Abstract
NMR spectroscopy is an emerging analytical tool for measuring complex drug product qualities, e.g., protein higher order structure (HOS) or heparin chemical composition. Most drug NMR spectra have been visually analyzed; however, NMR spectra are inherently quantitative and multivariate and thus suitable for chemometric analysis. Therefore, quantitative measurements derived from chemometric comparisons between spectra could be a key step in establishing acceptance criteria for a new generic drug or a new batch after manufacture change. To measure the capability of chemometric methods to differentiate comparator NMR spectra, we calculated inter-spectra difference metrics on 1D/2D spectra of two insulin drugs, Humulin R® and Novolin R®, from different manufacturers. Both insulin drugs have an identical drug substance but differ in formulation. Chemometric methods (i.e., principal component analysis (PCA), 3-way Tucker3 or graph invariant (GI)) were performed to calculate Mahalanobis distance (DM) between the two brands (inter-brand) and distance ratio (DR) among the different lots (intra-brand). The PCA on 1D inter-brand spectral comparison yielded a DM value of 213. In comparing 2D spectra, the Tucker3 analysis yielded the highest differentiability value (DM = 305) in the comparisons made followed by PCA (DM = 255) then the GI method (DM = 40). In conclusion, drug quality comparisons among different lots might benefit from PCA on 1D spectra for rapidly comparing many samples, while higher resolution but more time-consuming 2D-NMR-data-based comparisons using Tucker3 analysis or PCA provide a greater level of assurance for drug structural similarity evaluation between drug brands.
Keywords: Mahalanobis distance, distance ratio, PCA, Tucker3, graph invariant
INTRODUCTION
Industry practice for drug development and regulatory agency chemistry review require analytical similarity assessments between the reference listed drug (RLD) and its generic or biosimilar product (1–6). The physicochemical characterizations can be analytically challenging for complex drug products. The drugs can be complex mixtures (e.g., heparin, (7,8) glatiramer acetate (9), or conjugated estrogens (10)) from synthetic or biological processes. Some are also complex because of their large molecular size (e.g., protein therapeutics (11, 12)) or their formulation (liposomes, emulsions, or extended release coated forms (13)).
Higher order structure (HOS) and chemical composition similarity are critical quality attributes (CQAs) for many of the drugs described above. Recently, owing to the development of new NMR hardware, solution state 1- and 2-dimenstional (1/2D) NMR spectroscopy has become more practical for complex drug analysis, especially for injectable solutions where NMR can be non-invasively applied on the formulated form of the drug products (2,3,6,7,14–20). NMR spectra can be used to identify changes in chemical composition and HOS through characteristic peak patterns (i.e., peak positions, intensities, and line-widths). Simple 1D spectra may be collected in rapid fashion and heteronuclear 2D spectra, although requiring longer experimental time, have the advantages of resolving peaks and identifying finer details of molecular structure where changes may have occurred. While 1D NMR spectra of complex drugs have been analyzed using principal component analysis (PCA) (21,22), chemometric approaches for 2D spectral comparison have been hampered by difficulties in developing algorithms and software capable of supporting large data sets (i.e., 2048 × 2048 spectra yield 4,194,304 points) that require regularization to align multiple 2D spectra sets. Recently, PCA methods have been developed to compare 2D spectra of the protein therapeutic filgrastim (6) and polymeric low molecular weight heparin products (4). Thus, for protein or carbohydrate-based drugs, NMR peak locations or chemical shifts directly reflect chemical composition and any folded secondary and tertiary structure elements. In addition to the well-known PCA, other chemometric methods like the 3-way Tucker3 method (23–25) and sequential nearest neighbor (SNN) graph invariant (GI) (26) can be applied to 2D NMR spectra analysis. The SNN-GI was applied to filgrastim where 2D 1H-1H NOESY spectra were compared using the method (1). To our knowledge, Tucker3 method has not been used for any drug product NMR spectral analysis (27), and the SNN-GI method has not been applied to any 2D heteronuclear NMR spectral comparisons. A robust and systematic evaluation of these chemometric approaches has not been reported.
Therefore, herein both 1D 1H and 2D heteronuclear 1H-13C NMR spectra on the two differently formulated human insulin injection dosage forms were analyzed. Both drug products contained the identical regular insulin at the same strength of U-100. The differences in insulin HOS across brands (inter-brand) were compared by the scale-invariant Mahalanobis distance (28,29). This work represents an example of deriving a quantitative similarity comparison (i.e., a potential similarity metric) between an RLD and a generic or biosimilar drug product. In addition, a new distance ratio (DR), quantifying the variation among several lots of the same brand (intra-brand), was calculated to represent the normal range of differences or comparability metrics found in a given drug product. The results showed the Tucker3 analysis on 2D spectra yielded the best differentiability among the tested methods; and when lot-to-lot variation of Novolin R® was evaluated, the advantage of higher resolution 2D NMR spectral data over lower resolution 1D spectral data for comparisons were not as substantial as expected from the higher resolution data.
MATERIALS AND METHODS
NMR Spectroscopy
Insulin drug products were directly used as NMR samples. Briefly, 0.5 mL of insulin U-100 was mixed with 30 μL of D2O and loaded into a 5-mm 541-PP-7 NMR tube (Wilmad, Vineland, NJ). The protein concentration for all NMR samples was ~0.6 mM as indicated on the formulation labels. All NMR spectra were collected on a Bruker Ascend™ 850 MHz spectrometer equipped with a 5-mm TCI Z-gradient cryogenic probe. The experimental probe air temperature was set at 25°C and chemical shifts were internal or indirectly referenced (30). The carrier frequencies were set at 4.79 and 19.5 ppm for 1H and 13C nuclei, respectively. A1D 1H pulse sequence using the 3919 Watergate water suppression was used. The acquisition scans were composed of 32 k complex data points and 128 scans were averaged. The total 1D experimental time was 8 min. The spectral width was 14 ppm. The free induction decays (FIDs) were apodized with a Gaussian window function at 1 Hz line broadening and zero filled to 64 k before Fourier transformation. All 1D spectra were normalized to the summed spectrum area at an intensity value of 10,000 in MestReNova 10.0.1 (Mestrelab Research). For 2D data, spectral widths of 1H and 13C dimensions were 14 and 17 ppm, respectively. The number of complex data points collected along 1H and 13C dimensions were 2048 and 280, respectively. The multiplicity separated MS-HSQC pulse sequence was used (31). The total experimental time for each 2D spectrum was 7 h on these insulin drug samples at natural abundance (1.1% 13C). The data processing was performed using NMRPipe (32).
Principal Component Analysis
The 1D 1H spectra, excluding the regions of 1.15–1.25, 2.2–2.4, and 3.2–7.3 ppm, were binned at 0.01 ppm resolution, resulting in a total of 961 bins or observations. Spectral intensities were summed within each bin. The new summed intensities were subject to integrity check and Pareto scaling. The principal component analysis (PCA) was performed on all 24 spectra. The software MestReNova 10.0.1 (Mestrelab Research) was used for all data processing and PCA.
For 2D 1H-13C spectra, the core methyl region from 0 to 1.5 ppm along 1H and 12–27 ppm along 13C axes were evaluated and the methyl peak for ethanol was excluded from analysis. To compare spectral peak intensities at common 2D coordinates of all 24 spectra, the NMR data points were interpolated at pre-defined grid points, (6) which are coordinates at grid sizes of 0.005 and 0.05 ppm along 1H and 13C dimensions, respectively. The linear-interpolation function “meshgrid” within MATLAB 9.0 (The MathWorks Inc.) was employed. The 2D data matrix was then converted to a 1D array by appending each row elements sequentially. The 1D array consists of 83,979 (= 279 × 301) data points. The 1D array was further cleaned by removing intensities below S/N of 8, which resulted in 1044 data points in each array. Prior to PCA, each data point was mean subtracted and normalized by standard deviation. PCA was performed on the normalized 1D arrays using statistics toolbox of MATLAB 9.0.
Tucker3 Analysis
The Tucker3 analysis can only be performed on arrayed 2D spectra (24). The same set of 24 regularized 2D spectral matrices (279 × 301) in PCA analysis was arrayed to form a 3-way matrix (24 × 279 ×3 01), corresponding to modes A, B, and C. The 3-way matrix served as input for Tucker3 analysis using an R package. The outputs include a core 3D array (H), three component matrices (A, B, and C), and three diagonal matrices (La, Lb, and Lc) of each mode, explained in the R-package manual (https://cran.r-project.org/web/packages/ThreeWay/ThreeWay.pdf) and Kiers et al. (33). The diagonal elements in the La matrix (24 × 24) were treated as “intrinsic eigenvalues” in the R-package. The PC scores, PC1 and PC2, were calculated from both the first two columns in component matrix A and the first two diagonal values in matrix La. The corresponding R codes, adopted from the T3funcrep in ThreeWay library, were attached in supplementary material.
Graph Invariant
Within the same core methyl regions of the 2D 1H-13C NMR spectra defined before for PCA, a total of N (N = 24) cross-peaks with the S/N over 8 were picked using Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco). Ideally, N equals the number of methyl groups within a protein. The cross-peaks were sorted according to their intensities such that peak P1 to PN will have the highest to lowest intensities, respectively. The N × N distance matrix D was calculated as , where Δ13Cij and Δ1Hij are chemical shift differences between peaks Pi and Pj along the 13C and 1H dimensions, respectively. For the graph invariant method, the averaged SNN distances (ASD) were calculated for each value of number of graph peaks (NGP), ranging from 2 to N. For each NGP value, the distance matrix D was reduced to the size of NGP × NGP, retaining the stronger peaks with indices of 1 to NGP. Within the reduced matrix, for each row i, the shortest distance (graph edge) from elements Dij, j = 1, 2, 3…i−1, was chosen. All the shortest distances of the reduced NGP × NGP matrix were averaged to yield the value of ASD for each NGP. For every spectrum, the graph invariant curve of ASD versus NGP was calculated (Fig. S1).
Mahalanobis Distance
Mahalanobis distance (DM) was used as a metric for inter-brand comparisons (28). The PCA and Tucker3 scores, PC1 and PC2, were used as coordinates for DM calculations. SNN-GI curves were reduced to PCs as well. Then for each method, PC1/2 scores were stored as 2-element observation vectors Ha[i], Na[i], Hb[i] and Nb[i], where H and N refer to Humulin R® and Novolin R®, respectively, a and b refer to duplicated measurements, and i ranges from 1 to 6 to index lot numbers. The MATLAB function Mahalanobis was used. Equation (1) below describes the calculation used to derive Mahalanobis distances (DM),
| (1) |
where S = (SH + SN)/2 and SH and SN are the sample covariance matrices of and , respectively. Here, and (Eqs. (2) and (3)) are the mean vectors of H and N, respectively.
| (2) |
| (3) |
Distance Ratio
The same PC1/2 scores used in Mahalanobis distance were used for distance ratio (DR) calculations. For intra-brand variability, the squared distances between each measurement to the center (mean) of all the intra-brand measurements were calculated and averaged; the value was then normalized by the averaged square distances between the duplicated measurements to yield DR. In Eq. (4), ‖x‖ is the Euclidean norm; Ha, Hb, and can be replaced with Na, Nb, and to yield DR results for Novolin R® datasets.
| (4) |
RESULTS
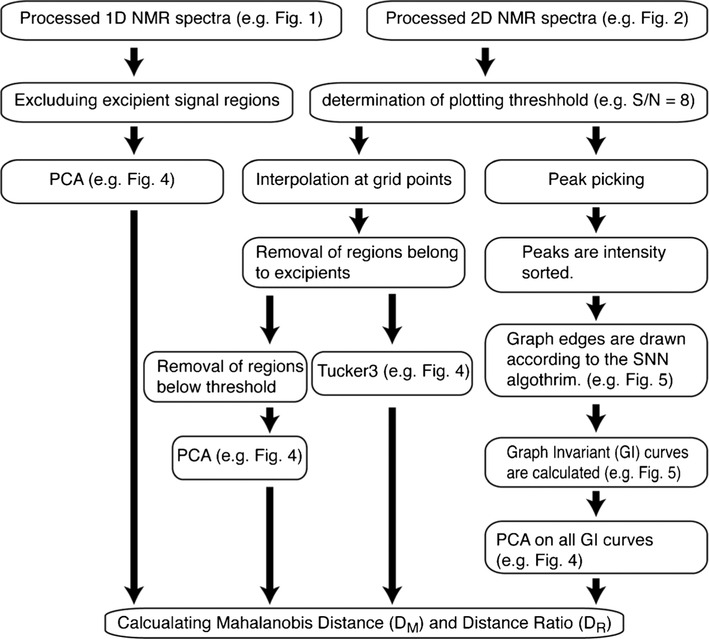
In this study, 1D 1H (Fig. 1) and 2D 1H-13C heteronuclear NMR spectra (Fig. 2) were collected on 12 insulin drug products; 6 lots each of Humulin R® or Novolin R® (Table S1). All NMR experiments were performed in duplicate to result in a total of 24 sets of 1D and 2D spectra for chemometric method analysis. To obtain a measure of the difference between the spectra of the two closely related comparator insulin brands, three orthogonal chemometric methods of PCA, Tucker3, or SNN-GI were applied (Fig. 3). All methods were unsupervised and no prior knowledge of NMR peak assignments was required.
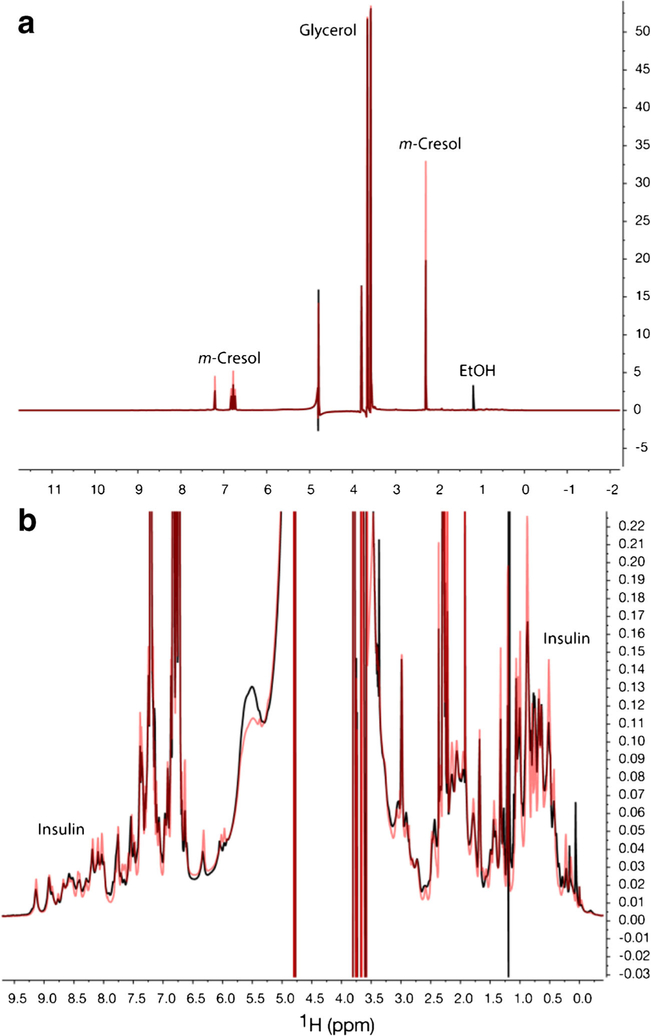
Fig. 1.
1D 1H NMR spectra of Humulin R® (black) and Novolin R® (red). The full spectra (a) showed the domination of excipient signals. The vertically zoomed spectra (b) showed resonances from insulin
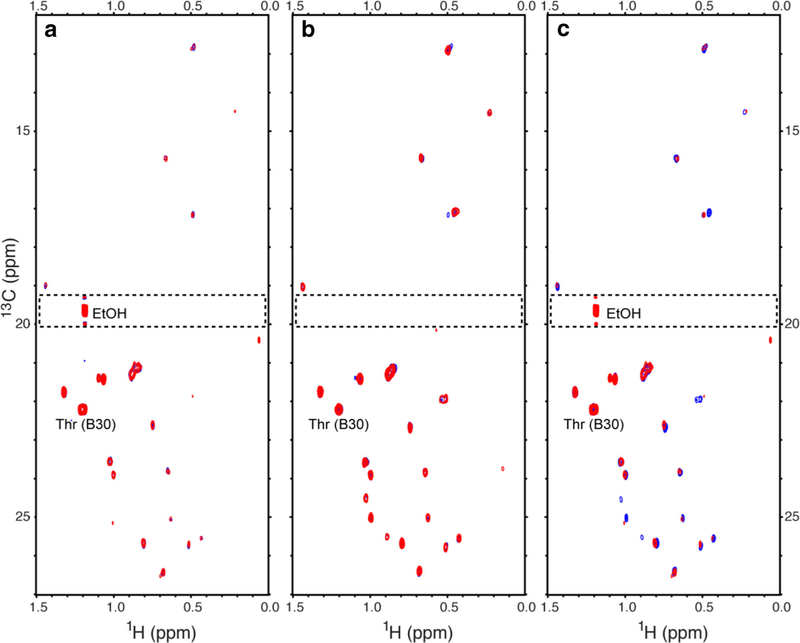
Fig. 2.
Plots of the 0 to 1.5 ppm proton and 13 to 27 ppm carbon region (methyl region) of 1H-13C multiplicity-separated (MS)-HSQC spectra for Humulin R® and Novolin R®. The overlaid spectra are HumulinR2 (blue) and HumulinR4 (red) (left panel), NovolinR1 (blue) and NovolinR3 (red) (middle panel), and HumulinR4 (red) and NovolinR1 (blue) (right panel). All spectra were plotted at a threshold of S/N of 8. The dashed rectangular regions, including the methyl peak from residual solvent ethanol, were excluded for PCA, Tucker3, and GI evaluation procedures. The strongest insulin peak was the annotated methyl resonance (Thr (B30)).
Fig. 3.
Schematic flow chart of PCA, Tucker3, and SNN-GI chemometric analysis methods on NMR spectra
PCA on 1D Spectra
In 1D 1H spectra, excipient signals from m-cresol and glycerol dominated spectra for both Humulin R® and Novolin R® (Fig. 1a). Residual process solvent signals from ethanol were observed in Humulin R®. Differences in insulin signals between the two formulations, after vertically zooming, can be visually observed (Fig. 1b). The peak line shape in Novolin R® spectrum was sharper than Humulin R®, indicating fast exchange among different HOS forms in Novolin R® (34). Based on visualization, 1D NMR was sensitive enough to observe insulin HOS differences found in the different formulations.
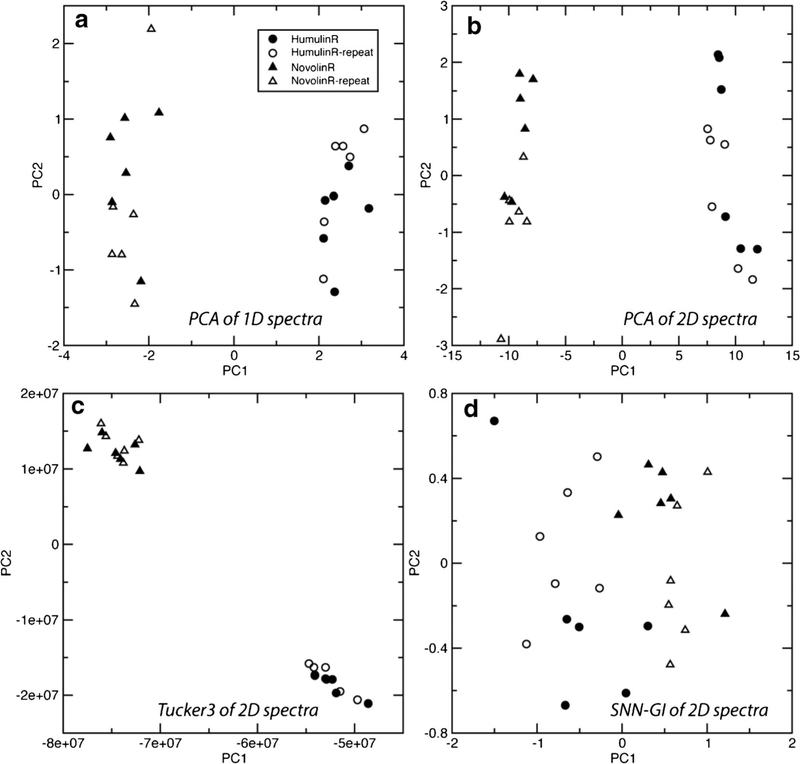
The PCA was performed directly on 24 1D spectra with insulin only resonance regions. The PC1 (66%) and PC2 scores (12%) totally account for 78% of the spectral variability (Fig. 4a, Table S2). PC1 components were sufficient to separate the two brands. The bins at chemical shifts of 0.76 and 0.87 ppm, methyl peaks, had the largest PC1 loading values. PC2 components differentiated intra-brand inter-lot spectra and there were no clear peaks dominant in the PC2 loading.
Fig. 4.
PCA score plots from chemometric methods of PCA on 1D spectra (a), PCA on 2D spectra (b), Tucker3 on 2D spectra (c), and PCA of SNN-GI curves (d)
PCA on 2D Spectra
For 2D heteronuclear NMR, a recently developed multiplicity-separated heteronuclear (1H-13C) single quantum correlation (MS-HSQC) experiment was applied, which generates clean spectra of methyls (CH3) (31). The MS-HSQC spectra (Fig. 2) enhanced the data comparison by selectively observing methyl peaks from insulin for HOS comparison. All methyl peaks detected were from human insulin except for one from the residual solvent ethanol methyl resonance (δ13C of 19.6 ppm and δ1H of 1.19 ppm) observed in all Humulin R® spectra (dashed boxes in Fig. 2). A visual inspection showed the inter-lot intra-brand spectra (Fig. 2a, b), overlaid very well. In addition, most observable peaks of inter-brand spectra aligned well, except for some missing peaks from Humulin R® HSQC spectra, e.g., resonances around δ13C of 25 ppm and δ1H of 1 ppm (Fig. 2c). In all spectra, the methyl resonances of Thr-30b showed the strongest intensity (Fig. 2). This strong intensity was attributed to the location of the residue in the C-terminal flexible portion of the peptide (35).
The PCA method of Ghasriani et al., which used interpolation for 2D 1H-15N HSQC NMR spectra, was used for this study (6). The PC1 (92%) and PC2 scores (2%) totally account for 94% of the spectral variability (Fig. 4b, Table S3). The largest PC1 loadings were in bins with 1H/13C chemical shifts of 1.22/22.2 and 0.795/25.7 ppm, corresponding to 2 methyl peaks from Thr-30b and Leu-13a, respectively (Fig. 2) (36). The largest PC2 loadings were in bins with 1H/13C chemical shift of 1.21/22.2 and 1.19/22.2 ppm, corresponding to the strong Thr-30b as well (Fig. 2). Similar to PCA of 1D spectra, both brands could be differentiated using the PC1 component and the loading had peaks from Leu-13a. The intra-brand or duplication variations were reflected along PC2 and the loading was from the strong Thr-30b peak, indicating that insulin concentration differences dominated PC2 (Fig. 4b).
Tucker3 on 2D Spectra
The Tucker3 analyses on 2D spectra were reported in NMR metabolomics (37) and relaxation studies (38). In this case, without unfolding the 2D spectra into 1D array prior to PCA, the entire set of 24 2D spectra or matrices were aligned as a 3-way array and decomposed using Tucker3 method. The first two diagonal values in the resulting diagonal matrix were the largest elements. These elements accounted for 82.5 and 4.9% of the spectral variability. Principal scores (Fig. 4c and Table S4), similar to PCA, can cluster spectra for comparison. The largest spectral component in PC1 was from the Leu-6b peak at 1H/13C chemical shifts of 0.895/25.65 ppm. Due to chemical exchange differences, the Leu-6b peak showed strong and weak intensity in Novolin R® and Humulin R®, respectively. The Tucker3 method appeared to weighs spectral features of all peaks more evenly, rather than emphasizing the strongest peaks in the spectra.
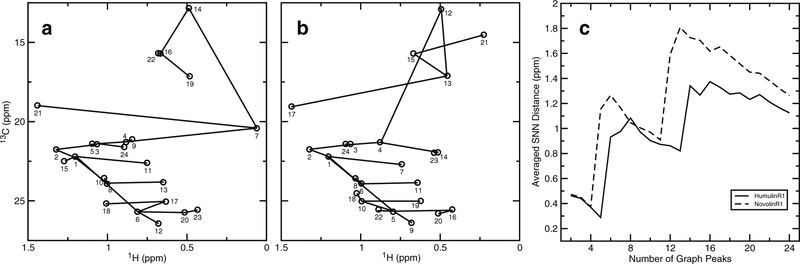
Graph Invariant on 2D Spectra
Among various mathematic representations of the graph invariant methods (26,39), the sequential nearest neighbor graph invariant (SNN-GI) approach was reported to compare 2D NMR spectra of filgrastim (1,26). However, the previous GI values were dominated by the much larger peak intensity values (1). Here, the SNN-GI approach was modified to remove the peak intensity from the invariant calculations because the peak intensity had been taken into account during the initial peak-sorting step (Fig. 3). In these spectra, the most intense peak was always that of the methyl proton carbon pairs of Thr-30b (Fig. 2), which was labeled as peak #1 in all graphs (Fig. 5). As shown in Fig. 5a, b, when NGP value reached 24, all peaks were connected with 23 graph edges. The GI curve was composed of ASD values as a function of NGP (Fig. 5c). Though the SNN-GI curve differences can be visualized, when all 24 spectra were graphed (Fig. S1), the difference was not significant enough to differentiate the two brands of insulin. PCA on 24 GI curves was performed to quantify the curve differences. The PC1 (82%) and PC2 scores (10%) accounted for 92% of the SNN-GI curve variability (Fig. 4d, Table S5).
Fig. 5.
The SNN-GI results from 2D 1H-13C HSQC NMR spectra. The SNN graphs of HumulinR1 (a) and NovolinR1 (b) and the corresponding GI curves (c) are shown. Each graph contains 24 nodes (peaks) and 23 edges (distances). The peaks (circles) were intensity sorted. The averaged SNN distance (ASD) values were calculated for the number of graph peaks ranging from 2 to 24 (c)
Quantifying Inter-Brand Spectra Difference
In computing the inter-brand spectra difference, Mahalanobis distances (DM) (a measure of separation in standard deviation) are considered between Humulin R® and Novolin R®. The Mahalanobis distances can only be calculated from PCA scores and as such, the 24 GI curves (Fig. S1) were reduced to principle components (Fig. 4d). The results showed Tucker3 analysis on the 3-way array composed of aligned 2D NMR spectra yielded the largest value of 305 in DM (Table I); therefore, Tucker3 was the most sensitive (i.e., had the largest dynamic range) of the methods compared for differentiating NMR spectral features between the two drug products. The PCA of 2D and 1D NMR spectra were slightly less differentiating with DM values of 255 and 213, respectively. The Mahalanobis distance from GI curves was 40, which was significantly smaller than the PCA and Tucker3 methods.
Table I.
Quantified NMR Spectra Difference Results
| NMR spectra | Chemometric method | Mahalanobis distances (DM) | Distance ratio (DR) | |
|---|---|---|---|---|
| Humulin R® | Novolin R® | |||
| 1D | PCA | 213 | 0.529 | 1.26 |
| 2D | PCA | 255 | 0.829 | 0.622 |
| Tucker3 | 305 | 4.37 | 1.38 | |
| SNN-GI | 40.4 | 0.449 | 0.414 | |
Quantifying Intra-Brand Spectra Difference
For intra-brand comparison, a new distance ratio (DR) metric was developed. DR was calculated using the same PC1 and PC2 scores (Fig. 4) used in DM calculations. The larger the DR value, the more differentiation between inter-lot spectra is observed and, thus, the better the method reproducibility. The Tucker3 method on 2D spectra was the best for identifying intra-brand inter-lot differences with the highest DR values of 4.37 and 1.38 for Humulin R® and Novolin R®, respectively (Table I). The GI method yielded the lowest DR values (below 0.5) for both insulin drugs. Therefore, similar to the inter-brand comparisons, the GI method on 2D spectra were not performing as well as the other approaches in differentiating intra-brand inter-lot variation. The PCA-2D method worked better than the PCA-1D method for Humulin R®, with a higher DR value of 0.829. Surprisingly, the PCA-1D method worked better for Novolin R® with a higher DR value of 1.26, which was close to Tucker3 value of 1.38.
CONCLUSION AND DISCUSSION
Here, we have described the three chemometric methods, PCA, Tucker3 and SNN-GI, to quantify NMR spectral differences between insulin drug products, Humulin R® and Novolin R®, and among different lots of each brand. The drug products have the same API in the same dosage strength U-100 and, therefore, the present inter-brand difference measure can be considered as a HOS analytical similarity comparison between an RLD and a pseudo-generic or -biosimilar formulation. The chemometric methods of Tucker3 on 2D NMR spectra yielded the best differentiability for inter-brand spectra similarity comparisons based on the largest Mahalanobis distance value. The SNN-GI method yielded the worst distance scores among all tested methods. The differentiability is a measure of the sensitivity of NMR/chemometric as an analytical tool to identify HOS difference due to formulation differences. The results demonstrated the Tucker3 and PCA methods, yielding DM values in the range of 200–300, were more sensitive and robust for this purpose than the graph method as implemented here. Practically, an acceptable DM value will be product and NMR spectrum dependent, and can be calculated using bootstrap sampling method on multiple lots of the innovator products.
The sensitivity of NMR to differentiate intra-brand inter-lot HOS difference was evaluated quantitatively as well. The comparison is relevant for analytical comparability testing within brand before and after manufacture changes. To take into account NMR method reproducibility in distance ratio DR calculations, spectral differences between duplicates were used to normalize inter-lot variations. Again, the Tucker3 analysis on 2D NMR spectra was the best differentiator and the GI method was the least differentiating, based on the DR value comparisons. Unexpectedly, when the drug sample had sharper lines and higher S/N, i.e., Novolin R®, 1D NMR with PCA showed better inter-lot differentiability than 2D NMR with PCA; the DR scores between PCA-1D and Tucker3–2D were also close. Given the fact that 1D 1H NMR experiment takes only minutes to acquire, high-throughput screening using 1D NMR and PCA could be practical for complex therapeutic comparability testing.
The failure of GI method on 2D spectra to differentiate 2D NMR spectra might be related to the graph invariant calculation (1). The current published GI method used the averaged inter-peak distance as a single graph invariant, which caused a significant dimension reduction and spectral variation removal. Though Tucker3 analysis on arrayed 2D spectra is highly differentiating, for ease of use and throughput, 1D NMR may be considered as a practical choice as a quality control analytical method for comparability test within a brand. In addition, 2D NMR spectral data (which takes longer to acquire but has higher resolution) with Tucker3 or classical PCA may be more suitable for the purposes of biosimilarity assessment and complex generic drug development.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the reviewer for pointing us to the Tucker3 method. We thank Prof. P.M. Kroonenberg for helpful discussion on Tucker3 application.
FUNDING INFORMATION
Support for this work from the US FDA CDER Critical Path Award is gratefully acknowledged.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1208/s12249-017-0911-1) contains supplementary material, which is available to authorized users.
Publisher's Disclaimer: Disclaimer This article reflects the views of the author and should not be construed to represent U.S. FDA’s views or policies.
REFERENCES
- 1.Zuperl S, Pristovsek P, Menart V, Gaberc-Porekar V, Novic M. Chemometric approach in quantification of structural identity/similarity of proteins in biopharmaceuticals. J Chem Inf Model. 2007;47(3):737–43. [DOI] [PubMed] [Google Scholar]
- 2.Poppe L, Jordan JB, Rogers G, Schnier PD. On the analytical superiority of 1D NMR for fingerprinting the higher order structure of protein therapeutics compared to multidimensional NMR methods. Anal Chem. 2015;87:5539–45. [DOI] [PubMed] [Google Scholar]
- 3.Poppe L, Jordan JB, Lawson K, Jerums M, Apostol I, Schnier PD. Profiling formulated monoclonal antibodies by H-1 NMR spectroscopy. Anal Chem. 2013;85(20):9623–9. [DOI] [PubMed] [Google Scholar]
- 4.Guerrini M, Rudd TR, Mauri L, Macchi E, Fareed J, Yates EA, et al. Differentiation of generic Enoxaparins marketed in the United States by employing NMR and multivariate analysis. Anal Chem. 2015;87(16):8275–83. [DOI] [PubMed] [Google Scholar]
- 5.Kozlowski S, Woodcock J, Midthun K, Sherman RB. Developing the nation’s biosimilars program. New Engl J Med. 2011;365(5):385–8. [DOI] [PubMed] [Google Scholar]
- 6.Ghasriani H, Hodgson DJ, Brinson RG, McEwen I, Buhse LF, Kozlowski S, et al. Precision and robustness of 2D-NMR for structure assessment of filgrastim biosimilars. Nat Biotechnol. 2016;34(2):139–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keire DA, Buhse LF, Al-Hakim A. Characterization of currently marketed heparin products: composition analysis by 2D-NMR. Anal Methods-Uk. 2013;5(12):2984–94. [Google Scholar]
- 8.Ye HP, Toby TK, Sommers CD, Ghasriani H, Trehy ML, Ye W, et al. Characterization of currently marketed heparin products: key tests for LMWH quality assurance. J Pharmaceut Biomed. 2013;85:99–107. [DOI] [PubMed] [Google Scholar]
- 9.Rogstad S, Pang E, Sommers C, Hu M, Jiang XH, Keire DA, et al. Modern analytics for synthetically derived complex drug substances: NMR, AFFF-MALS, and MS tests for glatiramer acetate. Anal Bioanal Chem. 2015;407(29):8647–59. [DOI] [PubMed] [Google Scholar]
- 10.Levy MJ, Boyneii MT, Rogstad S, Skanchy DJ, Jiang XH, Geerlof-Vidavsky I. Marketplace analysis of conjugated estrogens: determining the consistently present steroidal content with LC-MS. AAPS J. 2015;17(6):1438–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arzhantsev S, Vilker V, Kauffman J. Deep-ultraviolet (UV) resonance Raman spectroscopy as a tool for quality control of formulated therapeutic proteins. Appl Spectrosc. 2012;66(11):1262–8. [DOI] [PubMed] [Google Scholar]
- 12.Hmiel LK, Brorson KA, Boyne MT. Post-translational structural modifications of immunoglobulin G and their effect on biological activity. Anal Bioanal Chem. 2015;407(1):79–94. [DOI] [PubMed] [Google Scholar]
- 13.Korang-Yeboah M, Rahman Z, Shah D, Mohammad A, Wu SY, Siddiqui A, et al. Impact of formulation and process variables on solid-state stability of theophylline in controlled release formulations. Int J Pharm. 2016;499(1–2):20–8. [DOI] [PubMed] [Google Scholar]
- 14.Panjwani N, Hodgson DJ, Sauve S, Aubin Y. Assessment of the effects of pH, formulation and deformulation on the conformation of interferon alpha-2 by NMR. J Pharm Sci-Us. 2010;99(8):3334–42. [DOI] [PubMed] [Google Scholar]
- 15.Jin X, Kang S, Kwon H, Park S, Heteronuclear NMR. As a 4-in-1 analytical platform for detecting modification-specific signatures of therapeutic insulin formulations. Anal Chem. 2014;86(4):2050–6. [DOI] [PubMed] [Google Scholar]
- 16.Aubin Y, Jones C, Freedberg DI. Using NMR spectroscopy to obtain the higher order structure of biopharmaceutical products. Biopharm Int. 2010;Supplement:28–34. [Google Scholar]
- 17.Aubin Y, Hodgson DJ, Thach WB, Gingras G, Sauve S. Monitoring effects of excipients, formulation parameters and mutations on the high order structure of filgrastim by NMR. Pharm Res. 2015;32:3365–75. [DOI] [PubMed] [Google Scholar]
- 18.Aubin Y, Gingras G, Sauve S. Assessment of the three-dimensional structure of recombinant protein therapeutics by NMR fingerprinting: demonstration on recombinant human granulocyte macrophage-colony stimulation factor. Anal Chem. 2008;80(7):2623–7. [DOI] [PubMed] [Google Scholar]
- 19.Arbogast LW, Brinson RG, Marino JP. Mapping monoclonal antibody structure by 2D (13)C NMR at natural abundance. Anal Chem. 2015;87(7):3556–61. [DOI] [PubMed] [Google Scholar]
- 20.Amezcua CA, Szabo CM. Assessment of higher order structure comparability in therapeutic proteins using nuclear magnetic resonance spectroscopy. J Pharm Sci-Us. 2013;102(6):1724–33. [DOI] [PubMed] [Google Scholar]
- 21.Chen K, Long DS, Lute SC, Levy MJ, Brorson KA, Keire DA. Simple NMR methods for evaluating higher order structures of monoclonal antibody therapeutics with quinary structure. J Pharmaceut Biomed. 2016;128:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zang QD, Keire DA, Buhse LF, Wood RD, Mital DP, Haque S, et al. Identification of heparin samples that contain impurities or contaminants by chemometric pattern recognition analysis of proton NMR spectral data. Anal Bioanal Chem. 2011;401(3):939–55. [DOI] [PubMed] [Google Scholar]
- 23.Kiers HAL. Towards a standardized notation and terminology in multiway analysis. J Chemom. 2000;14(3):105–22. [Google Scholar]
- 24.Kroonenberg PM, Basford KE, Gemperline PJ. Grouping three-mode data with mixture methods: the case of the diseased blue crabs. J Chemom. 2004;18(11):508–18. [Google Scholar]
- 25.Tucker LR. Some mathematical notes on 3-mode factor analysis. Psychometrika. 1966;31(3):279. [DOI] [PubMed] [Google Scholar]
- 26.Randic M, Novic M, Vracko M. Novel characterization of proteomics maps by sequential neighborhoods of protein spots. J Chem Inf Model. 2005;45(5):1205–13. [DOI] [PubMed] [Google Scholar]
- 27.Bro R Review on multiway analysis in chemistry—2000–2005. Crit Rev Anal Chem. 2006;36(3–4):279–93. [Google Scholar]
- 28.Rencher AC. Methods of multivariate analysis. 2nd ed. Hoboken: Wiley-Interscience; 2003. [Google Scholar]
- 29.Brereton RG. The Mahalanobis distance and its relationship to principal component scores. J Chemom. 2015;29(3):143–5. [Google Scholar]
- 30.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, et al. H-1, C-13 and N-15 chemical-shift referencing in biomolecular NMR. J Biomol NMR. 1995;6(2):135–40. [DOI] [PubMed] [Google Scholar]
- 31.Chen K, Freedberg DI, Keire DA. NMR profiling of biomolecules at natural abundance using 2D H-1-N-15 and H-1-C-13 multiplicity-separated (MS) HSQC spectra. J Magn Reson. 2015;251:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe—a multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6(3):277–93. [DOI] [PubMed] [Google Scholar]
- 33.Kiers HAL, Kroonenberg PM, Tenberge JMF. An efficient algorithm for Tuckals3 on data with large numbers of observation units. Psychometrika. 1992;57(3):415–22. [Google Scholar]
- 34.Patil SM, Keire DA & Chen K AAPS J. 2017. 10.1208/s12248-017-0127-z. [DOI] [PMC free article] [PubMed]
- 35.Keller D, Clausen R, Josefsen K, Led JJ. Flexibility and bioactivity of insulin: an NMR investigation of the solution structure and folding of an unusually flexible human insulin mutant with increased biological activity. Biochemistry. 2001;40(35):10732–40. [DOI] [PubMed] [Google Scholar]
- 36.Chang XQ, Jorgensen AMM, Bardrum P, Led JJ. Solution structures of the R-6 human insulin hexamer. Biochemistry. 1997;36(31):9409–22. [DOI] [PubMed] [Google Scholar]
- 37.Dyrby M, Baunsgaard D, Bro R, Engelsen SB. Multiway chemometric analysis of the metabolic response to toxins monitored by NMR. Chemometr Intell Lab. 2005;76(1):79–89. [Google Scholar]
- 38.Pedersen HT, Bro R, Engelsen SB. Towards rapid and unique curve resolution of low-field NMR relaxation data: trilinear SLICING versus two-dimensional curve fitting. J Magn Reson. 2002;157(1):141–55. [DOI] [PubMed] [Google Scholar]
- 39.Randic M A graph theoretical characterization of proteomics maps. Int J Quantum Chem. 2002;90(2):848–58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.