Abstract
Design strategies have evolved over the past few years as a means to accelerate the drug development process so that the right therapies can be delivered to the right patients. Basket, umbrella and adaptive enrichment strategies represent a class of novel designs for testing targeted therapeutics in oncology. Umbrella trials include a central infrastructure for screening and identification of patients, and focus on a single tumor type or histology with multiple sub-trials each testing a targeted therapy within a molecularly defined subset. Basket trial designs offer the possibility to include multiple molecularly defined sub-populations, often across histology or tumor types, but included in one cohesive design to evaluate the targeted therapy in question. Adaptive enrichment designs offer the potential to enrich for patients with a particular molecular feature predictive of benefit for the test treatment based on accumulating evidence from the trial. This session will aim to discuss the fundamentals of these design strategies, the underlying statistical framework, the logistical barriers with implementation and ultimately, the interpretation of the trial results. New statistical approaches, extensive multi-disciplinary collaboration and state of the art data capture technologies are needed to implement these strategies in practice. Logistical challenges to implementation arising from centralized assay testing, requirement of multiple specimens, multidisciplinary collaboration, infrastructure requirements etc. will also be discussed. The session will present these concepts in the context of the National Cancer Institute’s precision medicine initiative trials: MATCH, ALCHEMIST, Lung MAP, as well as other trials such as FOCUS4.
Introduction
The traditional drug development paradigm of Phase I for establishing the safety profile, followed by Phase II for efficacy signal, followed by Phase III for establishing definitive clinical benefit is challenged by the use of targeted therapeutics and incorporation of biomarker assessment for medical treatment [1]. Biomarkers are a critical component of targeted therapies as they can be used to identify patients who are more likely to benefit from a particular treatment. In this context of personalized medicine, a Phase I study tests the methods of assessment of marker alteration in normal and tumor tissue samples and guides in the determination of cut points, and preliminary assessment of efficacy within molecularly defined subsets; a “Phase II” study includes careful retrospective assessment of the marker to establish clinical value, and Phase III trials are confirmatory in nature that validate the marker (and companion diagnostic) through large prospective randomized controlled trials (RCT) in a multi-center setting [2]. The fundamental challenge for development of new cancer therapeutics is therefore to be able to identify and assess activity in molecularly defined patient subsets starting from early phase trials in order to predict which patients will respond to a new agent/regimen.
Design strategies have evolved in the past few years as a means to accelerate this new drug development process so that the right therapies can be delivered to the right patients. Basket, umbrella and adaptive enrichment strategies represent a class of novel designs for testing targeted therapeutics in oncology [3]. This session will aim to address the following, using examples of ongoing or completed trials to present the concepts:
Enrichment or targeted trial design strategies - when are they appropriate?
Use of umbrella trials in improving clinical trial efficiency: impact of different drugs on different mutations (molecular subtypes) within a single tumor type.
Role of Basket trials in screening agents efficiently to identify the “exceptional” responders: impact of a single drug across multiple tumor types and/or histologic subtypes harboring the same mutation (molecular profile).
These design strategies challenge the historical paradigm of drug development. New statistical approaches, extensive multi-disciplinary collaboration and state of the art data capture technologies are needed to implement these strategies in practice. Logistical challenges to implementation arising from centralized assay testing, requirement of multiple specimens, multidisciplinary collaboration, infrastructure requirements etc. will also be discussed using illustrative examples where appropriate.
Enrichment Trial Designs
The enrichment design, also called targeted design, has become the most commonly used phase III design for co-development of a new cancer drug and a companion diagnostic. Patients are screened with the diagnostic test and those who are considered “test positive” are eligible for the clinical trial. Eligible patients are randomized to receive either the test drug or an appropriate control regimen. In some cases the randomization may be between the test drug and standard chemotherapy, or between standard chemotherapy alone versus standard chemotherapy plus the test drug. When there is no standard chemotherapy the randomization may be between the test drug and best supportive care. See Figure 1 for a schematic of the enrichment trial design strategy.
Figure 1:
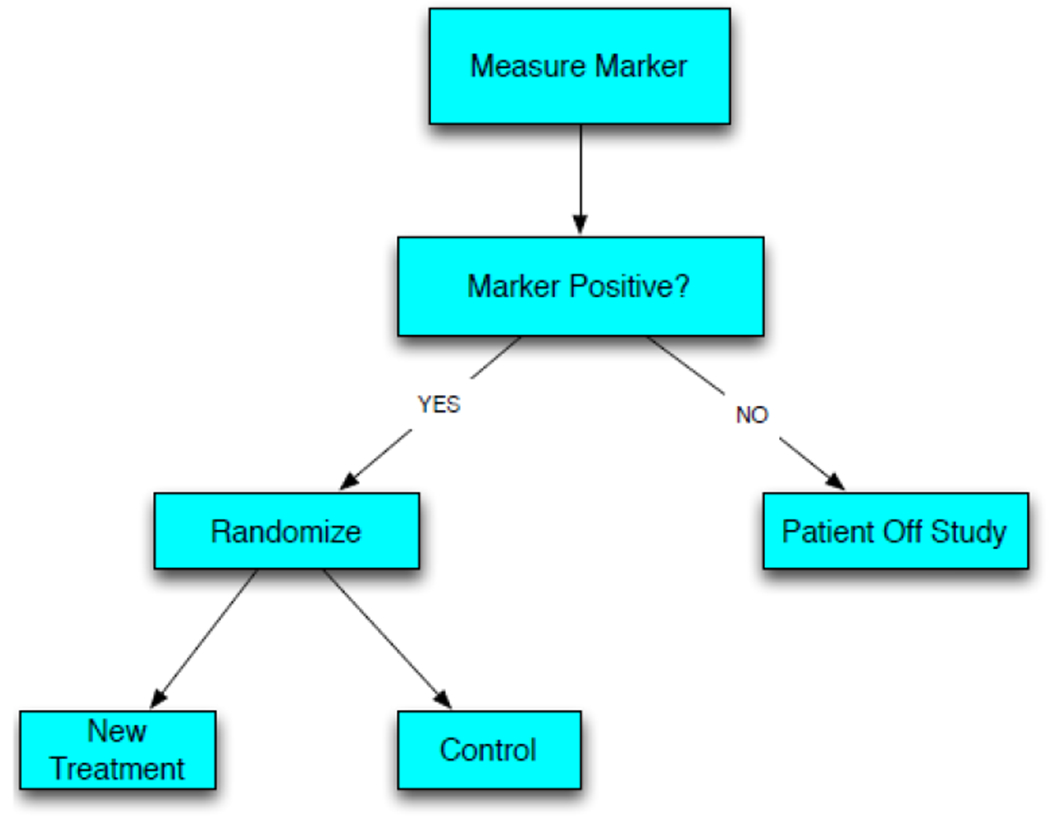
Schematic representation of the Enrichment Trial Design Strategy
Patients who are not marker positive are not eligible for the study. Consequently the design is best suited to settings where there is a strong biological rationale or phase II data indicating that marker negative patients are unlikely to benefit from the test drug. This was the case for the pivotal trials of vemurafenib for melanoma where the companion diagnostic was a sequencing test for a point mutation in the kinase domain of the BRAF gene [4]. It was also the case for crizotinib where the companion diagnostic was an assay for a translocation that activates the ALK kinase gene [5]. One of the first uses of the enrichment design in oncology was for the pivotal trial of trastuzumab in women with metastatic breast cancer [6]. The companion diagnostic in that case was over-expression of the HER2 protein or amplification of the HER2 gene. In these cases there was a close linkage between the target of the test drug and the diagnostic test measuring a genomic alteration known to constitutively activate an oncogene target of the test drug. Because kinase inhibitors generally have multiple targets, such drugs may be active for some patients who are test negative. The proportion of test negative patients who benefit from such a drug and the degree of benefit are generally much less than the treatment effect of the drug for test positive patients. Hence, use of the enrichment design is appropriate. Test negative patients can later be the focus of a separate clinical trial of the same drug after the drug is demonstrated to benefit those for whom it is designed to benefit. The test negative patients can be spared the toxicity of a drug from whom few are expected to benefit.
One of the main benefits of the enrichment design is efficiency; that is, reduced sample size requirements [7]. The sample size of a clinical trial can generally be expressed in terms of the significance level, the statistical power and the treatment effect to be detected with that power. For example, in comparing two treatments with regard to survival, the number of end-point events, D, that needs to be observed can be written as:
| (1) |
where, for a two-tailed 5% significance level, kα=1.96 and for power 90% kβ=1.28. The hazard ratio HR is a measure of the expected difference in the survival curve for the test treatment group and the control group. Broad eligibility clinical trials are frequently sized to detect 25% to 33% reductions in the hazard of death. Those reductions in hazard correspond to hazard ratios of 0.75 or 0.67 respectively. Using the natural logarithm in equation (1) gives a D value of 508 total events for HR=.75. That is, to have 90% power for detecting a 25% reduction in hazard using a two-sided significance level of 5% requires observing 508 events. If by the time of the final analysis only 25% of the patients are expected to fail, then in order to observe 508 events one needs to accrue 2032 patients. If, on the other hand, we only need to have 90% power for detecting a 33% reduction in hazard, i.e. a hazard ratio of 0.67, then expression (1) indicates that we only need to observe 262 events instead of 508. So a slight change in the size of the treatment effect to be detected can lead to a large reduction in the sample size requirement.
If we pre-select patients as in an enrichment design so that we expect an even larger reduction in hazard, say 50%, then expression (1) indicates that we only need to observe 88 events. Targeting a larger treatment effect by enriching the population of the clinical trial to exclude patients unlikely to benefit from the test treatment can thus dramatically reduces the number of randomized patients required. However, the number of patients screened with the diagnostic test in order to obtain the required number of randomized patients may still be substantial.
To use the enrichment design one should have strong biological rationale or phase II data that test negative patients are very unlikely to benefit from the treatment similarly to the test positive patients. One should also have an analytically validated test for use for selecting patients in the phase III clinical trial. Over-expression of the EGFR protein was thought to be a predictive biomarker for identifying patients who could benefit from erlotinib but it was not used as an eligibility factor. It later turned out that mutation in the kinase domain of EGFR was the appropriate biomarker for erlotinib in NSCLC, not over-expression of the protein. Hence, it was useful that the initial phase III studies did not employ an enrichment design with EGFR over-expression as a selection factor. Another case in which the enrichment design was not used came in the development of the monoclonal antibodies panitumamab and cetuximab against EGFR in the treatment of advanced colorectal cancer [8]. Over-expression of EGFR was not used to restrict eligibility and it later turned out that the appropriate biomarker was KRAS mutation. Patients with mutated KRAS did not benefit from the antibodies because their tumors were not driven by EGFR activation, but patients with wild type KRAS did benefit from the antibodies. So the enrichment design should be used primarily when the mechanism of the biomarker in the disease is well understood. It is also useful to have phase II data confirming that the understanding is correct.
Extensions of Enrichment Designs
Run-in Designs
The enrichment design has been most effectively used with genomic alterations of the drug target as the diagnostic test. Gene expression profiles have rarely been useful as predictive biomarkers for identifying which patients benefit from a specific drug therapy although they sometimes have been useful prognostic indicators. For some drugs, such as anti-angiogenic agents and immunological agents it has been difficult to identify pre-treatment predictive biomarkers. Hong and Simon [9] showed how the enrichment design can sometimes be used after a short run-in phase on the test drug. During the run-in phase a pharmacodynamic response, immunologic response or early tumor response is measured. The patients are randomized after the run-in phase to either continuing the test drug or to switching to a control regimen. The biomarker measured during the run-in can be used to restrict eligibility to the randomization. A variation of this approach is to use the control regimen during the run-in phase and to only randomize patients who do not have a strong initial response to the control. This was used successfully in a clinical trial of dose-dense intensification of chemotherapy for patients with poor-prognosis germ-cell tumors [10].
Adaptive Enrichment Designs
A second extension of the enrichment design is the adaptive enrichment approach described by Simon and Simon [11]. For example, one may have a companion diagnostic test to use with the drug, but a cut-point for test positivity may not have been adequately determined based on phase II trials. With the adaptive enrichment design, one starts without using the test result to limit eligibility. Interim analyses adaptively restrict eligibility to increase the cut-point of the test and exclude from eligibility the patients who do not appear to benefit from the test drug based on interim data for a short-term endpoint. The adaptive enrichment approach can also be used when there are multiple candidate biomarkers.
Umbrella Trial Designs
Having a national network of clinical sites doing molecularly targeted clinical trials using a common genomic screening platform is an important defining characteristic of the Umbrella Design and is essential for effectively conducting targeted clinical trials in an increasingly stratified set of tumors. The umbrella infrastructure usually has the flexibility to add (or drop/modify) sub trials of molecularly targeted drugs and companion diagnostics based on accumulating evidence from the ongoing trial (and newly emerging data). The umbrella design is essentially two or more enrichment designs linked thru a common patient-screening infrastructure. Patients are screened for a specific set of biomarkers and assigned to a biomarker-driven sub-study (targeted design) if it is determined that they have one of the target biomarkers. See Figure 2 for a schematic representation of this design.
Figure 2:
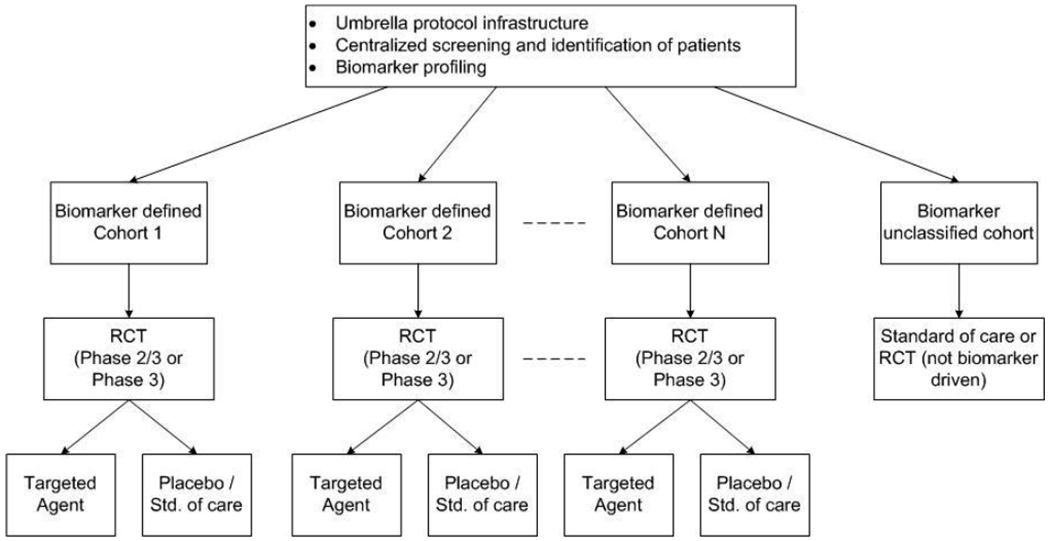
Schematic representation of the Umbrella Trial Design
The Lung MAP (Master Protocol-Phase II/III Biomarker-Driven Master Protocol for Second Line Therapy of Squamous Cell Lung Cancer, ) study in squamous lung cancer is an example of an umbrella protocol that is currently open at over hundreds of sites in the USA. Patients with advanced squamous lung cancer with one previous treatment have their tumors screened for genetic alterations in over 200 genes using a standardized sequencing platform. As a result of this tumor characterization, patients are recommended to one of 5 sub-trials within the umbrella framework. Four of the five clinical trials are enrichment trials with eligibility limited to those patients whose tumor harbors a specified genomic alteration in a gene which is a target of the test drug for that clinical trial. The fifth clinical trial is for patients whose tumor characterizations do not qualify them for one of the other four clinical trials. Each of the component clinical trials is a pivotal phase 2/3 clinical trial that can lead to the approval of a targeted drug with the genomic screening platform as companion diagnostic. Of note, a substantial amount of work is needed to analytically validate an assay to qualify it as a companion diagnostic. With the umbrella framework, this work qualifies the same platform for a large number of potential drugs.
The Phase 2/3 design allows for early termination if the test treatment is not promising relative to the internal control based on progression-free survival (PFS) or may proceed to a Phase 3 trial with PFS and overall survival (OS) as co-primary endpoints. The design within each sub-study is standardized such that the phase II interim analysis is to occur upon the observation of 55 progression events and the final phase III analysis is to occur upon the observation of 256 deaths (provided the study proceeds to Phase III). These are based on a phase II design with 90% power and 10% 1-sided type I error to detect a two-fold increase in median PFS and a phase III design with 90% power and 1-sided 2.5% type I error to detect a 50% increase in median OS.
The ALCHEMIST (Adjuvant Lung Cancer Enrichment Marker Identification and Sequencing Trial) clinical trial uses the Umbrella Design to identify and screen patients with the EGFR (~15% prevalence) and ALK (~5% prevalence) mutations in early stage resected non-squamous non-small cell lung cancer (NSCLC). The surgical specimen will be tested centrally through a CLIA certified laboratory for specific genomic alterations in the ALK or EGFR genes. If an ALK-EML4 rearrangement is found, the patient will be referred to a double blind randomized clinical trial comparing crizotinib to placebo adjuvant therapy. Patients with an activating mutation in EGFR will be referred to a double blind randomized clinical trial comparing erlotinib to placebo as adjuvant therapy. In the setting of these two parallel clinical trials studying resected NSCLC in rare molecularly defined subsets of patients, a central protocol infrastructure for the identification of potentially eligible patients and for central screening for appropriate genotypes is valuable. Furthermore, ALCHEMIST includes collection of tumor and blood specimens at baseline for genomic discovery efforts, and at the time of disease recurrence (optional tumor collection) to understand mechanisms of drug resistance and genomic evolution. Such advanced genomic analysis, performed on a clinically uniform and well characterized NSCLC population, will allow for new insights into the relationship between tumor biology and clinical outcome. Thus, the ALCHEMIST screening platform consists of three integrated protocols: ALCHEMIST-Screening (A151216; ), ALCHEMIST-EGFR (A081105; ), and ALCHEMIST-ALK (E4512; ).
An estimated 8000 patients will be screened through A151216 to facilitate accrual to the two sub-studies: ALCHEMIST-EGFR and ALCHEMIST-ALK. Both sub trials are harmonized in terms of primary endpoint (OS), as well as with regard to many of the trial logistics (eligibility criteria, follow-up schedule during treatment, long term follow-up etc.). The ALCHEMIST-EGFR trial has a target accrual of 410 patients, with centrally confirmed EGFR mutation status, in order to have at least 85% power to detect a hazard ratio of 0. 67 in favor of the erlotinib arm using a one-sided type I error rate of 5%. The ALCHEMIST-ALK trial has a target accrual of 378 patients, with centrally confirmed ALK status, in order to have at least 80% power to detect a 33% reduction in the OS hazard rate of 0.0105 to 0.0070 in favor of the crizotinib arm using a one-sided type I error rate of 5%. As with the Lung MAP protocol, ALCHEMIST is set up to accommodate the addition of sub trials of targeted therapeutics in this setting if and when they are ready to be tested in a Phase III setting.
Another umbrella trial example is the FOCUS4 Master Protocol (Molecular selection of therapy in colorectal cancer: a molecularly stratified randomized controlled trial program) [12]. It is an integrated protocol with parallel, molecularly stratified, randomised comparisons of maintenance therapies for patients with advanced or metastatic colorectal cancer after receiving 1st -line chemotherapy. It includes randomized phase 2/3 trials following the principles of the MAMS (Multi arm Multi stage [13, 14]) design for 4 predefined molecular cohorts. Similar to Lung MAP, for patients whose biomarker panel results are unclassifiable or for those whose molecular cohort is temporarily not open for recruitment, and for any patients unable or unwilling to travel, a concurrent non-biomarker driven trial comparing capecitabine against no treatment is available. Four pre-specified interim analyses are planned for each trial: Stage I (assess safety), Stage II (assess lack of benefit), Stage III (assess efficacy for PFS) and Stage IV (assess efficacy for OS). Stages I and II mirror a conventional phase 2 trial design and Stages III and IV are similar to a phase 3 trial design paradigm.
Basket Trial Designs
The discovery of driver oncogenes and the corresponding successful development of therapies targeting those genetic abnormalities have dramatically impacted clinical research, placing great focus on the biological causes of dramatic responses to therapy among populations that do not derive treatment benefit overall. The first observation of this phenomenon was the discovery that imatinib effectively targets the BCR-ABL tyrosine kinase in chronic myeloid leukemia; this was subsequently followed by the discovery that EGFR mutations in non-small cell lung cancer confer dramatic responses to treatment with gefitinib [15–18]. When an ‘exceptional responder’ is observed, often referred to as the n-of-one phenomenon, further investigation of the tumor biology and the therapeutic mechanism of action may lead to formally testing a predefined hypothesis that patients whose tumor harbor a particular genetic aberration respond to a specified targeted agent in the context of a prospective clinical study. Basket trials have come to be known as an efficient way of screening experimental therapeutics across multiple patient populations in early phase drug development. Basket trials generally assign treatments to patients based on the molecular alterations their tumors contain, regardless of the histologic type of the tumor. Like umbrella trials, there may be a common genetic screening platform, especially if the cohorts defined by the study are histology-independent and defined only by the presence of a single molecular aberration; however the term ‘basket trial’ has also been used to describe studies in which patients are assigned to cohorts by their cancer type. Whether cohort eligibility criteria are histology- or mutation-specific, these trials are generally conducted within the context of a single protocol. Figure 3 gives a generic representation of a Basket trial design schema. There is perceived efficiency in running multiple cohorts in this way, since conducting a stand-alone study within each cohort separately would be exponentially more resource-intensive.
Figure 3:
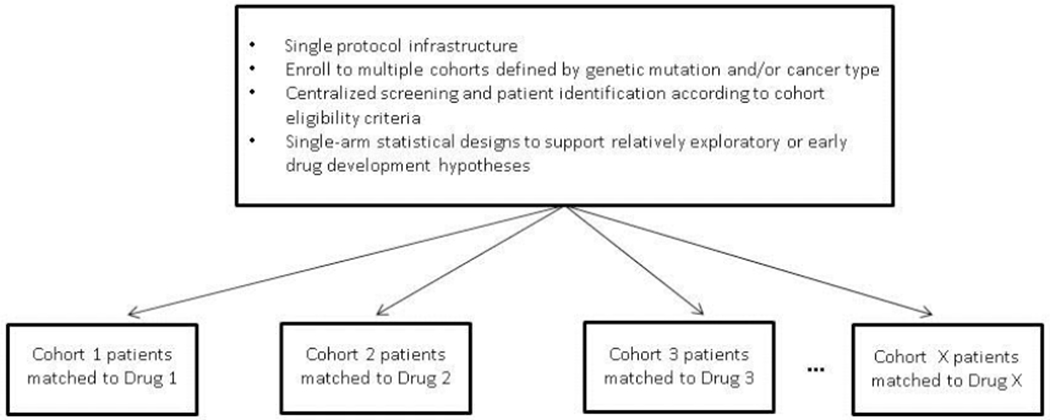
Schematic representation of the Basket Trial Design
Basket trials are not without their limitations, however. Paramount to their conduct is a strong scientific rationale for the marker-drug pairing, as well as reliable assay development for the marker of interest. Genomic variants have unknown or differential variability across tumor types [19], and there is often uncertainty about whether a particular mutation in a tumor of a particular histologic type should be considered actionable for treatment with a given drug. Resolving these uncertainties, however, is the reason for doing the study. These trials often study cancers so rare that it may be impossible to study them in a randomized setting, and usually require coordination and participation from multiple institutions in order to meet accrual objectives [20]. They can depend heavily on tumor availability for genetic screening, which can be difficult to obtain or to ethically justify in certain types of cancer. This design also depends on the availability of a sufficient number of drugs targeting diverse de-regulated pathways.
Basket trials are discovery trials and there has been uncertainty about how to design such trials statistically. Consequently, in some basket trials, justification of sample sizes has been absent despite increased interest in efficacy endpoints such as response, and the amendment process is frequently used to expand cohorts to sample sizes that would otherwise be sufficient for the proper conduct of a randomized phase II or III study [21, 22]. The practice of not justifying sample sizes is particularly worrisome since it counters the intent of minimizing the number of patients exposed to toxic or ineffective drugs, and in light of this investigators should consider early stopping rules and provide clearly defined criteria for study completion [21]. Additional steps should also be taken to acknowledge the impact of patient and tumor heterogeneity on the numerical variability of results and the multiple comparisons issues that arise due to excessive testing of subgroups during the conduct of these trials.
The Molecular Analysis for Therapy Choice (MATCH) study is an example of a basket trial being led by the ECOG-ACRIN Cancer Research Group, along with the National Cancer Institute, and is slated to activate within the National Clinical Trials Network this year [23]. As many as 3000 patients with refractory solid tumors or lymphoma and for whom no standard treatment prolonging survival exists will be screened with the goal of accruing approximately 35 patients (31 eligible) in as many as 20-25 biomarker-defined subgroups. Targeted agents for this trial were selected if (1) a drug is FDA-approved for a malignant indication and there is a molecular abnormality that can serve as a valid predictive marker; (2) the drug is investigational but met a clinical endpoint (PFS, tumor response) in any malignancy and has evidence of target inhibition and has evidence of a predictive molecular marker; and (3) the drug is investigational but has demonstrated clinical activity in any malignancy and evidence of target inhibition, and has evidence of a predictive molecular marker.
Patients on MATCH are assigned to histology-independent subgroups and treated with the agents matched to their tumor’s identified molecular abnormality. If a patient’s tumor harbors more than one molecular abnormality identified as actionable for treatment in MATCH, then treatment will be assigned based on the mutation detected in a higher percent of tumor cells; if this percentage is within 15% for two or more aberrations in >1 pathway, then the patient will be assigned to the treatment with fewer patients enrolled, or if equally enrolled the patient will be randomized between the treatments under consideration.
The primary endpoint of each cohort is objective response rate and 6-month PFS. Objective response will be compared to a historical control of 5% while 6-month PFS will be compared to a historical control of 15%. This design provides 92% power to test the alternative hypothesis that a drug confers an OR rate of 25% and 89% power that a drug provides a 6-month PFS rate of 38% while controlling the type I error rate at the one-sided 0.05 level. If the objective response rate is ≥ 5/31 (16%) or progression-free survival at 6 months is ≥ 9/31 (29%), then an agent will be considered promising and worthy of further testing. For each arm, there is a futility rule provided as a guideline; an interim analysis will be performed after 15 eligible patients have been enrolled. If no response is observed among them and the 6-month PFS is ≤ 2/15 (13%), the analysis result will be presented to the steering committee for further guidance on whether the arm should be stopped early.
Summary
Precision medicine’s fundamental assumption is that using the genetic makeup of the tumor and the genotype of the patient will enable targeted therapeutics to improve clinical outcomes [3]. While there have been notable successes with this approach, our methods for matching drugs to tumors is still rudimentary and numerous challenges remain to be addressed adequately. These include an understanding of the interaction of signaling pathways, the clonal evolution and heterogeneity of tumors, ability to obtain tumor biopsies (often multiple and over time), technical limitations with assays, centralized molecular testing, adequate resources and infrastructure for a quick turnaround of biomarker results to make these designs feasible, and effective multidisciplinary collaborations. Nevertheless, genomic technology has already become a part of routine clinical practice. Enrichment, Umbrella and Basket trial designs are gaining popularity as they present novel strategies to accelerate the drug development process so that the right therapies can be delivered to the right patients quickly.
Key Points.
Oncology is increasingly understood at the molecular level; therapeutic research has largely shifted from a focus on cytotoxic agents to newer targeted drugs that inhibit specific cancer cell growth and survival mechanisms.
Novel clinical trial design strategies that challenge the historical paradigm of drug development are needed to accelerate the drug development process so that the right therapies can be delivered to the right patients.
Patient selection and enrichment approaches are becoming increasingly relevant; however sufficient biologic rationale, understanding of the mechanism of action of the drug, assay characteristics and validated cut points are needed to support the enrichment strategy.
Umbrella trials incorporate a central infrastructure for screening and identification of patients with a focus on a single tumor type or histology; multiple sub-trials testing targeted therapeutics within molecularly defined subsets are embedded within the umbrella framework.
Basket trial designs offer the possibility to include multiple molecularly defined sub-populations, across histologic subtypes or tumor types, in one cohesive design to evaluate the targeted therapy in question.
References
- 1.Mandrekar SJ, Sargent DJ. Drug designs fulfilling the requirements of clinical trials aiming at personalizing medicine. Chin Clin Oncol. 2014. June 1;3(2):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elizabeth M, Hammond H, Taube SE. Issues and Barriers to Development of Clinically Useful Tumor Markers: A Development Pathway Proposal. Seminars in Oncology 2002; 29(3): 213–221. [DOI] [PubMed] [Google Scholar]
- 3.Andre F, Mardis E, Salm M, Soria JC, Siu LL, Swanton C. Prioritizing targets for precision cancer medicine. Ann Oncol. 2014;25(12):2295–303. [DOI] [PubMed] [Google Scholar]
- 4.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AMM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA, for the BRIM-3 Study Group. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. N Engl J Med 2011; 364:2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim D-W, Nakagawa K, Seto T, Crinó L, Ahn M-J, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu Y-L, Thomas M, O’Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Jänne PA. Crizotinib versus Chemotherapy in Advanced ALK-Positive Lung Cancer. N Engl J Med 2013; 368:2385–2394 [DOI] [PubMed] [Google Scholar]
- 6.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE Jr., Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–84. [DOI] [PubMed] [Google Scholar]
- 7.Simon R and Maitournam A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clinical Cancer Research 10:6759–63, 2005. [DOI] [PubMed] [Google Scholar]
- 8.Karapetis CS, Khambata-Ford S, Jonker DJ et al. ; K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med ;359(17):1757–65 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Hong F and Simon R. The run-in phase III design with post-treatment predictive biomarkers. Journal of the National Cancer Institute 106:1628–33, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon N and Simon R. Adaptive enrichment designs for clinical trials. Biostatistics, doi: 10.1093/kxt010, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fizazi K et al. Personalised chemotherapy based on tumour marker decline in poor prognosis germ-cell tumours (GETUG 13): a phase 3, multicentre, randomized trial. Lancet Oncology doi: 10.1016/S1470-2045(14)70490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan R, Maughan T, Crook A, Fisher D, Wilson R, Brown L, Parmar M. Evaluating many treatments and biomarkers in oncology: a new design. J Clin Oncol. 2013. December 20;31(36):4562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Royston P1, Parmar MK, Qian W. Novel designs for multi-arm clinical trials with survival outcomes with an application in ovarian cancer. Stat Med 2003; 22(14):2239–56. [DOI] [PubMed] [Google Scholar]
- 14.Parmar MKB, Barthel FM-S, Sydes M, Langley R, Kaplan R, Eisenhauer E, Brady M, James N, Bookman MA, Swart A-M, Royston P. Speeding up the Evaluation of New Agents in Cancer. J Natl Cancer Inst 2008;100:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Druker BJ, Sawyers CL, Kantarjian H, et al. : Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344(14):1038–1042. [DOI] [PubMed] [Google Scholar]
- 16.Druker BJ, Talpaz M, Resta DJ, et al. : Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1031–1037. [DOI] [PubMed] [Google Scholar]
- 17.Paez JG, Janne PA, Lee JC, et al. : EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. [DOI] [PubMed] [Google Scholar]
- 18.Lynch TJ, Bell DW, Sordella R. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. [DOI] [PubMed] [Google Scholar]
- 19.Sleijfer S, Bogaerts J, Siu LL. Designing transformative clinical trials in the cancer genome era. J Clin Oncol. 2013;31(15):1834–1841. [DOI] [PubMed] [Google Scholar]
- 20.Willyard C ‘Basket studies’ will hold intricate data for cancer drug approvals. Nat Med, 2013; 19:655. [DOI] [PubMed] [Google Scholar]
- 21.Dahlberg SE, Shapiro GI, Clark JW, Johnson BE. Evaluation of statistical designs in phase I expansion cohorts: the Dana-Farber/Harvard Cancer Center experience. JNCI J Natl Cancer Inst (2014) 106 (7): dju163 doi: 10.1093/jnci/dju163 [DOI] [PubMed] [Google Scholar]
- 22.Menis J, Hasan B, Besse B. New clinical research strategies in thoracic oncology: clinical trial design, adaptive, basket and umbrella trials, new end-points and new evaluations of response. Eur Respir Rev, 2014; 23(133):367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conley BA, Doroshow JH. Molecular Analysis for Therapy Choice: NCI MATCH. Semin Oncol, 2014; 41(3), 297–299. [DOI] [PubMed] [Google Scholar]


