Abstract
Endolysosome entrapment is one of the key barriers to the therapeutic use of biologic drugs that act intracellularly. The screening of prospective nanoscale endosome-disrupting delivery technologies is currently limited by methods that are indirect and cumbersome. Here, we statistically validate Galectin 8 (Gal8) intracellular tracking as a superior approach that is direct, quantitative, and predictive of therapeutic cargo intracellular bioactivity through in vitro high-throughput screening and in vivo validation. Gal8 is a cytosolically dispersed protein that, when endosomes are disrupted, redistributes by binding to glycosylation moieties selectively located on the inner face of endosomal membranes. The quantitative redistribution of a Gal8 fluorescent fusion protein from the cytosol into endosomes is demonstrated as a real-time, livecell assessment of endosomal integrity that does not require labeling or modification of either the carrier or the biologic drug and that allows quantitative distinction between closely related, endosome-disruptive drug carriers. Through screening two families of siRNA polymeric carrier compositions at varying dosages, we show that Gal8 endosomal recruitment correlates strongly (r = 0.95 and p < 10−4) with intracellular siRNA bioactivity. Through this screen, we gathered insights into how composition and molecular weight affect endosome disruption activity of poly[(ethylene glycol)-b-[(2-(dimethylamino)ethyl methacrylate)-co-(butyl methacrylate)]] [PEG-(DMAEMA-co-BMA)] siRNA delivery systems. Additional studies showed that Gal8 recruitment predicts intracellular bioactivity better than current standard methods such as Lysotracker colocalization (r = 0.35, not significant), pH-dependent hemolysis (not significant), or cellular uptake (r = 0.73 and p < 10−3). Importantly, the Gal8 recruitment method is also amenable to fully objective high-throughput screening using automated image acquisition and quantitative image analysis, with a robust estimated Z′ of 0.6 (whereas assays with Z′ > 0 have high-throughput screening utility). Finally, we also provide measurements of in vivo endosomal disruption based on Gal8 visualization (p < 0.03) of a nanocarrier formulation confirmed to produce significant cytosolic delivery and bioactivity of siRNA within tumors (p < 0.02). In sum, this report establishes the utility of Gal8 subcellular tracking for the rapid optimization and high-throughput screening of the endosome disruption potency of intracellular delivery technologies.
Keywords: endosomal escape, high-throughput screening, siRNA delivery, pH-responsive drug carriers
Graphical Abstract:

Intracellular delivery of biomacromolecules, such as proteins, peptides, and nucleic acids like mRNA, antisense oligodeoxynucleotides (ASO/ODN), miRNA, and siRNA, has high but relatively unrealized therapeutic potential. With the exception of vaccines, there have been only six U.S. Food and Drug Administration approved biomacromolecular drugs with intracellular mechanisms of action to date: five antisense or splice-modifying ODNs1,2 and one siRNA lipid nanoparticle formulation.3 Beyond the systemic pharmacokinetic barriers facing biomacromolecular drugs,4 the primary barrier to intracellular bioavailability is endolysosomal sequestration and degradation following endocytosis.5,6 Thus, efficient endosome-disrupting drug carrier formulations, often leveraging the progressive acidification that occurs as endosomes mature, are highly sought as research tools and for enabling clinically successful intracellular-acting biomacromolecular therapeutics.7 Most work in this field leverages lipids, polymers, viruses, cell-penetrating peptides, or photothermal energy to achieve endosome disruption through pH-sensitive conformation and solubility changes, counter-ion-induced osmotic swelling, thermally induced endosomal disruption, or pH-triggered unmasking of hydrophobic elements that disrupt endosomal lipid bilayer membranes.8–13
Despite numerous papers optimizing carrier-mediated drug delivery, methods to directly measure endosomal disruption are elusive. Quantitative Galectin-8 (Gal8) tracking in live cells overcomes multiple shortcomings of alternate experimental methods. This work quantitatively compares the measurement of Gal8 recruitment to other commonly used assays and establishes that it is sensitive and high-throughput. Combinatorial polymer and lipid chemistry is commonly used to create large libraries, and this assay is amenable to adoption as a component of the high-throughput screening pipeline for parallel measurement of the endosome disruptive potency of candidate formulations.
Although endosomal escape efficiency is a key characteristic of intracellular carriers, it is a relatively rare event14,15 that is difficult to directly measure, especially in a high-throughput capacity. The most commonly used approaches do not measure endosomal membrane integrity, instead relying on tracking pH-sensitive fluorophores attached to the biologic cargo or imaging colocalization of fluorescently labeled cargo with acidotropic dyes (e.g., Lysotracker) that accumulate in endolysosomal compartments.16–20 One limitation of these approaches is that sensitivity limitations of microscopy often require supra-therapeutic dosing for robust colocalization imaging analyses.21 Moreover, dependency on fluorophores (often bulky, hydrophobic organic dyes) is, in general, not ideal because they can alter cellular uptake and intracellular trafficking of the cargo,22 are subject to photobleaching, can be quenched when densely packed into a carrier system,23 and are susceptible to pH- and solvation-dependent fluctuations in quantum efficiency.22,24 Furthermore, many pH-responsive, endosomolytic carriers purposefully incorporate basic moieties that buffer pH in the endolysosomal environments.18,25,26 With these “proton sponge” formulations, carrier-mediated pH buffering can potentially alter signal from pH-sensitive and acidotropic fluorophores independent of whether the endosome has been physically disrupted. The use of a fluid-phase tracer (e.g., fluorescent dextrans) can also be used to track endosomal disruption; however, the endocytosis and trafficking of these tracer macromolecules may not match the biologic drug in question. For instance, it has been noted that there are differences between cytosolic release of calcein, a small fluid-phase tracer and fluorescent oligonucleotides loaded into polyplexes that traffic to the nucleus.15 Rather than measuring lack of colocalization with endosomes, another relevant approach for some classes of cargo is to measure colocalization with the ultimately targeted intracellular compartment (for example, the nucleus for plasmids or ODNs).27 Furthermore, trafficking studies using microscopy are ideally performed on live cells because fixation artifacts for biologic drugs such as peptides and nucleic acids can be misleading.28 Additionally, fluorescent microscopists are often biased toward choosing exposure and other image settings that detect the brightest areas of a cell (endosomes), which may obscure low-level cytosolic fluorescence. A pivotal study by Gilleron et al. showed that as little as 2% of siRNA cargo escaping endosomes can cause maximal gene silencing; therefore, functionally significant escape may be associated with cytosolic concentrations of molecules below the lower limit of typical fluorescent microscopic detection, especially under settings optimized for visualization of cargo-packed endolysosomal vesicles.14
The two methods that provide robust confirmation of endosomal disruption are transmission electron microscopy (TEM) and cellular fractionation, but these methods are not amenable to rapid, high-throughput analysis. TEM facilitated by electron dense labels (e.g., gold) provides a robust way to assess endosomal membrane integrity and escape, but TEM samples preparation is extremely time-consuming, cannot be obtained on live cells, provides only a very small field of view per imaging field, and can be difficult to interpret for nonexperts.14,29 Cell fractionation can be performed in two formats. In a semipermeabilization method, carefully timed weak detergents are applied to lyse the plasma membrane but not endolysosomal membranes. Cytosolic components are extracted, and the cytosolic drug is compared to the organelle-bound drug.30,31 In the mechanical separation method, cells are mechanically homogenized and separated by gradient ultracentrifugation, and the drug is quantified in each fraction.32,33 Both types of cell fractionation require experimental validation to identify and assess purity of extracted fractions, typically by Western blot, and require modification of the drug with radiolabels or fluorophores for quantification purposes. Both TEM and cellular fractionation can provide robust readouts but are labor intensive and low-through-put.29,30
To develop a live-cell, high-throughput-amenable endosomal escape screening assay that does not require chemical modification of the carrier or cargo, we sought to develop and validate Gal8 recruitment for direct visualization of endosomal disruption by drug carriers. Gal8 is a β-galactoside carbohydrate-binding protein described by Hadari et al. and expressed in a variety of tissues.34 Thurston et al. subsequently discovered the role of Gal8 in innate immunity, in which it functions to detect disrupted endosomes due to high-affinity binding with glycans selectively found on the inner leaflet of endosomal membranes.35 More recently, groups have used Galectin reporters and high-speed microscopy to track adenoviral entry into cells, showing that viral entry to cytosol is preceded by Galectin-3 redistribution.36 Wittrup et al. reported that the appearance of Gal8 positive spots temporally coincides with the cytosolic delivery of fluorescently labeled siRNA from lipid nanoparticles,37 and we reported the use of Gal8 recruitment to quantitatively assess kinetics of polymer-mediated endosome disruption for intracellular peptide delivery.29
Here, we focused on establishing quantitative correlations between carrier-mediated Gal8 recruitment levels and intracellular biologic cargo bioavailability and activity. Establishing the range and nature of this correlation is vital for verifying whether Gal8 recruitment imaging can be utilized to predict intracellular bioavailability of carrier formulations. Similar high-throughput and automated microscopy methods are already used in the high-throughput screening of compounds, genes, RNAi, and CRISPR-mediated gene knockouts.38–42 Establishing a suitable assay for assessing endosome disruption could become a powerful component of the pipeline for screening and understanding the structure-function properties among combinatorial chemistry-derived libraries of endosome-disrupting polymers and lipids, a highly popular carrier optimization approach.43–46 Herein, we screen level of Gal8 recruitment for two series of siRNA formulations (a composition series and a molecular weight, MW, series) of rationally designed polymers applied at different doses. This small library contains varied formulations with a range from negligible to very powerful pH-dependent endosome disruptive activity. This study design allows us to both validate the correlation between Gal8 imaging and intracellular cargo bioavailability (based on a luciferase knockdown readout) and also provides insights on how composition and molecular weight (MW) impact endosome disruption for our library. We also seek to compare the predictive power of the Gal8 recruitment imaging as a measure of intracellular bioavailability to other standard methods such as pH-dependent hemolysis profiling, Lysotracker colocalization analysis, and measurements of total cellular internalization. Finally, we also provide a proof of concept study for application of Gal8 recruitment imaging to visualize carrier-mediated endosome escape in vivo in an orthotopic breast tumor model. Collectively, these experiments and statistical analysis are designed to validate the use of Gal8 recruitment as a platform to rapidly screen and optimize intracellular drug delivery systems.
RESULTS AND DISCUSSION
Polymer Library Synthesis and Chemical Characterization.
To validate Gal8 recruitment as a predictor of intracellular bioavailability, we synthesized two series of polymers of varied composition and molecular weight to develop a library with widely varied endosome disruption potency. Reversible addition–fragmentation chain transfer (RAFT) was used to polymerize two series of diblock poly[(ethylene glycol)-b-[(2-(dimethylamino)ethyl methacry-late)-co-(butyl methacrylate)]] [PEG-(DMAEMA-co-BMA)] polymers (Figure S1). These polymers (general structure shown in Figure 1A) are composed of a constant 5000 g/mol PEG nanopolyplex (NP) corona-forming block and a second NP core-forming block of copolymerized DMAEMA (siRNA complexing, pH-responsive, and cationic) and BMA (core-stabilizing, hydrophobic, membrane interactive). This series of polymers extends recent work looking at siRNA NPs (si-NPs) with balanced cationic and hydrophobic core compostion20,43,47–50 and allows us to systematically examine the functional significance of the p(DMAEMA-co-BMA) block molecular weight, which our group has not previously studied.
Figure 1.
Polymer library construction and characterization. (A) Generalized structure for PEG-b-(DMAEMA-co-BMA) polymers. (B) The composition library consists of six polymers, ranging from 0% BMA to 75% BMA. Each polymer is composed of a 5000 g/mol PEG block and a second random copolymer block with an average MW of approximately 22 000 g/mol. (C) The MW library consists of seven polymers. Each contains a 5000 g/mol PEG block and a second random copolymer block composed of an approximately 50:50 ratio of DMAEMA to BMA monomers and with MW ranging from 11 300 to 47 500 g/mol. (D) Gel permeation chromatography traces show that all polymers in the composition library have similar, low polydispersity and approximately equivalent MW. (E) Gel permeation chromatography traces show that all the candidates in the MW library have well-controlled polydispersity and have a wide range of MW values. (F) Plot of log acid dissociation constant (pKa) shows that composition significantly impacts pKa; 75B is omitted due to insolubility under assay conditions; line of best fit: pKa = 7.36–0.019 x (x expressed in percent BMA). (G) Plot of log acid dissociation constant (pKa shows that MW has only a modest effect on pKa; line of best fit: pKa = 6.71 – (9.6 × 10−6) x (x expressed in grams per mole).
The first set of polymers with varied p(DMAEMA-co-BMA) block composition was designed to investigate the effects of the ratio of hydrophobe to cation on endosomal escape (Figure 1B and Table 1). These polymers consisted of constant si-NP corona-forming block (5000 g/mol PEG) and constant si-NP core-forming block molecular weight (average of 22 000 g/mol) but varied mole percentages (mol %) of hydrophobic BMA and cationic DMAEMA monomers ranging from 0 to 75 mol % BMA (Table 1). The second set of polymers was synthesized with a constant ratio of DMAEMA to BMA (50:50) but with varied total molecular weight of the p(DMAEMA-co-BMA) block (Figure 1C and Table 2). The MW library consisted of 7 polymers synthesized from an equimolar feed ratio of BMA and DMAEMA with total PEG-(DMAEMA-co-BMA) diblock polymer molecular weights ranging from 16 300 to 52 500 g/mol, named 50B-S, 50B-M, 50B-L, 50B-XL, 50B-2XL, 50B-3XL, and 50B-4XL. As a result of the RAFT-controlled free radical polymerization method, all resultant polymers have low polydispersity indices and controlled molecular weights as measured by gel permeation chromatography (Figure 1D,E), with expected compositions as measured by 1H nuclear magnetic resonance (NMR; Tables 1 and 2).
Table 1.
Polymer Characteristics in the Composition Library
| name | diblock MW (NMR)a | units of DMAEMA | units of BMA | DMAEMA content (%) | BMA content (%) |
|---|---|---|---|---|---|
| 0B | 28 600 | 150 | 0 | 100.0 | 0.0 |
| 25B | 29 000 | 118 | 38 | 75.6 | 24.4 |
| 40B | 28 300 | 93 | 61 | 60.4 | 39.6 |
| 50B | 26 100 | 71 | 70 | 50.4 | 49.6 |
| 60B | 26 500 | 60 | 85 | 41.1 | 58.9 |
| 75B | 24 400 | 34 | 99 | 25.4 | 74.6 |
Diblock MW refers to number-average total polymer molecular weight (Mn), including 5000 g/mol PEG “corona” block, in addition to the “core” p(DMAEMA-co-BMA) block.
Table 2.
Polymer Characteristics in the 50B MW Library
| name | diblock MW (NMR)a | units of DMAEMA | units of BMA | DMAEMA content (%) | BMA Content (%) |
|---|---|---|---|---|---|
| 50B-S | 16 300 | 39 | 36 | 52.0 | 48.0 |
| 50B-M | 23 000 | 63 | 57 | 52.5 | 47.5 |
| 50B-L | 27 500 | 75 | 75 | 50.0 | 50.0 |
| 50B-XL | 31 900 | 90 | 90 | 50.0 | 50.0 |
| 50B-2XL | 36 300 | 105 | 104 | 50.2 | 49.8 |
| 50B-3XL | 45 200 | 137 | 131 | 51.1 | 48.9 |
| 50B-4XL | 52 500 | 160 | 157 | 50.5 | 49.5 |
Diblock MW refers to number average total polymer molecular weight (Mn), including 5000 g/mol PEG “corona” block, in addition to the “core” p(DMAEMA-co-BMA) block.
To elucidate the relative effects of polymer composition on pH-dependent behavior, the logarithmic acid dissociation constant (pKa) was measured for all polymers via acid–base titration (Figure 1F,G). Composition had a strong effect on pKa, with increasing hydrophobic BMA mol % reducing the pKa of the protonatable amines on DMAEMA by 18.9 × 10−3 units per additional percent BMA (Figure 1F). In the MW library, core block molecular weight had only a modest effect on pKa with increasing molecular weights decreasing the pKa by 9.6 × 10−3 units per 103 g/mol (Figure 1G).
Composition Library Cytotoxicity and Gene Silencing Bioactivity.
The composition polymer library was screened for cytotoxicity and model gene knockdown in MDA-MB-231 cells that stably expressed firefly luciferase under a constitutive promoter. The successful delivery of siRNA targeting luciferase reduces the bioluminescence of luciferin treated cells, and data are normalized to bioluminescence of cells treated with dose-matched formulations loaded with a negative control, nontargeting siRNA. The 25B and 40B polymers formulated into si-NPs loaded with negative control nucleic acids were both significantly cytotoxic, while other compositions did not create cytotoxicity (Figure 2A). This is consistent with pH-dependent hemolysis data shown subsequently in this report that polymers that are membrane-disruptive at physiologic pH are generally cytotoxic. Without sufficient hydrophobicity to drive NP assembly and stability, the hydrophobic BMA-containing block becomes exposed to and disrupts outer cell membranes. The si-NPs should be finely tuned to become destabilized and expose the core block only under low-pH conditions in the endosomal environmental to avoid nonspecific cell-membrane-disruption-mediated cytotoxicity. In other words, the polymers containing lower percentages of BMA are sufficiently hydrophobic to interact with and disrupt lipid bilayer membranes but contain insufficient hydrophobic content to stably assemble (and block DMAMA-co-BMA block outer cell membrane interactions) at physiologic, extracellular pH.
Figure 2.
Composition library screening: visualization of Gal8 recruitment correlates with the most bioactive (50B) polymeric carrier composition. (A) Cytotoxicity of formulations loaded with a negative control siRNA (100 nM siRNA). Viability levels were normalized to phosphate-buffered saline (PBS)-treated cells; 25B and 40B caused statistically significant cytotoxicity. Overall p < 0.001 by ANOVA; single asterisks indicate significant toxicity by Dunnett’s post hoc testing vs PBS. (B) Bioactivity based on the silencing of luciferase activity (luciferase signal in cells treated with 100 nM luciferase siRNA normalized to cells treated with negative control siRNA with the same formulation. Overall p < 0.0001 by ANOVA; single asterisks indicate significant gene silencing by Dunnett’s post hoc testing vs PBS. (C) Schematic of intracellular Gal8 recruitment; in cells with intact endosomes, Gal8 is dispersed in the cytoplasm without access to intraendosomal glycans (blue circles). When endosomal membranes are disrupted, Gal8-YFP binds to these glycans, concentrating into bright, punctated fluorescent spots. (D) Gal8 confocal micrographs of nontoxic polymers confirms 50B induces robust Gal8 response (white arrow). Negligible Gal8 recruitment is detected for other polymeric carriers.
In the luciferase model gene knockdown studies, 25B, 40B, and 50B polymers showed significant gene silencing activity using a 100 nM siRNA dose (Figure 2B). This shows that 50B, containing equimolar cationic DMAEMA and hydrophobic BMA, is the only polymer of this library that is nontoxic and bioactive, confirming earlier studies.20 We carried forward the nontoxic formulations (0B, 50B, 60B, and 75B) into Gal8 recruitment experiments and hypothesized that, because it showed potent gene silencing, the 50B formulation would show the greatest Gal8 recruitment, as measured qualitatively by the appearance of punctate fluorescent spots (Figure 2C). These initial experiments had nearly binary results. No Gal8 recruitment was observed for 0B or 75B, while a minimal level of Gal8 recruitment was seen for 60B. Robust Gal8 recruitment, visualized as disappearance of diffuse cellular fluorescence and appearance of bright, punctate fluorescent spots was observed for 50B (Figure 2D). These initial experiments validated 50B as the best composition for the basis of a second library with varied molecular weight due to its combination of minimal toxicity and efficient siRNA delivery, which correlated with gene knockdown data. The desirable results produced using polymers with an equally balanced ratio of DMAEMA to BMA are consistent with several prior in vitro and in vivo experiments.20,47–50
MW Library Cytotoxicity and Gene Silencing Bioactivity.
We next sought to screen a broader series of 50B polymers with varied p(DMAEMA-co-BMA) block molecular weight because our previous, undocumented observations had suggested that the molecular weight of this block is an important driver of intracellular delivery, but this variable has not been carefully studied. Therefore, we synthesized a well-defined library of 50B polymers over a broad molecular weight range (Table 2) with the intention of yielding a series of NP formulations with varied levels of activity that would enable a robust Gal8 recruitment versus bioactivity correlation analyses. We characterized this polymer library for toxicity at a range of siRNA doses using the highly sensitive CellTiter Glo assay. It was found that no polymers were toxic at the lowest dose of 12.5 nM siRNA, while there was minor but statistically significant cytotoxicity at the highest polymer doses for the two largest MW p(DMAEMA-co-BMA) core blocks (Figure S2). A pair of polymers showed statistically significant toxicity; 50B-3XL exhibited 85% viability at 50 nM siRNA, and 50B-4XL exhibited 76% and 86% viability at 50 and 25 nM siRNA, respectively.
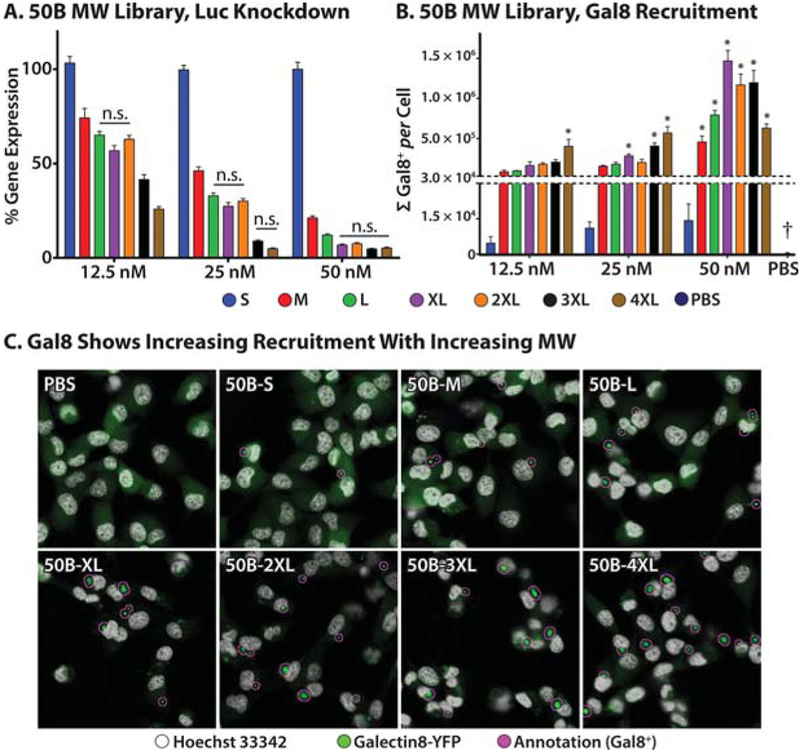
We then characterized this polymer library for luciferase knockdown activity relative to scrambled controls in luciferase expressing MDA-MB-231 cells. All polymers except 50B-S showed statistically significant gene silencing (Figure 3A). Because we had observed that the knockdown effect was saturated for several polymers at the 50 nM dose, we extended this study to assess lower doses. At the lowest dose of 12.5 nM, we could more sensitively detect the correlation between knockdown activity and molecular weight, with the largest-molecular-weight 50B-4XL formulation producing 74% reduction in luciferase activity at this relatively low dose. These data were statistically significant for polymer molecular weight, dose, and interaction thereof when tested by 2-way ANOVA (p < 10−4, all), with polymer molecular weight accounting for 69% of statistical variation. These data confirm that the MW polymer library provides a solid foundation for the correlation of endosomal escape and bioactivity across a series of polymers with structural similarity but varied levels of bioactive cargo delivery.
Figure 3.
MW library screening: gene knockdown level and Gal8 recruitment correlate. (A) Dose-dependent bioactivity for MW library. Overall p < 0.0001 for MW (59% of variation), dose (24%), and interaction (6%) by two-way ANOVA. For each dose, all polymers are statistically different from all other polymers by Tukey’s multiple comparisons test except within groups denoted “n.s.”. (B) Quantification of Gal8 recruitment confocal images reveals that Gal8 recruitment increases with increasing siRNA dose and increasing polymer MW. PBS-treated cells had near-zero response, highlighted with a dagger. Overall p < 0.0001 for MW (32% of variation), dose (35%), and interaction (25%). Single asterisks indicate polymers identified as significant hits by Dunnet’s comparison test. (C) Representative confocal micrographs of Gal8 recruitment at 50 nM siRNA dose. The images have been false-colored; white represents Hoechst staining; green represents Gal8; magenta represents automated annotation of Gal8 positive fluorescent spots.
Validation of Gal8 Recruitment Assay Using the MW Library.
To produce sufficient data points to assess the correlation between Gal8 recruitment and functional biomacromolecule delivery, Gal8 recruitment (Figure 3B) was assayed and quantified for all si-NPs at multiple doses. To robustly and objectively quantify Gal8 recruitment, automated methods were developed using MATLAB to identify and quantify Gal8-positive spots normalized to total cell number within each imaging frame. Images of recruited, punctate Gal8 spots were obtained for the MW library (Figure 3C), which were automatically annotated (Figure 3C, magenta) using MATLAB. Image processing algorithms are detailed in the Supporting Information and in Figure S3.
All 50B MW series polymers except 50B-S produced striking Gal8 recruitment. Increasing polymer MW increased Gal8 recruitment, with larger-MW polymers producing larger degrees of Gal8 recruitment at each dose tested. These data were statistically significant for polymer molecular weight, dose, and interaction thereof when tested by 2-way ANOVA (p < 10−4, all), with polymer molecular weight, dose, and interaction accounting for 32, 36, and 26% of statistical variation, respectively, suggesting that this method is more sensitive to polymer dosing than even gene-knockdown as a readout.
To assess the suitability of this assay, the signal-to-noise ratio (S/N), signal-to-background ratio (S/B), and Z-factor score51 were calculated using the highest response data (50 nM, 50B-XL) as an estimated reference positive control. S/N was calculated to be 2840, S/B to be 6078, and Z′ factor to be 0.61. In describeing the Z′ factor for assessing the quality of high-throughput screening (HTS), Zhang et al describe a value of Z′ ≥ 0.5 as “an excellent assay,” with a high band of separation between the negative control data and identified hits, whereas any assay with a positive Z′ factor “can be used for HTS”. In terms of user setup, the data in Figure 3B can notably be contained on a one-well plate, and by leveraging automated fluorescent confocal microscopy, these measurements can be acquired in less than 1 h of microscope time.
Cross-Validation against Standard Methods.
We next cross-validated the ability of Gal8 recruitment to serve as a predictor of intracellular bioactivity relative to other widely used standard endosomal-escape and drug-delivery characterization assays. A robust statistical cross-validation allows the direct comparison of the sensitivity of Gal8 recruitment against other methods of measuring endosomal escape. The 50B system is a well-suited polymer system for validating this technique because it has been well-characterized previously20,43,47–50 and is here characterized with varying molecular weights.
Hemolysis.
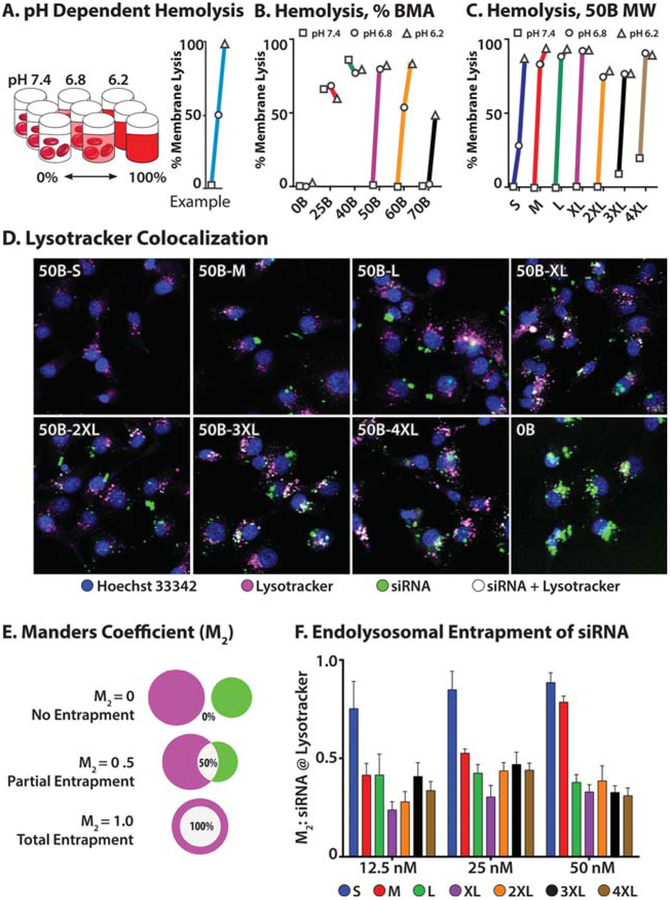
To assess pH-dependent membrane disruptive behavior, we used a hemolysis assay, which is commonly employed as a surrogate marker for pH-dependent endosome membrane disruption.52 Washed human red blood cells were incubated with si-NPs in buffer at pH values of 7.4, 6.8, and 6.2, corresponding to physiologic pH and progressively acidifying early endosomes. Intact red blood cells were removed by centrifugation, and the released hemoglobin within the supernatant was measured by absorbance (Figure 4A). As a control, we repeated hemolysis of the composition library (Figure 4B). No hemolysis was observed for purely cationic 0B without any hydrophobic BMA incorporation, whereas 25B and 40B exhibit hemolysis at all pH, consistent with their observed high cytotoxicity (Figure 2A). Only 50B exhibits no hemolysis at pH 7.4 and high hemolysis at pH 6.8. This “switch-like” hemolysis has been hypothesized to be predictive of bioactivity.30,52,53 Hemolysis results from the MW library showed that the smallest-MW polymer, 50B-S, produced weak hemolysis at pH 6.8, which may explain its low bioactivity throughout these studies. The largest two polymers had modest hemolysis at pH 7.4, again in agreement with the observation of mild cytotoxicity for 50B-3XL and 4XL at higher doses (Figure S2). All 50B polymers produced switch-like, pH-dependent behavior, with only minor differences in the hemolysis profiles for 50B-M through 50B-2XL, corresponding to MW from 23 000 to 36 300 g/mol. Taken together, these data suggest that overall pKa, relative hydrophobic content, and molecular weight all cooperate to dictate polymer pH-dependent membrane disruptive activity as measured by the hemolysis assay. A pair of primary conclusions from these data are that switch-like hemolysis profiles (i.e., low hemolysis at pH 7.4 with high hemolysis at pH 6.8) are necessary for endosomal disruption and that hemolysis at pH 7.4 is predictive of cytotoxicity. Hemolysis is useful for ascertaining crude pH-responsive profiles and for the prediction of cytotoxicity. However, desirable performance by hemolysis assay does not necessarily predict potent bioactivity because it does not have the sensitivity to discriminate differences in the 50B MW library that have a broad range of bioactivity levels. In fact, hemolysis at pH 7.4 (which is also indicative of cytotoxicity) was the only hemolysis outcome with statistically significant correlation to bioactivity (Supplemental Table 1).
Figure 4.
MW library pH-dependent hemolysis and Lysotracker colocalization. (A) Overview of red blood cell hemolysis assay; red blood cells are incubated with si-NPs at pH 7.4, 6.8, or 6.2. Intact red blood cells are separated by centrifugation; the relative amount hemoglobin released from disrupted red blood cells is measured in the supernatant by absorbance. (B) Hemolysis profiles for composition series reveal switch-like hemolysis profiles for 50B from pH 7.4 to 6.8, whereas 60B and 75B require increasingly acidic conditions for hemolysis. No lysis was observed for 0B, and 25B and 40B caused hemolysis at all pHs. pH legend: □, 7.4; ○, 6.8; △, 6.2. (C) Hemolysis profiles for MW series reveal switch-like hemolysis profiles for all 50B polymers; however, 50B-S has reduced hemolysis at pH 6.8, while 50B-3XL and 50B-4XL have observable hemolysis at pH 7.4. pH legend: □, 7.4; ○, 6.8; △, 6.2. (D) Representative false-color confocal micrographs of lysotracker microscopy at 50 nM siRNA dose; blue represents Hoechst nuclear staining, magenta represents Lysotracker staining, green represents Alexa-488-siRNA, and white represents colocalization of siRNA with Lysotracker. (E) Calculation of Manders coefficient of colocalization overlap; where siRNA and Lysotracker are not colocalized, a value of 0 is obtained, but complete colocalization results in a value of 1.0. (F) Quantification of siRNA colocalization with Lysotracker reveals that the majority of siRNA delivered using 50B-S is localized to endolysosomes, whereas less than half of siRNA is colocalized with Lysotracker for all larger polymers.
Lysotracker Colocalization Imaging.
Next, we correlated knockdown activity with Lysotracker colocalization, a commonly employed method to assess endosomal entrapment. Acidic, membrane-bound vesicles are stained using the acidotropic dye Lysotracker, while the cargo is tracked using a covalently attached fluorophore (in this case, Alexa-488, which has been shown to minimally alter cellular uptake;22 see Figure 4D). We used blinded, automated analysis to calculate the Manders Coefficient54 of overlap to qualitatively and quantitatively assess si-NP entrapment in endolysosomes (Figure 4E). At a 50 nM siRNA dose, Lysotracker colocalization matches expected values, with smaller-molec-ular-weight 50B polymers being more strongly colocalized with endolysosomal staining than larger ones. At lower doses, however, the decreased signal from the siRNA fluorophore reduces quality of localization (Figure 4F). In short, the Lysotracker colocalization data support the hypothesis that larger MW 50B polymers more efficiently escape endosomes. Statistical testing by 2-way ANOVA measured p < 10−4 for polymer molecular weight and p < 0.05 for interaction between polymer and dose. Polymer MW accounted for 65% of data set variation, with little measured dose dependency effect. One of the main shortcomings in using Lysotracker colocalization for endosome-buffering polymers is exemplified by the data on 0B polymer (Figure 4D). The 0B polymer is highly cationic, resulting in high cellular uptake and endosomal buffering capacity but minimally membrane disruptive, generating negligible endosomal escape activity and consequently little bioactivity (Figure 2B). The Manders coefficient for 0B is 0.22, in all likelihood suggesting via artifact that little endolysosomal colocalization exists. Presumably, Lysotracker is unable to accumulate in vesicles that contain 0B due to the polymer’s high tertiary amine content and associated pH-buffering capacity. These data support the contention that neither high cellular uptake nor a lack of Lysotracker colocalization ensures intracellular bioactivity, in agreement with our previous observations.43,49 Furthermore, we confirm earlier reports that accurate Lysotracker colocalization data requires siRNA doses higher than those that confer near-maximal gene knockdown,14 as the 12.5 and 25 nM doses of siRNA produce strong knockdown but have poor signal in Lysotracker colocalization analyses. This is because the amount of siRNA that reaches the cytoplasm necessary to achieve efficient gene knockdown is lower than the limit of detection of many microscopy setups.
Transmission Electron Microscopy.
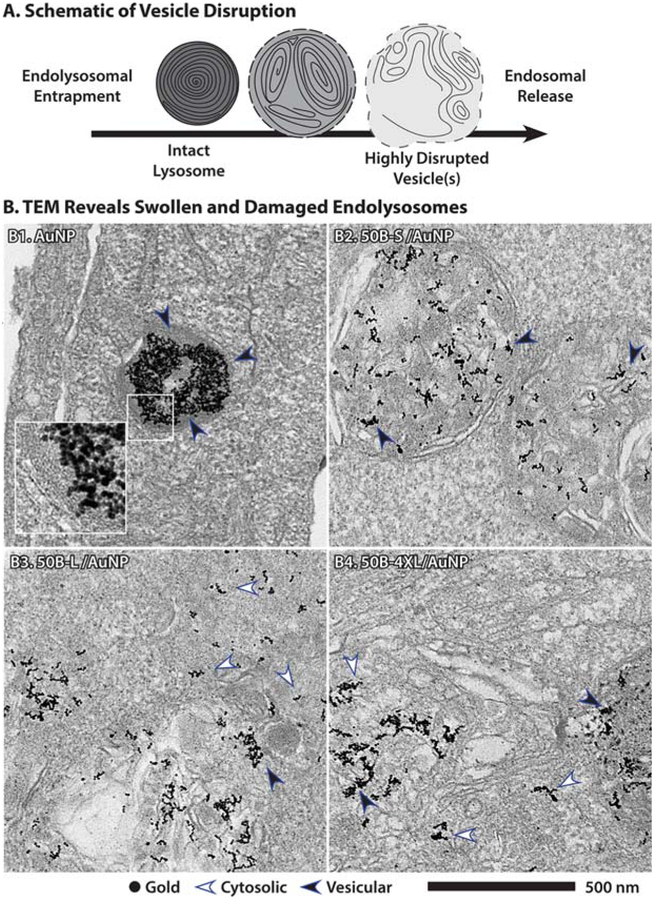
Endosomal disruption and escape was also visualized directly using TEM for the smallest (50B-S), an intermediate (50B-L), and the largest (50B-4XL) MW polymers with the goal of validating the Gal8 recruitment data. The si-NPs were formulated with 10 nm diameter gold nanoparticles (AuNPs)29 and applied to cells to enable visualization with TEM imaging. We hypothesized that, based on the Gal8 recruitment data, free AuNP and 50B-S/AuNP would localize to intact lysosomes and that 50B-L and 50B-4XL si-NPs would cause disruption and escape from endosomal membranes (Figure 5A), as we previously reported for endosome-disruptive poly(propylacrylic acid). The free AuNPs accumulated in electron dense, multilayered lysosomal structures (Figures 5B1 and S4). In the sample treated with 50B-S (Figures 5B1 and S5), the AuNP were found in membrane-bound multivesicular bodies, which supports literature reports of late endosomes or autolysosomes,55 but nearly all gold was found within membrane bound structures. For the 50B-L (Figures 5B3 and S6) and 50B-4XL (Figures 5B4 and S7) polymers that robustly triggered recruitment of Gal8, lysosomal structures appeared disrupted, and the AuNP label was apparent both in the cytosol (white arrowheads) and in structures that may retain a membrane (black arrowheads). Membranes, such as those seen encapsulating AuNP and 50B-S/AuNP, were not apparent, were visibly damaged in cells treated with si-NPs formulated with 50B-L and 50B-4XL, or both. These data confirm that pH-responsive 50B polymers disrupt the endo/lysosomal system and release large molecules and even 10 nm gold NPs to the cytosol.
Figure 5.
Transmission electron microscopy confirmation of endosomal disruption as predicted by Gal8 recruitment. (A) Endolysosomes are multilamellar, electron-dense structures that become more swollen with weakly membrane active polymers, while highly membrane active polymers more fully disrupt endosomal membranes and induce a fragmented, multivesicular phenotype with incomplete or discontinuous membranes. (B) Transmission electron microscopy shows AuNPs traffic to electron-dense lysosomes, B1, where they are fully enclosed by a lipid bilayer (black arrow), while 50B-S induces a swollen vesicle phenotype, B2, although the membrane remains intact. Endosome disruptive polymers 50B-L and 50B-4XL induce widespread loss of endosomal membrane and release AuNPs into cytoplasm (white arrows), although some AuNP remains trapped in membranes (black arrows).
Correlation Analysis.
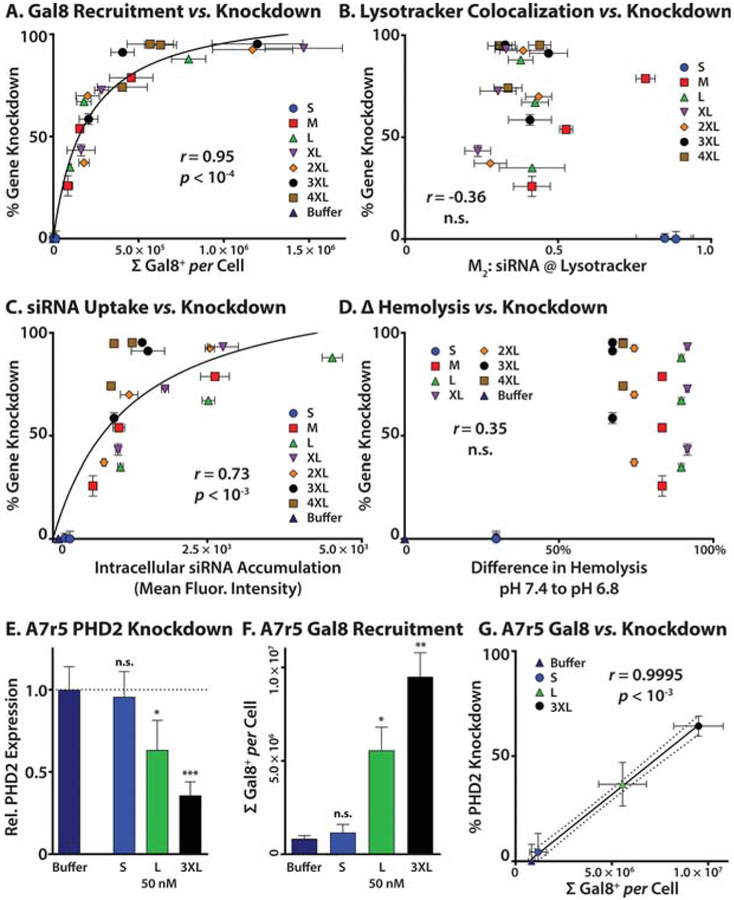
Finally, to statistically compare Gal8 recruitment against a variety of predictive assays used to screen biomacromolecular delivery technologies, we plotted the data from Figure 3B (Gal8 recruitment), Figure 4F (Lysotracker colocalization), and Figure S8 (cellular uptake), against dose-matched luciferase knockdown data (Figure 3A) to quantitatively assess the correlation of these measures to intracellular bioactivity using Spearman’s method (Figure 6A–C). For the hemolysis data set, we plotted the difference in hemolysis from pH values from 7.4 to 6.8 at 15 μg/mL against luciferase knockdown at all three doses because human red blood cell hemolysis was only performed at one concentration of polymer. Because this assay is performed on red blood cells in a buffered solution rather than inside an endosome, dose matching is not straightforward. Gal8 recruitment was found to have the highest correlation to luciferase knockdown, with r = 0.953 and p < 10−4 (Figure 6A). Measured Lysotracker colocalization was found to have very poor correlation to overall knockdown data, with nonsignificant correlation (Figure 6B; r = −0.36 and p = 0.10). Cellular uptake as measured by flow cytometry (Figure S8) was found to have intermediate correlation for the MW library, with r = 0.732 and p < 10−3 (Figure 6C). In addition, the change in hemolysis from pH 7.4 to 6.8 had very low predictive value (Figure 6D; r = 0.04 and p = 0.96) and does not clearly predict which polymers will have the highest in-cell endosome disruption capacity. The hemolysis assay is good at ruling out cytotoxic formulations and true negative hits; however, what appear to be ideal endosomolysis profiles can be see with “false positives” that do not generate high levels of in-cell activity, and the assay is not sensitive enough to discriminate between formulations with varied levels of efficacy. Furthermore, molecular weight of the polymers has statistically significant correlation to knockdown at all three doses tested, validating our hypothesis that increasing p(DMAEMA-co-BMA) si-NP core-forming block molecular weight increases gene knockdown activity (Figure S9, r > 0.9 and p < 0.01).
Figure 6.
Validation of Gal8 recruitment as a predictor of bioactivity relative to common standard delivery and endosome escape readouts. (A) Gal8 recruitment (x axis) and luciferase gene knockdown data (y axis) from MDA-MB-231 cells were plotted, and a hyperbolic fit was calculated. Correlations were calculated by Spearman’s method; r = 0.95 and p < 10−4. Gal8 recruitment and knockdown measurements are dose-matched (12.5, 25, and 50 nM) and include buffer-treated control. For additional discussion of these data, see Technical Note 1 in the Supporting Information. (B) Lysotracker colocalization as measured by Manders coefficient (x axis) and luciferase gene knockdown data (y axis) from MDA-MB-231 cells were plotted. Correlations were calculated by Spearman’s method; correlation was not statistically significant. Manders coefficients and knockdown measurements are dose-matched (12.5, 25, and 50 nM). (C) siRNA cellular uptake as measured by mean fluorescence intensity by flow cytometry (x axis) and luciferase gene knockdown data (y-axis) from MDA-MB-231 cells were plotted, and a hyperbolic fit was calculated. Correlations were calculated by Spearman’s method; r = 0.73 and p < 10−3. Cell uptake and knockdown measurements are dose-matched (12.5, 25, and 50 nM) and include buffer-treated vehicle control. (D) The difference in hemolysis response from pH 7.4 to 6.8 (x axis) from human erythrocytes and luciferase gene knockdown data (y axis) from MDA-MB-231 cells were plotted. Correlations were calculated by Spearman’s method for hemolysis data for each dose of knockdown data individually; despite the increased likelihood of type I errors, correlation at no individual dose was statistically significant. Hemolysis data are for 15 μg/mL polymer concentration, and knockdown data are at 0 (vehicle control), 12.5, 25, and 50 nM siRNA. (E) Relative PHD2 expression from A7r5 cells treated with si-NPs at 50 nM were measured via quantitative reverse-transcription polymerase chain reaction and calculated as 2−ΔΔCT. Treatment with 50B-L or 50B-3XL induced statistically significant reductions in PHD2 mRNA levels. Single and triple asterisks represent p < 0.05 and 0.001, respectively. (F) Gal8 recruitment measurements from A7r5 cells treated with 50 nM siNPs show that 50B-L and 50B-3XL induce statistically significant Gal8 recruitment. Single and double asterisks represent p < 0.05 and 0.01, respectively. (G) Gal8 recruitment (x axis) and PHD2 gene knockdown data (100 × [1 – 2−ΔΔCT]; y axis) were plotted. Because saturation was not observed in either Gal8 recruitment or gene knockdown data, a linear fit was calculated, and correlations were calculated by Pearson’s method; r = 0.995 and p < 10−3. Gal8 recruitment and knockdown measurements are dose-matched (50 nM) and include buffer-treated control. For additional discussion of these data, see Technical Note 1 in the Supporting Information.
Next, we wanted to confirm the predictive value of Gal8 recruitment imaging in a different cell type and for a therapeutically relevant, non-model gene. To do so, we focused on a non-cancer A7R5 rat vascular smooth muscle cells and the gene prolyl hydroxylase domain-containing protein 2 (PHD2) with a validated rat siRNA, which we have been interested in studying as a target relevant for pro-angiogenic wound healing therapy.56–58 For this study, we used the quantitative reverse-transcription polymerase chain reaction (qPCR) method to measure PHD2 mRNA levels following treatment with siNPs formulated with 50B-S, 50B-L, and 50B-3XL polymers to get a range of endosomolytic potency properties based on the MDA-MB-231 studies. In this study, we saw the expected trends between polymer molecular weight and gene knockdown (Figure 6E) and between polymer molecular weight and Gal8 recruitment level (Figure 6F). In agreement with the model gene observations in MDA-MB-231 cells, there was a statistically significant correlation (r = 0.995 and p < 0.001) between Gal8 recruitment level and percent gene knockdown (Figure 6G). As an added confirmation, we also found that knockdown of the model gene luciferase in the A7R5 cells followed the expected trend with regard to polymer molecular weight (Figure S10). These results confirm the more general application of Gal8 as predictive of gene silencing across multiple polymers, doses, cell types, and genes.
Gal8 Recruitment in Tumors in Vivo.
As a further extension of this technique, we sought proof of concept of Gal8 imaging for in vivo visualization and measurement of endosome disruption. To show that Gal8 recruitment occurs in vivo and that it corresponds to in vivo bioactivity, we used an optimized si-NP formulation based on our previous work.47,50,58 This system consists of the same 50B core as our in vitro library combined with an inert, zwitterionic corona chemistry and a hydrophobized siRNA to confer additional stability and pharmacokinetic benefits. To validate the utility of the Gal8 system in vivo, we characterized both in vivo luciferase knockdown and in vivo Gal8 recruitment in an orthotopic breast cancer model. Orthotopic MDA-MB-231 tumors carrying either firefly luciferase or Gal8-YFP constructs were introduced into the mammary fat pads of nude mice, and si-NPs were injected via the tail vein.
Gal8 imaging revealed that tumors from mice treated with si-NPs exhibited a significant increase in Gal8-associated endosomal disruption of 4.58-fold relative to mice treated with phosphate-buffered saline (PBS) injections (Figures 7A and S11A). In a separate cohort of mice, we measured, as expected, a statistically significant reduction in luciferase activity for the si-NPs formulated with luciferase targeting siRNA relative to the scrambled control (Figure 7B). These measurements indicate that Gal8 imaging is able to measure polymer-mediated endosomal disruption, which correlates to cytosolic siRNA delivery to tumor cells in vivo. These data are notable as providing visualization and measurement of endosomal disruption mediated by in vivo nanocarrier treatments performed on fresh, unfrozen, intact tissue without exogenous fluorophores or acidotropic dyes. However, a small number of confocal micrographs of tumors from PBS-treated animals showed detectable Gal8 recruitment, probably associated with cell stress response to the tumor microenvironment. For example, one extreme outlier within the PBS group, which was not excluded from analysis, showed Gal8 recruitment that was 348-fold higher than median PBS tumor. Likewise, Gal8 imaging of tumors from animals treated with si-NPs was highly variable, likely due to known heterogeneities in tumor NP penetration59–61 (Figure S11B). It is increasingly recognized that many types of cancer have dysregulated increased levels of autophagy to promote survival under stressful conditions,62,63 possibly leading to enhanced basal Gal8 recruitment in tumors in vivo relative to in vitro contexts. Furthermore, the Gal8 response observed was weaker than anticipated given the luciferase knockdown results, potentially due to suboptimal time points chosen. Multiple events with independent kinetics occur simultaneously in vivo: distribution by the vascular system, tissue and cellular uptake, endosomal escape, mRNA degradation by RISC, and endogenous degradation of luciferase protein. Gal8 imaging only provides a snapshot of endosomal disruption at one time point. Timing for the experiment could likely be improved, especially by using advanced techniques such as intravital microscopy or tumor window models, which would increase statistical power by providing same tumor controls and longitudinal sampling in a single tumor. This experiment is best understood as providing proof of concept that interesting information can be inferred about the real-time, in vivo status of endosomal integrity, although these studies are complicated by pharmacokinetics and potential false positives from baseline levels of autophagy. Despite these limitations, intratumoral Gal8 imaging shows strong promise as a technique for assessing endosomal disruption by nanocarriers in animal experiments, and there may be opportunity to improve in vivo application of this methodology.
Figure 7.
In vivo proof of concept for utilization of Gal8 imaging for visualization and measurement of endosome disruption within tumor tissue following the systemic delivery of endosome-disruptive si-NPs. (A) si-NPs induce significant endosomal disruption; n = 8 tumors per group, single asterisks indicate p < 0.05 by unpaired two-sided Mann-Whitney test. (B) si-NPs delivering luciferase targeting siRNA significantly reduce luciferase signal relative to scrambled sequence control; n =10 tumors per group, single asterisks indicate p < 0.02 by unpaired two-sided Student’s t test. (C) Representative images of saline (left) and si-NP treated tumors (right). Disrupted endosomes as identified algorithmically are marked with magenta circles.
CONCLUSIONS
One of the major limitations in developing improved intracellular drug delivery systems has been the difficulty of directly assessing active endosomal escape. Existing methods are prone to artifacts, typically require fluorophore labeling of the biologic cargo, are often indirect and thus not representative of the extent of endosomal damage, and are not amenable to high-throughput analysis. Here, we validated Gal8 recruitment as a measure of endosomal disruption that circumvents limitations of current standard techniques and correlated this endosomal disruption information to siRNA bioactivity. By validating the technique with a well-characterized series of different polymer compositions, we show that endosome disrupting polymers that facilitate intracellular biologic cargo activity result in increased Gal8 recruitment. This technique was then used to study the effects of 50B MW on endosome disruption. We show that Gal8 recruitment has a higher correlation to functional bioactivity for a library of endosome-escaping siRNA delivery polymers than alternative methods of predicting bioactivity for drug carriers, such as Lysotracker colocalization, hemolysis, or cellular uptake, and that this method further obviates the need for expensive, exogenous fluorophores to track cargo localization and uptake. This study also provided insights into the strong correlation between polymer molecular weight and endosome escape and bioactivity within the 50B MW polymer library. Furthermore, we extended this technique to orthotopic breast tumors in vivo, showing that Gal8-visualized, nanocarrier-mediated endosomal disruption also correlates with in vivo gene knockdown activity in the tissues of treated animals.
One under-appreciated aspect within the drug delivery field is that endosomal disruption ipso facto induces autophagy of damaged endosomes as a consequence of Gal8 recruitment.29,35,37 Gal8 signaling to NDP5264,65 leads to the formation of phagophores capturing these damaged organelles. Phagophores then mature into autophagosomes, which fuse with lysosomes.66 The steps downstream of endosomal disruption are poorly understood in the context of drug carriers, and additional efforts should focus on understanding and modulating the timing and mechanisms at play between disruption and ultimate recontainment of the damaged endosome, presumably in the time period when cargo escape can occur.
We believe that the quantitative Gal8 imaging technique will show broader utility beyond siRNA-focused library screens for silencing of model genes such as luciferase. For siRNA, activity and mechanistic (endosome disruption) screens completed in parallel are expected to provide a deeper level of structure–function insight and lead to better paths for carrier structural optimization relative to screening for activity alone. However, this assay may in fact find its greatest utility in screening formulations of biologic drugs whose delivery is more difficult to assay using high-throughput reporter (e.g., luciferase or GFP) methods. For example, the development of therapeutic peptides, enzyme replacement therapies, blocking antibodies, and nonviral gene editing systems have all been hampered to a degree by inefficient screening techniques.10,16,30,67–69 Our vision for the system is that Gal8 recruitment activity can be used as an early “down-selection criteria” in HTS screening of candidate delivery system libraries, especially when a specific parameter of structure or formulation is being investigated (e.g., overall MW, as we investigated here). In conjunction with additional, orthogonal HTS assays (e.g., toxicity), a large number of experimental groups can be reduced to a manageable number (“hits”, occurring at the intersection of top performers in both Gal8 and toxicity screening) for use in more intensive assays that provide additional information about intracellular pharmacokinetics, intracellular trafficking, or bioactivity. Indeed, our studies here show the best performing polymers also perform best in the Gal8 recruitment screen, although this predictive power is not perfect.
Despite its potentially broad applicability, there are some obvious limitations of the Gal8 recruitment imaging method. First, it requires the engineering of cell lines to express a transgene construct and requires a microscope equipped with a software-controlled stage, multiwell or 96-well plate mounts, and appropriate excitation and emission filters. While we used a typical Nikon confocal system here, this method should be amenable to other high-throughput and plate-reader-based microscopy systems, as Gal8 recruitment produces puncta that can be resolved with a relatively low-power 2× objective. Furthermore, cells must be very carefully handled in conjunction with this assay, and the use of inappropriate wash buffers may induce Gal8 recruitment independent of carrier-driven endosomal rupture, creating false-positive signals. Careful use of vehicle controls is a viable way to be sure to have an accurate baseline. Finally, while the system is amenable to multiple polymers, doses, and cell types, assay conditions such as time point and wash buffers may need to be optimized to account for differences in polymer characteristics or cell-type specific differences in mechanisms of endocytosis, endosomal trafficking, and endosomal disruption. In sum, imaging and quantification of Gal8 recruitment allows high-throughput, real-time, live-cell, and fluorophore-free measurements of active endosomal escape. This method provides a powerful tool for rapidly optimizing and elucidating structure–function relationships for libraries of biologic nanomedicine candidates.
MATERIALS AND METHODS
Materials.
All chemical synthesis reagents were obtained from Sigma-Aldrich and used as received unless otherwise noted. 2-(Dimethylamino)ethyl methacrylate (DMAEMA) and butyl methacrylate (BMA) monomers were passed twice through a basic alumina gravity column prior to use to remove inhibitors. 2,2-Azobis(2-methylpropionitrile) (AIBN) was recrystallized twice from methanol. Titration reagents, hydrochloric acid (HCl) solution (certified 0.0995–0.1005 N), and sodium hydroxide (NaOH) solution (certified 0.0995–0.1005 N), were obtained from Fisher Scientific. Poly[(ethylene glycol)-b-[(2-(dimethylamino)ethyl methacrylate)-co-(butyl methacrylate)]] polymers were synthesized using RAFT polymerization as previously reported.20,47 The macro-chain transfer reagent was synthesized by coupling 5000 g/mol PEG to 4-Cyano-4-(ethylsulfanylthiocarbonyl) sulfanylvpentanoic acid (ECT) using DCC-DMAP coupling. Macro-CTA synthesis and polymer synthesis are shown as Figure S1 and previously reported.20,50,70 An optimized in vivo polymer, poly[2-(methacryloxyethyl) phosphorylcholine] (PMPC)-b-(DMAEMA-co-BMA) was synthesized, characterized, and purified as previously described47 with a degree ofpolymerization of 75 for the each of DMAEMA, BMA, and 2-(methacryloxyethyl) phosphorylcholine. PD10 desalting columns (GE Healthcare) were used for the final purification of polymers according to manufacturer’s instructions. Oligonucleotide sequences were used as previously reported from Integrated DNA Technologies or Sigma-Aldrich.50 Alexa 488 labeled dsDNA were used for Lysotracker and cell uptake experiments. LysoTracker Red DND-99 was obtained from ThermoFisher Scientific.
Polymer Characterization.
Polymers were characterized using 1H nuclear magnetic resonance spectroscopy (Bruker, 400 MHz). Units of DMAEMA and BMA were calculated by integrating these peaks relative to the PEG peak; number average molecular weight (Mn) was calculated by adding the mass from these components and is referred to using the acronym MW throughout the manuscript. Polydispersity was evaluated with DMF mobile-phase gel permeation chromatography (GPC, Agilent Technologies), dissolved at 10 mg/mL in DMF containing 0.1 M LiBr.
pKa Measurement.
The pKa was measured for all polymers via acid-base titration using an MPT Titrino automatic titrator (Metrohm, Switzerland). Polymers were dissolved in 0.1 N HCl to a final concentration of 5 mg/mL and titrated to the basic end point with 100 μL additions of 0.1 N NaOH. The pKawas calculated as the half-neutralization point of each titration.
Nanoparticle Complexation.
Desalted, lyophilized polymers were dissolved in 100% ethanol at 33.3 mg/mL, diluted 1:10 into pH 4.0 citric acid buffer (10 mM), and stored at −20 °C until further use. Polymers were complexed with siRNA at an N/P ratio of 10 at pH 4.0 for 0.5 h and then neutralized with 5-fold excess pH 8.0 phosphate buffer (10 mM). The product was syringe filtered (0.45 μm) and then diluted into appropriate buffer for siNP characterization studies or DMEM containing 10% FBS for in vitro cellular studies. The naming scheme used for the composition library corresponds to the mole percent BMA in the feed for the p(DMAEMA-co-BMA) block of the polymer. The naming scheme used for the MW library corresponds to the relative MW of the p(DMAEMA-co-BMA) block, where S, M, L, XL, 2XL, 3XL, and 4XL correspond to small, medium, large, extra-large, double extra-large, triple extra-large, and quadruple extra-large, respectively.
Dynamic Light Scattering to Measure Nanoparticle Size.
Si-NPs were prepared at 100 μg/mL in PBS − /−. Hydrodynamic radius was measured using dynamic light scattering (Zetasizer Nano ZS, Malvern Instruments).
Cell Culture.
Human epithelial breast cancer cells (MDA-MB-231) were obtained from ATCC. To generate cells stably expressing luciferase and GFP, HEK-239-T cells from ATCC were transfected with pGreenFire1-CMV (constitutive firefly luciferase, GFP, and puromycin resistance) plasmid and packaging plasmids pMDLg/pRRE, pRSV-Rev, and pMD2.G with lipofectamine to generate psueodotyped lentiviral particles which were applied to MDA-MB-231s, selected with puromycin, and sorted using fluorescence activated cell sorting as previously reported.50 Gal8 retrovirus was generated using HEK 293-T cell transfected with Gal8-YFP, pUMVC, and pCMV-VSV-G. Luciferase transduced cells (Luc-MDA-MB-231) were obtained following fluorescence activated cell sorting (BD LSR II Flow Cytometer, San Jose, CA) for GFP+ cells after a 2 week puromycin selection. Gal8-YFP transduced cells (Gal8-MDA-MB-231) were obtained by 1 week selection with blasticidin, followed by single clonal expansions obtained through the limiting dilution method in blasticidin containing media; clonal populations were used to ensure consistent expression of YFP constructs. Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 0.1% gentamicin. All cell culture reagents were obtained from Gibco. Both MDA-MB-231 engineered cell lines were assessed for flow cytometry and were found to have minimally altered levels of cellular uptake of si-NPs (Figure S12).
Gal8 Recruitment Assays.
Gal8-MDA-MB231 cells were plated in Nunc Lab-Tek 8-well chambered coverglass (Nunc, Thermo Fisher Scientific Inc.) or Corning 96 Well Half-Area High-Content Imaging Glass-Bottom Microplates (Corning Inc.; product no. 4580) at a density of 6250 cells per square centimeter. Cells were left to adhere and proliferate for 24 h before being treated siRNA formulations. After a 17 h treatment, media was aspirated and replaced with warm FluoroBrite DMEM supplemented with 25 mM HEPES, 10% FBS, and Hoechst 33342. Images were acquired with Nikon C1si+ confocal microscope system on a Nikon Eclipse Ti-0E inverted microscope base, Plan Apo VC 20× objective, Galvano scanner, and 408/488/543 dichroic mirror. PerfectFocus was used to maintain focus between wells. The PerfectFocus offset was set to the optimal focal plane of the nuclear stain (blindly, without looking at the Gal8 layer) in combination with the well-scanning mode in ND Acquisition to acquire images, removing microscopist bias. A software-controlled motorized stage moved the plate between images. For additional information regarding microscopy systems for reproduction of this assay, please see Technical Note 2 in the Supporting Information. Images were exported to lossless multipage TIF from Nikon NIS-Elements AR version 4.30.01. Exported images were then analyzed in bulk using a blinded, automated MATLAB script, which is detailed below and has been made available via the FigShare platform.71 To improve print clarity, images were brightened via linear scaling to the max pixel intensity value per channel of the top 0.01% of Gal8 pixels or top 1% of Hoechst pixels using MATLAB. While these enhancements were made for print, all quantification was done on raw images.
Gal8 Image Processing.
A MATLAB script was written to identify Gal8 positive spots (Figure S2A–C). A binary mask was generated containing Gal8-positive puncta, which were used to integrate underlying Gal8 intensities on unprocessed images and normalized to the number of nuclei.
The Gal8 mask was generated by using a tophat filter on the YFP fluorescence channel to perform localized background subtraction to suppress diffuse cytosolic fluorescence (Figure S2D). This was followed by simple thresholding to identify Gal8 pixels (Figure S2E) and subsequent image “morphological opening” using a 3 × 3 pixel plus-sign-shaped structuring element to restrict positive pixels to puncta (Figure S2F). From this, a “visual check” layer was generated by the exclusive or (“XOR”) of the mask and its dilation, generating a “halo” around identified spots (Figure S2G) to allow the manual verification of algorithm detection success. The Gal8 mask (Figure S2F) was applied to the unprocessed Gal8 image and the result was integrated, quantifying total Gal8 present within the puncta. This sum was divided by cell number, which was determined by the following image analysis method: a simple threshold was applied to the Hoechst channel (Figure S2I), followed by an image opening to separate proximal nuclei (Figure S2J) and segmenting using a watershed transformation (Figure S2K). A rainbow colormap was applied to the image, and the result was exported (Figure S2L), which served as verification that nuclei were properly quantified. Note that contiguous nuclei (e.g., during mitosis) were counted as one cell. Fluorescent composite images (Figure S2A), as well as three method verification images were exported. The first verification image circles identified Gal8 spots (Figure S2M); the second verification image shows identified puncta as magenta overlays (Figure S2N); the third verification image identifies nuclear number (Figure S2L). Gal8 recruitment code has been made available by the authors via the FigShare platform at https://doi.org/10.6084/m9.figshare.7066472.71
Lysotracker Colocalization Image Processing.
Wild type MDA-MB-231 cells were seeded at a density of 6250 cells per square centimeter in half-area 96-well imaging plates as for Gal8 assays. Cells were treated with Alexa488-labeled dsDNA loaded si-NPs at indicated doses. After treatment for 24 h, media was replaced with Lysotracker Red (Invitrogen Life Technologies) containing media (75 nM) and incubated 1 h. Before imaging, cells were washed, and the media was replaced with Fluorobrite DMEM supplemented with 10% FBS and 25 mM HEPES. Images were analyzed using an automated MATLAB script for Manders Coefficient as originally described.54 The code to calculate Manders Coefficient of Overlap has been made available by the authors on the FigShare platform, accessible at https://doi.org/10.6084/m9.figshare.7066490.72
siRNA Cell Internalization Flow Cytometry Studies.
Wild-type MDA-MB-231 cells were seeded in triplicate in 12-well plates at a density of 60 000 cells per well and left to grow for 24 h, before being treated with si-NPs formulated with Alexa-488 labeled dsDNA at indicated doses and incubated for 16 h. Cells were washed with PBS, treated with 0.05% trypsin, and centrifuged at 500g for 5 min. The supernatant was removed, and cells were resuspended in 300 μL of PBS. Single-cell suspensions were analyzed using a BD LSR Fortessa flow cytometer, gated according to forward scatter and side scatter. Mean fluorescence intensities (MFI) were calculated using FlowJo (FlowJo LLC).
pH-Dependent Membrane Disruption Hemolysis assays.
Hemolysis with red blood cells was performed as previously described.52 Briefly, blood was donated by an anonymous human donor using a protocol approved by the Vanderbilt institutional review board (IRB). Red blood cells (RBCs) were isolated and washed by centrifugation and then diluted into buffered saline solutions at pH 7.4, 6.8, or 6.2 representative of extracellular, early endosomal, and late endosomal environments. si-NPs at concentration of 15 ug/mL polymer were incubated with RBCs in each buffer for 1 h at 37 °C. RBCs were centrifuged, and the optical absorption of the supernatant at 450 nm was used to measure the hemoglobin released compared to PBS (negative control) and 1% Triton X-100 (positive control) treated RBCs.
Luciferase Knockdown siRNA Bioactivity Assay.
Luc-MDA-MB-231 cells were plated in black-walled 96-well plates at a density of 67 000 cells per square centimeter. The cells adhered overnight and were then treated with the si-NPs containing siRNA targeting luciferase or nontargeting negative control siRNA for 24 h. Media containing 150 μg/mL luciferin was added at 24 and 48 h time-points, and bioluminescence was measured using an IVIS 200 Series imaging system (Xenogen Corporation).20
Cell Viability.
For the composition library toxicity studies, Luc-MDA-MB-231s were treated with PBS or si-NPs loaded with control nucleic acids. Luciferase activity was measured to assess intact cells as for luciferase knockdown studies. The ratio of luminescence of scrambled sequence si-NP treated cells to PBS treated cells at 24 h post-treatment was used as a measure of viability. For the MW library, the more sensitive CellTiter Glo assay (Promega), which measures retained cellular ATP, was used according to manufacturer protocol to measure dose-dependent and polymer-dependent toxicity effects. Wild-type MDA-MB-231 cells were plated in black-walled 96-well plates at a density of 67 000 cells per square centimeter and allowed to adhere overnight and were then treated with the si-NPs containing nontargeting negative control siRNA for 24 h before CellTiter Glo assay components were added to cells. Luminescence was read using an IVIS 200 Series imaging system.
Transmission Electron Microscopy.
MDA-MB-231 cells were plated in 100 mm dishes. Gold labeled si-NPs were formulated at 50 nM siRNA loaded into 50B-S, 50B-L, or 50B-4XL and loaded with anionic surface functionalized 10 nm colloidal gold (Sigma-Aldrich catalog no. 752584) as previously reported.29 Cells were treated for 24 h and washed with 0.1 M cacodylate buffer prior to fixation and processing. Samples were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer at pH 7.4 and 37 °C for 1 h and then stored at 4 °C overnight. Samples were submitted to the Vanderbilt Cell Imaging Shared Resource Electron Microscopy Center, where they were washed in 0.1 M cacodylate buffer and then incubated 1 h in 1% osmium tetraoxide at RT and washed with 0.1 M cacodylate buffer. Subsequently, the samples were dehydrated through a graded ethanol series, followed by three exchanges of 100% ethanol and two exchanges of pure propylene oxide (PO). Dehydrated samples were infiltrated with 25% Epon 812 resin and 75% PO for 0.5 h at room temperature (RT) followed by infiltration with Epon 812 resin and PO [1:1] for 1 h at RT and subsequent infiltration with fresh Epon 812 resin and PO [1:1] overnight at RT. The samples were subsequently infiltrated with resin for 48 h and then allowed to polymerize at 60 °C for 48 h. Samples were cut into 500–1000 nm thick sections using a Leica Ultracut microtome. Thick sections were contrast stained with 1% toluidine blue and imaged with a Nikon AZ100 microscope to locate cells. Ultrathin (70–80 nm) sections were cut and collected on 300-mesh copper grids and then poststained with 2% uranyl acetate followed by Reynolds’ lead citrate. Thin samples were imaged on a Philips/FEI Tecnai T12 electron microscope. For publication, TEM images were contrast enhanced and sharpened in Adobe Photoshop CS6 using the “Levels” tool, setting the 50% gray point to the center of the intensity distribution. Images were then sharpened using the “Smart Sharpen” filter, with “Amount: 100%” and “Radius: 5 px” to “Remove: Gaussian Blur.” This procedure was used on all images.
qPCR Studies.
qPCR was performed using TaqMan assay reagents and primers to quantify expression of PHD2. Total RNA was isolated purified from cultured A7r5 rat cells with Qiazol and RNeasy spin columns (Qiagen). RNA was reverse-transcribed (1 μg) with an iScript Reverse Transcription Supermix kit (Bio-Rad Laboratories). Relative expression of PHD2 was assessed as 2−ΔΔCT, normalized to housekeeping gene GAPDH (assay IDs: Rn00710295_m1, Rn01775763_gl; Applied Biosystems). siRNA was used as previously described.56–58
In Vivo Studies.
Athymic nude mice (catalog no. 002019 Nu/J) were ordered from Jackson Laboratories. All procedures were carried out under Vanderbilt University IACUC-approved protocols. Mice were injected with 106 MDA-MB-231 cells suspended in 1:1 Dulbecco’s modified Eagle medium (DMEM)-Matrigel into the mammary fat pad. MDA-MB-231 cells stably expressing firefly luciferase or Gal8-YFP were used for knockdown or Gal8 experiments, respectively. In vivo formulations of (PMPC)-b-(DMAEMA-co-BMA) with 50 mol % BMA and 50 mol % DMAEMA were made according to our previously published methods47 using palmitoylated siRNA.48,50,58 For luciferase studies, mice (5 per group, n = 10 tumors) were injected with 1 mg/kg siRNA against luciferase or scrambled control siRNA via the tail vein. To measure luminescence, mice were injected with 15 mg/kg luciferin intraperitoneally and imaged the day of and at 24 h after siRNA treatment using an IVS imaging system (Caliper Life Sciences). For Gal8 studies, mice were injected with in vivo formulations of polyplex at 1 mg/kg siRNA or an equal volume saline injection. Tumors were explanted into ice-cold PBS following euthanasia 18 h post-injection. The tumors were bisected with a razor blade and immediately subjected to confocal imaging. A total of 7 to 11 images were taken per tumor and averaged; each plotted point represents the average data for one tumor, and 8 tumors were analyzed per group.
Statistical Methods.
Data were analyzed and plotted using Microsoft Excel, Prism GraphPad 6, and MathWorks MATLAB R2016a. Lines of best fit for Figure 1 were generated through a linear fit in GraphPad; both lines have statistically significant nonzero slopes (p < 0.05). Luciferase activity and toxicity data for the composition library were tested using one-way ANOVA in GraphPad with Dunnett’s post hoc testing against PBS. For the MW library, cytotoxicity, luciferase activity, Manders coefficient, and Gal8 recruitment were tested using two-way ANOVA with no sample matching in GraphPad Prism. Where noted, post hoc analysis was performed using Tukey’s multiple comparisons test to calculate corrected p values for multiple comparisons, while Dunnett’s post hoc test was used for significant differences relative to control in GraphPad Prism. All 2-way ANOVA data presented are statistically significant at the data set level (p < 0.001) for polymer, dose, and interaction thereof. For Figure 6A–C, data were plotted using dose matched data sets; for Figure 6D, knockdown data at the 3 doses were plotted against hemolysis. Spearman correlation coefficients and twotailed p values were generated using GraphPad Prism; for hemolysis data, this calculation was done for each dose individually against hemolysis and as a whole. For Figure 6, hyperbolic lines of best fit were calculated using GraphPad Prism; best fit models did not converge for panels B and D of Figure 6. Correlations were calculated independently of lines of best fit. For in vivo experiments (Figure 7), p values were calculated using two-sided t tests without pairing for luciferase knockdown data, while Gal8 recruitment data were analyzed with two-sided Mann–Whitney testing without pairing, which was chosen due to statistically significant deviation of negative control data from normal distribution by testing with the D’Agostino and Pearson omnibus normality test (p < 0.05) as calculated in GraphPad Prism. The threshold of statistical significance, α, was set to 0.05 throughout this work. Signal-to-noise, signal-to-background, and Z′-factor calculations were carried out as described by Zhang et at.51 using the maximal response polymer condition (50 nM, 50B-XL) as an estimated positive control for the range of the assay.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the United States National Institutes of Health grant nos. R01HL122347, R01EB019409, and R01CA224241 and Department of Defense grant no. CDMRP W81XWH-14-1-0298 to C.L.D.; National Science Foundation Graduate Research Fellowship to K.V.K., T.A.W., M.A.J., and S.K.B. (grant nos. 0909667 and 1445197); and Vanderbilt SyBBURE Searle Undergraduate Research Program funding to S.C.D. Dynamic light scattering was conducted at the Vanderbilt Institute of Nanoscale Sciences and Engineering. Flow cytometry experiments were performed in the Vanderbilt University Medical Center Flow Cytometry Shared Resource, supported by the Vanderbilt Ingram Cancer Center (grant no. P30 CA68485) and the Vanderbilt Digestive Disease Research Center (grant no. DK058404). Electron microscopy was performed were performed through the use of the VU Cell Imaging Shared Resource (supported by NIH grant nos. CA68485, DK20593, DK58404, DK59637 and EY08126). We are thankful for electron microscopy assistance and expertise from J. Williams and for gifts of plasmids from F. Randow and R. Weinberg.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b05482.
Additional details on the polymer synthesis scheme, molecular weight library, the image processing pipeline, experimental controls and cells, cellular uptake, polymer weight and Gal8 correlations, spatial heterogeneity, stably integrated genetic constructs, correlation and statistics, lines of best fit, and microscopy requirements (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Stein CA; Castanotto D FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther 2017, 25, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Akcea Therapeutics Press Release. Akcea and Ionis Receive FDA Approval of TEGSEDI™ (inotersen) for the Treatment of the Polyneuropathy of Hereditary Transthyretin-Mediated Amyloidosis in Adults. https://ir.akceatx.com/news-releases/news-release-details/akcea-and-ionis-receive-fda-approval-tegseditm-inotersen (accessed Oct 25, 2018).
- (3).Mullard A FDA Approves Landmark RNAi Drug. Nat. Rev. Drug Discovery 2018, 17, 613–613. [DOI] [PubMed] [Google Scholar]
- (4).Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet 2014, 15, 541–555. [DOI] [PubMed] [Google Scholar]
- (5).Dowdy SF Overcoming Cellular Barriers for RNA Therapeutics. Nat. Biotechnol 2017, 35, 222–229. [DOI] [PubMed] [Google Scholar]
- (6).Whitehead KA; Langer R; Anderson DG Knocking Down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discovery 2009, 8, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Pack DW; Hoffman AS; Pun S; Stayton PS Design and Development ofPolymers for Gene Delivery. Nat. Rev. Drug Discovery 2005, 4, 581–593. [DOI] [PubMed] [Google Scholar]
- (8).Varkouhi AK; Scholte M; Storm G; Haisma HJ Endosomal Escape Pathways for Delivery of Biologicals. J. Controlled Release 2011, 151, 220–228. [DOI] [PubMed] [Google Scholar]
- (9).Belting M; Sandgren S; Wittrup A Nuclear Delivery of Macromolecules: Barriers and Carriers. Adv. Drug Delivery Rev. 2005, 57, 505–527. [DOI] [PubMed] [Google Scholar]
- (10).Akishiba M; Takeuchi T; Kawaguchi Y; Sakamoto K; Yu HH; Nakase I; Takatani-Nakase T; Madani F; Graslund A; Futaki S Cytosolic Antibody Delivery by Lipid-Sensitive Endosomolytic Peptide. Nat. Chem 2017, 9, 751–761. [DOI] [PubMed] [Google Scholar]
- (11).Li H; Nelson CE; Evans BC; Duvall CL Delivery of Intracellular-Acting Biologics in Pro-Apoptotic Therapies. Curr. Pharm. Des 2011, 17, 293–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kim H; Lee D; Kim J; Kim T; Kim WJ Photothermally Triggered Cytosolic Drug Delivery via Endosome Disruption Using a Functionalized Reduced Graphene Oxide. ACS Nano 2013, 7, 6735–6746. [DOI] [PubMed] [Google Scholar]
- (13).Sonawane ND; Szoka FC; Verkman AS Chloride Accumulation and Swelling in Endosomes Enhances DNA Transfer by Polyamine-DNA Polyplexes. J. Biol. Chem 2003, 278, 44826–44831. [DOI] [PubMed] [Google Scholar]
- (14).Gilleron J; Querbes W; Zeigerer A; Borodovsky A; Marsico G; Schubert U; Manygoats K; Seifert S; Andree C; Stoter M; Epstein-Barash H; Zhang L; Koteliansky V; Fitzgerald K; Fava E; Bickle M; Kalaidzidis Y; Akinc A; Maier M; Zerial M Image-Based Analysis of Lipid Nanoparticle-Mediated siRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat. Biotechnol 2013, 31, 638–646. [DOI] [PubMed] [Google Scholar]
- (15).Vermeulen LMP; Brans T; Samal SK; Dubruel P; Demeester J; De Smedt SC; Remaut K; Braeckmans K Endosomal Size and Membrane Leakiness Influence Proton Sponge-Based Rupture of Endosomal Vesicles. ACS Nano 2018, 12, 2332–2345. [DOI] [PubMed] [Google Scholar]
- (16).Berguig GY; Convertine AJ; Shi J; Palanca-Wessels MC; Duvall CL; Pun SH; Press OW; Stayton PS Intracellular Delivery and Trafficking Dynamics of a Lymphoma-Targeting Antibody-Polymer Conjugate. Mol. Pharmaceutics 2012, 9, 3506–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Medina-Kauwe LK; Xie J; Hamm-Alvarez S Intracellular Trafficking of Nonviral Vectors. Gene Ther. 2005, 12, 1734–1751. [DOI] [PubMed] [Google Scholar]
- (18).Tamura A; Oishi M; Nagasaki Y Enhanced Cytoplasmic Delivery of siRNA Using a Stabilized Polyion Complex Based on PEGylated Nanogels with a Cross-Linked Polyamine Structure. Biomacromolecules 2009, 10, 1818–1827. [DOI] [PubMed] [Google Scholar]
- (19).Sakurai Y; Hatakeyama H; Sato Y; Akita H; Takayama K; Kobayashi S; Futaki S; Harashima H Endosomal Escape and the Knockdown Efficiency of Liposomal-siRNA by the Fusogenic Peptide ShGALA. Biomaterials 2011, 32, 5733–5742. [DOI] [PubMed] [Google Scholar]
- (20).Nelson CE; Kintzing JR; Hanna A; Shannon JM; Gupta MK; Duvall CL Balancing Cationic and Hydrophobic Content of PEGylated siRNA Polyplexes Enhances Endosome Escape, Stability, Blood Circulation Time, and Bioactivity in Vivo. ACS Nano 2013, 7, 8870–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Basha G; Novobrantseva TI; Rosin N; Tam YYC; Hafez IM; Wong MK; Sugo T; Ruda VM; Qin J; Klebanov B; Ciufolini M; Akinc A; Tam YK; Hope MJ; Cullis PR Influence of Cationic Lipid Composition on Gene Silencing Properties of Lipid Nanoparticle Formulations of siRNA in Antigen-Presenting Cells. Mol. Ther 2011, 19, 2186–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hughes LD; Rawle RJ; Boxer SG Choose Your Label Wisely: Water-Soluble Fluorophores Often Interact with Lipid Bilayers. PLoS One 2014, 9, e87649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Swiecicki JM; Thiebaut F; Di Pisa M; Gourdin-Bertin S; Tailhades J; Mansuy C; Burlina F; Chwetzoff S; Trugnan G; Chassaing G; Lavielle S How to Unveil Self-Quenched Fluorophores and Subsequently Map the Subcellular Distribution of Exogenous Peptides. Sci. Rep 2016, 6, 20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gruber HJ; Hahn CD; Kada G; Riener CK; Harms GS; Ahrer W; Dax TG; Knaus HG Anomalous Fluorescence Enhancement of Cy3 and Cy3.5 versus Anomalous Fluorescence Loss of Cy5 and Cy7 upon Covalent Linking to IgG and Noncovalent Binding to Avidin. Bioconjugate Chem. 2000, 11, 696–704. [DOI] [PubMed] [Google Scholar]
- (25).Behr J The Proton Sponge: A Trick to Enter Cells the Viruses Did Not Exploit. Chim. Int. J. Chem 1997, 2, 34–36. [Google Scholar]
- (26).Martens TF; Remaut K; Demeester J; De Smedt SC; Braeckmans K Intracellular Delivery of Nanomaterials: How to Catch Endosomal Escape in the Act. Nano Today 2014, 9, 344–364. [Google Scholar]
- (27).Zelphati O; Szoka FC Mechanism of Oligonucleotide Release from Cationic Liposomes. Proc. Natl. Acad. Sci. U. S. A 1996, 93, 11493–11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Richard JP; Melikov K; Vives E; Ramos C; Verbeure B; Gait MJ; Chernomordik LV; Lebleu B Cell-Penetrating Peptides. A Reevaluation of the Mechanism of Cellular Uptake. J. Biol. Chem 2003, 278, 585–590. [DOI] [PubMed] [Google Scholar]
- (29).Kilchrist KV; Evans BC; Brophy CM; Duvall CL Mechanism of Enhanced Cellular Uptake and Cytosolic Retention of MK2 Inhibitory Peptide Nano-Polyplexes. Cell. Mol. Bioeng 2016, 9, 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Evans BC; Hocking KM; Kilchrist KV; Wise ES; Brophy CM; Duvall CL Endosomolytic Nano-Polyplex Platform Technology for Cytosolic Peptide Delivery to Inhibit Pathological Vasoconstriction. ACS Nano 2015, 9, 5893–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Liu X; Fagotto F A Method to Separate Nuclear, Cytosolic, and Membrane-Associated Signaling Molecules in Cultured Cells. Sci. Signaling 2011, 4, pl2. [DOI] [PubMed] [Google Scholar]
- (32).Shi J; Chou B; Choi JL; Ta AL; Pun SH Investigation of Polyethylenimine/DNA Polyplex Transfection to Cultured Cells Using Radiolabeling and Subcellular Fractionation Methods. Mol. Pharmaceutics 2013, 10, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cohen RN; van der Aa MAEM; Macaraeg N; Lee AP; Szoka FC Quantification of Plasmid DNA Copies in the Nucleus after Lipoplex and Polyplex Transfection. J. Controlled Release 2009, 135, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Hadari YR; Paz K; Dekel R; Mestrovic T; Accili D; Zick Y Galectin-8. A New Rat Lectin, Related to Galectin-4. J. Biol. Chem 1995, 270, 3447–3453. [DOI] [PubMed] [Google Scholar]
- (35).Thurston TLM; Wandel MP; Von Muhlinen N; Foeglein A; Randow F Galectin 8 Targets Damaged Vesicles for Autophagy to Defend Cells against Bacterial Invasion. Nature 2012, 482, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Maier O; Marvin SA; Wodrich H; Campbell EM; Wiethoff CM Spatiotemporal Dynamics of Adenovirus Membrane Rupture and Endosomal Escape. J. Virol 2012, 86, 10821–10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wittrup A; Ai A; Liu X; Hamar P; Trifonova R; Charisse K; Manoharan M; Kirchhausen T; Lieberman J Visualizing Lipid-Formulated siRNA Release from Endosomes and Target Gene Knockdown. Nat. Biotechnol 2015, 33, 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Perlman ZE; Slack MD; Feng Y; Mitchison TJ; Wu LF; Altschuler SJ Multidimensional Drug Profiling by Automated Microscopy. Science 2004, 306, 1194–1198. [DOI] [PubMed] [Google Scholar]
- (39).Mitchison TJ Small-Molecule Screening and Profiling by Using Automated Microscopy. ChemBioChem 2005, 6, 33–39. [DOI] [PubMed] [Google Scholar]
- (40).Pepperkok R; Ellenberg J High-Throughput Fluorescence Microscopy for Systems Biology. Nat. Rev. Mol. Cell Biol 2006, 7, 690–696. [DOI] [PubMed] [Google Scholar]
- (41).Conrad C; Gerlich DW Automated Microscopy for High-Content RNAi Screening. J. Cell Biol 2010, 188, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).de Groot R; Luthi J; Lindsay H; Holtackers R; Pelkmans L Large-Scale Image-Based Profiling of Single-Cell Phenotypes in Arrayed CRISPR-Cas9 Gene Perturbation Screens. Mol. Syst Biol 2018, 14, e8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Werfel TA; Jackson MA; Kavanaugh TE; Kirkbride KC; Miteva M; Giorgio TD; Duvall C Combinatorial Optimization of PEG Architecture and Hydrophobic Content Improves Ternary siRNA Polyplex Stability, Pharmacokinetics, and Potency in Vivo. J. Controlled Release 2017, 255, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Siegwart DJ; Whitehead KA; Nuhn L; Sahay G; Cheng H; Jiang S; Ma M; Lytton-Jean A; Vegas A; Fenton P; Levins CG; Love KT; Lee H; Cortez C; Collins SP; Li YF; Jang J; Querbes W; Zurenko C; Novobrantseva T; et al. Combinatorial Synthesis of Chemically Diverse Core-Shell Nanoparticles for Intracellular Delivery. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 12996–13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Green JJ; Langer R; Anderson DG A Combinatorial Polymer Library Approach Yields Insight into Nonviral Gene Delivery. Acc. Chem. Res 2008, 41, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Akinc A; Zumbuehl A; Goldberg M; Leshchiner ES; Busini V; Hossain N; Bacallado SA; Nguyen DN; Fuller J; Alvarez R; Borodovsky A; Borland T; Constien R; de Fougerolles A; Dorkin JR; Narayanannair Jayaprakash K; Jayaraman M; John M; Koteliansky V; Manoharan M; et al. A Combinatorial Library of Lipid-like Materials for Delivery of RNAi Therapeutics. Nat. Biotechnol 2008, 26, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Jackson MA; Werfel TA; Curvino EJ; Yu F; Kavanaugh TE; Sarett SM; Dockery MD; Kilchrist KV; Jackson AN; Giorgio TD; Duvall CL Zwitterionic Nanocarrier Surface Chemistry Improves siRNA Tumor Delivery and Silencing Activity Relative to Polyethylene Glycol. ACS Nano 2017, 11, 5680–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Jackson MA; Bedingfield SK; Yu F; Stokan ME; Miles RE; Curvino EJ; Hoogenboezem EN; Bonami RH; Patel SS; Kendall PL; Giorgio TD; Duvall CL Dual carrier-cargo hydrophobization and charge ratio optimization improve the systemic circulation and safety of zwitterionic nano-polyplexes. Biomaterials 2018, 192, 245–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Beavers KR; Werfel TA; Shen T; Kavanaugh TE; Kilchrist KV; Mares JW; Fain JS; Wiese CB; Vickers KC; Weiss SM; Duvall CL Porous Silicon and Polymer Nanocomposites for Delivery of Peptide Nucleic Acids as Anti-MicroRNA Therapies. Adv. Mater 2016, 28, 7984–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Sarett SM; Werfel TA; Chandra I; Jackson MA; Kavanaugh TE; Hattaway ME; Giorgio TD; Duvall CL Hydrophobic Interactions between Polymeric Carrier and Palmitic Acid-Conjugated siRNA Improve PEGylated Polyplex Stability and Enhance in Vivo Pharmacokinetics and Tumor Gene Silencing. Biomaterials 2016, 97, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zhang JH; Chung TDY; Oldenburg K R A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol Screening 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- (52).Evans BC; Nelson CE; Yu SS; Beavers KR; Kim AJ; Li H; Nelson HM; Giorgio TD; Duvall CL Ex Vivo Red Blood Cell Hemolysis Assay for the Evaluation of pH-Responsive Endosomolytic Agents for Cytosolic Delivery of Biomacromolecular Drugs. J. Visualized Exp 2013, 73, 50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Evans BC; Hocking KM; Osgood MJ; Voskresensky I; Dmowska J; Kilchrist KV; Brophy CM; Duvall CL MK2 Inhibitory Peptide Delivered in Nanopolyplexes Prevents Vascular Graft Intimal Hyperplasia. Sci. Transl Med. 2015, 7, 291ra95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Manders EMM; Verbeek FJ; Aten JA Measurement of Co-Localization of Objects in Dual-Colour Confocal Images. J. Microsc 1993, 169, 375–382. [DOI] [PubMed] [Google Scholar]
- (55).Klumperman J; Raposo G The Complex Ultrastructure of the Endolysosomal System. Cold Spring Harbor Perspect. Biol 2014, 6, a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Nelson CE; Kim AJ; Adolph EJ; Gupta MK; Yu F; Hocking KM; Davidson JM; Guelcher SA; Duvall CL Tunable Delivery of siRNA from a Biodegradable Scaffold to Promote Angiogenesis in Vivo. Adv. Mater 2014, 26, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Martin JR; Nelson CE; Gupta MK; Yu F; Sarett SM; Hocking KM; Pollins AC; Nanney LB; Davidson JM; Guelcher SA; Duvall CL Local Delivery of PHD2 siRNA from ROS-Degradable Scaffolds to Promote Diabetic Wound Healing. Adv. Healthcare Mater 2016, 5, 2751–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Sarett SM; Kilchrist KV; Miteva M; Duvall CL Conjugation of Palmitic Acid Improves Potency and Longevity of siRNA Delivered via Endosomolytic Polymer Nanoparticles. J. Biomed. Mater. Res., Part A 2015, 103, 3107–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Jain RK; Stylianopoulos T Delivering Nanomedicine to Solid Tumors. Nat. Rev. Clin. Oncol 2010, 7, 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Yuan F; Dellian M; Fukumura D; Leunig M; Berk DA; Torchilin VP; Jain RK Vascular Permeability in a Human Tumor Xenograft: Molecular Size Dependence and Cutoff Size. Cancer Res. 1995, 55, 3752–3757. [PubMed] [Google Scholar]
- (61).Yuan F; Leunig M; Huang SK; Berk DA; Papahadjopoulos D; Jam RK Microvascular Permeability and Interstitial Penetration of Sterically Stabilized (Stealth) Liposomes in a Human Tumor Xenograft. Cancer Res. 1994, 54, 3352–3356. [PubMed] [Google Scholar]
- (62).Mathew R; Karantza-Wadsworth V; White E Role of Autophagy in Cancer. Nat. Rev. Cancer 2007, 7, 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).White E The Role for Autophagy in Cancer. J. Clin. Invest 2015, 125 (1), 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Li S; Wandel MP; Li F; Liu Z; He C; Wu J; Shi Y; Randow F Sterical Hindrance Promotes Selectivity of the Autophagy Cargo Receptor NDP52 for the Danger Receptor Galectin-8 in Antibacterial Autophagy. Sci. Signaling 2013, 6, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Kim BW; Beom Hong S; Hoe Kim J; Hoon Kwon D; Song HK Structural Basis for Recognition of Autophagic Receptor NDP52 by the Sugar Receptor Galectin-8. Nat. Commun 2013, 4, 1613. [DOI] [PubMed] [Google Scholar]
- (66).Feng Y; He D; Yao Z; Klionsky DJ The Machinery of Macroautophagy. Cell Res. 2014, 24, 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Gupta B; Levchenko TS; Torchilin VP Intracellular Delivery of Large Molecules and Small Particles by Cell-Penetrating Proteins and Peptides. Adv. Drug Delivery Rev 2005, 57, 637–651. [DOI] [PubMed] [Google Scholar]
- (68).Lim SI; Lukianov CI; Champion JA Self-Assembled Protein Nanocarrier for Intracellular Delivery of Antibody. J. Controlled Release 2017, 249, 1–10. [DOI] [PubMed] [Google Scholar]
- (69).Lonn P; Kacsinta AD; Cui X-S; Hamil AS; Kaulich M; Gogoi K; Dowdy SF Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci. Rep 2016, 6, 32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Convertine AJ; Benoit DSW; Duvall CL; Hoffman AS; Stayton PS Development of a Novel Endosomolytic Diblock Copolymer for siRNA Delivery. J. Controlled Release 2009, 133, 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Kilchrist KV Galectin 8 Recruitment Analysis MATLAB Software. FigShare 2018, DOI: 10.6084/m9.figshare.7066472. [DOI] [Google Scholar]
- (72).Kilchrist KV Manders Coefficient of Overlap Calculation Analysis MATLAB Software. FigShare 2018, DOI: 10.6084/m9.fig-share.7066490. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.