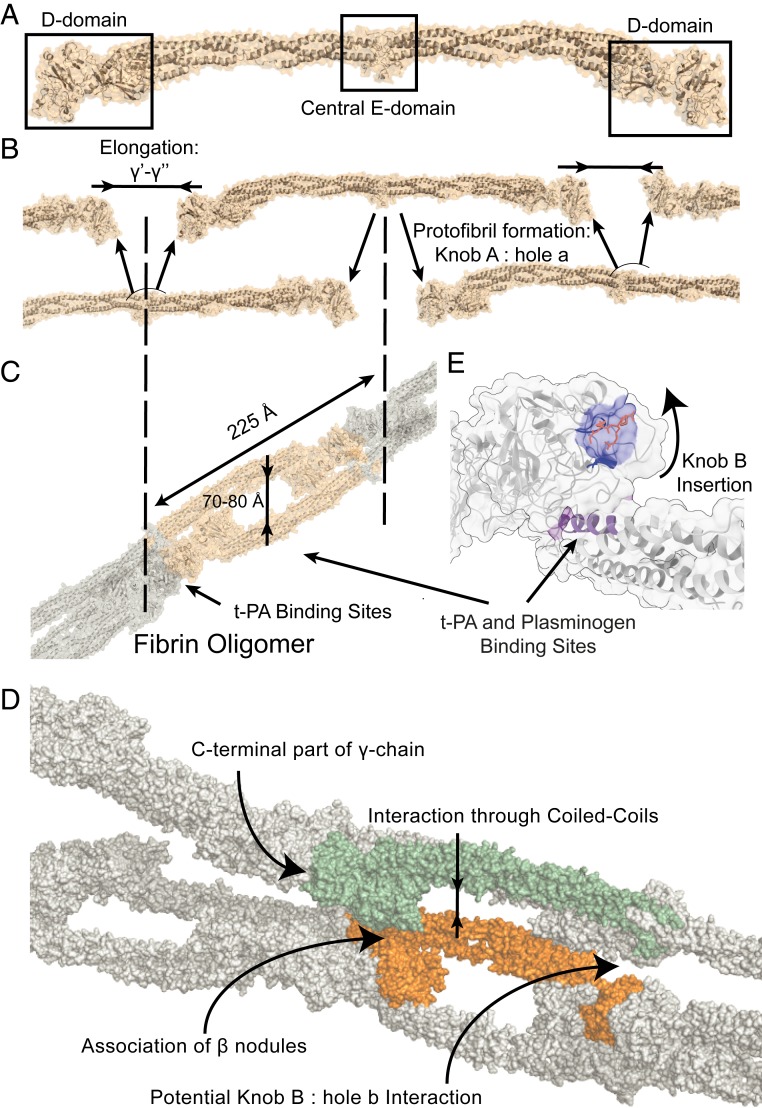
Fibrin clot formation is a multistage process that can be affected by numerous factors, including modifications of the initial protein building blocks, cellular environment, physiological properties of the environment, and the presence or absence of blood flow (1–3). Clot formation starts with the release of fibrinopeptides A and B from the N termini of the α- and β-chains of the large fibrinogen protein (Fig. 1A). Fibrinopeptide release is followed by the A:a knob-hole interaction of the parallel fibrin monomers, and oligomers are formed through simultaneous linear elongation through the sewing of the γ-nodules with the next fibrin monomer (4–6) (Fig. 1B). Oligomers polymerize further into protofibrils (Fig. 1C), after which lateral aggregation follows (Fig. 1D), a process about which much less is known. Protofibrils have successfully been visualized with microscopy methods (7, 8), and their packing into fibrin fibers with a periodicity of 225 Å has been well documented (9, 10) although this so far provided only a low-resolution view. This final, three-dimensional scaffold is formed through bilateral or trimolecular junction mechanisms (11, 12), and some potential sites involved in lateral aggregation are shown in Fig. 1D. Concurrently with the polymerization process, covalent cross-linking of fibrin mediated by factor XIIIa takes place (13), which strengthens the final structure with naturally occurring disulfide bonds (14) [for a detailed review about clot formation, see Weisel and Litvinov (15)]. The clots are finally degraded by plasmin cleavage upon formation of a ternary complex combining the clot, tissue plasminogen activator (t-PA), and plasminogen (Fig. 1 C and E), followed by conversion of plasminogen into plasmin (16). To date, it has been difficult to capture the molecular structure of fibrin within fully formed clots with traditional structural biology techniques, such as crystallography or electron microscopy (EM), and structural models are solely available for smaller disconnected parts. To illustrate, a high-resolution crystal structure was reported (17); however, this structure lacks domains important for lateral aggregation like the αC-terminal, βN-terminal, and γC-terminal domains.
Fig. 1.
Structural model of fibrin clot formation. (A) Structural model of the fibrin monomer (α2β2γ2), after cleavage of the fibrinopeptides from fibrinogen (based on PDB: 3ghg). (B) Fibrin monomers form double-stranded oligomers through linear elongation and via the well-described A:a knob-hole interaction. (C) Double-stranded protofibrils, which laterally aggregate with a linear periodicity of 225 Å while maintaining cavities of 70 to 80 Å. The sites potentially involved in clot degradation through plasminogen cleavage are highlighted. (D) Schematic representation of laterally aggregating protofibrils. Suggested interaction sites for lateral aggregation of the protofibrils are highlighted, including interaction through laterally aggregating coiled coils, association of β-nodules, and the potential site of the knob b:hole B interaction. Part of the first protofibril that is involved in lateral aggregation is shown in orange. The second half of the full protofibril, which includes the domain in green, is omitted, and the γC-terminal nodule is highlighted for clarity. (E) The potential function of knob B upon insertion into hole B. Cryptic sites important for clot degradation become accessible for formation of a ternary complex between clot, t-PA, and plasminogen.
Cross-linking mass spectrometry (XL-MS) has emerged as a powerful tool for studying protein–protein interactions. It is capable of extracting protein structure distance information from any protein (18) and has been successfully applied to elucidate the structure of elongated proteins (19), as well as tissue samples (20). The technique utilizes small reagents with two reactive moieties that form a covalent bond between two amino acids in close proximity. Upon application to, and proteolytic digestion of, protein–protein complexes, four distinct peptide products are formed: nonmodified, monolinked, loop-linked, and cross-linked peptides. The first three products consist of single peptides in various forms that yield no or limited structural information. The fourth product consists of two peptides bound together by the cross-linking reagent; this yields valuable distance information for protein tertiary structure (the two peptides originate from the same protein) or for protein quaternary structure (the two peptides originate from different proteins). Identification of the two peptides by mass spectrometry allows for localization of the cross-link within the proteins at the residue level. We apply XL-MS utilizing the reagent DSSO (a lysine–lysine reactive cross-linking reagent) to study fibrin clots extracted from platelet-poor plasma and uncovered 284 unique distance constraints. This dataset was combined with structural modeling (protein structure prediction and protein–protein docking; combined with XL-MS, this approach is termed integrative structural modeling) to illuminate dark areas on the fibrin(ogen) structure. Our study, supported by biochemical evidence, resolves 78% (previously ∼66%, counted by amino acids present in the full protein sequence) of the total fibrin structure after processing, validating 92% of the distance constraints in our dataset. To validate the final structure, we investigated 22 mutations reported to lead to impaired clot formation. For the complete set, our model provides explanations at the molecular level (Movie S1). Additionally, previous studies suggested that albumin hinders fibrinolysis by interfering with activation or hindering access to cleavage sites of clot degradation enzymes (21). From our data, the interaction interface between albumin and fibrin clots can be uncovered, from which it can clearly be demonstrated that albumin indeed interferes with fibrinolysis by sterically hindering access to sites involved in plasminogen activation and plasmin cleavage. All data and structures have been made available through the PRIDE and PDB-Dev repositories (22, 23).
Materials and Methods
For fibrinogen, we highlight the residue numbering as provided by the UniProt database: i.e., based on the numbering in the nascent form of the individual chains, including the signal- and fibrinopeptides.
Fibrin Clot Formation, Purification, and Sample Preparation.
Deidentified plasma samples were obtained after informed consent and in accordance with the ethics board of Sanquin (Amsterdam, The Netherlands). Three samples of blood extracted from one voluntary donor, citrated to prevent coagulation, were treated separately to induce clot formation and consecutively to cross-link the structure. After a series of centrifugation steps at 120, 2,000, and 10,000 × g, clot formation in 1 mL of platelet-poor plasma was initiated by addition of 16 mM CaCl2 (Sigma-Aldrich) and 9 pM tissue factor (Innovin, Siemens Healthcare Diagnostics). The final mixtures were heated to 37 °C for 5 min, the resulting clots were washed with cross-linking buffer containing 20 mM Hepes, 150 mM NaCl, and 1.5 mM MgCl2 (all Sigma-Aldrich) adjusted to pH 7.8 with NaOH (Sigma-Aldrich) and with no reagent present. This buffer and sample preparation are designed to preserve the native conformation of the proteins, and therefore the clot, while washing unspecifically bound proteins. Cross-linking buffer was aspirated from the clots, which were consequently filtered with a molecular weight cutoff (MWCO) filter from Amicon (10 kDa; Sigma-Aldrich), Microcon (30 kDa; Merck Millipore), and one sample was processed without prior filtering. After these steps, cross-linking buffer supplemented with 2 mM DSSO (18) (Thermo Fisher Scientific) was pushed by centrifugal force through the filters (10 min, 6000 × g for Amicon and 14,000 × g for Microcon) in four repetitions to maintain an ample supply of fresh nondegraded cross-linking reagent. The cross-linking reaction was then quenched by addition of a 20 mM Tris⋅HCl (Sigma-Aldrich) solution at pH 8.5, and the cross-linked clots were then snap frozen in liquid nitrogen.
The snap-frozen cross-linked clots were processed as previously described with minor adjustments (18). Briefly, clots were homogenized on a bead-mill device (Retsch) for 5 min at 120 Hz. Then, the samples were treated with protein deglycosylation mix II (NEBB) overnight, attempting to fully remove both N- as well as O-linked glycans, followed by acetone-cold precipitation to clean up the protein mixture. As deglycosylation is applied after the cross-linking reaction, the removal of the glycans will not impact the distance information derived from the cross-links, while improving the identification performance. Proteins were resuspended in a solution containing 1% SDC and 10 mM TCEP with 40 mM CAA (Sigma-Aldrich) as reduction and alkylating agents and heated to 37 °C for 1 h. The resulting solution was diluted with 50 mM ammonium bicarbonate (Sigma-Aldrich) and digested by a combination of LysC (Wako) and Trypsin (Promega). The final peptide mixtures were desalted with BioSelect solid-phase extraction C18 columns (300-Å pore size material; Vydac) and fractionated with an Agilent 1200 HPLC pump system (Agilent) coupled to an Opti-LYNX trap column (Optimize Technologies) and strong cation exchange (SCX)-separation column (PolyLC), resulting in 25 fractions per fibrin clot. For the serum experiments, human serum from an anonymous healthy donor was provided by Sanquin Research (Amsterdam, The Netherlands). The whole blood was collected in a 9-mL Vacuette tube (Greiner Bio-One) containing Z Serum Clot Activator and then was left undisturbed at room temperature for 30 to 60 min. The clotted material was removed by centrifugation at 1,800 × g for 20 min at room temperature, and the serum was transferred as a 1-mL aliquot to a clean 1.5-mL Eppendorf tube, snap frozen in liquid nitrogen, and stored at −80 °C until further analysis. Proteins were processed in a similar fashion as the clots, excluding homogenization, deglycosylation, and fractionation.
Liquid Chromatography with Mass Spectrometry and Data Analysis.
Each fraction was separated with an Agilent 1290 Infinity UHPLC system (Agilent) on a 50-cm analytical column packed with C18 beads coupled online to an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). For the serum experiments, the Orbitrap HF-X (Thermo Fisher Scientific) was used with the same acquisition parameters. Details of the full setup, separation gradient, data acquisition methods, and data analysis for cross-linking experiments were previously described (18). Briefly, each SCX fraction was separated on a UHPLC 1290 system (Agilent Technologies, Santa Clara, CA) fitted with a 50-cm C18 analytical column (bead-size 2.7 µm) over a 3-h gradient coupled on-line to an Orbitrap Lumos mass spectrometer (Thermo Scientific, San Jose, CA). The MS acquisition method was configured with the following settings: survey MS1 Orbitrap scan at 60,000 resolution, automatic gain control (AGC) target of 4e5 ions, and maximum inject time of 50 ms; MS2 Orbitrap scan at 30,000 resolution, AGC target of 5e4 ions, and maximum inject time of 100 ms for detection of DSSO signature peaks; and four MS3 Ion Trap scans at AGC target of 3e4 ions, maximum inject time of 90 ms for sequencing selected signature peaks (representing the individual peptides). The resulting raw files were analyzed with the Proteome Discoverer software suite version 2.3.0.522 (Thermo Fisher Scientific) with the incorporated XLinkX node for analysis of cross-linked peptides. Data were searched against the reviewed Homo Sapiens UniProt database (20,230 entries, version 2018_01, downloaded from UniProt) with the maximum FDR rate set to 1%; further standard settings were used. We were able to use the full human proteome due to the gas-phase cleavable characteristics of DSSO, essentially liberating the individual peptides for mass analysis and preventing the need for complex searches where each peptide needs to be combined with every other peptide to fit a single mass. For the shotgun proteomics experiments, the same approach was used, with the following modifications: Only unfractionated samples were analyzed, and the resolution setting for the MS2 Orbitrap scan was reduced to 15,000. Quantification was performed with the iBAQ (intensity-Based Absolute Quantification) algorithm in MaxQuant version 1.6.2.3 (24). For analysis of the specificity of human serum albumin (HSA) binding, quantification of detected precursor ions was performed across all of the available cross-linked fractions. From the 1,205 unique cross-link pair identifications, we retained only those detected in two out of the three samples, resulting in 451 unique peptide residue pairs (from 697 spectral matches), 284 of which (from 437 spectral matches) provide distance constraints for the fibrin(ogen) chains/subunits and their interacting proteins. As each sample is treated slightly differently prior to cross-linking, the selected cross-links represent the core interaction points (i.e., amino acids in the protein that are connected to many different locations/proteins). Furthermore, as the sample is solid-phase, which decreases the space between atoms when compared to solution, combined with the mechanical forces applied during clot purification, we define the maximum cross-linker distance as 43 Å. This is in line with previously published studies applying solid-phase cross-linking (20) and cross-link–driven docking (25).
Modeling and Docking of Albumin and Individual Fibrinogen Domains into Fibrin.
Predictions of the location of individual domains were performed with the web services provided by ThreaDomEx (26) and Robetta (27). Additional template searches were performed with HHalign-Kbest (28). Each domain was modeled separately with I-TASSER (29), Robetta (27), and RaptorX (30). A highly detailed description of these modeling procedures, allowing for replication of the results, is provided in SI Appendix, Note S1. The resulting structures were scored with z-DOPE (31), Errat (32), Procheck (33), QMEAN (34), ProQ2 (35), and the final model of each domain chosen based on a combination of available scores (best or second best in at least three out of five scores), combined with previously reported biochemical features of the modeled domain to ensure biological validity. Tables with the scores are provided in SI Appendix, Note S1. The detected intralinks within each domain were used for validation of the resulting model (apart from α432–491, where the cross-links were not used in the validation procedure).
Interlinks and overlength intralinks (potentially arising from the interaction between two copies of the same subunit) were clustered with DisVis (36) to determine collections of cross-links pointing to each location. Clusters generated with DisVis are provided as SI Appendix, Figs. S4–S8. For docking laterally aggregating protofibrils, we took the available fibrinogen template (PDB: 3ghg) as a basis as it represents the building block of a protofibril, and we split this template from a hexamer to a single trimer. Trimers were submitted to CPORT (37) to highlight active residues, which were included in the docking procedure. Docking was performed with HADDOCK (38) and ClusPro (39). Highly detailed descriptions of each docking run, allowing for replication of the results, are provided in SI Appendix, Notes S2 and S3. Obtained structures were visualized with pyMol (40) and ChimeraX (41).
Analysis of Mutation Sites.
Conservation analysis of the generated structures was performed with the Consurf webserver (42), with standard settings using 300 sequences from the Uniref-90 database (43) and modeled structures as PDB input. Mapping of the conservation sites and relevant tables can be found in SI Appendix, Fig. S11 and Datasets S1–S7. Combined Annotation Dependent Depletion (CADD) (44) scores were calculated with the web version of ANNOVAR (45), with the reported mutations as input (for further details, see http://wannovar.wglab.org/; higher scores are more likely to have detrimental effects). A CADD score represents an aggregation of multiple scoring methods operating at the nucleotide level, describing the effect of a genetic mutation on the protein of interest and therefore serves as an effective parameter to highlight deleterious, functional, and/or disease-causative variants. For those residues with reported mutations, the side chain backbone-dependent rotamers were generated with pyMol, and the conformation with the lowest strain values and no clashes within the structure was selected for final analysis.
Results
So far, studies aimed at deciphering the high-resolution structure of fibrin assemblies have used a combination of atomic force microscopy and molecular dynamics simulations (46, 47). These studies provide invaluable insights into the structural organization of fibrin oligomers; however, to circumvent technological limitations, they were not performed on full clots. A recent mass spectrometry-based study mapped endogenously cross-linked peptide species, suggesting a more compact organization of protein blocks than so far assumed (48) and, importantly, highlights the potential of mass-spectrometry based investigations of fibrin clots. Here, we employ in situ cross-linking mass spectrometry on purified clots extracted from plasma specifically to delve into the mechanisms underlying lateral aggregation (SI Appendix, Fig. S2).
Initial Data Check.
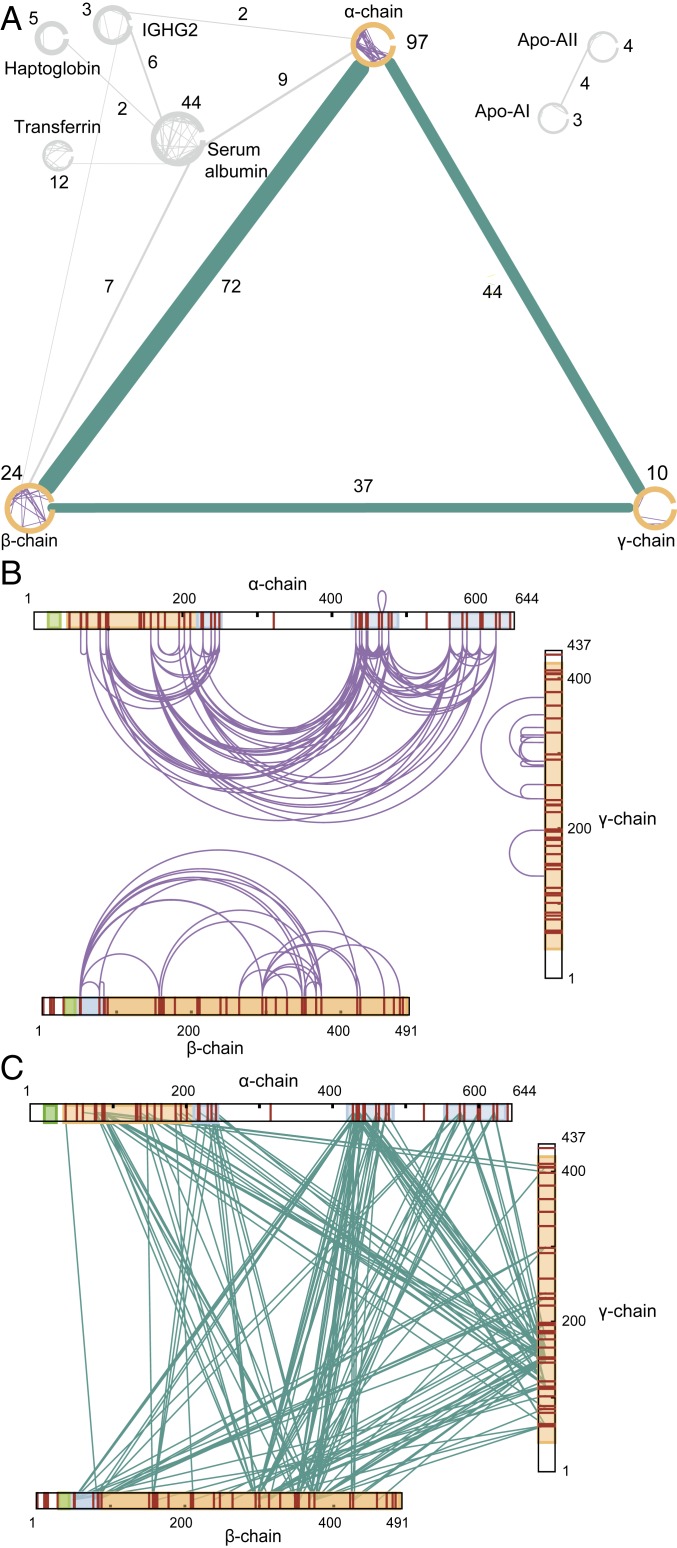
To gain insight into the sample purity and data quality of our XL-MS measurements, we first visualized the detected cross-links as a network (49). We only retained proteins with at least one intra- and one interlink (Fig. 2A), excluding proteins for which insufficient structural information was detected. The core of the resulting network is built up from the fibrinogen α- (97 intralinks; 116 interlinks to the other chains), β- (24 intralinks; 109 interlinks to the other chains), and γ-chain (10 intralinks; 81 interlinks to the other chains). In addition to cross-links between the fibrin(ogen) subunits, we also detect cross-links involving serum albumin, transferrin, haptoglobin, immunoglobulin Gs (IgGs), and apolipoproteins apolipoprotein A-I (apo A-I) and apolipoprotein A-II (apo A-II). In total, three interlinks were detected between fibrin(ogen) and Ig while, for HSA, 44 intralinks and 16 interlinks to the α- and β-chains of fibrin(ogen) were detected. Excitingly, a very large degree of the sequence of fibrin(ogen) subunits were covered by cross-links (Fig. 2 B and C). As anticipated, for two lysine-poor regions on the α-chain, no cross-links were detected.
Fig. 2.
Overview of cross-linking MS data. (A) Cross-links detected in two out of three samples. Only proteins with at least 1 inter- and 1 intralink are depicted. (B) XL-MS intra- and (C) interlinks detected on the sequences of the fibrin(ogen) chains. Cleaved off fibrinopeptides A and B are shown in green, and domains with reported PDB structures are shown in orange. Domains for which additional structural models are presented in this study are shown in blue.
Mapping Detected Restraints on Available Structures.
For structural validation, we mapped the detected intralinks of serum albumin on the existing crystal structure, for which we find 90.9% of restraints within the set distance constraint (PDB: 1uor) (SI Appendix, Table S1). For fibrin(ogen), the detected inter- and intralinks were mapped on existing crystal structures of the fibrinogen hexamer and fibrin (4, 17). For the first partial structure of fibrin (PDB: 1fzc), a total of 43 restraints could be mapped, of which 81% were within the allowed distance constraint. For the assembled α, β, and γ trimer (PDB: 3ghg), used as the basis for this study, a total of 86 cross-links could be mapped, of which 58% were within the allowed distance constraint (SI Appendix, Table S2). The lower percentage of cross-links within the defined distance cutoff indicates that, upon assembly of the full protofibril, likely some degree of structural rearrangement takes place.
Illuminating Dark Areas on the Fibrinogen Structure.
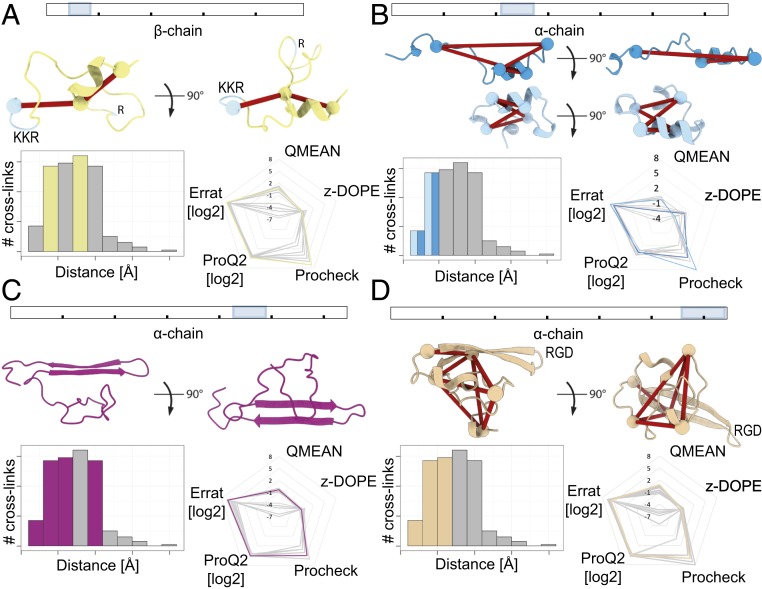
The available structures for fibrinogen and fibrin lack several domains, or, as we term them, dark areas. To define those dark areas amenable to integrative structural modeling supported by our XL-MS results, we separated the chains into domains for which currently no structural information is available (Table 1). In total, we identified four suitable domains. 1) For the N-terminal domain of the β-chain at residue 21–53 (UniProt: 51–83), 29 inter- and two intralinks were detected. This domain was previously reported to bind the anticoagulant heparin (50), and, even though a heparin-binding pocket is not strictly defined, some structural characteristics are present (51, 52). A total of 18 models were chosen for manual inspection. By enforcing the exposure of a heparin-binding site and the restraints imposed by the intralinks, a high scoring conformation could be selected (Fig. 3A and SI Appendix, Table S2, Beta_within). 2) The α-chain domain at residue 201–230 (UniProt: 220–249) was detected with four intra- and 19 interlinks. From a total of 15 models, we selected two models with different conformations (folded and elongated) that score high on all metrics and satisfy the distance constraints of the detected intralinks well (in both cases, four of four valid restraints) (SI Appendix, Table S2, Alpha1). As the two versions are likely present (see next paragraph), we selected both (Fig. 3B). 3) For the α-chain domain at residue 413–472 (UniProt: 432–491) (part of the αC-terminal domain), we detected 69 inter- and 20 intralinks. This domain was previously reported (15) to be involved in fibrin clot formation by interacting with other fibrin copies, making it unclear for the detected cross-links whether they arise from intra- or intermolecular contacts. As such, the cross-links for this domain were excluded from the modeling procedures. A structure for the bovine form was previously reported (PDB: 2jor) (53), which we used for homology modeling, with no distance restraints but with forced disulfide bond formation between αCys’461 (UniProt: 442) and αCys’491 (UniProt: 472). We were successful in mapping all 20 detected restraints within the set distance constraint for the best scoring of 15 models (Fig. 3C and SI Appendix, Table S2, Alpha3a_Within). 4) For the α-chain domain at residue 539–601 (UniProt: 558–620), we detected 50 inter- and seven intralinks. Upon activation of this domain, three lysines are hypothesized to interact with glutamine residues, either between different molecules or with the flexible region of the αC-terminal domain, forming β-sheets (54). From 16 manually inspected models, we chose the highest scoring conformation with an exposed integrin-binding RGD motif secured by β-sheets (Fig. 3D and SI Appendix, Table S2, Alpha3b_within).
Table 1.
Overview of fibrinogen domains
| Fibrinogen chain | Residue numbers of the nascent chains (and according to UniProt sequence) | Comment | Available structure | Submitted to structure prediction | Involved in detected cross-links |
| α | (1–19) | Signal peptide | No | No | No |
| 1–15(20–35) | Fibrinopeptide A | No | No | No | |
| 17–26 (36–45) | No structural info | No | No | No | |
| 27–230 (46–249) | α-chain coiled coil | Partially, 46–219 (PDB: 3ghg) | Yes, 220–249 | Yes | |
| 231–412 (250–431) | Includes highly flexible parts | No | No | No | |
| 413–472 (432–491) | Part of N-term subdomain (interactive domain) | Full template (PDB: 2jor, based on the structure of bovine domain) | Yes | Yes | |
| 473–538 (492–557) | Includes highly flexible parts | No | No | No | |
| 539–601 (558–620) | Part of C-term subdomain (RGD-containing domain) | No | Yes | Yes | |
| 602–610 (621–629) | No structural info | No | No | No | |
| 611–625 (630–644) | Cleaved during processing | No | No | No | |
| β | (1–30) | Signal peptide | No | No | No |
| 1–14 (31–44) | Fibrinopeptide B | No | No | No | |
| 15–20 (45–50) | No structural info | No | No | No | |
| 21–53 (51–83) | Part of knob B (heparin-binding domain) | No | Yes | Yes | |
| 58–456 (88–486) | β-chain coiled coil and knob | Yes (PDB: 3ghg) | No | Yes | |
| γ | (1–26) | Signal peptide | No | No | No |
| 1–12 (27–38) | No structural info | No | No | No | |
| 13–394 (39–420) | γ-chain coiled coil and nodule | Yes (PDB: 3ghg) | No | Yes | |
| 395–424 (421–450) | No structural info | No | No | No |
Fig. 3.
Structural models of individual domains. Each panel shows the location within the protein sequence of the relevant domain (Top), the structural model of the relevant domain with mapped cross-links (Middle), the histogram with the overall distance distribution of the mapped cross-links in gray and the distances of the cross-links mapped onto the domain in color (Bottom Left), and the radial graph with the scores for all of the obtained models (Bottom Right). On the radial graphs, each corner represents the individual scoring algorithm: log2 of ProQ2 and Errat scores, QMEAN, z-DOPE, and the number of passed evaluations for the Procheck score; further to the outside is better. The score distributions for the selected structural model are highlighted in the color selected for the domain. (A) The β-chain N terminus, residues 21–53 (UniProt: 51–83), with amino acid motief KKR highlighted. (B) Part of the α-chain domain 1, residues 201–230 (UniProt: 220–249). Here, we have chosen two border conformations for our structural investigations. All relevant restraints belong to the same bins on the histograms, and, therefore, those bins are highlighted with two colors (light blue for the top conformation and dark blue for the bottom conformation). (C) The N-terminal subdomain of the α-chain C terminus, residues 413–471 (UniProt: 432–491). (D) The C-terminal subdomain of the α-chain C terminus, residues 539–601 (UniProt: 558–620), with amino acid motief RGD highlighted.
The domains at residues UniProt: 1–35 on the α-chain, UniProt: 1–44 on the β-chain, and UniProt: 1–26 on the γ-chain were excluded as they harbor signal- and/or fibrinopeptides, which were also not detected in our shotgun proteomics experiments (SI Appendix, Fig. S3). As these are only present in the UniProt sequence, these locations are only provided as UniProt. Residues 17–26 (UniProt: 36–45) on the α-chain, 15–20 (UniProt: 45–50) and 55–57 (UniProt: 85–87) on the β-chain, and 1–12 (UniProt: 27–38) and 395–411 (UniProt: 421–437) on the γ-chain were omitted, as we detected no structural information for these regions. The domain 231–412 (UniProt: 250–431) on the α-chain (previously reported to be highly flexible) (9) was also excluded, as we did not detect any structural information, while the region before α231 (UniProt: 250) represents the naturally occurring fibrinogen truncated variants (i.e., fragment X) (55). The domain at residue 473–538 (UniProt: 492–557) on the α-chain (part of the αC-terminal domain), but preceding the domain we uncovered in point 4 above, was excluded as we did not detect structural information; this domain was reported to potentially form beta-sheets upon activation by FXIII (54), which could not be confirmed from our data. These preceding regions are considered as highly flexible linkers in our model. We also excluded the α-chain C-terminal domain at residues 602–625 (UniProt: 621–644) as we also did not detect structural information for this domain, in line with previous reports that this region is partially removed following translation (56). In our shotgun proteomics experiment, we detect peptides from the elongated 420-kDa version of the α-chain; however, these peptides do not comprise more than 0.3% of the total detected intensity, and we therefore do not expect them to play an important role within the clot (SI Appendix, Fig. S3). In total, we improve the fibrin(ogen) structure coverage, supported by biochemical evidence, from 66 to 78%.
Assembly of the Fibrin Scaffold.
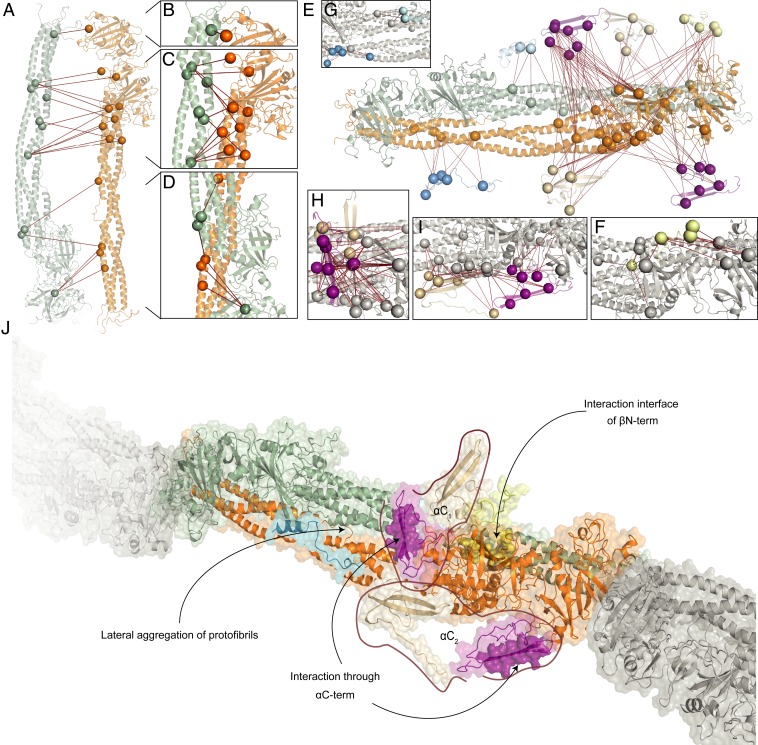
Our initial attempts to map the detected restraints on an existing model of protofibrils (i.e., a dimerized trimer) (47) were unsuccessful. None of the distance restraints fit with the structural model due to, e.g., the predicted cavity of ∼70 Å (15). Combined with our experimental setup ensuring that a purified whole clot was investigated, we concluded that our XL-MS results can best be assigned to laterally aggregating protofibrils. Trying to predict the conformation of laterally aggregating protofibrils of fibrin monomers is, however, challenging as these monomers are highly flexible (57). We resolved this by driving the assembly of the final repeating unit by a combination of our XL-MS results and interaction interface analysis by DisVis and CPORT for scaffold assembly and finally protein–protein docking with HADDOCK (Fig. 4). As the initial fibrinogen template possesses C2 symmetry, we split the hexamer into pairs of identical trimers and use these as parts of two distinct laterally aggregating protofibrils (colored in green and orange in Fig. 1D). CPORT analysis shows that most of the potential interaction sites are localized on discrete parts of the molecule and are not shared between the trimers. From our DisVis analysis, we identified two major clusters of overlength intralinks between the fibrin(ogen) chains (cluster I: 24; cluster II: 7 and 5 unclustered cross-links) (SI Appendix, Fig. S4). With the set of cross-links from cluster I, protein docking with HADDOCK on the two copies of the fully assembled fibrin(ogen) trimer was performed to create the structure of two distinct laterally aggregating protofibrils that acts as the basis for the next steps (Fig. 4A). After docking, 18 of 24 cross-links were within the defined distance constraint (Fig. 4 B–D and SI Appendix, Table S2, Dimer). For all of those restraints, at least one involved residue is located prior to or within the flexible region. Interestingly, five out of the six overlength cross-links and all cross-links from cluster II together with five unclustered restraints involve residues βLys’265 (UniProt: 295) and βLys’344 (UniProt: 374) on the β-nodule, which, together with the predicted interaction interfaces from DisVis, suggests they arise from the antiparallel alignment of the β-nodules.
Fig. 4.
Assembly of the full fibrin clot model. (A) The docked structures of two distinct protofibrils; for clarity, we show only part of the first laterally aggregating protofibril in green and part of the second in orange. All of the restraints used for docking are shown in red. (B–D) Zoom-in to cross-link-rich regions on the assembled structural model with the zoom on Top (B), Middle (C), and Bottom (D) parts of the docked scaffold. (E) Placement of the structural models of previously not-resolved domains that are generated in this work. (F–H) Zoom-in to cross-link-rich regions on the assembled model with the scaffold (gray): (F) βN-termini (yellow), (G) α-chain residues 201–230 (UniProt: 220–249) in folded (light blue) and elongated (blue) conformations, (H) α-Interactive domain cluster I (purple, top) and RGD-binding domain cluster II (beige, top), and (I) α-Interactive domain cluster II (purple, bottom) and RGD-binding domain cluster I (beige, bottom). (J) The final model of fibrin with the highlighted interaction interfaces with the color scheme described above. Each copy of α-interactive and -RGD domains is grouped as they would belong to the same fibrinogen molecule.
To place the modeled domains onto the final structure, we performed HADDOCK docking with the interlinks from each of the four domains as the final filtering step (Fig. 4E). 1) From 29 detected interlinks for the β-chain N terminus, eight cross-links are used later for placement of the αC terminus domains, and the remaining 21 are used for placement of the β-chain N terminus onto the dimer. All 21 cross-links are identified as true positives by the DisVis analysis (SI Appendix, Fig. S5). After docking, 16 of 21 cross-links were within the set distance constraint (Fig. 4F and SI Appendix, Table S2, Beta_withDimer); the overlength cross-links are, however, located on a flexible region in the β-chain and can be explained from this perspective. 2) For the α-chain domain at residue 201–230 (UniProt: 220–249), 23 interlinks were detected to the other chains. Both the elongated as well as the folded conformation of this domain were fused to αHis’219 (UniProt: 219), with additional refinement to optimize placement accuracy and side-chain placement (Fig. 4G). After docking, 19 of 23 interlinks for the elongated conformation and 4 of 23 for the folded conformation were within the set distance constraint (SI Appendix, Table S2, Alpha1a_Elongated and Alpha1a_Folded). As the folded conformation hides accessible lysines, the imbalance of explained interlinks was anticipated. 3) The C terminus of the α-chain was previously suggested to be split into an N- and C-terminal subdomain (54); this is in line with our domain predictions (Table 1). For the α-chain domain at residue 413–442 (UniProt: 432–461) (N-terminal subdomain), 69 interlinks were detected to the α-chain; the large number of interlinks was expected due to its highly interactive nature (54). From our DisVis analysis, we detected three clusters of interlinks (cluster I: 46, cluster II: 19, and cluster III: 4) (SI Appendix, Fig. S6), indicating this domain is localized in three distinct positions in our sample. Cluster I is situated in front of the β nodule (Fig. 4H, purple residues). After docking 44 of 46 interlinks are within the set distance constraint (SI Appendix, Table S2, Alpha3a_01). Cluster II is placed under the β- and γ-chain nodules (Fig. 4I, purple residues). After docking, 19 of 19 interlinks are within the set distance constraint (SI Appendix, Table S2, Alpha3a_02). Cluster III is situated in the middle of the coiled-coil region of the aggregated protofibrils (SI Appendix, Table S2, Alpha3a_Central), which we hypothesize arises from a transitional state toward the active form (9). We note, however, that, with four cross-links, this is an observation of relatively low confidence and we do not pursue this further. 4) For the α-chain domain at residue 539–601 (UniProt: 558–620) (C-terminal subdomain), 50 interlinks were detected, of which 17 are to the indistinguishable N-terminal subdomains and therefore not included in the docking procedure. The remaining cross-links are connected to the other chains and therefore provide localization information for the C-terminal subdomain. Fitting with point 3 above, from our DisVis analysis, we also detect two clusters of interlinks (cluster I: 21, cluster II: 12) (SI Appendix, Fig. S7), indicating that both this domain as well as the N-terminal subdomain are localized in two distinct positions in our samples. After docking, 18 of 21 interlinks for cluster I (Fig. 4I, beige; SI Appendix, Table S2, Alpha3b_01) and 7 of 12 interlinks for cluster II were within the set distance constraint (Fig. 4H, beige; SI Appendix, Table S2, Alpha3b_02). Supporting this placement, the 17 discarded cross-links are within the set distance constraint when mapped between the N- and C-terminal subdomains after docking (SI Appendix, Table S2, Alpha3a_3b). Based on this observation, we suggest that each C-terminal subdomain can be matched to its respective N-terminal subdomain, the combinations of which we term αC1 and αC2 (Fig. 4J). Outliers from each docking run were mapped on the alternative subunit (where applicable), resulting in two additional valid restraints. As most of the docked regions are part of the regions containing the nodules that are not flexible, per se, we postulate that, even with the alternative conformations within the clot, the placement of the docked domains will be preserved.
Interaction Interface of Albumin and Fibrin Clots.
As the fibrin trimer is present in two copies, a total of 31 cross-linked pairs (of 16 detected) can be mapped, for which we used a structure of HSA without a bound ligand (PDB: 1uor). DisVis analysis uncovered three distinct clusters of 17, 9, and 4 cross-links, respectively (SI Appendix, Fig. S8). One nonclustered restraint, as well as restraints from cluster III, represent the mirrored location of clusters I and II, respectively, and were not used in docking. The selected docked model, obtained with restraints from cluster I, places albumin inside the cavity on the structural model of the fibrin clot (Fig. 5 A, left HSA molecule). It is possible to map 11 out of 17 restraints within the defined distance cutoff (Fig. 5B). The selected model, obtained with restraints from cluster II, places albumin on the exposed part of the fibrin clot (Fig. 5 A, right HSA molecule) with six valid restraints out of nine (Fig. 5C). Located relatively close to both the distinct protofibrils, this placement, however, interferes with certain conformations of the fibrin coiled-coil regions, which is in line with fewer detected restraints in comparison to cluster I. In conclusion, from 16 detected cross-links, it was possible to validate 13 (SI Appendix, Table S3), and 3 are outliers which can be placed on another model docked from cluster I. While this model possesses higher balanced scores, in total, it satisfies only nine restraints and therefore was not selected for further modeling steps (SI Appendix, Note S3).
Fig. 5.
Interaction Interfaces between the fibrin clot and HSA. (A) Overview of the detected cross-links between albumin and the fibrin clot. (B and C) Placement of albumin according to cluster I and cluster II restraints on the fibrin clot. Only validated restraints are shown. (D) Binding delays fibrin degradation by hindering access to sites of ternary complex formation and plasmin cleavage with a zoom-in of (E) cluster I and (F) cluster II.
Discussion
Fibrin clot formation has received a large amount of interest over the years, and, at a molecular level, many details have been resolved through various structural biology techniques. For example, from the generated partial protein structures, details on how fibrin oligomers are formed were readily uncovered (4–6, 58). These oligomers elongate and form double-stranded protofibrils, which also laterally aggregate to form thick fibers and intricate lattices. The molecular details of this process are less understood as some of the protein regions involved in lateral aggregation have so far been difficult to investigate. We applied a combination of XL-MS and structural modeling to uncover these dark regions and shed light on the lateral aggregation mechanism. This approach is not hampered by protein complexity and/or size and flexibility although it provides structural information of low resolution. From the presented dataset of 284 distance constraints (where a distance constraint is a unique combination of two protein positions connected by the cross-linker, regardless of missed cleavages or PTMs), it was not only possible to validate structural predictions, validating 92% of the detected distance constraints, but also unambiguously place them on the full structure. Because XL-MS is not limited to the proteins of interest, we also uncover interactors (like HSA and IgG). Based on sufficient amounts of detected interprotein cross-links, we were able to dock HSA on our final fibrin model and get insights into the role of HSA during fibrin clot degradation. As the used docking software uses the distance constraints only as a final filtering step and we ensured that the structural prediction software did not include these data where possible, the distance constraints provided by XL-MS constitute an independent biochemical validation for this structure.
Mapping Known Mutations.
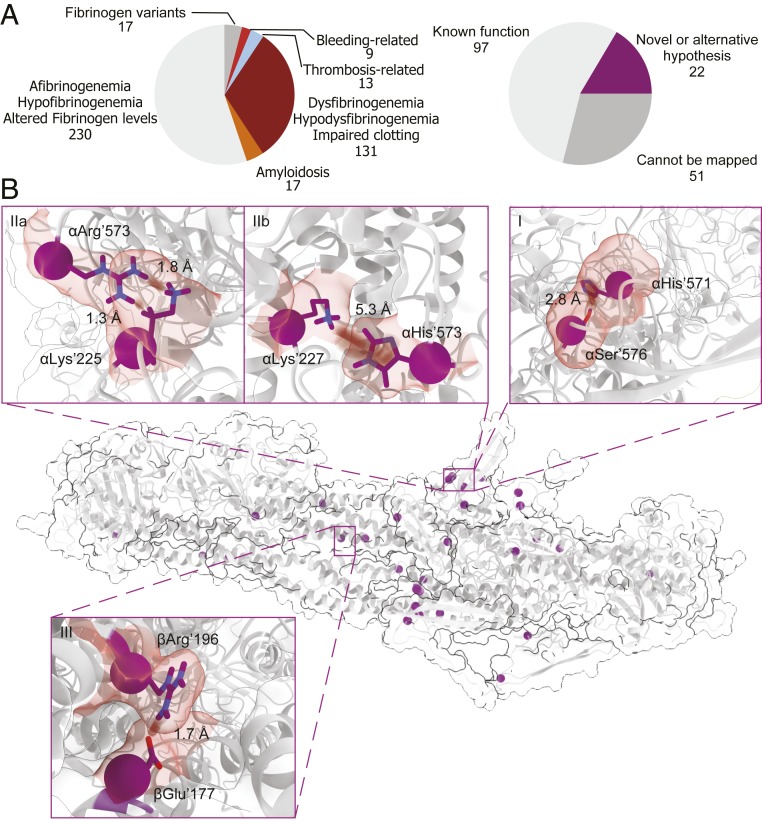
The assembled structural model is only useful if it provides insights into the molecular details explaining how mutations result in impaired clot formation (Fig. 6). From a list of 417 known mutations, we focused on 174 mutations potentially related to altered clot structure, including 22 mutations for which our structural model provides insights (Fig. 6A). From the final list of 170 mutations, 57% could be explained with existing models, and 30% were not possible to map on our structure [four had no reported sites; 24 are located on the α-chain, before residue 27 (UniProt: 46), and two on the β-chain, before residue 21 (UniProt: 51), both of which are part of signal peptides and fibrinopeptides; 16 are located on flexible domains, and, finally, five arise from frameshift mutations on the β- and γ-chain C-terminal domains]. Additionally, we examined an overlapping prediction list of 304 mutations with calculated CADD scores (44), for which we include a conservation score (42) (SI Appendix, Fig. S11 and Datasets S1–S7). For all of the 18 selected mutation sites, it was possible to uncover molecular details explaining impaired clot formation (SI Appendix, Table S4). Here, we discuss in more detail a subset of three mutations (Fig. 6B and Table 2). 1) For αPro’552 (UniProt: 571) mutated to histidine (CADD score: 17.0), we find the potential for a hydrogen-π interaction with the highly conserved αSer’557 (UniProt: 576) (Fig. 6 B, I). Previously, this mutation was linked to amyloidosis (59), hypothesized to occur through beta-sheet extension in the region α561–584 (UniProt: 580–603) (60, 61). Interaction to αSer’557 observed in our structure confirms that the histidine introduces additional stiffness in the domain, causing extensive beta-sheet formation in the region. The high conservation score of αSer’557 additionally indicates an important role for this residue in normal clots, which, with the hydrogen-π interaction, will be reduced or even abolished. 2) For αArg’554 (UniProt: 573), three debilitating mutations were so far reported (62–64). Even though this residue is only moderately conserved (score 6 out of 9), from our structure, we find that αArg’554 from the RGD-containing domain in αC1 is involved in an H–H bond to the highly conserved residue αLys’206 (UniProt: 225) in the folded conformation of the α201–230 (UniProt: 220–249) domain. The moderate conservation score likely arises from the replacement of arginine with serine or threonine in other organisms, which are also capable of forming an H–H bond to αLys’206. This bond makes it structurally important for the correct folding of α539–601 (UniProt: 558–620), and its loss alone potentially disturbs correct clot formation (Fig. 6 B, IIa). Mutation to cysteine (CADD score: 18.2; fibrinogen Dusart) is associated with impaired clot formation by the involvement of the cysteine with albumin complexes through S–S bonds (62). Cysteine is a polar residue also capable of forming H–H bonds, and this substitution does not necessarily affect the structure directly. Mutation to histidine (CADD score: 13.4; fibrinogen San Diego V), also associated with impaired clot formation, has so far not satisfactorily been explained. From our model, we find the potential of αHis’554 to form a cation-π interaction with αLys’208 (UniProt: 227) (Fig. 6 B, IIb). This lysine is involved in the correct folding of the α201–230 domain, and it might disturb the dynamic equilibrium between the elongated and folded forms. Loss of this equilibrium affects the correct placement of these subunits that are directly involved in lateral aggregation to αC1 and αC2. This fits well with previous reports that clots with decreased turbidity values and relatively fewer laterally aggregated protofibrils are found for fibrinogen San-Diego V(61). The role of the mutation to leucine (CADD score: 13.7) remains elusive, but we hypothesize that this affects the flexibility of the region in α539–601, resulting in beta-sheet extension. 3) For βArg’166 (UniProt: 196) mutated to cysteine (CADD score: 28.9; fibrinogen Longmont), we find, in our model, that the arginine is crucial for lateral aggregation of protofibrils through the coiled-coil region by the formation of a salt bridge to βGlu’147 (UniProt: 177) on the parallel coil (Fig. 6 B, III). Its loss disrupts this process, supported by the finding that polymerization is not normalized after removal of albumin–fibrinogen complexes (65).
Fig. 6.
Mapping known mutations on the final structural model of fibrin clots. (A) Selection of previously reported mutations. (B) Locations of selected mutation sites (purple). (I–III) Several highlighted mutations. (I) Showcases the αHis’552 (UniProt: 571) in interaction with αSer’557 (UniProt: 576). (IIa) Shows the role of αArg’554 (UniProt: 573). (IIb) Shows an additional interaction with αLys’208 (UniProt: 227) created by its substitution to αHis’554 (UniProt: 573). (III) Depicts salt bridge formation between βArg’166 (UniProt: 196) and βGlu’147 (UniProt: 177) from two laterally aggregated protofibrils, which is disrupted when βArg’166 (UniProt: 196) is mutated to Cys.
Table 2.
Analyzed fibrinogen mutation sites
| Gene | Amino acid change (UniProt) | Associated to/source | CADD score | Conservation score | Potential effect | Conservation scores of affected residues |
| FGA | P552H (571) | Amyloidosis (59) | 17.0 | 4 | Change of hydrophobic Pro’552 (UniProt: 571) to polar histidine—affects flexibility through, e.g., π–π stacking with Phe'551 (UniProt: 570) or cation-π interaction with Arg'554 (UniProt: 573). | αPhe’551–1 (UniProt: 570) |
| The histidine gives rise to a H-π interaction with Ser'557 (UniProt: 576) and interferes with an important structural function implied by its high conservation score. | αSer’557–9 (UniProt: 576) | |||||
| Alternatively, H-π interaction with Glu’221 (UniProt: 240) is also possible. Another rotamer provides the potential for π–π stacking with Tyr'582 (UniProt: 601), fixing an otherwise flexible loop into beta-sheet extension. | αTyr’582–1 (UniProt: 601) | |||||
| R554C, R554H, R554L (573) | Dysfibrinogenemia (62, 63), amyloidosis (64) | 18.2, 13.4, 13.7 | 6 | In αC1, αArg’554 (UniProt: 573) is involved in a H–H bond with the side chain of αLys'206 (UniProt: 225) in the folded conformation. In other organisms, the Arg often changed to Ser or Thr, also capable of maintaining this interaction. Within αC2, this mutation has the potential to form a salt bridge with αGlu’428 (UniProt: 447). | αLys’206–9 (UniProt: 225) | |
| Mutation to His provides the basis for π–π stacking to the neighboring αPhe’551 (UniProt: 570) and potentially a cation-π interaction to αLys'227. | αGlu’428–6 (UniProt: 447) | |||||
| Amyloidosis-causing substitution to Leu might cause the extension of α561–584 (UniProt: 580–603) beta-sheet. | αPhe’551–1 (UniProt: 570) | |||||
| αLys’208–1 (UniProt: 227) | ||||||
| αArg’554–6 (UniProt: 573) | ||||||
| FGB | R166C (196) | Dysfibrinogenemia (65) | 28.9 | 6 | In addition to the formation of albumin complexes, βArg’166 (UniProt: 196) is crucial for lateral aggregation through a salt bridge with βGlu’147 (UniProt: 177). In some organisms, this residue is changed to lysine, which allows salt bridge formation as well. | βGlu’158–8 (UniProt: 177) |
Interaction of Fibrin Clots with Serum Albumin.
There are several proteins which were previously reported as noncovalently bound to fibrin clots (21, 66, 67), and almost all of them are presented in our shotgun proteomics data (Dataset S9). After filtering our cross-linking dataset, we only retain restraints for albumin, apolipoproteins, and immunoglobulins (Dataset S8). Although we detected apolipoproteins in our dataset, there is no intermolecular connection to fibrin. In the case of immunoglobulins, the amount of detected restraints is relatively low, and all are focused on a single Ig residue, making prediction of the interaction interface extremely difficult. In the case of albumin, 16 intermolecular restraints were detected, allowing placement of albumin on fibrin clots with high confidence (Fig. 5 A–C). Before proceeding, we evaluated the specificity of this binding by estimating albumin content in both human serum as well as in our clot samples (SI Appendix, Fig. S9 A and B). Generally, albumin is several folds more abundant than fibrinogen in plasma (68). This situation alters drastically in our clot samples where only a 1.75-fold excess of albumin is observed (SI Appendix, Fig. S2B). We also examined the presence of albumin in human serum, and it is still the most abundant protein, with several folds excess over the remaining proteins. Further confirmation of the specificity of the interaction was derived from the detected abundances of albumin intra- and intermolecular cross-links to the fibrin clot (SI Appendix, Fig. S9C). As it is generally harder to form an interlink, it is expected that interlinks have lower detected abundances. In our data, intra- and interlinks, however, have the same abundance distributions, further confirming the specificity of the interaction at the indicated stoichiometry.
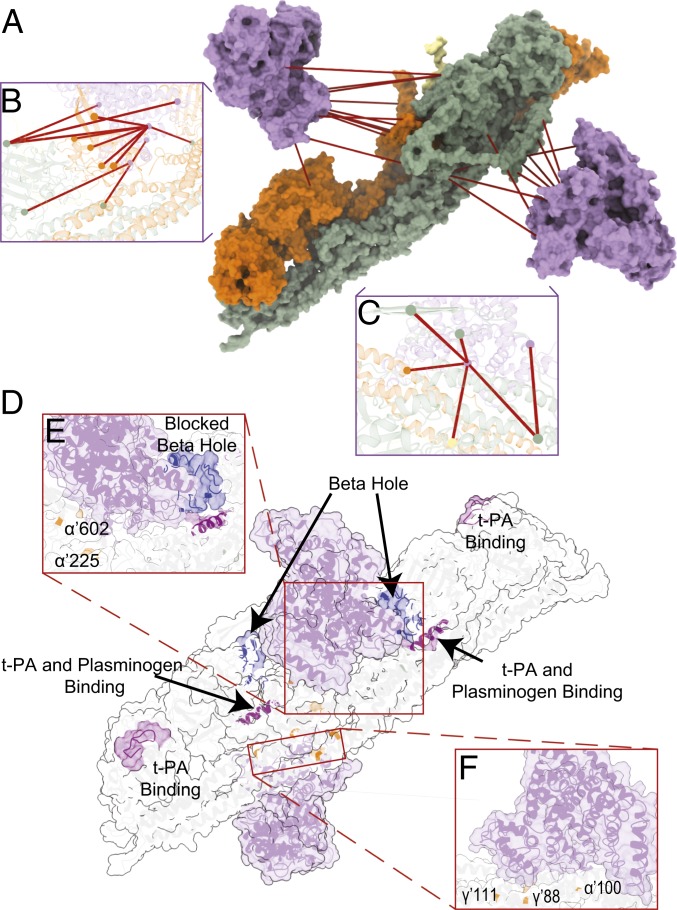
Fibrin clots are cleaved by plasmin, which is formed upon activation of plasminogen. Activation takes place after formation of a ternary complex comprising of t-PA, plasminogen, and fibrin. The first cleavage points of plasmin are located at three sites on the αC-terminal domain (69). Then so-called “D-dimers” are formed, which consists of two connected D-domains and 1 E-domain (16, 70). Products of fibrin degradation can be further cleaved into smaller pieces, with the cleavage on multiple sites (69, 71). Two sites on fibrinogen are known to bind either t-PA or both t-PA and plasminogen. Previously, it was suggested that, upon clot formation, a structural rearrangement takes place that makes both sites active (72). More specifically, site α’148–160 (UniProt: 167–179) is normally buried inside the structure and exposed after insertion of knob B into hole b (73). In our model, the placement of albumin suggested by cluster I blocks hole B (Fig. 5 D and E). This placement interferes with knob B insertion and therefore blocks the site for ternary complex formation. It was also reported that αC-termini contain cryptic t-PA and plasminogen high-affinity binding sites that are active only in fibrin clots. However, while the exact location of these sites remains elusive, we predict these sites to be present between α’425 (UniProt: 444) and α’600 (UniProt: 619). In our structural model, access to a large part of αC1 is hindered by albumin. In our model, albumin both interferes with ternary complex formation and limits access of biomolecules like plasminogen to a large part of αC1 and the plasmin cleavage sites it harbors (Fig. 5D and SI Appendix, Table S5). Combined, this interferes with cleavage and clearance of the clot, which fits well with a previously reported decrease of fibrinolysis rates at elevated albumin concentrations (21). In addition, apart from interfering with ternary complex formation, albumin hinders access to a number of plasmin cleavage sites (Fig. 5D and SI Appendix, Table S5). The location suggested by cluster I ensures that albumin covers site α’583 (UniProt: 602) from αC1 (Fig. 5E), which has been reported to be cleaved by plasmin (69). The location suggested by cluster II ensures that albumin binding interferes with cleavage by blocking plasmin cleavage sites αArg’104 (UniProt: 123) and γLys’62 (UniProt: 88) completely (71) (Fig. 5F). Another cleavage site is located on γLys’85 (UniProt: 111); however, cleavage here is known to be less efficient, which is in line with our structural model where access to that site is hindered by the laterally aggregating regions within the fibrin clot. In total, we docked two albumin molecules to the structural model of a fibrin clot. If we take into account all 31 possible cross-linked peptide pairs between the fibrin clot and HSA, we could also mirror the model from cluster II. Such a placement results in binding of three albumin molecules to six copies of each fibrin(ogen) protein (SI Appendix, Fig. S10). This is indeed very close to our copy-number estimation (SI Appendix, Fig. S2B), which suggests a ratio of 1.76 to 1 of albumin to fibrin(ogen)s.
Binding of Fibrin Clots to Endothelial Cells and Thrombocytes.
Excitingly, our structural model also provides a detailed molecular basis for the binding sites for platelets and endothelial cells. The α-chain domain at residues 539–601 (UniProt: 558–620) contains a functional RGD sequence fixed in place by beta-sheets. The folding of this domain and its placement within the final structural model are biochemically supported by seven intra- and 50 interlinks. This provides an excellent starting point for further structural investigations. Several important binding events are associated with precisely this spot due to the affinity to integrin receptors exposed on the cellular membrane (74–76). In the case of platelet binding, this RGD motif competes with the γ-chain, and the interaction between fibrin and integrin αIIbβ3 was previously proposed as a therapeutic target to destabilize thrombi (77, 78). We envision that XL-MS can be used to study the interactions made by both platelets as well as endothelial cells to provide a molecular picture for these interactions. Potentially, from such an unbiased screen, more information in terms of other receptors and/or other locations on the full fibrin clot structure can be obtained. Importantly, though, from the detailed structural information, this will open up avenues for the development of approaches for the treatment of thrombosis and/or hemophilia.
Supplementary Material
Acknowledgments
We thank all members of the A.J.R.H. group for their helpful contributions, especially Kelly Dingess for support with the video. We acknowledge financial support by the large-scale proteomics facility Proteins@Work (Project 184.032.201) embedded in the Netherlands Proteomics Centre and supported by the Netherlands Organization for Scientific Research (NWO). This work is part of the research program NWO TA with Project 741.018.201, which is partly financed by the NWO; additional support came through the European Union Horizon 2020 program INFRAIA project Epic-XS (Project 823839).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.R.E. is a guest editor invited by the Editorial Board.
Data deposition: The RAW mass spectrometry data and all associated output files used in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (identifier PXD011680). The final structural model has been deposited into the PDB-Dev database (identifier PDBDEV_00000030).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911785117/-/DCSupplemental.
References
- 1.Weisel J. W., Nagaswami C., Computer modeling of fibrin polymerization kinetics correlated with electron microscope and turbidity observations: Clot structure and assembly are kinetically controlled. Biophys. J. 63, 111–128 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feghhi S., Sniadecki N. J., Mechanobiology of platelets: Techniques to study the role of fluid flow and platelet retraction forces at the micro- and nano-scale. Int. J. Mol. Sci. 12, 9009–9030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neeves K. B., Illing D. A. R., Diamond S. L., Thrombin flux and wall shear rate regulate fibrin fiber deposition state during polymerization under flow. Biophys. J. 98, 1344–1352 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everse S. J., Spraggon G., Veerapandian L., Riley M., Doolittle R. F., Crystal structure of fragment double-D from human fibrin with two different bound ligands. Biochemistry 37, 8637–8642 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Kostelansky M. S., Betts L., Gorkun O. V., Lord S. T., 2.8 A crystal structures of recombinant fibrinogen fragment D with and without two peptide ligands: GHRP binding to the “b” site disrupts its nearby calcium-binding site. Biochemistry 41, 12124–12132 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Litvinov R. I., et al. , Polymerization of fibrin: Direct observation and quantification of individual B:b knob-hole interactions. Blood 109, 130–138 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chernysh I. N., Nagaswami C., Weisel J. W., Visualization and identification of the structures formed during early stages of fibrin polymerization. Blood 117, 4609–4614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allan P., Uitte de Willige S., Abou-Saleh R. H., Connell S. D., Ariëns R. A. S., Evidence that fibrinogen γ′ directly interferes with protofibril growth: Implications for fibrin structure and clot stiffness. J. Thromb. Haemost. 10, 1072–1080 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Weisel J. W., Medved L., The structure and function of the alpha C domains of fibrinogen. Ann. N. Y. Acad. Sci. 936, 312–327 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Yermolenko I. S., Lishko V. K., Ugarova T. P., Magonov S. N., High-resolution visualization of fibrinogen molecules and fibrin fibers with atomic force microscopy. Biomacromolecules 12, 370–379 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Mosesson M. W., DiOrio J. P., Siebenlist K. R., Wall J. S., Hainfeld J. F., Evidence for a second type of fibril branch point in fibrin polymer networks, the trimolecular junction. Blood 82, 1517–1521 (1993). [PubMed] [Google Scholar]
- 12.Fogelson A. L., Keener J. P., Toward an understanding of fibrin branching structure. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 81, 051922 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Standeven K. F., et al. , Functional analysis of fibrin gamma-chain cross-linking by activated factor XIII: Determination of a cross-linking pattern that maximizes clot stiffness. Blood 110, 902–907 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Mosesson M. W., Siebenlist K. R., Meh D. A., The structure and biological features of fibrinogen and fibrin. Ann. N. Y. Acad. Sci. 936, 11–30 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Weisel J. W., Litvinov R. I., Fibrin formation, structure and properties. Subcell. Biochem. 82, 405–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosesson M. W., Fibrinogen and fibrin structure and functions. J. Thromb. Haemost. 3, 1894–1904 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Kollman J. M., Pandi L., Sawaya M. R., Riley M., Doolittle R. F., Crystal structure of human fibrinogen. Biochemistry 48, 3877–3886 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Klykov O., et al. , Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc. 13, 2964–2990 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Armony G., et al. , Cross-linking reveals laminin coiled-coil architecture. Proc. Natl. Acad. Sci. U.S.A. 113, 13384–13389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavez J. D., et al. , Chemical crosslinking mass spectrometry analysis of protein conformations and supercomplexes in heart tissue. Cell Syst. 6, 136–141.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Sain-van Der Velden M. G. M., Smolders H. C., Van Rijn H. J. M., Voorbij H. A. M., Does albumin play a role in fibrinolysis by its inhibition of plasminogen activation? Fibrinolysis Proteolysis 14, 242–246 (2000). [Google Scholar]
- 22.Klykov O., et al. , Missing regions within the molecular architecture of human fibrin clots structurally resolved by XL-MS and integrative structural modeling. PRIDE Repository. https://www.ebi.ac.uk/pride/archive/projects/PXD011680. Deposited 4 May 2019. [DOI] [PMC free article] [PubMed]
- 23.Klykov O., et al. , Missing regions within the molecular architecture of human fibrin clots structurally resolved by XL-MS and integrative structural modeling. PRIDE Repository. https://pdb-dev.wwpdb.org/static/cif/PDBDEV_00000030.cif. Deposited 20 March 2019. [DOI] [PMC free article] [PubMed]
- 24.Schwanhäusser B., et al. , Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Kastritis P. L., et al. , Capturing protein communities by structural proteomics in a thermophilic eukaryote. Mol. Syst. Biol. 13, 936 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., et al. , ThreaDomEx: A unified platform for predicting continuous and discontinuous protein domains by multiple-threading and segment assembly. Nucleic Acids Res. 45, W400–W407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D. E., Chivian D., Baker D., Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J., Picord G., Tuffery P., Guerois R., HHalign-Kbest: Exploring sub-optimal alignments for remote homology comparative modeling. Bioinformatics 31, 3850–3852 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Yang J., et al. , The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Källberg M., et al. , Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen M.-Y., Sali A., Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colovos C., Yeates T. O., Verification of protein structures: Patterns of nonbonded atomic interactions. Prorein Sci. 2, 1511–1519 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M., AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Benkert P., Tosatto S. C. E., Schomburg D., QMEAN: A comprehensive scoring function for model quality assessment. Proteins 71, 261–277 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Ray A., Lindahl E., Wallner B., Improved model quality assessment using ProQ2. BMC Bioinformatics 13, 224–236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Zundert G. C. P., Bonvin A. M. J. J., DisVis: Quantifying and visualizing accessible interaction space of distance-restrained biomolecular complexes. Bioinformatics 31, 3222–3224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Vries S. J., Bonvin A. M. J. J., CPORT: A consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One 6, e17695 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Zundert G. C. P., et al. , The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Xia B., Vajda S., Kozakov D., Accounting for pairwise distance restraints in FFT-based protein-protein docking. Bioinformatics 32, 3342–3344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrodinger L., The PyMOL Molecular Graphics System (Version 1.8). https://pymol.org/2/support.html. Accessed 23 December 2019.
- 41.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashkenazy H., et al. , ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzek B. E., Wang Y., Huang H., McGarvey P. B., Wu C. H.; UniProt Consortium , UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rentzsch P., Witten D., Cooper G. M., Shendure J., Kircher M., CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhmurov A., et al. , Structural basis of interfacial flexibility in fibrin oligomers. Structure 24, 1907–1917 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhmurov A., et al. , Atomic structural models of fibrin oligomers. Structure 26, 857–868.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt L. R., et al. , Mass spectrometry-based molecular mapping of native FXIIIa cross-links in insoluble fibrin clots. J. Biol. Chem. 294, 8773–8778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Graaf S. C., Klykov O., van den Toorn H., Scheltema R. A., Cross-ID: Analysis and visualization of complex XL-MS-driven protein interaction networks. J. Proteome Res. 18, 642–651 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Odrljin T. M., Shainoff J. R., Lawrence S. O., Simpson-Haidaris P. J., Thrombin cleavage enhances exposure of a heparin binding domain in the N-terminus of the fibrin beta chain. Blood 88, 2050–2061 (1996). [PubMed] [Google Scholar]
- 51.Mulloy B., Linhardt R. J., Order out of complexity–protein structures that interact with heparin. Curr. Opin. Struct. Biol. 11, 623–628 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Muñoz E. M., Linhardt R. J., Heparin-binding domains in vascular biology. Arterioscler. Thromb. Vasc. Biol. 24, 1549–1557 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burton R. A., Tsurupa G., Hantgan R. R., Tjandra N., Medved L., NMR solution structure, stability, and interaction of the recombinant bovine fibrinogen alphaC-domain fragment. Biochemistry 46, 8550–8560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsurupa G., et al. , On the mechanism of αC polymer formation in fibrin. Biochemistry 51, 2526–2538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorkun O. V., Henschen-Edman A. H., Ping L. F., Lord S. T., Analysis of A α 251 fibrinogen: The α C domain has a role in polymerization, albeit more subtle than anticipated from the analogous proteolytic fragment X. Biochemistry 37, 15434–15441 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Weisel J. W., Fibrinogen and fibrin. Adv. Protein Chem. 70, 247–299 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Köhler S., Schmid F., Settanni G., The internal dynamics of fibrinogen and its implications for coagulation and adsorption. PLoS Comput. Biol. 11, e1004346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Litvinov R. I., Gorkun O. V., Owen S. F., Shuman H., Weisel J. W., Polymerization of fibrin: Specificity, strength, and stability of knob-hole interactions studied at the single-molecule level. Blood 106, 2944–2951 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gillmore J. D., et al. , Diagnosis, pathogenesis, treatment, and prognosis of hereditary fibrinogen A alpha-chain amyloidosis. J. Am. Soc. Nephrol. 20, 444–451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yazaki M., Yoshinaga T., Sekijima Y., Kametani F., Okumura N., Hereditary fibrinogen Aα-chain amyloidosis in Asia: Clinical and molecular characteristics. Int. J. Mol. Sci. 19, E320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garnier C., et al. , VLITL is a major cross-β-sheet signal for fibrinogen Aα-chain frameshift variants. Blood 130, 2799–2807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koopman J., et al. , Molecular basis for fibrinogen Dusart (A α 554 Arg–>Cys) and its association with abnormal fibrin polymerization and thrombophilia. J. Clin. Invest. 91, 1637–1643 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris T. A., et al. , High prevalence of dysfibrinogenemia among patients with chronic thromboembolic pulmonary hypertension. Blood 114, 1929–1936 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benson M. D., Liepnieks J., Uemichi T., Wheeler G., Correa R., Hereditary renal amyloidosis associated with a mutant fibrinogen α-chain. Nat. Genet. 3, 252–255 (1993). [DOI] [PubMed] [Google Scholar]
- 65.Lounes K. C., et al. , The impaired polymerization of fibrinogen Longmont (Bbeta166Arg–>Cys) is not improved by removal of disulfide-linked dimers from a mixture of dimers and cysteine-linked monomers. Blood 98, 661–666 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Talens S., Leebeek F. W. G., Demmers J. A. A., Rijken D. C., Identification of fibrin clot-bound plasma proteins. PLoS One 7, e41966 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galanakis D. K., Anticoagulant albumin fragments that bind to fibrinogen/fibrin: Possible implications. Semin. Thromb. Hemost. 18, 44–52 (1992). [DOI] [PubMed] [Google Scholar]
- 68.Geyer P. E., et al. , Plasma proteome profiling to assess human health and disease. Cell Syst. 2, 185–195 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Henschen A., McDonagh J., “Blood coagulation” in Blood Coagulation, Zwaal R. F. A., Hemker H. C., Eds. (Elsevier Science Publishers, Amsterdam, ed. 1, 1986), pp. 171–241. [Google Scholar]
- 70.Cesarman-Maus G., Hajjar K. A., Molecular mechanisms of fibrinolysis. Br. J. Haematol. 129, 307–321 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Walker J. B., Nesheim M. E., The molecular weights, mass distribution, chain composition, and structure of soluble fibrin degradation products released from a fibrin clot perfused with plasmin. J. Biol. Chem. 274, 5201–5212 (1999). [DOI] [PubMed] [Google Scholar]
- 72.Yakovlev S., et al. , Conversion of fibrinogen to fibrin: Mechanism of exposure of tPA- and plasminogen-binding sites. Biochemistry 39, 15730–15741 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Doolittle R. F., Pandi L., Binding of synthetic B knobs to fibrinogen changes the character of fibrin and inhibits its ability to activate tissue plasminogen activator and its destruction by plasmin. Biochemistry 45, 2657–2667 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Cheresh D. A., Human endothelial cells synthesize and express an Arg-Gly-Asp-directed adhesion receptor involved in attachment to fibrinogen and von Willebrand factor. Proc. Natl. Acad. Sci. U.S.A. 84, 6471–6475 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asakura S., et al. , Fibroblasts spread on immobilized fibrin monomer by mobilizing a beta1-class integrin, together with a vitronectin receptor alphavbeta3 on their surface. J. Biol. Chem. 272, 8824–8829 (1997). [DOI] [PubMed] [Google Scholar]
- 76.Suehiro K., Gailit J., Plow E. F., Fibrinogen is a ligand for integrin alpha5beta1 on endothelial cells. J. Biol. Chem. 272, 5360–5366 (1997). [DOI] [PubMed] [Google Scholar]
- 77.Adelman B., Gennings C., Strony J., Hanners E., Synergistic inhibition of platelet aggregation by fibrinogen-related peptides. Circ. Res. 67, 941–947 (1990). [DOI] [PubMed] [Google Scholar]
- 78.Höök P., et al. , Strong binding of platelet integrin αIIbβ3 to fibrin clots: Potential target to destabilize thrombi. Sci. Rep. 7, 13001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.