We thank Drs. Hartl and Gao for pointing out the difference in read lengths between the cancer and normal samples (1). Indeed, Illumina discontinued their 50-base pair (bp) sequencing kits after we finished sequencing the cancer samples, forcing us to switch to 75-bp sequencing kits on the normal samples. The different sequencing kits did incur small changes of sequence alignment metrics. While acknowledging the possibility that this may bias the assessment of cancer vs. normal discrimination performance of our method, we believe such a possibility is substantially ameliorated by the following considerations.
First, a recent study reported that “with the exception of 25 bp reads, there is little difference for the detection of differential expression regardless of the read length” (2). In line with this report, the absolute difference of mismatch rates (0.5% vs. 0.8%) between the 50-bp and 75-bp reads is much smaller than the typically used threshold (4%) of the mapping software (STAR). Furthermore, among the sequence alignment metrics Hartl and Gao list (1), only the rates of uniquely mapped reads directly affect the quantification of extracellular RNA (exRNA) levels. However, the rates of uniquely mapped sequences contributed to sample-to-sample variation in PC1, which is independent of the observed cancer–normal difference (PC2) (second row in figure 1B of ref. 1).
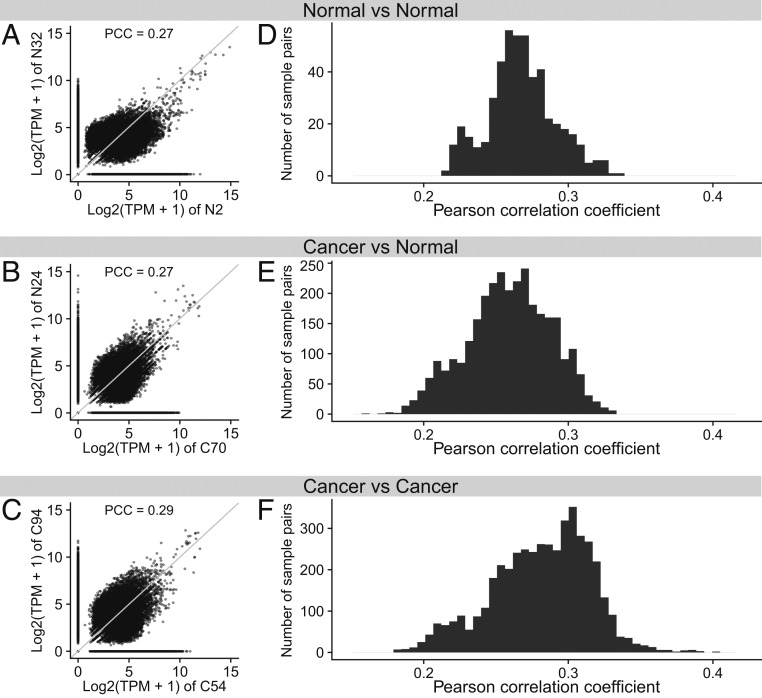
Second, we cannot identify a batch difference in the distributions of the measured exRNA levels: “The distributions of TPM exhibited little difference between any 2 cancer samples or between a cancer sample and a normal sample (Fig. 4A and SI Appendix, Fig. S9). Thus, every sample contains a similar proportion of highly expressed exRNAs, regardless of the threshold for calling highly expressed exRNAs” (3). Furthermore, the pairwise differences of the normal samples in exRNA levels nearly completely overlapped with the between-batch pairwise differences (Fig. 1). Taken together, read length is unlikely to affect the quantification of exRNAs or bias our cancer–normal comparison, which was completely based on exRNA quantification.
Fig. 1.
Genome-wide comparisons of exRNA levels within and between the 2 batches. Pairwise difference is measured by 1-correlation. (A–C) Scatter plots of log2-transformed transcripts per million (TPMs) of all genes between 2 normal samples (A), a cancer (x axis) and a normal sample (y axis) (B), and 2 cancer samples (C). (D–F) Histograms of pairwise Pearson correlation coefficients (PCCs) between every pair of normal samples (D), every cancer–normal pair (E), and every pair of cancer samples (F), based on the log2-transformed TPMs of all genes.
However, the aforementioned considerations cannot dwarf our caution that “this study did not rule out all possible confounding factors that may contribute [to] the separation of cancer and normal samples” and “future studies … are needed, preferably as double-blind prospective trials” (3).
Finally, the read-length difference does not affect other major findings in our paper. Specifically, Small Input Liquid Volume Extracellular RNA Sequencing (SILVER-seq) can reproducibly produce exRNA sequencing libraries from different aliquots of the same serum sample. exRNA levels measured by SILVER-seq correlated with those measured by standard exRNA sequencing methods. SILVER-seq detected fragments of tissue-specific RNAs in human sera. SILVER-seq–derived exRNA levels reflected chronological age. SILVER-seq–based classifiers exhibited good discrimination power between recurrent and nonrecurrent cancer samples.
Footnotes
The authors declare no competing interest.
References
- 1.Hartl C., Gao Y., Clarifying the effect of library batch on extracellular RNA sequencing. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.1916312117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chhangawala S., Rudy G., Mason C. E., Rosenfeld J. A., The impact of read length on quantification of differentially expressed genes and splice junction detection. Genome Biol. 16, 131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Z., et al. , Extracellular RNA in a single droplet of human serum reflects physiologic and disease states. Proc. Natl. Acad. Sci. U.S.A. 116, 19200–19208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]