Abstract
Blastocystis is a food and water borne intestinal parasite commonly identified in humans and many other animals worldwide. Of the nine potentially zoonotic subtypes of Blastocystis, seven have been reported in bird species. However molecular studies of Blastocystis subtype diversity in birds are limited. In this study, fecal samples from 109 domestic and captive wild birds from Minas Gerais, Brazil were tested for the presence of Blastocystis subtypes using PCR and next generation amplicon sequencing of a fragment of the small subunit ribosomal RNA (SSU rRNA) gene. Birds from 11 orders and 38 species from both local markets and bird conservation facilities were sampled. Blastocystis was present in 14.7% of samples, and eight subtypes, six previously reported (ST5, ST6, ST7, ST10, ST14, ST24) and two novel subtypes (named ST27 and ST28), were identified. The most commonly identified subtypes were ST7 and ST6 identified in 10 (62.5%) and 6 (37.5%) of 16 Blastocystis positive samples. At least one of the three zoonotic subtypes identified (ST5, ST6, and ST7) was found in 81.3% of Blastocystis positive samples. Infection with multiple Blastocystis subtypes was common and identified in 62.5% of positive samples. This study is the first to use next generation amplicon sequencing to characterize Blastocystis subtype diversity in birds. The findings presented here confirm that birds may serve as reservoirs of zoonotic subtypes of Blastocystis and that the role of birds in transmission of Blastocystis to humans requires further study.
Keywords: Blastocystis, Brazil, Birds, Subtypes, Mixed infection, Next generation amplicon sequencing (NGS)
Highlights
-
•
Blastocystis was detected in 14.7% of the birds.
-
•
Eight subtypes, including three potentially zoonotic and two potentially novel subtypes, were identified.
-
•
Zoonotic subtypes ST6 and ST7 were the most commonly found subtypes in Blastocystis positive birds.
-
•
Multiple Blastocystis subtypes were common in infected birds (62.5% of the positive samples).
-
•
Birds could contribute to the transmission of zoonotic Blastocystis subtypes to humans.
1. Introduction
Blastocystis is an intestinal protist parasite which is frequently reported in humans and other animals. Infection has been associated with a wide range of symptoms including diarrhea, abdominal pain, and chronic hives with asymptomatic infection/colonization being most common (Tan, 2008). Blastocystis transmission likely occurs via the ingestion of cysts from feces either through direct contact or ingestion of contaminated food or water. Blastocystis has been found in both food and water samples (Caradonna et al., 2017; Kolören and Karaman, 2019; Zahedi et al., 2019).
The genus Blastocystis constitutes a genetically diverse group of organisms divided into subtypes based on polymorphism in the small subunit ribosomal RNA (SSU rRNA) gene. There are currently 26 proposed subtypes of Blastocystis named ST1 to ST26 (Alfellani et al., 2013; Maloney et al., 2019a; Zhao et al., 2017). While the degree of host specificity of each of these subtypes remains unresolved, there are nine subtypes which have been reported in both humans and animals, ST1-8 and ST12, indicating the potential for zoonotic transmission of this parasite (Ramírez et al., 2016; Stensvold and Clark, 2016). Zoonotic transmission is further supported by evidence from both pig and chicken farms where Blastocystis subtypes found in animals were also found in caretakers or staff (Greige et al., 2018; Li et al., 2007; Wang et al., 2014; Yan et al., 2007). Both pigs and caretakers were found to have ST5 in reports from Australia and China (Li et al., 2007; Wang et al., 2014; Yan et al., 2007). Similarly, samples from chickens and staff in a Lebanon slaughterhouse were both found to have ST6 (Greige et al., 2018). Thus, the role of animals in human infection requires more study.
Studies on the prevalence and subtype diversity of Blastocystis in bird species are limited. However, the data that exist indicate birds may be host to several potentially zoonotic subtypes including ST1, ST2, ST4, ST5, ST6, ST7, and ST8 (Cian et al., 2017; Deng et al., 2019; Ramírez et al., 2014; Roberts et al., 2013; Valença-Barbosa et al., 2019). In this study, the presence of Blastocystis subtypes was determined in fecal samples from domestic and captive wild birds from 11 orders and 38 species collected in the state of Minas Gerais (Brazil) using PCR and next generation amplicon sequencing. This study provides important information on the subtype diversity of Blastocystis in birds and will aid in determining their role in the transmission of Blastocystis to humans and other animals.
2. Material and methods
2.1. Source of specimens
From October 2013 to September 2014, 109 fecal specimens were collected from captive birds at the Brazilian Institute of the Environment and Renewable Natural Resources (IBAMA) (n = 15), a private aviary (n = 6), a school of falconry (n = 10), and 14 local markets (n = 78) in Uberlândia and Belo Horizonte in the state of Minas Gerais, Brazil. The birds included 11 orders and 38 species (Table 1). At the time of sampling, all birds examined appeared in good health and no diarrhea was observed. To collect the samples, the animals were placed in individual sanitized cages and fresh feces were collected from the bottom of the cages. Fecal specimens were placed into sterile polystyrene tubes with records of the date, location, identification number, and species, and transferred in isothermal boxes to the Parasitology Laboratory of Federal University of Uberlândia (UFU) and held at −20 °C until DNA extraction.
Table 1.
Blastocystis prevalence and subtypes in domestic and wild captive birds in Uberlândia and Belo Horizonte in the state of Minas Gerais (Brazil).
| Location | Order | Scientific name (common name) | No. of samples examined | No. of Blastocystis positives | Blastocystis prevalence (%) | Subtype(s) | |
|---|---|---|---|---|---|---|---|
| Uberlândia | School of Falconry | Falconiformes |
Falco femoralis (Aplomado Falcon) |
6 | 0 | 0 | |
| Strigiformes |
Bubo virginianus (Great Horned Owl) |
1 | 0 | 0 | |||
|
Tyto alba (American Barn Owl) |
2 | 0 | 0 | ||||
| Accipitriformes |
Buteo albicaudatus (White-tailed Hawk) |
1 | 0 | 0 | |||
| Aviary | Psittaciformes |
Agapornis nigrigenis (Black-cheeked Lovebird) |
1 | 1 | 100 | ST14 | |
|
Agapornis roseicollis (Peach-faced Lovebird) |
1 | 0 | 0 | ||||
| Nymphicus hollandicus (Cockatiel) | 3 | 0 | 0 | ||||
| Piciformes |
Ramphastos toco (Toco Toucan) |
1 | 0 | 0 | |||
| Market 1 | Anseriformes |
Cairina moschata (Muscovy Duck) |
2 | 0 | 0 | ||
| Market 2 | Anseriformes |
Anser cygnoides (Swan Goose) |
2 | 0 | 0 | ||
| Anas querquedula (Garganey) | 1 | 1 | 100 | ST7, ST10, ST27, ST28 | |||
| Galliformes |
Pavo cristatus (Indian Peafowl) |
1 | 1 | 100 | ST27, ST28 | ||
|
Meleagris gallopavo (Turkey) |
1 | 0 | 0 | ||||
| Psittaciformes | Aratinga leucophthalma (White-eyed Parakeet) | 1 | 0 | 0 | |||
| Melopsittacus undulatus (Budgerigar) | 1 | 0 | 0 | ||||
| Struthioniformes |
Struthio camelus (Ostrich) |
2 | 2 | 100 | ST5, ST10, ST24 ST5 |
||
| Market 3 | Anseriformes |
Anser cygnoides (Swan Goose) |
1 | 1 | 100 | ST7 | |
|
Aix galericulata (Mandarin Duck) |
1 | 0 | 0 | ||||
|
Anas platyrhynchos (Wild Duck) |
1 | 1 | 100 | ST7 | |||
|
Cairina moschata (Muscovy Duck) |
1 | 0 | 0 | ||||
| Galliformes |
Numida meleagris (Helmeted Guineafowl) |
2 | 2 | 100 | ST6, ST7 | ||
| Market 4 | Passeriformes |
Serinus canaria (Wild Canary) |
8 | 0 | 0 | ||
| Anseriformes |
Cairina moschata momelanotus (Muscovy Duck) |
1 | 1 | 100 | ST7, ST14 | ||
| Galliformes |
Numida meleagris (Helmeted Guineafowl) |
1 | 0 | 0 | |||
| Market 5 | Psittaciformes | Melopsittacus undulatus (Budgerigar) | 1 | 0 | 0 | ||
| Galliformes |
Coturnix coturnix (Quail) |
1 | 0 | 0 | |||
| Market 6 | Anseriformes | Callonetta leucophrys (Ringed Teal) | 1 | 0 | 0 | ||
|
Aix galericulata (Mandarin Duck) |
1 | 0 | 0 | ||||
| Market 7 | Galliformes |
Coturnix coturnix (Quail) |
1 | 1 | 100 | ST6, ST7 | |
| Market 8 | Galliformes |
Coturnix coturnix (Quail) |
1 | 0 | 0 | ||
| Market 9 | Galliformes |
Coturnix coturnix (Quail) |
1 | 0 | 0 | ||
|
Numida meleagris (Helmeted Guineafowl) |
1 | 0 | 0 | ||||
| Market 10 | Galliformes |
Coturnix coturnix (Quail) |
1 | 0 | 0 | ||
| Market 11 | Galliformes |
Numida meleagris (Helmeted Guineafowl) |
1 | 0 | 0 | ||
| Belo Horizonte | IBAMA | Psittaciformes |
Amazona aestiva (Blue-fronted Parrot) |
3 | 0 | 0 | |
| Aratinga leucophthalma (White-eyed Parakeet) | 1 | 0 | 0 | ||||
| Passeriformes |
Saltator similis (Green-winged Saltator) |
9 | 0 | 0 | |||
|
Saltatricula atricollis (Black-throated Saltator) |
1 | 0 | 0 | ||||
| Schistochlamys ruficapillus (Cinnamon Tanager) | 1 | 0 | 0 | ||||
| Market 12 | Anseriformes |
Cairina moschata (Muscovy Duck) |
2 | 0 | 0 | ||
| Anas querquedula (Garganey) | 1 | 0 | 0 | ||||
| Columbiformes |
Columba livia (Rock Pigeon) |
2 | 0 | 0 | |||
| Galliformes |
Phasianus colchicus (Pheasant) |
2 | 1 | 50 | ST6 | ||
| Market 13 | Galliformes |
Pavo cristatus (Indian peafowl) |
2 | 1 | 50 | ST27, ST28 | |
|
Phasianus colchicus (Pheasant) |
7 | 2 | 28.6 | ST6, ST7 | |||
| Casuariiformes |
Dromaius novaehollandiae (Emu) |
1 | 0 | 0 | |||
| Psittaciformes |
Pyrrhura perlata (Crimson-bellied Parakeet) |
3 | 0 | 0 | |||
|
Pyrrhura roseifrons (Rose-fronted Parakeet) |
1 | 0 | 0 | ||||
|
Psittacula krameri (Ringneck Parakeet) |
4 | 0 | 0 | ||||
|
Pionus menstruus (Blue-headed Parrot) |
1 | 0 | 0 | ||||
|
Aratinga jandaya (Jandaya Parakeet) |
2 | 0 | 0 | ||||
|
Ara chloropterus (Red-and-green Macaw) |
1 | 0 | 0 | ||||
|
Amazona aestiva (Blue-fronted Parrot) |
2 | 0 | 0 | ||||
|
Pyrrhura cruentata (Blue-throated Parakeet) |
1 | 0 | 0 | ||||
| Market 14 | Passeriformes |
Serinus canaria (Wild Canary) |
2 | 0 | 0 | ||
|
Sicalis flaveola (Saffron Finch) |
2 | 0 | 0 | ||||
| Sporophila caerulescens (Double-collared Seedeater) | 2 | 0 | 0 | ||||
| Psittaciformes | Melopsittacus undulatus (Budgerigar) | 1 | 0 | 0 | |||
| Anseriformes |
Anser cygnoides (Swan Goose) |
1 | 0 | 0 | |||
| Galliformes |
Phasianus colchicus (Pheasant) |
1 | 1 | 100 | ST7 | ||
| Total | 109 | 16 | 14.7 | ST5, ST6, ST7, ST10, ST14, ST24, ST27, ST28 | |||
2.2. DNA extraction
Genomic DNA was extracted directly from individual fecal specimens using the QIAamp Stool Mini Kit (Qiagen GmbH, Hilden, Germany) per the manufacturer's instruction with minor modifications. Modifications included the addition of 0.3 g of zirconia beads (Stratech Scientific, Luton, U.K.) to 0.2 g of feces and 1.4 ml lysis buffer (Mclauchlin et al., 1999), then mixture was heated at 95 °C for 5 min followed by vigorous shaking (2 rounds of 15 min) to facilitate the rupture of parasites. The nucleic acid was eluted in 150 μl of AE buffer to increase the quantity of DNA recovered.
2.3. Detection by PCR, NGS amplicon library preparation, and bioinformatic analysis
To detect Blastocystis, a ca. 500 base pair fragment of the Blastocystis SSU rRNA gene, which contains a variable region suitable for subtyping, was amplified by PCR (Santín et al., 2011). PCR products were analyzed using a QIAxcel (Qiagen, Valencia, CA). All positive samples were used to conduct next generation amplicon sequencing and libraries were prepared as previously described (Maloney et al., 2019b). Briefly, all positive samples were amplified by PCR using primers ILMN_Blast505_532F and ILMN_Blast998_1017R. These primers amplify a region of the SSU rRNA gene and are identical to Blast505_532F/Blast998_1017R (Santín et al., 2011), except for containing the Illumina overhang adapter sequences on the 5′ end. Final libraries were quantified using the Quant-iT dsDNA Broad-Range Assay Kit (ThermoFisher, Waltham, MA) on a SpectraMax iD5 (Molecular devices, San Jose, CA) prior to normalization. A final pooled library concentration of 8 pM with 20% PhiX control was sequenced using Illumina MiSeq 600 cycle v3 chemistry (Illumina, San Diego, CA). Paired end reads were processed and analyzed with an in-house pipeline that uses the BBTools package v38.22 (Bushnell, 2014), VSEARCH v2.8.0 (Rognes et al., 2016), and BLAST+ 2.7.1. After removing singletons, clustering and the assignment of centroid sequences to operational taxonomic units (OTU) was performed within each sample at a 98% identity threshold. Only those OTUs with a minimum of 100 sequences were retained. All raw fastq files were deposited to the NCBI sequence read archive under the accession number PRJNA557715. The nucleotide sequences for unique sequences obtained in this study have been deposited in GenBank under the accession numbers MN472766-MN472838.
2.4. Cloning and Sanger sequencing of novel sequences
To support novel sequence designations PCR products from samples containing novel sequences were purified using Exonuclease I/Shrimp Alkaline Phosphatase (Exo-SAP-IT™, USB Corporation, Cleveland, OH), and sequenced in both directions using primers utilized for PCR screening in 10 μl reactions, Big Dye™ chemistries, and an ABI 3130 sequencer analyzer (Applied Biosystems, Foster City, CA). Sequence chromatograms of each strand were aligned and examined with Lasergene software (DNASTAR, Inc., Madison, WI). As the sequence traces of these specimens had the appearance of a mixed infections, the SSU rDNA product was cloned using the TOPO TA cloning kit (Invitrogen Corp. Carlsbad, CA). Transformants were screened by PCR and sequenced in both direction using M13 forward and reverse primers per the sequencing protocol previously described. Up to 8 clones per specimen were sequenced. Sanger and NGS sequences were compared to confirm the identity of novel sequences.
2.5. Phylogenetic analysis
All Sanger sequences and OTUs were assigned a Blastocystis subtype based on the best match by BLAST search in the GenBank database. Novel subtypes were assigned when 5% sequence divergence from the SSU rRNA of known subtypes was observed (Clark et al., 2013). Sequences obtained in this study as well as nucleotide sequences from Blastocystis subtypes previously identified (Supplementary Table 1) were aligned with the Clustal W algorithm using MEGA X (Kumar et al., 2018). Phylogenetic analyses were made by the Neighbor-Joining (NJ) method of Saitou and Nei (Saitou and Nei, 1987), and genetic distance calculated with the Kimura 2-parameter model using MEGA X (Kumar et al., 2018).
3. Results
Sixteen (14.7%) of the 109 domestic and captive wild bird specimens examined were Blastocystis positive by PCR (Table 1). Using next generation amplicon sequencing, a total of 3,073,032 paired end reads were generated from the 16 positive samples with an average of 192,065 reads per sample. After end trimming, quality filtering, and pair merging 655,725 reads remained. The removal of chimeric sequences left 543,620 merged reads which were used for operational taxonomic unit (OTU) generation. Clustering generated 77 Blastocystis OTUs among the 16 samples, and of those 65 were unique sequences (Table 2, Table 3; Supplementary Table 2).
Table 2.
Blastocystis subtypes identified in positive samples by next generation amplicon sequencing.
| Bird ID | Common name | Location | Subtypes (% of sample) |
|---|---|---|---|
| 32 | Ostrich | Uberlândia | ST5(10)/ST10a(28.4)/ST24(61.6) |
| 33 | Ostrich | Uberlândia | ST5(100) |
| 36 | Indian Peafowl | Uberlândia | ST27(80.8)/ST28(19.2) |
| 38 | Garganey | Uberlândia | ST7a(79.3)/ST10(7.4)/ST27(8.6)/ST28(4.7) |
| 63 | Pheasant | Belo Horizonte | ST6a(100) |
| 98 | Swan Goose | Uberlândia | ST7a(100) |
| 101 | Mallard | Uberlândia | ST7a(100) |
| 102 | Helmeted guineafowl | Uberlândia | ST6(0.5)/ST7a(95.5) |
| 103 | Helmeted guineafowl | Uberlândia | ST6a(2.4)/ST7a(97.6) |
| 105 | Muscovy duck | Uberlândia | ST7a(84.7)/ST14(15.3) |
| 132 | Indian Peafowl | Belo Horizonte | ST27(98.7)/ST28(1.3) |
| 133 | Pheasant | Belo Horizonte | ST6(34.4)/ST7a(65.6) |
| 135 | Pheasant | Belo Horizonte | ST6a(95.1)/ST7a(4.9) |
| 158 | Pheasant | Belo Horizonte | ST7a(100) |
| 174 | Black-cheeked Lovebird | Uberlândia | ST14(100) |
| 208 | Quail | Uberlândia | ST6(2)/ST7a(98) |
Denotes intra-subtype variability.
Table 3.
Blastocystis subtypes identified in domestic and captive wild birds from Minas Gerais (Brazil) including number of samples for which each subtype was identified and number of unique operational taxonomic units (OTUs) among the subtypes identified using next generation sequencing.
| Subtype | No. positive samples | % of positives samples | No. of unique OTUs |
|---|---|---|---|
| ST5 | 2 | 12.5 | 2 |
| ST6 | 6 | 37.5 | 6 |
| ST7 | 10 | 62.5 | 48 |
| ST10 | 2 | 12.5 | 4 |
| ST14 | 2 | 12.5 | 2 |
| ST24 | 1 | 6.3 | 1 |
| ST27 | 3 | 18.8 | 1 |
| ST28 | 3 | 18.8 | 1 |
There were no positive samples from captive birds at either IBAMA or the School of Falconry. Only one bird sample collected from the Uberlândia aviary was positive for Blastocystis. Birds at markets were more frequently positive for Blastocystis, and 19.2% (15) of the 78 market birds were Blastocystis positive. Of the 11 bird orders sampled, four had Blastocystis positive specimens, Galliformes (9), Anseriformes (4), Struthioniformes (2), and Psittaciformes (1) (Table 1).
Eight subtypes were identified in Blastocystis positive birds, six previously reported subtypes ST5, ST6, ST7, ST10, ST14, and ST24 and two proposed novel subtypes named ST27 and ST28. Nucleotide sequences of novel subtypes were obtained both by NGS and Sanger sequencing and cloning. The sequence similarity between these samples and any existing subtyped nucleotide sequence available in the GenBank database was approximately 90%. ST27 branches within the ST6 and ST9 clade, and ST28 branches within the ST15 and ST17 clade (Fig. 1). Both novel subtypes were identified in 3 birds, an Indian peafowl (#36) and a garganey (#38) from Uberlândia, and an Indian peafowl (#132) from Belo Horizonte (Table 2).
Fig. 1.
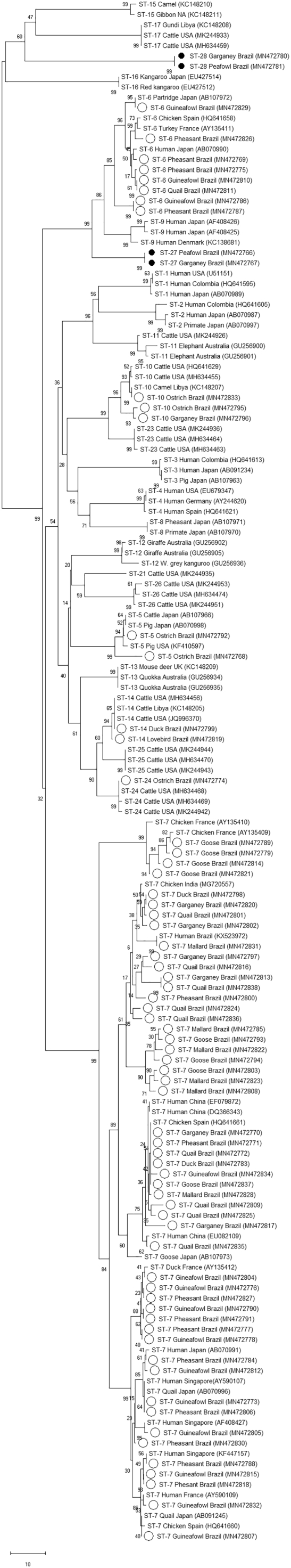
Phylogenetic relationships among Blastocystis subtypes identified in this study and known Blastocystis subtypes (Supplementary Table 1), was inferred by a neighbor-joining analysis of the SSU rRNA gene sequence, based on genetic distances calculated by the Kimura two-parameter model (MEGA X software). The analysis involved 149 nucleotide sequences. There was a total of 429 positions in the final dataset. Bootstrap proportions (in percent) are attached to the internal branches (1000 replicates). Reference sequences have host and location information with the GenBank accession number in parenthesis. Nucleotide sequences determined in this study are identified with circles before the subtype name. Black filled circles indicate novel subtypes.
Phylogenetic relationships among Blastocystis subtypes identified in this study and known Blastocystis subtypes (Supplementary Table 1), was inferred by a neighbor-joining analysis of the SSU rRNA gene sequence, based on genetic distances calculated by the Kimura two-parameter model (MEGA X software). The analysis involved 149 nucleotide sequences. There was a total of 429 positions in the final dataset. Bootstrap proportions (in percent) are attached to the internal branches (1000 replicates). Reference sequences have host and location information with the GenBank accession number in parenthesis. Nucleotide sequences determined in this study are identified with circles before the subtype name. Black filled circles indicate novel subtypes.
All birds with ST6, ST7, ST27 and ST28 infections belonged to orders Anseriformes and Galliformes (Table 1). Birds with ST14 infections belonged to orders Psittaciformes and Anseriformes while ST10 infections were identified in Anseriformes and Struthioniformes. ST5 and ST24 were only identified in order Struthioniformes.
The most frequently observed subtypes in this study were ST7 and ST6 found in 62.5% and 37.5% of positive samples, respectively (Table 1). Novel subtypes ST27 and ST28 were observed in 18.8% of positive samples and occurred as co-infections in all birds infected with these subtypes. ST5, ST10, and ST14 were observed in 12.5% of positive samples. ST24 was the only subtype observed in only a single sample.
Mixed subtype infections were observed in 62.5% of positive samples. Mixed ST6/ST7 infections were the most common mixed infections and were observed in 5 birds. Mixed infection with ST27/ST28 was observed in two birds. Combinations of ST5/ST10/ST24, ST7/ST10/ST27/ST28, and ST7/ST14 were all observed in single birds.
Sixty-five unique Blastocystis sequences were observed among the eight subtypes identified in this study. Most intra-subtype heterogeneity was found in ST7 which had 48 (74%) of the 65 unique sequences in this study (Table 3). ST6 had six unique sequences. ST10 had four unique sequences. ST14 and ST5 had two unique sequences. No intra-subtype heterogeneity was observed for ST24, ST27, or ST28, and these subtypes each had only one unique sequence (Table 3).
4. Discussion
Blastocystis is one of the most common parasites found in humans and many other animals worldwide, and wild and domestic birds have been identified as hosts of potentially zoonotic Blastocystis subtypes (Cian et al., 2017; Greige et al., 2018; Ramírez et al., 2014; Roberts et al., 2013; Tanizaki et al., 2005; Valença-Barbosa et al., 2019; Wang et al., 2018). Yet, molecular data on Blastocystis in birds in the Americas are limited. In this study, the subtypes of Blastocystis present in 11 orders and 38 species of both domestic and captive wild birds from Minas Gerais, Brazil were identified using PCR and next generation amplicon sequencing (NGS) to better understand the subtype distribution of Blastocystis in birds.
Of the 109 samples tested for Blastocystis, 14.7% (16) were positive. Of the 11 bird orders sampled, Blastocystis positive specimens were identified in orders Galliformes (9), Anseriformes (4), Struthioniformes (2), and Psittaciformes (1). It is noteworthy that with the exception of one tree bird all of the positive birds were ground and water birds. These findings could suggest that ground and water birds are more likely to be exposed to Blastocystis that tree birds, possibly because of a higher fecal exposure associated with their feeding habits for Galliformes and Struthioniformes, and with exposure to parasites present in surface waters for Anseriformes. However, tree birds were found to have a Blastocystis infection prevalence of 90% in a study of multiple Passeriformes species from Colombia (Ramírez et al., 2014). Therefore, it is also possible that other environmental factors play a role in the exposure and infection risk of birds.
The prevalence of Blastocystis in birds in this study (14.7%) is within the range of 2.2–34.6% that has been reported for Blastocystis in other studies from birds (Cordón et al., 2009; Bergamo Do Bomfim and Machado Do Couto, 2013; Cian et al., 2017; Roberts et al., 2013; Valença-Barbosa et al., 2019; Wang et al., 2018; Zhao et al., 2017). The only exception is the aforementioned study from wild bird species from Colombia which reported a prevalence of 90% (Ramírez et al., 2014). In a study of market birds from Brazil 34.6% were reported as Blastocystis positive using morphological diagnosis (Bergamo Do Bomfim and Machado Do Couto, 2013). In another study from Brazil which included both captive and wild birds, a Blastocystis prevalence of 21% was reported (Valença-Barbosa et al., 2019). Two studies of captive wild bird species in China found Blastocystis in 2.2% and 7% of samples (Wang et al., 2018; Zhao et al., 2017). A study of birds in French zoos found Blastocystis in 8.6% of samples birds (Cian et al., 2017). Blastocystis was reported in 23.4% of birds in a study of farm, bushland, and zoo birds from Australia (Roberts et al., 2013). Blastocystis was reported in 23.6% of birds examined in an ornithological garden in Spain (Cordón et al., 2009). While some of these studies were not designed to assess the true population level prevalence of Blastocystis in birds, it is clear from these reports that birds are frequently hosts of this parasite. It is also clear from these reports that the prevalence of Blastocystis varies widely and more work is needed to understand what factors may influence this variability.
ST6 and ST7 are the two subtypes most frequently identified in birds, however wild and domestic bird species have been reported to harbor other Blastocystis subtypes including ST1, ST2, ST4, ST5, ST6, ST7, ST8, ST10, and ST20 (Cian et al., 2017; Deng et al., 2019; Ramírez et al., 2014; Roberts et al., 2013; Valença-Barbosa et al., 2019; Wang et al., 2018; Zhao et al., 2017). Eight Blastocystis subtypes were identified in the birds in this study, ST5, ST6, ST7, ST10, ST14, ST24, and two novel subtypes with the proposed name designation of ST27 and ST28. ST5 which was identified in two ostriches in this study has been reported in ostriches from China, in ostriches and a greater rhea from French zoos, and in a chicken from Brazil (Cian et al., 2017; Valença-Barbosa et al., 2019; Zhao et al., 2017). The seemingly common occurrence of ST5 in ostriches and the ability of birds to host this subtype is noteworthy as ST5 has been found in both pigs and pig caretakers in China and Australia (Li et al., 2007; Wang et al., 2014; Yan et al., 2007). Thus, birds could also be considered a potential source of zoonotic ST5 infection. Furthermore, ostriches in this study were also positive for ST10 and ST24 and have been reported to host ST4, ST6, ST10, and ST20 in other studies indicating that ostriches may be host to both zoonotic and enzootic Blastocystis subtypes (Chandrasekaran et al., 2014; Roberts et al., 2013; Zhao et al., 2017). ST10, ST14, and ST24 which are most commonly reported in ruminants were identified in four samples in this study, ST10 and ST24 in an ostrich, ST10 in a garganey, and ST14 in a Muscovy duck and a black-cheeked lovebird, indicating that birds may be a source of transmission between livestock and wildlife (Fayer et al., 2012; Maloney et al., 2019a; Zhu et al., 2017). Whether subtypes common to ruminants can successfully colonize avian hosts remains to be demonstrated. The two novel subtypes, ST27 and ST28, were identified in three samples, two Indian peafowls and a garganey. These were the only subtypes identified in Indian peafowls in this study, and the only other molecular data from Indian peafowls reported an untypable subtype in a Indian peafowl from a French zoo (Cian et al., 2017). The sequence of this untypable subtype does not overlap with the region used for subtyping in this study, however like the ST28 described here it also branches with ST15 in a SSU rRNA gene cladogram (Cian et al., 2017). While limited, these data indicate that Indian peafowl may be the main host to these unique subtypes of Blastocystis.
Of the three potentially zoonotic subtypes identified in birds in this study, ST6 and ST7 were the most commonly found. In fact, one or both subytpes were identified in 68.8% (11) of the 16 Blastocystis positive samples. These subtypes have been reported in humans in South America and worldwide. ST6 and ST7 have been reported in humans from Colombia and Poland (Ramírez et al., 2016; Rudzińska et al., 2019). ST6 was reported in humans in Argentina, and ST7 was identified in children in Brazil (Oliveira-Arbex et al., 2018; Ramírez et al., 2016). The zoonotic transmission of ST6 was reported between chickens and humans working in a poultry slaughter house in Lebanon (Greige et al., 2018). As ST5, ST6, and ST7 were identified in birds in this and other studies, birds may serve as reservoirs of these subtypes and play a role in the transmission of these subtypes to humans. It should be noted that the four subtypes most prevalent in humans (ST1-ST4) were not observed in birds in this study, nor are these subtypes frequently reported in subtype surveys of avian hosts. However, as studies in birds are limited the role of birds as hosts and potential reservoirs of these subtypes remains to be fully elucidated.
Currently Blastocystis subtype designations are based solely on heterogeneity in the SSU rRNA gene. While heterogeneity in this gene is unlikely to directly translate to differences in host specificity or pathogenicity, intra-subtype variability could be an important factor influencing these infection outcomes. In this study, intra-subtype heterogeneity in the SSU rRNA gene was observed for ST5, ST6, ST7, ST10 and ST14, and multiple variants of ST6, ST7, and ST10 occurred in the same host (Table 2, Table 3). This variability was most pronounced in ST7 which had 48 unique sequences among the 10 ST7 positive samples. Heterogeneity in the SSU rRNA gene has been reported in the ST7 strain B isolate (Poirier et al., 2014). In fact, 17 non-identical copies of the gene were found in the whole genome sequence of this strain (Denoeud et al., 2011; Poirier et al., 2014). However, these non-identical copies were reported to have between 98.1 and 99.9% nucleotide sequence similarity (Poirier et al., 2014). As the similarity threshold for OTU cluster generation in the present study was 98%, it is unlikely that the amount of variability observed in ST7 is only due to copy level differences in the SSU rRNA gene of a single ST7 isolate. To date, no significant associations have been found between SSU rRNA gene sequence variants of a subtype and infection outcomes in a host. However, NGS represents a unique tool for exploring this type of variability since it provides greater depth of information about both intra and inter-subtype variability in an individual sample (Maloney et al., 2019b). Future studies utilizing this method may help to resolve some of these issues.
5. Conclusions
Although Blastocystis is commonly reported in both wild and domestic bird species worldwide, few molecular studies have sought to describe the subtype diversity of Blastocystis in birds. In this study the prevalence and subtype diversity of Blastocystis was described in domestic and captive wild birds from Brazil. Eight subtypes of Blastocystis, including three zoonotic and two novel subtypes, were identified. These results support the need for more research on the potential role of birds in the transmission of Blastocystis between humans and other domestic animals.
The following are the supplementary data related to this article.
Reference sequences for each Blastocystis subtype included in the alignment used to conduct phylogenetic analysis with their GenBank accession number, host, and location.
List of Blastocystis nucleotide sequences submitted to Genbank including for each information on subtype, host, and isolate identification name.
Acknowledgements
This study was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), process number 99999.009537/2014-07.
References
- Alfellani M.A., Taner-Mulla D., Jacob A.S., Imeede C.A., Yoshikawa H., Stensvold C.R., Clark C.G. Genetic diversity of Blastocystis in livestock and zoo animals. Protist. 2013;164:497–509. doi: 10.1016/j.protis.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Bergamo Do Bomfim T.C., Machado Do Couto M.C. Morphological diagnosis and occurrence of Blastocystis spp. obtained from the stool samples of domestic bird species commercialized in municipal markets. J. Parasitol. Vector Biol. 2013;5:20–26. [Google Scholar]
- Bushnell, B. 2014. BBMap download|SourceForge.net [WWW Document].
- Caradonna T., Marangi M., Del Chierico F., Ferrari N., Reddel S., Bracaglia G., Normanno G., Putignani L., Giangaspero A. Detection and prevalence of protozoan parasites in ready-to-eat packaged salads on sale in Italy. Food Microbiol. 2017;67:67–75. doi: 10.1016/j.fm.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran H., Kumar Govind S., Panchadcharam C., Bathmanaban P., Raman K., Thergarajan G. 2014. High Lipid Storage in Vacoular Forms of Subtype 6 Blastocystis sp. in ostrich. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cian, A., El Safadi, D., Osman, M., Moriniere, R., Gantois, N., Benamrouz-Vanneste, S., Delgado-Viscogliosi, P., Guyot, K., Li, L.-L., bastien Monchy, S., Noë, C., Poirier, P., line Nourrisson, C., Wawrzyniak, I., déric Delbac, F., phanie Bosc, S., Chabé, M., Petit, T., Certad, G., Viscogliosi, E., 2017. Molecular Epidemiology of Blastocystis sp. in Various Animal Groups From Two French Zoos and Evaluation of Potential Zoonotic Risk. 10.1371/journal.pone.0169659. [DOI] [PMC free article] [PubMed]
- Clark C.G., van der Giezen M., Alfellani M.A., Stensvold C.R. Recent developments in Blastocystis research. Adv. Parasitol. 2013;82:1–32. doi: 10.1016/B978-0-12-407706-5.00001-0. [DOI] [PubMed] [Google Scholar]
- Cordón G.P., Prados A.H., Romero D., Moreno M.S., Pontes A., Osuna A., Rosales M.J. Intestinal and haematic parasitism in the birds of the Almuñecar (Granada, Spain) ornithological garden. Vet. Parasitol. 2009;165(3–4):361–366. doi: 10.1016/j.vetpar.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Deng L., Yao J.X., Liu H.F., Zhou Z.Y., Chai Y.J., Wang W.Y., Zhong Z.J., Deng J.L., Ren Z.H., Fu H.L., Yan X., Yue C.J., Peng G.N. First report of Blastocystis in giant pandas, red pandas, and various bird species in Sichuan province, southwestern China. Int. J. Parasitol. Parasites Wildl. 2019;9:298–304. doi: 10.1016/j.ijppaw.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F., Roussel M., Noel B., Wawrzyniak I., da Silva C., Diogon M., Viscogliosi E., Brochier-Armanet C., Couloux A., Poulain J., Segurens B., Anthouard V., Texier C., Blot N., Poirier P., Ng G.C., Tan K.S., Artiguenave F., Jaillon O., Aury J.M., Delbac F., Wincker P., Vivarès C.P., El Alaoui H. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 2011;12:R29. doi: 10.1186/gb-2011-12-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayer R., Santin M., Macarisin D. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitol. Res. 2012;111:1349–1355. doi: 10.1007/s00436-012-2971-1. [DOI] [PubMed] [Google Scholar]
- Greige S., El Safadi D., Bécu N., Gantois N., Pereira B., Chabé M., Benamrouz-Vanneste S., Certad G., El Hage R., Chemaly M., Hamze M., Viscogliosi E. Prevalence and subtype distribution of Blastocystis sp. isolates from poultry in Lebanon and evidence of zoonotic potential. Parasites and Vectors. 2018:11. doi: 10.1186/s13071-018-2975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolören Z., Karaman Ü. Investigation of Blastocystis subspecies in water samples collected from Ordu province. Turkiye Parazitol Derg. 2019;43:111–117. doi: 10.4274/tpd.galenos.2019.5976. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K., Battistuzzi F.U. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-H., Zhou X.-N., Du Z.-W., Wang X.-Z., Wang L.-B., Jiang J.-Y., Yoshikawa H., Steinmann P., Utzinger J., Wu Z., Chen J.-X., Chen S.-H., Zhang L. Molecular epidemiology of human Blastocystis in a village in Yunnan province, China. Parasitol. Int. 2007;56:281–286. doi: 10.1016/j.parint.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Maloney J.G., Lombard J.E., Shivley C.B., Urie N.J., Santin M. Zoonotic and genetically diverse subtypes of Blastocystis in US pre-weaned dairy heifer calves. Parasitol. Res. 2019;118:575–582. doi: 10.1007/s00436-018-6149-3. [DOI] [PubMed] [Google Scholar]
- Maloney J.G., Molokin A., Santin M. Next generation amplicon sequencing improves detection of Blastocystis mixed subtype infections. Infect. Genet. Evol. 2019;73:119–125. doi: 10.1016/j.meegid.2019.04.013. [DOI] [PubMed] [Google Scholar]
- McLauchlin J., Pedraza-Díaz S., Amar-Hoetzeneder C., Nichols G.L. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 1999;37:3153–3158. doi: 10.1128/jcm.37.10.3153-3158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Arbex A.P., David É.B., Guimarães S. Blastocystis genetic diversity among children of low-income daycare center in Southeastern Brazil. Infect. Genet. Evol. 2018;57:59–63. doi: 10.1016/j.meegid.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Poirier P., Meloni D., Nourrisson C., Wawrzyniak I., Viscogliosi E., Livrelli V., Delbac F. Molecular subtyping of Blastocystis spp. using a new rDNA marker from the mitochondria-like organelle genome. Parasitology. 2014;141:670–681. doi: 10.1017/S0031182013001996. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Sánchez L.V., Bautista D.C., Corredor A.F., Flórez A.C., Stensvold C.R. Blastocystis subtypes detected in humans and animals from Colombia. Infect. Genet. Evol. 2014;22:223–228. doi: 10.1016/j.meegid.2013.07.020. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Sánchez A., Hernández C., Flórez C., Bernal M.C., Giraldo J.C., Reyes P., López M.C., García L., Cooper P.J., Vicuña Y., Mongi F., Casero R.D. Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. 2016;41:32–35. doi: 10.1016/j.meegid.2016.03.017. [DOI] [PubMed] [Google Scholar]
- Roberts T., Stark D., Harkness J., Ellis J. Subtype distribution of Blastocystis isolates from a variety of animals from New South Wales, Australia. Vet. Parasitol. 2013;196:85–89. doi: 10.1016/j.vetpar.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4 doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudzińska M., Kowalewska B., Wąż P., Sikorska K., Szostakowska B. Blastocystis subtypes isolated from travelers and non-travelers from the north of Poland – a single center study. Infect. Genet. Evol. 2019;75:103926. doi: 10.1016/j.meegid.2019.103926. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Santín M., Gómez-Muñoz M.T., Solano-Aguilar G., Fayer R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol. Res. 2011;109:205–212. doi: 10.1007/s00436-010-2244-9. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Clark C.G. Current status of Blastocystis: a personal view. Parasitol. Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Tan K.S.W. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008;21:639–665. doi: 10.1128/CMR.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizaki A., Yoshikawa H., Iwatani S., Kimata I. Infectivity of Blastocystis isolates from chickens, quails and geese in chickens. Parasitol. Res. 2005;96:57–61. doi: 10.1007/s00436-005-1326-6. [DOI] [PubMed] [Google Scholar]
- Valença-Barbosa C., Do Bomfim T.C.B., Teixeira B.R., Gentile R., Da Costa Neto S.F., Magalhães B.S.N., De Almeida Balthazar D., Da Silva F.A., Biot R., D'Avila Levy C.M., Santos H.L.C. Molecular epidemiology of Blastocystis isolated from animals in the state of Rio de Janeiro, Brazil. PLoS One. 2019;14 doi: 10.1371/journal.pone.0210740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Owen H., Traub R.J., Cuttell L., Inpankaew T., Bielefeldt-Ohmann H. Molecular epidemiology of Blastocystis in pigs and their in-contact humans in Southeast Queensland, Australia, and Cambodia. Vet. Parasitol. 2014;203:264–269. doi: 10.1016/j.vetpar.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Wang J., Gong B., Liu X., Zhao W., Bu T., Zhang W., Liu A., Yang F. Distribution and genetic diversity of Blastocystis subtypes in various mammal and bird species in northeastern China. Parasites and Vectors. 2018;11 doi: 10.1186/s13071-018-3106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Su S., Ye J., Lai X., Lai R., Liao H., Chen G., Zhang R., Hou Z., Luo X. Blastocystis sp. subtype 5: a possibly zoonotic genotype. Parasitol. Res. 2007;101:1527–1532. doi: 10.1007/s00436-007-0672-y. [DOI] [PubMed] [Google Scholar]
- Zahedi A., Greay T.L., Paparini A., Linge K.L., Joll C.A., Ryan U.M. Identification of eukaryotic microorganisms with 18S rRNA next-generation sequencing in wastewater treatment plants, with a more targeted NGS approach required for Cryptosporidium detection. Water Res. 2019;158:301–312. doi: 10.1016/j.watres.2019.04.041. [DOI] [PubMed] [Google Scholar]
- Zhao G.H., Hu X.F., Liu T.L., Hu R.S., Yu Z.Q., Yang W.B., Wu Y.L., Yu S.K., Song J.K. Molecular characterization of Blastocystis sp. in captive wild animals in Qinling Mountains. Parasitol. Res. 2017;116:2327–2333. doi: 10.1007/s00436-017-5506-y. [DOI] [PubMed] [Google Scholar]
- Zhu W., Tao W., Gong B., Yang H., Li Y., Song M., Lu Y., Li W. First report of Blastocystis infections in cattle in China. Vet. Parasitol. 2017;246:38–42. doi: 10.1016/j.vetpar.2017.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reference sequences for each Blastocystis subtype included in the alignment used to conduct phylogenetic analysis with their GenBank accession number, host, and location.
List of Blastocystis nucleotide sequences submitted to Genbank including for each information on subtype, host, and isolate identification name.


