Abstract
Background
Non-surgical cytological specimens are adequate not only for accurate histological subtyping but also for molecular profiling. A modified amplification refractory mutation system polymerase chain reaction (ARMS PCR), known as SuperARMS PCR, was improved by optimizing the primers designation, which provides a higher sensitivity and specificity approach for free plasma DNA detection. It is unclear whether SuperARMS PCR detects epidermal growth factor receptor (EGFR) mutations in cytology samples. The aim of this study was to compare the EGFR mutations detected by ARMS PCR and SuperARMS PCR in cytology samples derived from advanced non-small cell lung cancer (NSCLC) patients.
Methods
From March 2016 to March 2018, a total of 234 cytological samples were obtained from primary or metastatic lesions of NSCLC, including 144 fine-needle aspirations (FNAs), 36 endobroncheal ultrasonography (EBUS) FNAs, 36 transbronchial needle aspirations (TBNAs) and 18 pleural effusion (PLEs). EGFR mutations were simultaneously detected using an ADx-ARMS EGFR kit (Amoy Diagnostics CO., ltd., Xiamen, China) and an ADx-SuperARMS EGFR kit (Amoy Diagnostics CO., ltd., Xiamen, China). Digital droplet PCR (ddPCR) and next-generation sequencing (NGS) were further used to verify the EGFR mutant inconsistent samples.
Results
All of the 234 patients with advanced or recurrent NSCLC were diagnosed and assessed by two cytopathologists, and their EGFR mutation statuses were successfully detected by ARMS and SuperARMS. Importantly, the SuperARMS and ARMS methods showed a highly concordant result of 94.0% (220/234) (95%CI: 85.0, 95.0%). The positive rate of the SuperARMS was higher than the ARMS in the cytology samples for EGFR detection (46.2% vs. 40.2%). The specific EGFR mutation sites in 16 samples (6.8%) were not completely consistent between the SuperARMS and ARMS. A total of 14 patients showed EGFR mutations when detected by SuperARMS, but by ARMS there were EGFR wild-type. Two patients were detected as having one more EGFR mutation site by SuperARMS than by ARMS. ddPCR and NGS were used to further confirm the EGFR mutations in these inconsistent samples. Eight samples had the same mutation results as the SuperARMS, and 6 samples were not verified because the remaining DNA was insufficient. A total of 78 EGFR mutation patients received Tyrosine Kinase Inhibitor (TKI) treatment. The overall objective response rate (ORR) was 88.5% (69/78) for EGFR TKI treatment.
Conclusion
SuperARMS showed a high sensitivity and specificity for EGFR detection and thus, is expected to become a routine test in the clinic to be used as a widely available, easy-to-operate and sensitive method for EGFR mutation detection in liquid-based cytology samples.
Keywords: EGFR, SuperARMS, ARMS, NSCLC, Liquid-based cytology
Introduction
Lung cancer remains the most common cause of cancer death worldwide. Approximately 70% of patients with non-small cell lung cancer (NSCLC) come to clinical attention at an advanced stage [1]. Small biopsy or cytological specimens may be a good choice in these patients, while they have no surgical options. The types of non-surgical cytological samples include fine-needle aspiration (FNA), endobronchial ultrasound-guided (EBUS) biopsy, transbronchial needle aspiration (TBNA), bronchoscopic brush (BB) and pleural effusion (PLE), which are reliable for the diagnosis and staging of thoracic malignancy, especially of NSCLC [2–6].
Many investigators report that cytological material obtained with minimally invasive procedures is adequate not only for accurate histological subtyping but also for molecular profiling [7–9]. In fact, studies have demonstrated optimal results using a multitude of cytological samples for molecular tests [10–12]. In our previous study, epidermal growth factor receptor (EGFR) mutation detection was performed by ARMS in liquid-based cytology samples from patients with NSCLC and their paired tissue samples, and the results of the two groups were identical [13]. Cytology specimens provide high quality DNA for the evaluation of clinically relevant mutations. Research groups, such as Allegrini S and Malapelle U, have also directly used liquid-based cytology samples for EGFR and Kirsten rat sarcoma viral oncogene (KRAS) mutation detection with better results [14, 15]. Under an era of individualized targeted therapy for NSCLC, it is crucial, for guiding treatment decisions, to obtain informative cytological material for both diagnosis and molecular testing. In our lab, we have increasingly used cytology specimens for molecular testing when cytology material was the only specimen source available.
Amplification Refractory Mutation System (ARMS) is an improved polymerase chain reaction (PCR) system and is an important platform for the detection of genes that drive NSCLC. The AmoyDx EGFR 29 Mutations Detection Kit is approved by the China Food and Drug Administration (CFDA) for the clinical testing of EGFR mutations using tissue samples. Recently, a novel technique called SuperARMS, which is a modified version of ARMS that optimizes the primers designation, was shown to provide a high sensitivity and specificity approach for free plasma DNA detection. No studies have been performed to compare ARMS and SuperARMS for EGFR mutation using cytology specimens. Therefore, it is unclear whether SuperARMS can be used to detect EGFR mutations in cytology samples and if it improves the sensitivity compared with ARMS. Thus, we conducted the present study to compare the EGFR mutations detected by ARMS and SuperARMS PCR in cytology samples derived from advanced NSCLC patients.
Materials and methods
Cytology specimens and study design
From March 2016 to March 2018, a total of 234 patients with advanced NSCLC were retrospectively enrolled in this study at the Shanghai Pulmonary Hospital, Tongji University. All the cytological samples were obtained from primary or metastatic lesions of NSCLC and included 144 FNAs, 36 EBUS FNAs, 36 TBNAs and 18 PLEs. The imaging data were independently reviewed by the authors to evaluate their treatment responses according to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. Progression-free survival (PFS) was calculated from the date of the initiating tyrosine kinase inhibitors (TKI) treatment to a radiologic or clinical observation of the disease progression.
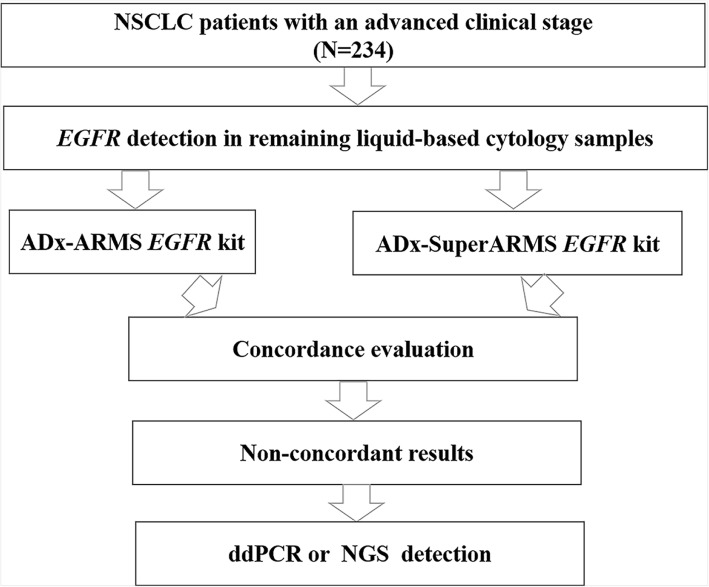
A flowchart describing the study design is presented in Fig. 1. The study protocol was approved by the Institute Review Board of the Shanghai Pulmonary Hospital.
Fig. 1.
A flowchart describing the study design
Abbreviations: NSCLC = non-small cell lung cancer; EGFR = epidermal growth factor receptor; NGS = next-generation sequencing.
Sample collection
The liquid-based cytology specimen preparations were handled according to the standard specimen processing protocols in our laboratory. Cytology samples from FNAs, EBUS FNAs, TBNAs and PLEs were taken by clinicians and sent to pathology department for Thinprep cytologic test (TCT) within 1 day. One ThinPrep slide was stained with hematoxylin and eosin (H&E) from the various cell types and was reviewed for morphologic evaluation by two cytopathologists. The remainder of the specimen liquid-based cytology specimens were rinsed in CytoLyt solution (Hologic) and were centrifuged to generate a cell pellet stored at 4 °C. If the H&E–stained smear was positive for NSCLC, we quantitated the number of tumor cells as more than 200 cells by the H&E–stained smear and observed the residual cell pellet, which was visible to the naked eye, to ensure that the specimen was adequate for EGFR molecular testing.
DNA extraction
The residual cell pellets were used for the DNA extraction, which was performed using a Tissue DNA Kit (Amoy Diagnostics Co., Xiamen, China), following the manufacturer’s instructions. The optical density of the extracted DNA samples was measured using a microplate spectrophotometer (Biotek). The A260/A280 value of all the samples was 1.8 to 2.1. The extracted DNA was used for EGFR molecular testing.
EGFR mutation detection
The EGFR mutations in the residual liquid-based cytology samples were simultaneously detected using an ADx-ARMS EGFR kitand an ADx-SuperARMS EGFR kit, according to the manufacturer’s instructions. Briefly, the DNA templates were added to the EGFR Reaction Mix, which included the primers, probes, dNTPs, buffer, Mg2+ and Taq DNA polymerases. The PCR reactions were performed on a Stratagene Mx3000P quantitative PCR (qPCR) system. After the PCR reaction was completed, the data interpretation was conducted according to results interpretation criteria of the EGFR mutation detection kit.
ddPCR detection
The ddPCR was performed according to the manufacturer’s instructions. Briefly, the mutant reaction solution was prepared, and then, the emulsified microdroplets were generated in the QX200TM Droplet Generator instrument and were put into a 96-well plate for amplification. The PCR reaction conditions were as follows: incubation at 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60°Cfor 60 s. After the PCR amplification, the 96-well plate was placed in the QX200 microdrop analyzer, and the data analyses were conducted using Quanta Soft analysis software.
NGS detection
DNA sequencing was carried out using a capture-based sequencing panel (Amoy Diagnostics CO., Ltd., Xiamen, China). The kit covered the targeted drug-related hot spot mutation regions of 10 genes, including EGFR et al. The liquid-based cytology DNA samples were sequenced with a NextSeq 500 (Illumina, Inc), with pair-end reads, with a reading length of PE150 and a sequencing depth of > 10,000x. The sequence data were analyzed using the AmoyDx NGS Data Analysis System ADXLC10 module (Amoy Diagnostics CO., Ltd., Xiamen, China).
Statistical analysis
The statistical analyses were performed with SPSS, version 17.0 (IBM, Armonk, NY). A two-sided tailed Fisher’s exact test was applied to the data for one subgroup with a count less than 5. p < 0.05 was considered significantly different.
Results
Patient characteristics
A total of 234 patients with advanced or recurrent NSCLC were retrospectively enrolled into the study. The patients’ clinical characteristics are listed in Table 1. The median age was 65 years (range, 31–85 years). The patients consisted of 79 women and 155 men. Eighty-four patients were smokers, 145 were never-smokers, and 5 were unclear smokers. Fifty-eight patients were classified as at stage III, and 176 were at stage IV. The liquid-based cytology sample types included: FNA from the lungs for 114 patients, FNA from the lymph nodes for 30 patients, EBUS for 36 patients, TBNA for 36 patients and PLE for 15 patients. All the specimens were confirmed by two cytopathologists, including 172 adenocarcinoma, 21 squamous and 41 NSCLC.
Table 1.
Clinical characteristics (n = 234)
| Factors | No.(%) |
|---|---|
| Sex | |
| Male | 155(48.7%) |
| Female | 79(48.7%) |
| Age, years | |
| Median | 65 |
| Range | 31–85 |
| Smoking history | |
| Never | 145(62.0%) |
| Former/current | 84(35.9%) |
| Unclear | 5(2.1%) |
| Pathology | |
| Adenocarcinoma | 172(73.5%) |
| Squamous | 21(9.0%) |
| NSCLC | 41(17.5%) |
| Tumor Stage | |
| III | 58(24.8%) |
| IV | 176(75.2%) |
| Cytological sample types | |
| FNA (lung) | 114(48.7%) |
| FNA (lymph node) | 30(12.8%) |
| EBUS FNA | 36(15.4%) |
| TBNA | 36(15.4%) |
| Pleural effusion | 18(7.7%) |
Comparisons of the SuperARMS and the ADx-ARMS for detecting EGFR in the cytological specimens
All 234 liquid-based cytology samples were successfully detected for their EGFR mutation status by ARMS and SuperARMS. The results are shown in Table 2. When tested by ARMS, 94 of 234 (40.2%) patients were identified to have an EGFR mutation. When using the SuperARMS, 108 of 234 (46.2%) patients were demonstrated to have an EGFR mutation. The positive rate of the SuperARMS was higher than the ARMS in the cytology samples for EGFR detection (46.2% vs. 40.2%). The two-sided tailed Fisher’s exact test showed that this was a significant difference (p < 0.05). The negative results and the positive results from the SuperARMS were in accordance with the ARMS results, at 90.0% (95% CI, 91.0,97.1%) and 100.0% (94/94), respectively. The concordance rate of the EGFR mutation status between SuperARMS and ARMS was 94.0% (220/234) (95%CI:85.0,95.0%). Fourteen patients showed an EGFR mutation by SuperARMS but were EGFR wild-type by ARMS. In addition, two patients were detected with one more EGFR mutation site by SuperARMS than by ARMS.
Table 2.
Comparison of EGFR mutation status using ARMS and SuperARMS (N = 234)
| ARMS | Total | |||
|---|---|---|---|---|
| Mt | Wt | |||
| SuperARMS | Mt | 94 | 14 | 108 |
| Wt | 0 | 126 | 126 | |
| Total | 94 | 140 | 234 | |
| PCR* | 100% (94/94) | |||
| NCR* | 90%(126/140)(95%CI:91.0,97.1%) | |||
| Overall agreement* | 94%(220/234)(95%CI:85.0,95.0%) | |||
*percentage [95% confidence interval (CI)];
Mt: mutation; Wt: wild type; PCR: positive coincident rate; NPV: negative coincident rate
SuperARMS for detecting specific EGFR mutation types
Out of total of 234 samples, 16 cases were discordant between SuperARMS and ARMS (Table 3). Among them, 14 (87.5%) cases were detected as positive for EGFR mutations only by SuperARMS, while the other two samples showed different mutations by these two methods. We performed ddPCR and NGS to further confirmed the inconsistent samples(n = 12/16, 4 samples were insufficient). 1 sample with EGFR 20-ins mutation (Sample ID:FK020) is beyond capability of ddPCR, so 11/12 samples were identified by ddPCR at first. Then we successful detected 5 sample using NGS (Table 3). 2 samples were negative for mutations。 2 samples had a special L858R mutation type (NM_005228.3: exon21:c.2573 T > G:p.L858R), which was only detected by NGS and SuperARMS. This observation indicates that SuperARMS and NGS methods can detect more sites of mutation than ARMS in the tested regions.
Table 3.
Sixteen Nonconcordant Cases of SuperARMS and ARMS for Cytology EGFR Detection
| Sample ID | ARMS | Super-ARMS | ddPCR | NGS | |
|---|---|---|---|---|---|
| FK008 | 19-DEL | 19-DEL/T790 M | 19-DEL/T790 M | NA | |
| FK020 | WT | 20-INS | NA | 20-INS | |
| FK055 | WT | L858R | WT | NE DNA | |
| FK062 | WT | L858R | WT | NM_005228.3:exon21:c.2573_2574delinsGT:p.L858R | |
| FK066 | WT | L858R | WT | NM_005228.3:exon21:c.2573 T > G:p.L858R | |
| FK080 | WT | 19-DEL | 19-DEL | NA | |
| FK108 | WT | L858R | L858R | NA | |
| FK129 | WT | L858R | NE DNA | NE DNA | |
| FK130 | WT | L858R | NE DNA | NE DNA | |
| FK135 | WT | 19-DEL | 19-DEL | NA | |
| FK147 | WT | 19-DEL | WT | WT | |
| FK166 | WT | L858R | WT | NE DNA | |
| FK190 | WT | 19-DEL | WT | WT | |
| FK206 | 19-DEL | 19-DEL/L858R | NE DNA | NE DNA | |
| FK220 | WT | 20-ins | NE DNA | NE DNA | |
| FK221 | WT | 19-DEL/L858R | 19-DEL/L858R | NA | |
*NE DNA: not enough DNA
EGFR mutation status and prediction of EGFR-TKI efficacy
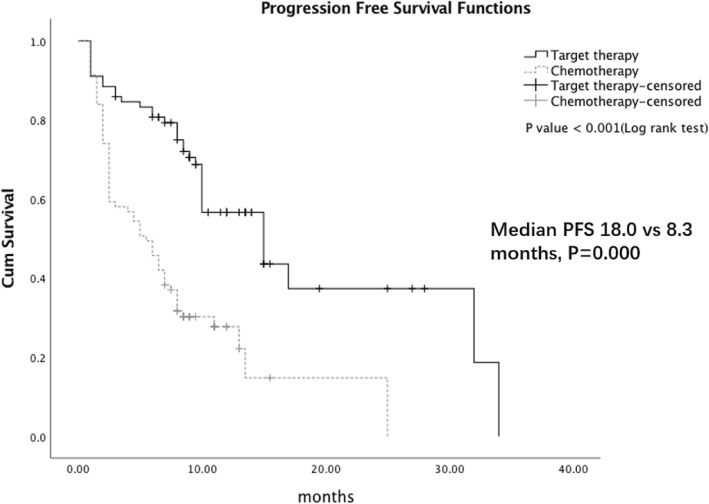
As shown in Table 4, out of the 94 positive EGFR mutation patients by ARMS, 77 patients received EGFR-TKI treatment. In another 14 patients with an EGFR mutation by SuperARMS, only 1 patient received EGFR-TKI treatment. The ORR was 88.5% (69/78) for EGFR TKI treatment. In patients with or without EGFR mutation, 81 received chemotherapy. The median PFS was significantly prolonged in the EGFR mutation patients compared with the EGFR wild-type patients (18.0 months vs 8.3 months, P < 0.01) (Fig. 2). These data suggested that EGFR mutation status in cytology samples detected by ARMS and SuperARMS is predictive of tumor response to EGFR-TKIs and survival results.
Table 4.
EGFR Mutation Status and TKIs treatment
| Sample No. | Positive No. | Negtive No. | Sensitive MT No. | targeted therapy No. | |
|---|---|---|---|---|---|
| ARMS | 234 | 94 | 140 | 92 | 77 |
| SuperARMS | 234 | 108 | 126 | 104 | 78 |
Fig. 2.
PFS in EGFR mutation patients compared with EGFR wild-type patients
Discussion
Currently, the molecular spectrum of tumor patients promotes the development of individualized treatment. Especially in patients with NSCLC, the mutation status of EGFR has great guiding significance for the use of TKIs [16]. Now more than ever, obtaining adequate material for the molecular testing of NSCLC is crucial. In the clinical setting, a cytological specimen may be the only material available for the determination of EGFR mutation status. Liquid-based cytology specimens have been used for molecular assays with optimal results, although rare large series have been published.
It has been reported that the number of tumor cells in the remaining liquid base cell sample varies significantly between the different samples, with limited sensitivity of the ARMS detection in low tumor cell content samples [17]. SuperARMS is a highly sensitive kit that detects 41 EGFR mutations in exons 18 to 21 in human plasma cfDNA samples. To the best of our knowledge, ours is the first study to evaluate the performance of SuperARMS in liquid-based cytology samples for EGFR detection.
The feasibility of conventional smears and the remaining liquid-based cytology samples for molecular detection has been evaluated in multiple laboratories [18–20]. Gilda da Cunha Santos’s team compared the nucleic acid quality of the remaining liquid-based cytology samples, smears and cell blocks for molecular detection, and found that well-preserved liquid-based cytology samples had a good nucleic acid yield and quality [21]. Maurizio Martini’s research team explained the role of cytological samples in tumor diagnosis, which is not only used in the diagnosis, but also in molecular testing, and a small amount of high-quality nucleic acid provides effective molecular genetic features [22]. Consistent with these reports in the literature, our liquid-based cytology samples had good DNA quality and or the EGFR gene status was successfully detected using SuperARMS and ARMS.
In this study, various cytological specimens, including FNA, EBUS FNA, TBNA and PLE, were all subjected to the EGFR mutational analysis using SuperARMS and ARMS, and 100% (234/234) of the samples were suitable for testing. Firstly, the SuperARMS and ARMS methods showed highly concordant results, at 94.0% (220/234) (95%CI: 85.0, 95.0%). Moreover, SuperARMS detected more mutations compared with ARMS. Our study showed that SuperARMS, as a highly sensitive molecular detection method, can be used for the detection of EGFR not only in plasma ctDNA, but also in cytological samples DNA.
The high sensitivity and specificity of SuperARMS has been demonstrated in a series of liquid biopsy studies [23, 24]. Our study confirmed that SuperARMS had high sensitivity and specificity in liquid-based cytology samples. The positive rate of SuperARMS was higher than ARMS in the cytology samples for EGFR detection (46.2% vs. 40.2%) (Table 2). Our study showed that SuperARMS was not only used in the blood, but also in liquid-based cytology samples. Is there a false positive result in the SuperARMS? We analyzed the reasons for the inconsistency of the results of the validation samples. The false positive results caused by the contamination of aerosol in the laboratory should be excluded. Notably that two samples had the special L858R mutation type (NM_005228.3: exon21:c.2573 T > G:p.L858R), which was detected by SuperARMS and NGS. The specific reason for this observation is that the SuperARMS and NGS methods detect more sites.
Many previous studies report that there is a significant correlation between EGFR mutation status and treatment for PFS [25, 26]. Consistent with these studies, our study revealed that patients with EGFR mutations, using targeted drugs as the first line of treatment, had a longer PFS compared to patients without EGFR mutations, using chemotherapy drugs as the first line of treatment, as detected by both ARMS and SuperARMS.
In conclusion, the SuperARMS had a high sensitivity and specificity for EGFR detection in liquid-based cytology samples, and these results were good predictors of the efficacy of EGFR-TKIs. Thus, the SuperARMS is expected to become a test that is used routinely in the clinic as a widely available, easy-to-operate and sensitive method for EGFR mutation detection in liquid-based cytology samples.
Acknowledgements
We thank all laboratory members in the Department of Pathology, Shanghai Pulmonary Hospital, for their kind help.
Ethical approval and consent to participate
The study was approved by the ethics committee of Shanghai Pulmonary Hospital (No. K17–040). Additional patient consent for this retrospective study was not required.
Abbreviations
- 20-ins
20 exon insertion mutation
- ARMS
amplification Refractory Mutation System
- BB
bronchoscopic brush
- EBUS
endobronchial ultrasound-guided
- EGFR
epidermal growth factor receptor
- FNA
fine-needle aspiration
- H&E
hematoxylin and eosin
- NSCLC
non-small cell lung cancer
- ORR
objective response rate
- PCR
polymerase chain reaction
- PFS
progression-free survival
- PLE
pleural effusion
- RECIST
response Evaluation Criteria in Solid Tumors
- TBNA
transbronchial needle aspiration
- TKI
tyrosine kinase inhibitor
Authors’ contributions
WW, ZY C, LK H and CY W designed the study. WW, ZY C, LP Z and WZ analyzed and interpreted the patient data. WW, ZY C and ZW performed the experiments. All authors read and approved the final manuscript.
Funding
This study was sponsored by “Shanghai Sailing Program” (17YF1415800).
Availability of data and materials
The raw data are available upon request.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wei Wu and Ziyang Cao contributed equally to this work.
References
- 1.Walker S. Updates in non-small cell lung cancer. Clin J Oncol Nurs. 2008;12(4):587–596. doi: 10.1188/08.CJON.587-596. [DOI] [PubMed] [Google Scholar]
- 2.Fielding D, Kurimoto N. Endobronchial ultrasound-guided transbronchial needle. Aspiration for diagnosis and staging of lung cancer. Clin Chest Med. 2018;39(1):111–123. doi: 10.1016/j.ccm.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Roh MH. The utilization of cytologic fine-needle aspirates of lung cancer for molecular diagnostic testing. J Pathol Transl Med. 2015;49(4):300–309. doi: 10.4132/jptm.2015.06.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias S, Liu QH, Frimpong B, et al. Role of the Endobronchial landmarks guiding TBNA and EBUS-TBNA in lung Cancer staging. Can Respir J. 2016;2016:1652178. doi: 10.1155/2016/1652178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinfort DP, Leong TL, Laska IF, et al. Diagnostic utility and accuracy of rapid on-site evaluation of bronchoscopic brushings. Eur Respir J. 2015;45(6):1653–1660. doi: 10.1183/09031936.00111314. [DOI] [PubMed] [Google Scholar]
- 6.Lococo F, Cesario A, Attili F, Chiappetta M, et al. Transoesophageal endoscopic ultrasound-guided fine-needle aspiration of pleural effusion for the staging of non-small cell lung cancer. Interact Cardiovasc Thorac Surg. 2013;17(2):237–241. doi: 10.1093/icvts/ivt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy-Chowdhuri S, Aisner DL, Allen TC, et al. Biomarker testing in lung carcinoma cytology specimens: a perspective from members of the pulmonary pathology society. Arch Pathol Lab Med. 2016;140(11):1267–1272. doi: 10.5858/arpa.2016-0091-SA. [DOI] [PubMed] [Google Scholar]
- 8.Ellison G, Zhu G, Moulis A, et al. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol. 2013;66(2):79–89. doi: 10.1136/jclinpath-2012-201194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei S, Lieberman D, Morrissette JJ, et al. Using “residual” FNA rinse and body fluid specimens for next-generation sequencing: an institutional experience. Cancer Cytopathol. 2016;124:324–329. doi: 10.1002/cncy.21666. [DOI] [PubMed] [Google Scholar]
- 10.Treece AL, Montgomery ND, Patel NM, et al. FNA smears as a potential source of DNA for targeted next-generation sequencing of lung adenocarcinomas. Cancer Cytopathol. 2016;124:406–414. doi: 10.1002/cncy.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Cunha Santos G, Saieg MA. Preanalytic specimen triage: smears, cell blocks, cytospin preparations, transport media, and cytobanking. Cancer Cytopathol. 2017;125(S6):455–464. doi: 10.1002/cncy.21850. [DOI] [PubMed] [Google Scholar]
- 12.Roy-Chowdhuri S, Chen H, Singh RR, et al. Concurrent fine needle aspirations and core needle biopsies: a comparative study of substrates for nextgeneration sequencing in solid organ malignancies. Mod Pathol. 2017;30(4):499–508. doi: 10.1038/modpathol.2016.228. [DOI] [PubMed] [Google Scholar]
- 13.Wu CY, Hou LK, Ren SX, et al. High feasibility of liquid-based cytological samples for detection of EGFR mutations in Chinese patients with NSCLC. Asian Pac J Cancer Prev. 2014;15:7885–7889. doi: 10.7314/APJCP.2014.15.18.7885. [DOI] [PubMed] [Google Scholar]
- 14.Allegrini S, Antona J, Mezzapelle R, et al. Epidermal growth factor receptor gene analysis with a highly sensitive molecular assay in routine cytologic specimens of lung adenocarcinoma. Am J Clin Pathol. 2012;138(3):377–381. doi: 10.1309/AJCPVAGIUC1AHC3Y. [DOI] [PubMed] [Google Scholar]
- 15.Malapelle U, de Rosa N, Rocco D, et al. EGFR and KRAS mutations detection on lung cancer liquid-based cytology: a pilot study. J Clin Pathol. 2012;65(1):87–91.12. doi: 10.1136/jclinpath-2011-200296. [DOI] [PubMed] [Google Scholar]
- 16.Juan O, Popat S. Treatment choice in epidermal growth factor receptor mutation-positive non-small cell lung carcinoma: latest evidence and clinical implications. Ther Adv Med Oncol. 2017;9(3):201–216. doi: 10.1177/1758834016687262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy-Chowdhuri S, Aisner DL, Allen TC, et al. Biomarker testing in lung carcinoma cytology specimens. Arch Pathol Lab Med. 2016;140:1267–1272. doi: 10.5858/arpa.2016-0091-SA. [DOI] [PubMed] [Google Scholar]
- 18.Betz BL, Roh MH, Weigelin HC, et al. The application of molecular diagnostic studies interrogating EGFR and KRAS mutations to stained cytologic smears of lung carcinoma. Am J Clin Pathol. 2011;136(4):564–571. doi: 10.1309/AJCP84TUTQOSUONG. [DOI] [PubMed] [Google Scholar]
- 19.Reynolds JP, Zhou Y, Jakubowski MA, et al. Next-generation sequencing of liquid-based cytology non-small cell lung cancer samples. Cancer Cytopathol. 2017;125(3):178–187. doi: 10.1002/cncy.21812. [DOI] [PubMed] [Google Scholar]
- 20.Doxtader EE, Cheng YW, Zhang Y. Molecular testing of non-small cell lung carcinoma diagnosed by Endobronchial ultrasound-guided Transbronchial fine-needle aspiration: the Cleveland Clinic experience. Arch Pathol Lab Med. 2019;143(6):670–676. doi: 10.5858/arpa.2017-0184-RA. [DOI] [PubMed] [Google Scholar]
- 21.da Cunha Santos G, Saieg MA. Preanalytic specimen triage: Smears, cell blocks, cytospin preparations, transport media, and cytobanking. Cancer Cytopathol. 2017;125(S6):455–464. doi: 10.1002/cncy.21850. [DOI] [PubMed] [Google Scholar]
- 22.Martini M, Capodimonti S, Cenci T, et al. To obtain more with less: Cytologic samples with ancillary molecular techniques-the useful role of liquid-based cytology. Arch Pathol Lab Med. 2018;142(3):299–307. doi: 10.5858/arpa.2017-0148-RA. [DOI] [PubMed] [Google Scholar]
- 23.Cui S, Ye L, Wang H, et al. Use of SuperARMS EGFR mutation detection kit to detect EGFR in plasma cell-free DNA of patients with lung adenocarcinoma. Clin Lung Cancer. 2018;19(3):e313–e322. doi: 10.1016/j.cllc.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Feng WN, Gu WQ, Zhao N, et al. Comparison of the SuperARMS and droplet digital PCR for detecting EGFR mutation in ctDNA from NSCLC patients. Transl Oncol. 2018;11(2):542–545. doi: 10.1016/j.tranon.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, Jin B, Chu T, et al. EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non-small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: a real-world study in China. Lung Cancer. 2016;96:87–92. doi: 10.1016/j.lungcan.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Sutiman N, Tan SW, Tan EH, et al. EGFR mutation subtypes influence survival outcomes following first-line Gefitinib therapy in advanced Asian NSCLC patients. J Thorac Oncol. 2017;12(3):529–538. doi: 10.1016/j.jtho.2016.11.2225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data are available upon request.