Significance
Carbon-13 MRI was used to assess exchange of hyperpolarized 13C label between injected [1-13C]pyruvate and the endogenous tumor lactate pool in breast cancer patients. Higher levels of 13C label exchange were observed in more-aggressive tumors, including all triple-negative cancers. The 13C label exchange correlated significantly with the expression of the transmembrane transporter mediating uptake of pyruvate into tumor cells and hypoxia inducible factor 1 (HIF1α), but no significant correlation with the expression of lactate dehydrogenase, the enzyme that catalyzes the exchange. The study has shown that 13C MRI can be used for metabolic imaging of breast cancer patients in the clinic, creating possibilities for noninvasive cancer monitoring in this patient group.
Keywords: metabolic imaging, magnetic resonance imaging, cancer metabolism, breast cancer
Abstract
Our purpose is to investigate the feasibility of imaging tumor metabolism in breast cancer patients using 13C magnetic resonance spectroscopic imaging (MRSI) of hyperpolarized 13C label exchange between injected [1-13C]pyruvate and the endogenous tumor lactate pool. Treatment-naïve breast cancer patients were recruited: four triple-negative grade 3 cancers; two invasive ductal carcinomas that were estrogen and progesterone receptor-positive (ER/PR+) and HER2/neu-negative (HER2−), one grade 2 and one grade 3; and one grade 2 ER/PR+ HER2− invasive lobular carcinoma (ILC). Dynamic 13C MRSI was performed following injection of hyperpolarized [1-13C]pyruvate. Expression of lactate dehydrogenase A (LDHA), which catalyzes 13C label exchange between pyruvate and lactate, hypoxia-inducible factor-1 (HIF1α), and the monocarboxylate transporters MCT1 and MCT4 were quantified using immunohistochemistry and RNA sequencing. We have demonstrated the feasibility and safety of hyperpolarized 13C MRI in early breast cancer. Both intertumoral and intratumoral heterogeneity of the hyperpolarized pyruvate and lactate signals were observed. The lactate-to-pyruvate signal ratio (LAC/PYR) ranged from 0.021 to 0.473 across the tumor subtypes (mean ± SD: 0.145 ± 0.164), and a lactate signal was observed in all of the grade 3 tumors. The LAC/PYR was significantly correlated with tumor volume (R = 0.903, P = 0.005) and MCT 1 (R = 0.85, P = 0.032) and HIF1α expression (R = 0.83, P = 0.043). Imaging of hyperpolarized [1-13C]pyruvate metabolism in breast cancer is feasible and demonstrated significant intertumoral and intratumoral metabolic heterogeneity, where lactate labeling correlated with MCT1 expression and hypoxia.
Breast cancer accounts for ∼25% of all cancer cases and is the leading cause of cancer death among women worldwide (1). Breast tumors show considerable heterogeneity, both within and between tumors, which partly accounts for the variable clinical course of the disease and response to treatment. Some of this heterogeneity is captured by hormone receptor expression and HER2 amplification/overexpression, which can be used to guide targeted treatment options. Genomic and transcriptional information can also indicate prognosis and may be used to select therapy pathways (2). Alterations in tumor metabolic pathways, which drive tumor growth, can influence treatment response but are not easily assessed in a routine clinical setting (3, 4). A clinical tool that measures spatial and temporal variations in tumor metabolism may further stratify patients in ways that are complementary to histological and molecular profiling.
A major metabolic change in most cancer types is a switch to aerobic glycolysis, known as the Warburg effect, which results in increased lactate formation (5, 6). Hyperpolarized 13C MRI (HP 13C MRI), which increases the MRI signal acquired from 13C-labeled substrates by more than 10,000-fold (7), is an emerging clinical tool that can be used to probe this altered metabolism. The most widely used hyperpolarized 13C-labeled substrate is [1-13C]pyruvate, which is the product of glycolysis, and lies at the intersection of several key metabolic pathways (8). Pyruvate is reduced to lactate in the reaction catalyzed by lactate dehydrogenase (LDH), which, in tumors, is predominantly the A isoform (LDHA) (9). The massive gain in sensitivity afforded by hyperpolarization means that the spatial distribution of intravenous (i.v.) injected hyperpolarized 13C-pyruvate and the hyperpolarized 13C-lactate formed from it, can be imaged in real time (10).
Preclinical studies have shown that the tumor metabolic phenotype revealed by hyperpolarized 13C-lactate labeling can reflect disease aggressiveness (11) and provide rapid assessment of treatment response, with multiple studies demonstrating an early reduction in 13C-lactate labeling following therapy (12, 13). MRI of hyperpolarized 13C-pyruvate metabolism has translated into the clinic with lactate labeling demonstrated in prostate cancer and brain tumors and preliminary evidence showing that it can be used for treatment response assessment in prostate cancer (7, 14, 15). The latter studies suggest that the technique could also be used to monitor the response of breast cancer patients to neoadjuvant treatment (NAT), which is used increasingly in routine patient management. NAT can be used to downstage the tumor, permitting breast-conserving surgery, improving operability, and allowing direct response assessment in situ. A complete pathological response (pCR) at surgery indicates a better prognosis, especially in estrogen receptor-negative breast cancer (16–19). Identifying effective treatments on an individual patient basis is essential to reduce both morbidity and cost, as ineffective NAT increases the time during which patients harbor chemoresistant tumor cells, thus increasing the risk of metastatic spread (20). Despite the increasing use of advanced multiparametric MRI in breast cancer patients undergoing NAT, there is an unmet need to develop more-specific and sensitive treatment response biomarkers to differentiate responders and nonresponders at an early time point. The aims of this study were, firstly, to demonstrate the feasibility of translating this imaging technique into the clinical breast cancer field; secondly, to metabolically phenotype breast cancers and evaluate which subtypes are most suitable for response assessment using HP 13C MRI; and. thirdly, to correlate lactate labeling with biological factors that may determine this labeling, such as expression of the monocarboxylate transporters, LDH, and markers of hypoxia and vascularity.
Methods
Patient Recruitment.
Local research ethics committee approval was obtained for this prospective study (National Research Ethics Service Committee East of England, Cambridge South, Research Ethics Committee number 15/EE/0378; National Institute for Health Research [NIHR] portfolio number 30388). Seven women diagnosed with invasive carcinoma of the breast measuring at least 1.5 cm in maximum diameter on ultrasound or mammogram were consented between November 2016 and June 2018.
Proton MRI.
Patients were imaged in a clinical 3T scanner (MR750; GE Healthcare). The MRI system inbuilt 1H body coil was used to acquire three-dimensional (3D) fast gradient echo scout images and, subsequently, T1-weighted axial and coronal fast spoiled gradient echo images, which were used to plan the 13C MRI. Following 13C MRI, diagnostic quality proton breast imaging was undertaken in the prone position in a dedicated eight-channel phased array receive-only breast coil (SI Appendix, Methods). For dynamic contrast-enhanced (DCE) MRI, a 3D fast spoiled gradient echo sequence with k-space data sharing was used (volume image breast assessment–time-resolved imaging of contrast kinetics [VIBRANT-TRICKS]) as described previously (21, 22) and reconstructed using an in-plane voxel-size of 0.68 × 0.68 mm (slice thickness = 1.4 mm). Gadobutrol (Gadovist; Bayer, Schering) was injected at 0.1 mmol/kg body weight and a flow rate of 3.0 mL/s followed by a 25-mL saline flush. In total, 48 VIBRANT-TRICKS volumes were acquired, over 8 min with a temporal resolution of 9.4 s. Contrast agent injection was started between phases 2 and 3.
Preparation and Injection of 13C-Pyruvate.
Hyperpolarization of samples containing 1.47 g of [1-13C]pyruvic acid (Sigma Aldrich) and 15 mM electron paramagnetic agent (EPA; Syncom) was performed in a clinical hyperpolarizer (SPINlab; 5T Research Circle Technology) by microwave irradiation at 139 GHz at ∼0.8 K for ∼3 h followed by rapid dissolution in 38 mL of superheated sterile water and filtration to remove EPA to a concentration below ≤3 μM. The filtered formulation was neutralized with a buffer solution (SI Appendix, Methods). Sample pH, temperature, pyruvate and EPA concentrations, polarization, and volume were measured by the SPINlab quality control (QC) module to ensure appropriate QC criteria were obtained. Then 0.4 mL/kg of ∼250 mM hyperpolarized pyruvate solution was injected at 5 mL/s using a power injector (Medrad) followed by a saline flush of 25 mL.
Carbon-13 MRI.
A dedicated eight-channel 13C breast coil (Rapid Biomedical) was used. A 13C-labeled 8 M urea sample (Sigma-Aldrich), positioned adjacent to the tumor-containing breast, was used to set the 13C transmit gain and center frequency. Images were acquired using a dynamic coronal Iterative Decomposition with Echo Asymmetry and Least squares estimation (IDEAL) spiral chemical shift imaging (CSI) sequence (SI Appendix, Methods and Table S1) (23). IDEAL spiral imaging data were reconstructed in MATLAB R2018b (The MathWorks, Inc.). Complex imaging data from the eight breast coil channels were combined as the square root of the sum of squares, with signal from each channel weighted by the maximal signal-to-noise ratio (SNR) of pyruvate. For analysis of metabolite ratios integrated over time, the complex data were summed over all time points prior to coil combination to minimize noise propagation. Images at the pyruvate, pyruvate hydrate, lactate, alanine, and bicarbonate resonance frequencies were reconstructed with an in-plane voxel size of 1.6 × 1.6 to 1.9 × 1.9 mm2 and a slice thickness of 30 mm.
Tumor regions of interest (ROIs) were generated by thresholding the sum of the summed lactate and pyruvate signals using a MATLAB program developed in-house. The threshold was chosen for each tumor individually so that, when comparing thresholded 13C images and the DCE images, the diameter of the ROI on the 13C images matched the maximum diameter of the tumor on the coronal DCE images at peak enhancement. This semiautomated approach was validated by comparing the results with those obtained using ROIs drawn manually. There was a good correlation between the lactate-to-pyruvate signal ratio (LAC/PYR) based on manual and thresholded ROIs (SI Appendix, Fig. S1). There were no significant differences between ROIs drawn in a semiautomated fashion and those drawn manually for LAC/PYR (P = 0.711), summed lactate SNR (SNRLAC) (P = 0.107), and summed pyruvate SNR (SNRPYR) (P = 0.097). To avoid potential bias introduced by drawing ROIs manually, the semiautomated thresholding approach was used for further analyses in this study. Estimation of noise (mean and SD) was performed using entire bicarbonate images, summed over the time course (or pyruvate hydrate, if there were visible artifacts in the bicarbonate image). The noise distribution in images of the different individual metabolites was expected to be the same, since all of the images were calculated from the same underlying set of multi-echo time data, which was confirmed empirically. Noise was best characterized from an entire image where spiral acquisition artifacts were absent. To generate the SNR for pyruvate and lactate (SNRmetabolite) within each tumor, the following formula was used:
Mean SIROI tumor is the mean signal intensity in the tumor ROI; mean and SD of SInoise were computed from the entire noise image series as described above. The factor of √2 accounts for the narrowed Rayleigh distribution of magnitude noise, with an approximate adjustment for the use of multiple receivers (24, 25). The summed metabolite SNR over the entire image time course is referred to as summed SNRPYR and summed SNRLAC. By dividing the latter by the former, the [1-13C]lactate/[1-13C]pyruvate SNR ratio (LAC/PYR) was calculated. As an alternative metric for assessing tumor metabolism of [1-13C]pyruvate, the forward exchange rate constant (kPL) from the most metabolically active voxel within each tumor was calculated, which was determined as the voxel with the highest time-averaged lactate signal. This approach was taken to maximize SNR, which is crucial for modeling of this rate constant. These results were compared with the calculated LAC/PYR (SI Appendix, Fig. S2). Based on the high variance of the fits of kPL in tumors that had low summed SNRLAC, the LAC/PYR was chosen as a more robust measure of pyruvate to lactate conversion.
DCE-MRI Analysis.
In-house−developed MATLAB software was used to generate B1 maps. MIStar (Apollo Medical Imaging) was used to generate B1-corrected T1 maps, to perform motion correction of the DCE-MRI data using a 3D affine model, and for pharmacokinetic modeling using the standard Tofts model (26). Tumor ROIs were drawn on the DCE-MRI data by a consultant radiologist specialized in breast imaging with 10 y of experience (V8.5.2 Pixmeo SARL; OsiriX). These ROIs were used to calculate tumor volumes and to extract voxel-wise pharmacokinetic parameters (Ktrans, kep, ve, and iAUC90) using in-house software written in MATLAB. Only voxels with an adequate goodness of fit were included in the analyses.
Immunohistochemistry and Quantification of Monocarboxylate Transporters 1 and 4.
For six patients, immunohistochemical (IHC) staining for the monocarboxylate transporters 1 and 4 (MCT1 and MCT4) was performed on formalin-fixed, paraffin-embedded tumor blocks using Leica’s Polymer Refine Detection System (DS9800) in combination with their Bond automated system (Leica Biosystems Newcastle Ltd) (SI Appendix, Methods). Insufficient tissue was available for analysis in one patient.
HALO v2.2.1870.15 (Indica Labs) and the area quantification v1 module were used for automated analysis of scanned sections (optical densities are provided in SI Appendix, Methods). Areas of weak, moderate, and strong staining were summed and divided by the total tissue area to obtain the percentage of positive tissue for MCT1 and MCT4 expression.
RNA Sequencing.
RNA from frozen tumor tissue sections from six patients was extracted using the QIAGEN miRNeasy Mini Kit (catalog no. 217004; QIAGEN; details in SI Appendix, Methods). RNA quantification was performed using Nanodrop technology (ThermoFisher Scientific). Assessment of the RNA integrity number was performed using a high-sensitivity RNA assay and a 2100 Bioanalyzer (Agilent Technologies).
RNA sequencing libraries were constructed using the TruSeq Stranded Total RNA Gold library preparation kit (Illumina). The libraries were sequenced as paired-end reads (2 × 75 cycles) on an HiSeq2500 platform to give a mean coverage of ×150. Postprocessing of the gene count data included normalization, scaling, and the correction of library preparation effects (details provided in SI Appendix, Methods).
Statistical Analysis.
The lactate signal is likely to be dominated by the intracellular compartment, particularly at early time points, whereas experiments in vitro and in vivo (27–29) have shown that, at later time points, there is substantial export of hyperpolarized 13C-labeled lactate into the extracellular space. The pyruvate signal is derived mainly from the extracellular pool, which includes intravascular pyruvate. Therefore, the summed SNRLAC lactate signal from the tumor and the LAC/PYR are likely to reflect vascular delivery of pyruvate, expression of the transporters mediating cellular pyruvate uptake (MCT1 and MCT4), and expression of LDHA, which catalyzes exchange of the hyperpolarized 13C label between pyruvate and lactate. The correlation of LAC/PYR, summed pyruvate signal (SNRPYR), and summed lactate signal (SNRLAC) with tumor volume and IHC markers of hypoxia (hypoxia-inducible factor 1-alpha [HIF1α]) were analyzed to assess the contributions of hypoxia, pyruvate uptake, and metabolism to the observed hyperpolarized 13C signals.
Statistical analyses were performed using R (version 3.4.0; R Foundation). Normal distribution of the data was assessed using the Shapiro−Wilk test for normality (threshold P ≥ 0.05). Correlations were assessed using Spearman’s rank correlation test including the rank correlation coefficient (rho) when data of at least one variable were nonnormal, and Pearson's correlation including the correlation coefficient R for data with normal distribution. Differences between measurements based on manually drawn versus thresholded ROIs were compared using Student’s t test for paired samples. All statistical tests were two-sided. P values below 0.05 were considered significant. No multiple testing correction was applied: Significant tests should be interpreted as exploratory rather than confirmatory.
Data Availability.
Transcriptomic data are deposited at the European Genome-phenome Archive (EGA ID EGAS00001004118; https://www.ebi.ac.uk/ega/studies/EGAS00001004118). Imaging raw data, and MATLAB scripts for data in this manuscript can be obtained from radiology-13c-mri-breast@lists.cam.ac.uk.
Results
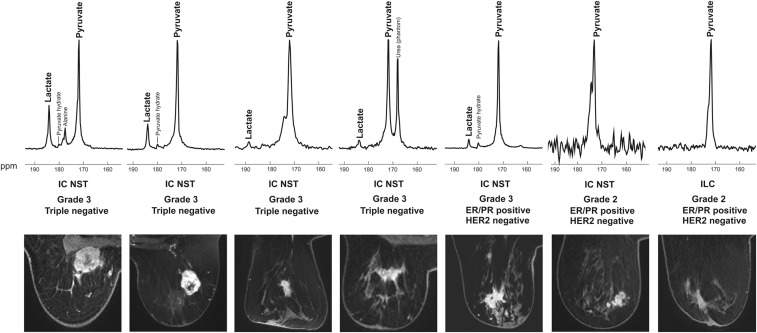
The feasibility of using HP 13C MRI in breast cancer was demonstrated in seven patients with a histopathological diagnosis of the disease. These included one grade 2 (G2) invasive lobular carcinoma (ILC), which was estrogen and progesterone receptorpositive (ER/PR+) and HER2/neu-negative (HER2−); two invasive carcinomas of no specific type (IC NST), which were ER/PR+ HER2− (one G2 and one G3); and four IC NST ER/PR− HER2− G3 (triple-negative breast cancer [TNBC]; ER and PR negativity defined as Allred score 0 to 3) (Fig. 1). Patient characteristics (age, body mass index, breast parenchymal density) are shown in SI Appendix, Table S2. No adverse effects were observed when the patients were monitored for 1.5 h after injection of the hyperpolarized agent. In all patients, HP 13C-lactate signal was observed exclusively in the tumors but not in other areas of the breast. Fibroglandular breast tissue demonstrated low HP 13C-pyruvate signal in some patients, whereas adipose breast tissue showed no signal. No other metabolite signals were observed in breast tissue.
Fig. 1.
Spectra and images from the patient cohort. (Top) Spectra from a coronal dynamic IDEAL spiral CSI slice covering each tumor; the spectra have been summed over all 15 time points. (Bottom) Axial slices from the equivalent DCE-MRI data at the time point of maximum tumor enhancement. Abbreviations: ppm, parts per million; ER, estrogen receptor; PR, progesterone receptor; HER2, HER2/neu.
Intratumoral and Intertumoral Metabolic Heterogeneity.
Intertumoral metabolic heterogeneity was observed with variation in the summed LAC/PYR, summed SNRPYR, and summed SNRLAC. The LAC/PYR ranged from 0.021 to 0.473 (mean ± SD, 0.145 ± 0.164), summed SNRPYR ranged from 6.2 to 74.3 (43.8 ± 25.8), and summed SNRLAC ranged from −0.1 to 22.3 (6.5 ± 7.8) (Fig. 2). Hyperpolarized lactate signal was observed in all of the G3 tumors (TNBC and IC NST). There was no discernable lactate signal in the two G2 tumors despite detectable pyruvate in all seven tumors (Fig. 1). In addition, there was significant variation in the LAC/PYR and summed SNRLAC within the TNBC subgroup (ranges 0.031 to 0.473 and 1.1 to 22.3, respectively).
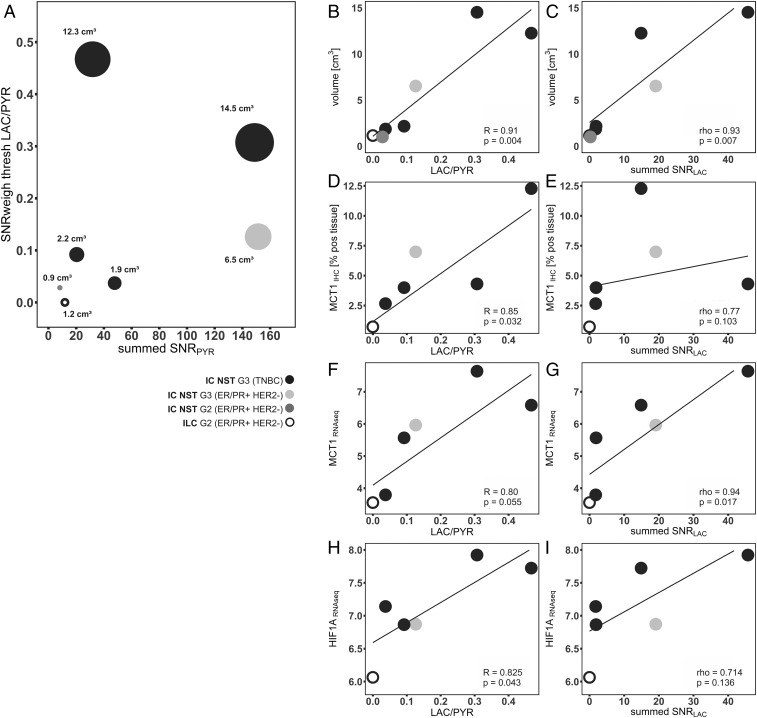
Fig. 2.
Correlation of hyperpolarized 13C MRI data with tumor volume and expression of the MCT1 and HIF1α. (A) Each patient is represented by an individual point, with the size of each circle proportional to the respective tumor size. (B and C) Correlation of LAC/PYR and summed SNRLAC with tumor volume. (D−G) Correlation of LAC/PYR and summed SNRLAC with expression of MCT1, determined by both (D and E) IHC and (F and G) RNA sequencing. (H and I) Correlation of LAC/PYR and summed SNRLAC with expression of HIF1α determined by RNA sequencing. Abbreviations: IHC [% pos tissue], percentage of formalin fixed paraffin embedded tissue positive for IHC staining; RNAseq, normalized expression based on RNA sequencing.
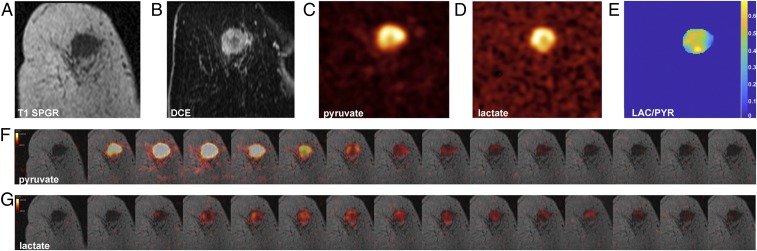
Intratumoral metabolic heterogeneity was observed in the largest TNBC. The summed SNRLAC and summed SNRPYR were higher in the tumor periphery than in the tumor core, which was similar to the pattern of rim-like contrast enhancement on DCE-MRI, suggesting that the rate of pyruvate delivery to the tumor has a significant influence on lactate labeling. (Fig. 3).
Fig. 3.
The 13C-pyruvate and 13C-lactate images acquired following i.v. injection of hyperpolarized [1-13C]pyruvate in a patient with TNBC. (A) Coronal T1 3D spoiled gradient echo (SPGR) image. (B) Coronally reformatted DCE image at peak enhancement after i.v. injection of a gadolinium-based contrast agent. (C) Summed hyperpolarized 13C-pyruvate and (D) summed hyperpolarized 13C-lactate images: area of low 13C-pyruvate and 13C-lactate signals in the center of the tumor, likely corresponding to an area with low enhancement on DCE. (E) LAC/PYR map showing intratumoral heterogeneity (background removed by thresholding). The dominant intratumoral heterogeneity was concordant between the DCE-MRI and HP 13C MRI images and represents decreased delivery of both the gadolinium-based contrast agent and 13C-pyruvate to the center of the tumor. (F and G) Dynamic hyperpolarized 13C-pyruvate and 13C-lactate images acquired over 15 time points after i.v. injection of hyperpolarized [1-13C]pyruvate (delay = 12 s; temporal resolution = 4 s).
Correlation of Lactate Labeling with Tumor Volume and MCT1 and HIF1α Expression.
The summed SNRLAC and LAC/PYR showed significant correlations with tumor volume (R = 0.974, P < 0.001 and R = 0.903, P = 0.005, respectively; Fig. 2 A–C). The LAC/PYR was also significantly correlated with the expression of MCT1 on IHC (R = 0.85, P = 0.032), and the summed SNRLAC was significantly correlated with MCT1 on RNA sequencing (R = 0.907, P = 0.013; Fig. 2 D–G). HIF1α expression determined by RNA sequencing was significantly correlated with the LAC/PYR (R = 0.83, P = 0.043; Fig. 2 H and I). However, a 42-gene RNA-based hypoxia signature that had been developed in breast cancer (30) did not correlate significantly with the LAC/PYR (R = 0.39, P = 0.442) or summed SNRLAC (rho = 0.23, P = 0.658; Fig. 2 F–I). There were no significant correlations between the LAC/PYR or summed SNRLAC and MCT4 expression, where this was determined by IHC (rho = 0.54, P = 0.297 and rho = 0.14, P = 0.803, respectively) or by RNA sequencing (R = 0.41, P = 0.420 and rho = −0.54, P = 0.297, respectively; SI Appendix, Fig. S4 A–D), nor with the expression of LDHA determined by RNA sequencing (R = 0.439, P = 0.383 and rho = 0.257, P = 0.658, respectively; SI Appendix, Fig. S4 E and F).
Correlation of 13C MRI with DCE-MRI.
A significant correlation was observed between ve and the LAC/PYR (R = 0.84, P = 0.035). However, this correlation was driven mainly by one tumor showing low ve and high LAC/PYR (SI Appendix, Fig. S3) and is thus unlikely to be representative of the entire cohort. No other significant correlations were observed between DCE parameters (Ktrans, kep, ve, or iAUC90) and the LAC/PYR, summed SNRPYR, and summed SNRLAC.
Discussion
Previous clinical studies have demonstrated 13C MRI with hyperpolarized [1-13C]pyruvate in human prostate cancer and in a range of brain tumors (7, 14, 15). Here we investigated the metabolism of hyperpolarized [1-13C]pyruvate in breast cancer patients and demonstrated the feasibility and safety of the technique, as well as significant intertumoral metabolic heterogeneity. Hyperpolarized 13C-lactate signal was observed in the summed spectra from all of the TNBCs and in all of the higher-grade (G3) tumors, whereas there was no discernable hyperpolarized 13C-lactate signal in the two lower-grade (G2) tumors. This is consistent with previous preclinical studies in prostate cancer, which have shown higher lactate signal in more-aggressive tumors, and suggests that increased lactate labeling can be used as a biomarker for aggressive disease (11). In addition, we observed intratumoral heterogeneity in lactate labeling in a single large triple-negative breast tumor, where the level of lactate labeling was correlated with delivery of a gadolinium-based contrast agent. Extensive preclinical studies have shown that hyperpolarized 13C-pyruvate metabolism is frequently modulated following treatment (31). The high levels of lactate labeling in TNBC, which commonly undergo NAT, would make them suitable for response assessment using this technique.
Previous studies showing that intertumoral metabolic heterogeneity is more pronounced than intratumoral metabolic heterogeneity (32) and that core biopsy samples can be used to reliably assess intertumoral differences (33–37) provided us with a rationale for comparing global imaging-based metrics (mean LAC/PYR and mean summed SNRLAC) with the results from IHC and RNA sequencing of single tumor biopsies. The strong correlation between the LAC/PYR with tumor volume, which is known to correlate with hypoxia (38), led us to investigate the contribution of hypoxia to the measured lactate signal. In tumors with a high LAC/PYR, there was a significant increase in HIF1α expression on RNA sequencing (R = 0.83, P = 0.043) (SI Appendix, Fig. S4), suggesting that hypoxia may account for the correlation between LAC/PYR and tumor volume.
Metabolic reprogramming of cancer cells is often the downstream effect of oncogene activation or deletion of tumor suppressor genes (39–41). MCT1, which imports pyruvate into cells, and LDHA, which catalyzes hyperpolarized 13C label exchange between the injected pyruvate and endogenous lactate pool, can be up-regulated by the transcription factors HIF-1α and c-Myc, either constitutively, such as following activation of the PI3K/Akt/mTOR pathway, or as a response to hypoxia (42). There was a significant correlation between MCT1 protein expression and HIF1α messenger RNA expression and tumors with high LAC/PYR and summed SNRLAC. The expression of MCT1 is elevated in glycolytic breast tumors, and high MCT1 expression predicts poor prognosis (43). The correlation between the LAC/PYR and the expression of MCT1, which, unlike MCT4, transports pyruvate in preference to lactate, can be explained by the influence of pyruvate transport on the rate of exchange. A number of studies have demonstrated that the tumor cell pyruvate transport can have a significant influence on 13C label exchange and the apparent exchange rate, including experiments on the effects of pyruvate concentration (44), cell lysis (45), modulations of LDH activity (45), and MCT1 inhibition (29), and a recent study in prostate cancer patients which analyzed MCT1 expression (46). Hyperpolarized 13C MRI was complementary to the DCE-MRI measurements, with a correlation between kep and the summed SNRPYR but no correlation between the other parameters derived from the DCE-MRI and the hyperpolarized 13C magnetic resonance spectroscopic imaging data. Since kep is related to tissue perfusion, the correlation with summed SNRPYR is to be expected. The lack of correlation between the other parameters suggests that the hyperpolarized [1-13C]pyruvate experiment reflects other aspects of tumor biology that are not captured by DCE-MRI. The combination of the two imaging strategies could be exploited for early response assessment, where quantitative DCE-MRI has demonstrated the potential to increase accuracy in the prediction of pCR compared to standard clinical MRI, but the results still depend on the molecular tumor subtype (47, 48). Response assessment using proton magnetic resonance spectroscopy (1H-MRS) has proven challenging in the breast, due to overlap of the lactate resonance with the intense lipid signals from adipose tissue (49). A recent multicenter study on the assessment of early treatment response in breast cancer using 1H-MRS showed very limited feasibility (50). Although uptake of the glucose analog 18F-FDG imaged with positron emission tomography is widely used in oncological imaging for cancer detection, staging, and response assessment, ionizing radiation should ideally be minimized in women of reproductive age, and it is not used routinely in the assessment of primary breast cancer. In addition, it reflects cellular uptake via the glucose transporters and phosphorylation by hexokinase, but it does not allow the assessment of the downstream metabolism that can be probed with hyperpolarized [1-13C]pyruvate.
This study, although in a relatively small cohort, demonstrates the feasibility and safety of hyperpolarized 13C MRI in patients with early breast cancer and that metrics obtained from 13C MRI measurements of hyperpolarized [1-13C]pyruvate metabolism are correlated with the molecular characteristics of the tumors. Increased hyperpolarized [1-13C]lactate signal in larger and more-aggressive tumors appears to be driven by hypoxia, through increased MCT1 expression.
Supplementary Material
Acknowledgments
This work was supported by a Wellcome Trust Strategic Award, Cancer Research UK (CRUK; Grants C8742/A18097, C19212/A16628, C19212/A911376, and C197/A16465), the Austrian Science Fund (Grant J4025-B26), the CRUK Cambridge Centre, the CRUK & Engineering and Physical Sciences Research Council Cancer Imaging Centre in Cambridge and Manchester, the Mark Foundation for Cancer Research and Cancer Research UK Cambridge Centre (Grant C9685/A25177), Addenbrooke’s Charitable Trust, the National Institute for Health Research Cambridge Biomedical Research Centre, Cambridge Experimental Cancer Medicine Centre, and Cambridge University Hospitals National Health Service Foundation Trust.
Footnotes
Competing interest statement: A research agreement is in place between GE Healthcare and K.M.B. and F.A.G.
This article is a PNAS Direct Submission. M.G.V.H. is a guest editor invited by the Editorial Board.
Data deposition: RNA sequencing data are deposited at the European Phenome-genome Database (EGA ID EGAS00001004118) under a controlled license policy. The Data Access Committee can be contacted via radiology-13c-mri-breast@lists.cam.ac.uk. Imaging raw data and MATLAB scripts described in this manuscript can be obtained from radiology-13c-mri-breast@lists.cam.ac.uk.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913841117/-/DCSupplemental.
References
- 1.Torre L. A., et al. , Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Paik S., et al. , A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R. A., The hallmarks of cancer. Cell 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Kim S., Kim D. H., Jung W.-H., Koo J. S., Metabolic phenotypes in triple-negative breast cancer. Tumour Biol. 34, 1699–1712 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Cappelletti V., et al. , Metabolic footprints and molecular subtypes in breast cancer. Dis. Markers 2017, 1–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson S. J., et al. , Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C]pyruvate. Sci. Transl. Med. 5, 198ra108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurhanewicz J., et al. , Hyperpolarized 13C MRI: Path to clinical translation in oncology. Neoplasia 21, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutta P., et al. , Evaluation of LDH-A and glutaminase inhibition in vivo by hyperpolarized 13C-pyruvate magnetic resonance spectroscopy of tumors. Cancer Res. 73, 4190–4195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golman K., in ’t Zandt R., Thaning M., Real-time metabolic imaging. Proc. Natl. Acad. Sci. U.S.A. 103, 11270–11275 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albers M. J., et al. , Hyperpolarized 13C lactate, pyruvate, and alanine: Noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 68, 8607–8615 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day S. E., et al. , Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat. Med. 13, 1382–1387 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Witney T. H., et al. , Detecting treatment response in a model of human breast adenocarcinoma using hyperpolarised [1-13C]pyruvate and [1,4-13C2]fumarate. Br. J. Cancer 103, 1400–1406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park I., et al. , Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies. Magn. Reson. Med. 80, 864–873 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aggarwal R., Vigneron D. B., Kurhanewicz J., Hyperpolarized 1-[13C]-pyruvate magnetic resonance imaging detects an early metabolic response to androgen ablation therapy in prostate cancer. Eur. Urol. 72, 1028–1029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennisi A., Kieber-Emmons T., Makhoul I., Hutchins L., Relevance of pathological complete response after neoadjuvant therapy for breast cancer. Breast Cancer Basic Clin. Res. 10, 103–106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rastogi P., et al. , Preoperative chemotherapy: Updates of national surgical adjuvant breast and bowel project protocols B-18 and B-27. J. Clin. Oncol. 26, 778–785 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Cortazar P., et al. , Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 384, 164–172 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) , Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: Meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 19, 27–39 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaidya J. S., et al. , Rethinking neoadjuvant chemotherapy for breast cancer. BMJ 360, j5913 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Kershaw L. E., Cheng H.-L. M., A general dual-bolus approach for quantitative DCE-MRI. Magn. Reson. Imaging 29, 160–166 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Bedair R., et al. , Effect of radiofrequency transmit field correction on quantitative dynamic contrast-enhanced MR imaging of the breast at 3.0 T. Radiology 279, 368–377 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Wiesinger F., et al. , IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1-13C]pyruvate. Magn. Reson. Med. 68, 8–16 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Gilbert G., Measurement of signal-to-noise ratios in sum-of-squares MR images. J. Magn. Reson. Imaging 26, 1678 (2007). Author reply in: J. Magn. Reson. Imaging26, 1679 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Constantinides C. D., Atalar E., McVeigh E. R., Signal-to-noise measurements in magnitude images from NMR phased arrays. Magn. Reson. Med. 38, 852–857 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tofts P. S., et al. , Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: Standardized quantities and symbols. J. Magn. Reson. Imaging 10, 223–232 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Keshari K. R., et al. , Hyperpolarized 13C-pyruvate magnetic resonance reveals rapid lactate export in metastatic renal cell carcinomas. Cancer Res. 73, 529–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koelsch B. L., et al. , Separation of extra- and intracellular metabolites using hyperpolarized 13C diffusion weighted MR. J. Magn. Reson. 270, 115–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrao E. M., et al. , Analysis of 13 C and 14 C labeling in pyruvate and lactate in tumor and blood of lymphoma-bearing mice injected with 13 C- and 14 C-labeled pyruvate. NMR Biomed. 31, e3901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye I. C., et al. , Molecular portrait of hypoxia in breast cancer: A prognostic signature and novel HIF-regulated genes. Mol. Cancer Res. 16, 1889–1901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comment A., Merritt M. E., Hyperpolarized magnetic resonance as a sensitive detector of metabolic function. Biochemistry 53, 7333–7357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gogiashvili M., et al. , Impact of intratumoral heterogeneity of breast cancer tissue on quantitative metabolomics using high-resolution magic angle spinning 1H NMR spectroscopy. NMR Biomed. 31, e3862 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Knuttel F. M., et al. , Meta-analysis of the concordance of histological grade of breast cancer between core needle biopsy and surgical excision specimen. Br. J. Surg. 103, 644–655 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Chen X., et al. , Preoperative core needle biopsy is accurate in determining molecular subtypes in invasive breast cancer. BMC Cancer 13, 390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Focke C. M., Decker T., van Diest P. J., Reliability of the Ki67-labelling index in core needle biopsies of luminal breast cancers is unaffected by biopsy volume. Ann. Surg. Oncol. 24, 1251–1257 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Lee J., et al. , Efficacy of an RNA-based multigene assay with core needle biopsy samples for risk evaluation in hormone-positive early breast cancer. BMC Cancer 19, 388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cha Y. J., et al. , Comparison of tumor-infiltrating lymphocytes of breast cancer in core needle biopsies and resected specimens: A retrospective analysis. Breast Cancer Res. Treat. 171, 295–302 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Dunst J., et al. , Tumor volume and tumor hypoxia in head and neck cancers. The amount of the hypoxic volume is important. Strahlenther. Onkol. 179, 521–526 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Ogrodzinski M. P., Bernard J. J., Lunt S. Y., Deciphering metabolic rewiring in breast cancer subtypes. Transl. Res. 189, 105–122 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Deberardinis R. J., Sayed N., Ditsworth D., Thompson C. B., Brick by brick: Metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18, 54–61 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elia I., Fendt S.-M., In vivo cancer metabolism is defined by the nutrient microenvironment. Transl. Cancer Res. 5, S1284–S1287 (2016). [Google Scholar]
- 42.Courtnay R., et al. , Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 42, 841–851 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Hong C. S., et al. , MCT1 modulates cancer cell pyruvate export and growth of tumors that co-express MCT1 and MCT4. Cell Rep. 14, 1590–1601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris T., Eliyahu G., Frydman L., Degani H., Kinetics of hyperpolarized 13C1-pyruvate transport and metabolism in living human breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 18131–18136 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witney T. H., Kettunen M. I., Brindle K. M., Kinetic modeling of hyperpolarized 13C label exchange between pyruvate and lactate in tumor cells. J. Biol. Chem. 286, 24572–24580 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granlund K. L., et al. , Hyperpolarized MRI of human prostate cancer reveals increased lactate with tumor grade driven by monocarboxylate transporter 1. Cell Metab., 10.1016/j.cmet.2019.08.024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fowler A. M., Mankoff D. A., Joe B. N., Imaging neoadjuvant therapy response in breast cancer. Radiology 285, 358–375 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Drisis S., et al. , Quantitative DCE-MRI for prediction of pathological complete response following neoadjuvant treatment for locally advanced breast cancer: The impact of breast cancer subtypes on the diagnostic accuracy. Eur. Radiol. 26, 1474–1484 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Bolan P. J., Magnetic resonance spectroscopy of the breast: Current status. Magn. Reson. Imaging Clin. N. Am. 21, 625–639 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Bolan P. J., et al. ; ACRIN Trial team ISPY-1 Investigators , MR spectroscopy of breast cancer for assessing early treatment response: Results from the ACRIN 6657 MRS trial. J. Magn. Reson. Imaging 46, 290–302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomic data are deposited at the European Genome-phenome Archive (EGA ID EGAS00001004118; https://www.ebi.ac.uk/ega/studies/EGAS00001004118). Imaging raw data, and MATLAB scripts for data in this manuscript can be obtained from radiology-13c-mri-breast@lists.cam.ac.uk.