Abstract
Colorectal cancer (CRC) can be classified into different types. Chromosomal instable (CIN) colon cancers are thought to be the most common type of colon cancer. The risk of developing a CIN-related CRC is due in part to inherited risk factors. Genome-wide association studies have yielded over 40 single nucleotide polymorphisms (SNPs) associated with CRC risk, but these only account for a subset of risk alleles. Some of this missing heritability may be due to gene-gene interactions. We developed a strategy to identify interacting candidate genes/loci for CRC risk that utilizes both linkage and RNA-seq data from mouse models in combination with allele-specific imbalance (ASI) studies in human tumors. We applied our strategy to three previously identified CRC susceptibility loci in the mouse that show evidence of genetic interaction: Scc4, Scc5 and Scc13. 525 SNPs from genes showing differential expression in the mouse and/or a previous role in cancer from the literature were evaluated for allele-specific imbalance in 194 paired human normal/tumor DNAs from CIN-related CRCs. 103 SNPs showing suggestive evidence of ASI (31 variants with uncorrected p-values < 0.05) were genotyped in a validation set of 296 paired DNAs. Two variants in SNX10 (SCC13) showed significant evidence of allelic selection after multiple comparisons testing. Future studies will evaluate the role of these variants in combination with interacting genetic partners in colon cancer risk in mouse and humans.
Keywords: colorectal cancer, allele-specific imbalance, colon cancer susceptibility loci, Scc4, Scc5, Scc13
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer-related death in the United States.1 Genetic alterations drive the transformation of normal colon epithelium into adenomas and ultimately malignant adenocarcinomas.2 Pathways such as chromosomal instability (CIN), microsatellite instability, and CpG island methylator phenotype lead to genomic alterations and thus promote tumorigenesis.3 Estimates suggest that as many as 80 to 85 percent of sporadic colorectal cancers demonstrate CIN, which is marked by an accelerated rate of gains or losses of whole or partial chromosomes.3 In CRC, CIN is typically coupled with mutational activation of proto-oncogenes like KRAS, inactivation of tumor suppressor genes like APC and TP53, and loss of heterozygosity at 18q.3 One mechanism by which CIN leads to activation and inactivation of oncogenes and tumor suppressor genes, respectively, is by altering gene dosage as a result of copy number alterations.
Array comparative genomic hybridization (aCGH) studies can be used to identify refined regions of somatic copy number alterations in human colorectal tumors. Such studies have led to the identification of oncogenes and tumor suppressor genes that play a role in the development and progression of this cancer.4, 5 One important limitation of this approach is the inability to detect allele-specific copy number alterations. Allelic imbalance arises when there is complete loss of one allele or copy number gain of one allele relative to the other.6 This phenomenon can be detected by comparing the proportion of one allele to the other in cells from an individual who is constitutively heterozygous at that locus. In a proportion of loci showing allelic imbalance, one allele may exhibit preferential copy number gain or loss compared to the other allele. This preferential, or allele-specific, imbalance (ASI) may implicate the presence of a susceptibility allele or resistance allele at that genomic locus. Indeed in previous studies, ASI has been observed among 40% of mouse skin cancer susceptibility loci and at human variants associated with colon cancer risk by genome-wide association studies (GWAS).7-10 Technologies such as next-generation sequencing and quantitative genotyping permit identification of relative gains or losses of alleles in tumor DNA samples compared to germline DNA from the same individual.
To date, the identification of low-penetrance CRC susceptibility variants has largely been accomplished by population-based case-control GWAS. While these studies have identified over 40 independent low-penetrance variants from different populations, the proportion of genetic risk that they explain is modest and incomplete.11 Given that heritable factors are estimated to account for 12–35% of the risk for developing CRC,12 it has been proposed that the remainder of risk comes from rare variants, copy number variants, gene-gene interactions, gene-environment interactions and parent-of-origin effects—though overestimates of heritability are also a possible explanation for the missing heritability.13
Gene-gene interaction studies have not identified convincing combinations of variants contributing to CRC susceptibility. One complementary strategy for identifying genetic regions contributing to CRC susceptibility via genetic interactions is to exploit genetic mapping of susceptibility loci using linkage analysis in mouse models. The identification of genetic interactions is more straightforward in mice due the high degree of homogeneity among inbred mouse strains and the long stretches of linkage disequilibrium in the mouse crosses used for linkage mapping. Approximately 20 quantitative trait loci for CRC susceptibility have been mapped by crossing mouse strains resistant to and susceptible to chemically induced colon cancer or by looking for modifiers of transgenic mutations such as ApcMin which cause spontaneous development of intestinal adenomas.14-23 Mouse models of chemically induced CRC follow a similar multistage disease progression and develop tumors with many of the same mutations observed in human CIN tumors.24 This similarity suggests that the genes controlling cancer susceptibility in mice will be relevant to humans.25
Additional analysis using mouse models has led to the identification of loci involved in genetic interactions.17, 18, 26 One colon cancer susceptibility locus, Scc1, has been mapped to mouse chromosome 2 and refined to the candidate gene Ptprj.27 In humans, PTPRJ functions as a tumor suppressor by negatively regulating growth-promoting receptor tyrosine kinases. Two murine Scc loci, Scc5 and Scc13, show synergistic interactions with the Ptprj locus to enhance risk.18 The Scc5 locus also demonstrates a reciprocal interaction with Scc4, wherein the risk associated with the allele at Scc5 is dependent on the allele present at Scc4.26 In our previous studies, we showed that single nucleotide polymorphisms (SNPs) in PTPRJ were associated with susceptibility to CRC.28 Given the importance of variants in PTPRJ in human CRC risk, we were interested in what variants drive susceptibility at Ptprj-interacting loci. Importantly, the genes responsible for risk at Scc4, Scc5 and Scc13 are unknown but are intriguing candidates for study in the context of human colon cancer.
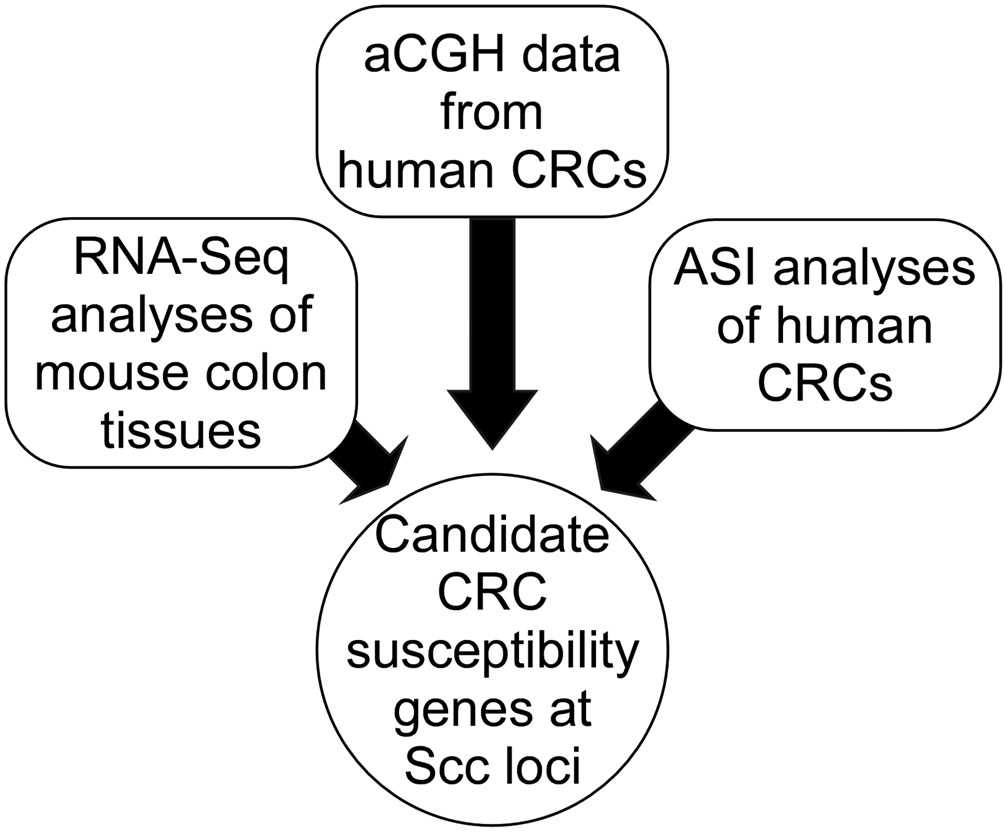
In the present study, we aimed to identify candidate CRC susceptibility genes and variants within the Ptprj-interacting Scc5 and Scc13 loci, as well as the Scc5-interacting locus Scc4. We employed a cross-species approach in which we integrated RNA-seq colon transcriptome data from the parental mouse lines used in the linkage studies with aCGH data and ASI mapping of human colon tumors (Figure 1).29 Here, we describe the results of using this approach to identify candidate genes for CRC susceptibility.
Figure 1: Experiment Design.
Data from three sources were used to identify candidate susceptibility genes for CRC. RNA-seq data from normal colons was generated from cancer susceptible STS/A and cancer resistant Balb/cHeA mice. aCGH data from human CRCs was used to confirm that SCC loci for study were showing copy number changes. Allele-specific imbalance (ASI) studies were completed on paired normal and colon tumor DNAs for the orthologous human loci to Scc4, Scc5 and Scc13.
Materials and Methods
Mouse Samples
All studies were approved by The Ohio State University Institutional Animal Care and Use Committee. Snap-frozen large intestines from 4–5 week old age-matched female Balb/cHeA and female STS/A mice were obtained from the Netherlands Cancer Institute. Colon specimens were homogenized in 1 mL Ribozol (Amresco, Solon, OH, USA) using medium power for 15-second pulses, followed by incubation on ice. Total RNA was isolated according to the Ribozol manufacturer’s protocol. RNA quantity and quality were assessed by NanoDrop-1000 and by Agilent Bioanalyzer. RNA samples used for library preparation had RNA integrity number greater than 8.
RNA-Seq and Analysis of Next-Generation Sequencing Data
The Illumina TruSeq RNA sample preparation kit was used to generate mRNA libraries from 2μg of total RNA isolated from the Balb/cHeA and STS/A colon specimens according to manufacturer’s recommended guidelines. Briefly, total RNA was selected for poly-A mRNA using poly-T oligo-attached magnetic beads. Poly-A-selected mRNA was next fragmented and primed for cDNA synthesis, reverse transcribed into first strand cDNA using SuperScript II reverse transcriptase (Life Technologies, Carlsbad, CA, USA), and subsequently synthesized into double-stranded cDNA using DNA polymerase I and RNase H supplied in the kit. Double-stranded cDNA fragments were subjected to an end repair process, the addition of a single ‘A’ base, and ligation of adapters. Finally, double-stranded cDNA was size selected, multiplexed and sequenced on an Illumina Genome Analyzer IIX.
Partek Genomic Suite (Partek, Inc., St. Louis, MO, USA) was used for performing differential expression analysis, alternative splicing analysis and SNP calling of the RNA-seq libraries. Each short sequence read was mapped to the corresponding position of the mouse genome (mm9 assembly) and reads not meeting the quality threshold were discarded. When performing differential expression analysis on the transcript level, the log likelihood ratio for each transcript was calculated among the samples using the number of reads that mapped to the transcripts according to the recommended RNA-seq analysis procedure by Partek (http://www.partek.com/Tutorials/microarray/User_Guides/RNASEQ.pdf). Next, p-values for each transcript were calculated via a chi-squared test. When performing the alternative-splicing quantification, a contingency table with two rows representing the samples and as many columns as the number of isoforms for each gene was created. Each entry in the table was estimated using an expectation/maximization algorithm. Then p-values were calculated by performing chi-squared statistics on the contingency table using the log likelihood ratio.
For the RNA-seq data, a base might not match the reference at the given position due to read errors, alignment errors or the presence of SNPs. Read errors were eliminated using the quality scores obtained by the sequencer. However, special care was taken when SNPs were identified and these were differentiated from potential alignment errors per Partek Genomic Suite recommendations (http://www.partek.com/Tutorials/microarray/User_Guides/NGS_Genotype_Likelihoods.pdf). For a possible SNP position, the likelihood of each genotype was calculated using the frequency of the bases. Finally, the log-odd-ratios were calculated and reported for the genotype with the maximum likelihood ratio. The RNA-seq data generated in this study are available in the Gene Expression Omnibus database (project identifier SRP056672).
Human Samples
Studies were approved by the Institutional Review Board at The Ohio State University. Study participants provided written informed consent for use of their tissues in research. Samples and their preparation have been previously described.10 Samples, all from Ohio CRC cases, consisted of a discovery set of 194 colon tumor/normal DNA pairs and a validation set of 296 colon tumor/normal DNA pairs. A pathologist confirmed all colon cancer diagnoses. To enrich for tumors showing CIN, samples were excluded if they were absent for any of the mismatch repair proteins, demonstrated microsatellite instability, were from the right side of the colon or showed a high degree of mucin.
Sources of DNA for this study included formalin-fixed paraffin-embedded (FFPE) tissue blocks, flash-frozen colon tissue, and blood samples. DNA isolation from FFPE tissues was performed by extraction of tissue from paraffin by xylene and ethanol washes, digestion by proteinase K treatment in lysis buffer, and purification by phenol/chloroform extraction and ethanol precipitation. DNA was assessed with a NanoDrop-1000 spectrophotometer for quantity and quality. Between 10 and 20ng of DNA were used for each genotyping reaction.
Choice of Tagging SNPs for study
SNPs tagging for linkage disequilibrium blocks at an R-squared cutoff of 0.80 within candidate genes were selected for inclusion in this study. The International HapMap Project “Annotate Tag SNP Picker” tool was used to select tagging SNPs. In total, 525 SNPs were assessed in the discovery sample set of human normal/tumor DNA pairs.
Sequenom Quantitative Genotyping and R-ratio calculations
Multiplexed primers for PCR amplification and allele-specific single-base extension reactions were designed using the Sequenom MassARRAY Assay Design 3.1 software (Supplemental Table 1). Mass spectrometry-based genotyping of paired tumor and normal DNA was performed using Sequenom MassARRAY iPlex Gold (Sequenom Inc., San Diego, CA, USA) according to the manufacturer’s protocol as described.10 Each 384-well Sequenom plate included four negative template controls (dH2O), two samples tested in duplicate, and four positive control DNAs.
As described previously, for all SNPs tested we scored preferential allelic imbalance by calculating the R-ratio for each normal/tumor DNA pair.10, 30, 31 The R-ratio represents the ratio of the two allele peak areas measured by the Sequenom MassARRAY iPLEX software in the normal heterozygous DNA divided by the ratio of the two allele peak areas in the paired tumor DNA (R-ratio = Normal(allele 1/allele 2) / Tumor(allele 1/allele 2)). For pairs in which the tumor was heterozygous for a SNP but the normal DNA from that individual failed to genotype, an average of the two normal alleles from all heterozygous normal samples at that SNP was used in place of the failed normal sample to calculate an R-ratio. Samples with R-ratio greater than 1.5 were deemed to have relative loss of the first allele (allele 1), while samples with R-ratio less than 0.67 were classified as showing relative loss of the second allele (allele 2).
Analysis of Allele-Specific Imbalance
The number of tumor samples from heterozygous individuals that showed relative loss of allele 1 (“allele 1 imbalance”) was compared to the number of tumor samples showing relative loss of allele 2 (“allele 2 imbalance”). A chi-squared test (df = 1) was used to assess the observed imbalances for statistically significant deviation from the expected 50:50 distribution of random allelic imbalances. SNPs with p-value < 0.10 were considered suggestive of preferential allelic imbalance and were therefore subjected to testing in the validation sample set to rule out false positives.
Validation Studies
Following statistical analysis of allele-specific imbalance in the discovery sample set, 103 variants with p-values < 0.10 were genotyped by Sequenom MassARRAY iPlex Gold in a replication sample set of 296 paired normal/tumor DNAs. The same quantitative genotyping protocol and statistical analyses used for the discovery sample set were employed with the validation sample set. Allele imbalance counts were combined for the discovery and validation sets and chi-squared analysis was conducted on the sum. Bonferroni correction was used to adjust for the number of statistical tests (n = 103).
Results
RNA-seq Analysis of CRC-Sensitive and CRC-Resistant Mouse Strains
The inbred mouse strains Balb/cHeA and STS/A differ in susceptibility to colon cancer when treated with carcinogens 1,2-dimethyl-hydrazine (DMH) or azoxymethane (AOM).24, 26 Balb/cHeA mice, the resistant strain, are reported to develop an average of 0.8–1.3 tumors per mouse and STS/A mice, the susceptible strain, develop an average of 8–18.4 tumors per mouse following DMH or AOM treatment, respectively.15, 22, 26, 32 Linkage analysis performed using recombinant inbred strains of Balb/cHeA by STS/A mice led to the identification of numerous quantitative trait loci (termed Susceptibility to colon cancer, or Scc, loci) that are linked to colon tumor formation.16, 17, 26, 27 The genes that underlie susceptibility at these murine QTLs may likewise play a role in human CRC susceptibility. Although specific polymorphisms identified in a mouse model of cancer susceptibility may not be conserved in human populations, it is likely that many genes that contribute to cancer susceptibility in the mouse serve similar roles in human disease.33 As a screening method to identify potential candidate CRC susceptibility genes of interest from our loci of interest (Scc4, Scc5 and Scc13), we performed RNA-seq from normal colon tissue of one female mouse per strain.
To identify SNPs, expression, and splicing pattern differences between the strains, the RNA-seq data for STS/A and Balb/cHeA was analyzed for the genes mapping within the three Scc loci of interest. Among the 119 genes within Scc4, 74 transcripts representing 55 genes showed different expression levels. At Scc5, 64 transcripts from 54 of the 137 annotated genes showed differential expression. Of the 185 genes at Scc13, 40 transcripts from 32 different genes showed differential expression. After eliminating genes that showed less than a 1.5-fold difference in expression and those that had very low expression (< 20 total reads in both strains), 95 genes expressed in the colon exhibited differential expression between the strains (Supplemental Table 2).
In addition to expression differences, numerous coding SNPs were identified between Balb/cHeA and STS/A (Supplemental Table 3). Non-synonymous variants were assessed for predicted disruption to protein structure and function using the in silico tools SIFT and PolyPhen-2. None of the amino-acid changing SNPs that differed between Balb/cHeA and STS/A were predicted to be damaging to protein structure or function. These findings do not rule out the possibility that the amino acid substitutions could have a moderate effect on protein function that could lead to changes in cancer susceptibility.
Array Comparative Genomic Hybridization Analysis of Human Colorectal Tumors
To determine if the human orthologous loci to Scc4, Scc5, and Scc13 showed copy number gains or losses, suggesting that these loci could be informative in ASI studies, we evaluated copy number data for the orthologous loci to the mouse Scc regions from published human aCGH studies as well as from our own aCGH studies.34-37 As the exact coordinates of the aCGH data were not available for most of the datasets, we looked at either whole chromosome arms or chromosome bands depending on the detail of the publicly available data. We observed a range of copy number aberrations depending on the locus and the study (Table 1). Mouse Scc13 correlates to human 4q25 and 7p14. Among our cohort of samples, 7 of 67 tumors (10.4%) showed loss of at least 50% of bacterial artificial chromosomes (BACs) mapping to the 4q25 band. In other published aCGH datasets, losses occurred at frequencies varying from 0% to 35%. Conversely, at the 7p14 locus, BACs exhibited gains in 16 of 67 tumors in our set (24%) and between 25% and 45% in other studies. In our samples, the BAC at 7p showing the highest frequency of genomic gains in these tumors (55%, or 37 of 67 tumors), CTB-111H21, encompasses a genomic segment at 7p14.3 containing the gene SCRN1. As this is not specifically mentioned, we do not know what the frequency of gain of this specific BAC is in the other published aCGH studies. As expected, 5q, where the APC tumor suppressor gene maps, shows frequent loss in CRCs across studies. Of particular note for the SCC5 locus, the BAC CTD-2202A14, which does not contain APC, is lost in 46% of our 67 tumors and contains several candidate genes from this study including the PTPRJ substrate PDGFRB. The SCC4 locus maps to human 2p25, which shows gains in 3–19% of tumors and losses in less than 10% of tumors.
Table 1:
Chromosomal losses and gains by aCGH at human SCC4, SCC5 and SCC13
| Arm | Gains/ Losses |
Our Cohort (n = 67) |
Nakao et al., 2004*36 (n=125) |
Jones et al., 2005*34 (n = 30) |
Lassman et al., 2007*37 (n = 22) |
Dyrso et al., 2011*35 (n = 40) |
|---|---|---|---|---|---|---|
| 2p | Gains | 13/67 (19%) |
10% | 3% | 15% | <10% |
| Losses | 0/67 (0%) |
<3% | 3% | 5% | <10% | |
| 5q | Gains | 1/67 (1.5%) |
<5% | 0% | 8% | <5% |
| Losses | 21/67 (31%) |
25% | 40% | <5% | 20% | |
| 4q | Gains | 2/67 (3%) |
<5% | 0% | 5% | <5% |
| Losses | 7/67 (10%) |
20% | 35% | 0% | 30% | |
| 7p | Gains | 16/67 (24%) |
35% | 45% | 25% | 43% |
| Losses | 0/67 (0%) |
<5% | 0% | 0% | 0% |
Percentages are approximations.
Sequenom Allele-Specific Imbalance Mapping
As there was evidence of genomic aberrations in greater than 15% of CRCs for most of the Scc equivalent regions, we next determined whether any of the orthologs of genes showing sequence or expression differences between Balb/cHeA and STS/A exhibited ASI in human colon tumors. We chose genes for Sequenom MassARRAY quantitative SNP genotyping in human tumors based largely on the mouse RNA-seq data, but we also included genes and/or SNPs from these loci that showed evidence in the literature as being associated with any type of cancer, colon biology, and/or a suggestion of being associated with CRC risk from previous GWAS. From these criteria, we identified 81 genes and intergenic regions of interest. We performed quantitative genotyping of 525 haplotype-tagging SNPs in DNAs in our discovery set of 194 normal/colon tumor pairs. These corresponded to 103 SNPs mapping to 18 genes at SCC4, 278 haplotype-tagging SNPs mapping to 34 genes in SCC5, and 144 SNPs from 29 genes at SCC13. Among these 525 SNPs, 74 SNPs showed evidence of ASI with a p-value of < 0.01 including 19 SNPs at SCC4, 28 at SCC5 and 27 at the SCC13 locus (Supplemental Table 4). As this was our discovery set, we set a generous cutoff of p-value < 0.10 for selection for subsequent validation studies, which resulted in 103 SNPs for further study (Supplemental Table 4).
For our validation set we evaluated the 103 SNPs in 296 normal/tumor pairs (Supplemental Table 5). When the validation data were combined with data from the discovery set, quantitative genotyping yielded 31 SNPs showing statistical significance with nominal p-values < 0.05 (Supplemental Table 6). Two SNPs in the gene SNX10 at SCC13 showed significant evidence of ASI after Bonferroni correction for multiple comparisons (n = 103) (Table 2). Genes with SNPs showing non-statistical evidence of ASI (adjusted p-values of < 0.2) include GRAMD3 and CEP120 at SCC5, EPAS1 at SCC4, and LANCL2 and SCRN1 at SCC13 (Supplemental Table 6). Though these genes do not meet statistical significance for ASI in our study, they could be considered as candidates for much larger studies with greater statistical power.
Table 2:
SNPs showing evidence of ASI after replication
| SNP ID | Tagged Gene (Locus) |
Alleles | Sample Set | Allele 1 Imbalance* |
Allele 2 Imbalance† |
Total Imbalance§ |
P-value‡ | Adjusted P-value∞ |
Potential Role in Cancer |
|---|---|---|---|---|---|---|---|---|---|
| rs1919935 |
SNX10 (SCC13) |
CT | Discovery | 18/67 (27%) | 6/67 (9%) | 24/67 (36%) | 0.01431 | Endosome trafficking of EGFR and PDGFR | |
| Validation | 22/109 (20%) | 4/109 (4%) | 26/109 (24%) | 0.00042 | |||||
| Combined | 40/176 (23%) | 10/176 (6%) | 50/176 (28%) | 0.00002 | 0.00228 | ||||
| rs2699814 |
SNX10 (SCC13) |
TA | Discovery | 5/89 (6%) | 19/89 (21%) | 24/89 (27%) | 0.00427 | Endosome trafficking of EGFR and PDGFR | |
| Validation | 6/136 (4%) | 16/136 (12%) | 22/136 (16%) | 0.03301 | |||||
| Combined | 11/225 (5%) | 35/225 (16%) | 46/225 (20%) | 0.00040 | 0.04143 | ||||
| rs6958331 |
LANCL2 (SCC13) |
CT | Discovery | 17/63 (27%) | 7/63 (11%) | 24/63 (38%) | 0.04123 | Positive regulator of Akt | |
| Validation | 14/93 (15%) | 2/93 (2%) | 16/93 (17%) | 0.00270 | |||||
| Combined | 31/156 (20%) | 9/156 (6%) | 40/156 (26%) | 0.00050 | 0.05193 | ||||
| rs6891155 |
CEP120 (SCC5) |
GA | Discovery | 26/98 (27%) | 11/98 (11%) | 37/98 (38%) | 0.01366 | Centriole assembly | |
| Validation | 26/153 (17%) | 12/153 (8%) | 38/153 (25%) | 0.02314 | |||||
| Combined | 52/251 (21%) | 23/251 (9%) | 75/251 (30%) | 0.00081 | 0.08365 | ||||
| rs4835907 |
GRAMD3 (SCC5) |
TA | Discovery | 8/89 (9%) | 21/89 (24%) | 29/89 (33%) | 0.01578 | Membrane-coupled processes | |
| Validation | 10/118 (8%) | 23/118 (19%) | 33/118 (28%) | 0.02364 | |||||
| Combined | 18/207 (9%) | 44/207 (21%) | 62/207 (30%) | 0.00096 | 0.09888 |
Relative loss of allele 1 compared to allele 2
Relative loss of allele 2 compared to allele 1
Total number of tumors with imbalance/total heterozygous samples (% of heterozygotes showing imbalance)
Chi-squared statistical test, df = 1
Bonferroni corrected p-value
Discussion
Here, we show that variants at loci orthologous to mouse Scc loci exhibit evidence of ASI in human colon tumors (Table 2). We identified two variants in the SNX10 gene that are candidates for colon cancer susceptibility based on data from the mice used in the original linkage analyses as well as our ASI studies (Table 2, Supplemental Table 6). Additional variants in GRAMD3, CEP120, LANCL2, SCRN1 and EPAS1 did not show statistically meaningful ASI, but some of them are interesting candidates given their published roles in cancer and relevance to cancer-related cellular processes (Table 2).
The gene SNX10 is a member of the sorting nexin family. This family plays a role in endocytosis, endosome sorting, and endosome signaling.38 To date this gene has not been implicated in any cancers. However, sorting nexins have been known to regulate the trafficking and signaling of such molecules as EGFR and PDGFR, both of which are substrates for PTPRJ.38 The PX domain of the sorting nexin protein binds to phosphatidylinositol-3-phosphate (PtdIns3P), which facilitates SNX protein localization to the membrane.38 It is possible that SNX10 binds to PtdIns3P and contributes to the endosomal trafficking of EGFR in colon epithelial cells, though this hypothesis has not been tested.
At the SCC5 locus, no genes showed significant ASI. Similarly, the SCC4 locus failed to reveal statistically significant ASI in our study. However, an intriguing candidate at the SCC4 locus is the EPAS1 gene, which is also known as HIF2α. This transcription factor regulates genes involved in angiogenesis, metabolism, and other processes involved in cellular adaptation to hypoxia.39 Constitutively activated Epas1 promotes colon carcinogenesis in the mouse by regulating the COX2/mPGES-1/PGE(2) pathway.40 One variant in EPAS1 emerged in a genome-wide association study of renal cell carcinoma, while a different variant in this gene showed an interaction with the well-validated CRC susceptibility variant rs6983267 at 8q24 in a prostate cancer association study.41,42, At this time, however, no SNPs in EPAS1 have been associated with CRC. While the tagging SNPs in EPAS1 that we tested for ASI did not achieve a significant p-value after multiple comparisons testing, it remains possible that variants in this gene contribute to CRC risk by some unknown interaction with variants in the SCC5 locus.
Our RNA-seq analysis revealed few coding variations between Balb/cHeA and STS/A (Supplemental Table 3). Most of the differences between the strains were in mRNA expression (Supplemental Table 2), suggesting that variants outside of the coding regions may play a critical role in the regulation of gene expression and contribute to CRC sensitivity by modifying gene expression. Such variants may include promoter SNPs, SNPs mapping to regulatory elements, variants in non-coding RNAs, or intronic SNPs that influence mRNA splicing. One limitation of the use of RNA-seq data to identify candidate sequence variants is the inability to sequence promoter or intergenic regions that may differ between the strains. However, if the variants driving the linkages are acting in cis to regulate gene expression, the gene expression differences can act as a surrogate for the regulatory variants. It is important to note, however, that our RNA-seq data is constrained to one mouse per strain, thereby necessitating the use of alternative methods to validate expression or coding differences. Targeted sequencing of promoter and regulatory regions that control the transcription of genes showing significant differential expression between the susceptible and resistant strains could reveal regulatory SNPs that contribute to the observed expression differences. As we used mRNA for our RNA-seq study, we were not able to capture any difference in microRNA or other non-coding RNAs that map to these regions.
Our aCGH data along with that described in the literature supports the exploration of the human orthologs of Scc4, Scc5, and Scc13 in CRC susceptibility (Table 1). We observed similar patterns of chromosome arm gains and losses as a number of other aCGH analyses of CRC tumors.34-37 We demonstrated gains in 19% of tumors at 2p (SCC4) and in 24% of tumors at 7p (SCC13). Interestingly, gains at 7p14.1 and 7p15.3 were observed in 60% and 35% of tumor genomes of patients with microsatellite stable hereditary nonpolyposis colorectal cancer, respectively.43 The BAC showing highest frequency of gain at 7p in our cohort maps to SCRN1, a gene which shows higher expression in colon tumor tissue compared to adjacent normal tissue and in which increased expression is correlated with poorer prognosis.44 In our cohort, losses were observed in 10% of tumors at 4q (SCC13) and 31% of tumors at 5q (SCC5). In addition to gains or losses that spanned multiple adjacent BAC clones or whole arms, we also identified a handful of single BAC clones showing frequencies of gains or losses greater than or approaching 35% (data not shown). Genes such as SCRN1, PDGFRB, and PRDM5 map within these focal regions of aberration, but no SNPs within these genes showed statistically significant evidence of ASI in our study.
There are limitations to this study. From work performed by our laboratory and others, we know that not all CRC susceptibility alleles demonstrate allele-specific gains or losses in tumors.9, 10 We may be missing interesting candidate genes or variants for future studies by prioritizing those that contain variants with ASI. Secondly, we primarily focused on testing tagging SNPs that map near or within genes. As many GWAS-identified SNPs for cancer are intergenic and are thought to alter regulatory elements of genes important in CRC development, we may be missing critical SNPs/regulatory elements important for cancer susceptibility.45 Finally, the identification of “causal” variants being selected for during tumorigenesis (i.e. drivers of ASI) is dependent in part on the allele-frequency and frequency at which a locus shows copy number gains and losses in tumors. Variants that have lower rates of heterozygosity and/or map in regions with less frequent gains and losses will require larger samples sizes than those used in this study. Thus, we were likely underpowered to detect variants showing ASI at SCC4 as it showed less frequent gains and losses relative to other loci. If a less stringent method of multiple comparisons adjustments were made, a number of additional variants from these loci could be considered as candidates.
To date, none of our candidate SNPs (or genes) has been identified from genome-wide association studies for CRC risk. This is not entirely unexpected as we anticipate that risk of these SNPs will be dependent upon their interacting loci and will not necessarily meet the stringent p-values for risk when considered independently. Interestingly some of the murine Scc and colon cancer susceptibility (Ccs) loci have been mapped within 3.3 cM of loci identified through GWAS of human CRC risk.46 A study by the EPICOLON consortium identified a SNP, rs954353, in the human orthologous locus to Scc15 that showed evidence of risk in their Phase I study.47 Finally, work by us and others suggest that variants and haplotypes in PTPRJ, the candidate gene for SCC1, may be important for colon and breast cancer risk.28, 48,49 As these variants were not replicated in larger studies, it will be important to look at them in the context of candidate interacting alleles at the human orthologs of Scc5 and Scc13.
In summary, we used a cross-species approach to identify potential candidate genes from mouse Scc loci. We identified multiple candidate genes showing mRNA expression differences between the strains of mice and identified two variants in the gene SNX10 showing statistically significant evidence of ASI in human tumors. Future studies will be necessary to determine (1) if these genes and the others showing suggestive evidence of ASI are important in the differences in colon cancer susceptibility between the strains, and (2) if these variants are involved in gene-gene interactions for human CRC susceptibility.
Supplementary Material
Supplemental Table 1: Primers for Sequenom MassARRAY quantitative SNP genotyping
Supplemental Table 2: Genes showing differential expression between Balb/cHeA and STS/A by RNA-seq
Supplemental Table 3: Coding SNPs identified in Balb/cHeA and STS/A by RNA-seq
Supplemental Table 4: Allele-specific imbalance data for 525 SNPs in discovery sample set
Supplemental Table 5: Allele-specific imbalance data for 103 SNPs in validation sample set
Supplemental Table 6: Allele-specific imbalance data for 103 SNPs in discovery sample set and validation sample set combined
Novelty/Impact: Only a portion of the estimated genetic variants contributing to colorectal cancer (CRC) risk have been identified. We used a cross-species approach combining data from mouse linkage studies of CRC susceptibility and RNA-seq analysis with allele-specific somatic changes in human tumors. Variants in the SNX10 gene that maps to the human equivalent of mouse Scc13 were found to show allele-specific imbalance in colon tumors, suggesting that variants in this region may be important in CRC.
Acknowledgments
Mouse tissue samples were obtained from Charlotte Pfauth of the Netherlands Cancer Institute. The OSU Tissue Procurement Shared Resource (CCTPSR) and the Cooperative Human Tissue Network aided in sample ascertainment. Jerneja Tomsic assisted with sample acquisition. The OSU Human Genetics Sample Bank processed normal genomic DNA for the validation studies. Brittany Price and the University of Chicago Sequenom core provided Sequenom MassARRAY support. The OSU CCC Genomics Shared Resource provided qPCR support.
This study was funded in part by the NIH/NCI (NCI CA134461 to A. E. Toland) and the Ohio State University Comprehensive Cancer Center Core grant (NCI CA16058). M. Gerber was funded by an OSU College of Medicine Systems and Integrated Biology training grant and a Pelotonia Graduate Fellowship. N. Schulz was funded by an OSU College of Medicine Medical Student Research Scholarship. The authors have no financial or personal conflicts of interest to disclose.
Abbreviations:
- aCGH
array comparative genomic hybridization
- ASI
allele-specific imbalance
- BAC
bacterial artificial chromosome
- CIN
chromosomal instability
- CRC
colorectal cancer
- GWAS
genome-wide association study
- Scc/SCC
Susceptibility to colon cancer locus
- SNP
single nucleotide polymorphism
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA 2014;64: 104–17. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61: 759–67. [DOI] [PubMed] [Google Scholar]
- 3.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology 2010;138: 2059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin ES, Tonon G, Sinha R, et al. Common and distinct genomic events in sporadic colorectal cancer and diverse cancer types. Cancer Res 2007;67: 10736–43. [DOI] [PubMed] [Google Scholar]
- 5.Xie T, G DA, Lamb JR, et al. A comprehensive characterization of genome-wide copy number aberrations in colorectal cancer reveals novel oncogenes and patterns of alterations. PLoS One 2012;7: e42001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mei R, Galipeau PC, Prass C, et al. Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome Res 2000;10: 1126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagase H, Mao JH, Balmain A. Allele-specific Hras mutations and genetic alterations at tumor susceptibility loci in skin carcinomas from interspecific hybrid mice. Cancer Res 2003;63: 4849–53. [PubMed] [Google Scholar]
- 8.Tuupanen S, Niittymaki I, Nousiainen K, et al. Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res 2008;68: 14–7. [DOI] [PubMed] [Google Scholar]
- 9.Niittymaki I, Tuupanen S, Li Y, et al. Systematic search for enhancer elements and somatic allelic imbalance at seven low-penetrance colorectal cancer predisposition loci. BMC Med Genet 2011;12: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerber MM, Hampel H, Schulz NP, et al. Evaluation of allele-specific somatic changes of genome-wide association study susceptibility alleles in human colorectal cancers. PLoS one 2012;7: e37672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K, Civan J, Mukherjee S, et al. Genetic variations in colorectal cancer risk and clinical outcome. World J Gastroenterol 2014;20: 4167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao S, Peters U, Berndt S, et al. Estimating the heritability of colorectal cancer. Human Mol Gen 2014;23: 3898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maher B Personal genomes: The case of the missing heritability. Nature 2008;456: 18–21. [DOI] [PubMed] [Google Scholar]
- 14.Jacoby RF, Hohman C, Marshall DJ, et al. Genetic analysis of colon cancer susceptibility in mice. Genomics 1994;22: 381–7. [DOI] [PubMed] [Google Scholar]
- 15.Moen CJ, van der Valk MA, Snoek M, et al. The recombinant congenic strains--a novel genetic tool applied to the study of colon tumor development in the mouse. MammGenome 1991;1: 217–27. [DOI] [PubMed] [Google Scholar]
- 16.Moen CJ, Groot PC, Hart AA, et al. Fine mapping of colon tumor susceptibility (Scc) genes in the mouse, different from the genes known to be somatically mutated in colon cancer. Proc Natl Acad Sci USA 1996;93: 1082–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Wezel T, Ruivenkamp CA, Stassen AP, et al. Four new colon cancer susceptibility loci, Scc6 to Scc9 in the mouse. Cancer Res 1999;59: 4216–8. [PubMed] [Google Scholar]
- 18.Ruivenkamp CA, Csikos T, Klous AM, et al. Five new mouse susceptibility to colon cancer loci, Scc11-Scc15. Oncogene 2003;22: 7258–60. [DOI] [PubMed] [Google Scholar]
- 19.Van Der Kraak L, Meunier C, Turbide C, et al. A two-locus system controls susceptibility to colitis-associated colon cancer in mice. Oncotarget 2010;1: 436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meunier C, Kwan T, Turbide C, et al. Genetic control of susceptibility to carcinogen-induced colorectal cancer in mice: the Ccs3 and Ccs5 loci regulate different aspects of tumorigenesis. Cell Cycle 2011;10: 1739–49. [DOI] [PubMed] [Google Scholar]
- 21.Eversley CD, Yuying X, Pearsall RS, et al. Mapping six new susceptibility to colon cancer (Scc) loci using a mouse interspecific backcross. G3 (Bethesda) 2012;2: 1577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu P, Lu Y, Liu H, et al. Genome-wide association and fine mapping of genetic loci predisposing to colon carcinogenesis in mice. Mol Cancer Res 2012;10: 66–74. [DOI] [PubMed] [Google Scholar]
- 23.Nnadi SC, Watson R, Innocent J, et al. Identification of five novel modifier loci of Apc(Min) harbored in the BXH14 recombinant inbred strain. Carcinogenesis 2012;33: 1589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg DW, Giardina C, Tanaka T. Mouse models for the study of colon carcinogenesis. Carcinogenesis 2009;30: 183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balmain A, Nagase H. Cancer resistance genes in mice: models for the study of tumour modifiers. Trends Genet 1998;14: 139–44. [DOI] [PubMed] [Google Scholar]
- 26.van Wezel T, Stassen AP, Moen CJ, et al. Gene interaction and single gene effects in colon tumour susceptibility in mice. Nat Genet 1996;14: 468–70. [DOI] [PubMed] [Google Scholar]
- 27.Ruivenkamp CA, van Wezel T, Zanon C, et al. Ptprj is a candidate for the mouse colon-cancer susceptibility locus Scc1 and is frequently deleted in human cancers. Nat Genet 2002;31: 295–300. [DOI] [PubMed] [Google Scholar]
- 28.Toland AE, Rozek LS, Presswala S, et al. PTPRJ haplotypes and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 2008;17: 2782–5. [DOI] [PubMed] [Google Scholar]
- 29.Ewart-Toland A, Balmain A. The genetics of cancer susceptibility: from mouse to man. Toxicol Pathol 2004;32 Suppl 1: 26–30. [DOI] [PubMed] [Google Scholar]
- 30.Dworkin AM, Ridd K, Bautista D, et al. Germline variation controls the architecture of somatic alterations in tumors. PLoS Genet 2010;6: e1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleming JL, Dworkin AM, Allain DC, et al. Allele-specific imbalance mapping identifies HDAC9 as a candidate gene for cutaneous squamous cell carcinoma. Int J Cancer 2014;134: 244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki R, Kohno H, Sugie S, et al. Strain differences in the susceptibility to azoxymethane and dextran sodium sulfate-induced colon carcinogenesis in mice. Carcinogenesis 2006;27: 162–9. [DOI] [PubMed] [Google Scholar]
- 33.Quigley D, Balmain A. Systems genetics analysis of cancer susceptibility: from mouse models to humans. Nat Rev Genet 2009;10: 651–7. [DOI] [PubMed] [Google Scholar]
- 34.Jones AM, Douglas EJ, Halford SE, et al. Array-CGH analysis of microsatellite-stable, near-diploid bowel cancers and comparison with other types of colorectal carcinoma. Oncogene 2005;24: 118–29. [DOI] [PubMed] [Google Scholar]
- 35.Dyrso T, Li J, Wang K, L et al. Identification of chromosome aberrations in sporadic microsatellite stable and unstable colorectal cancers using array comparative genomic hybridization. Cancer Genet 2011;204: 84–95. [DOI] [PubMed] [Google Scholar]
- 36.Nakao K, Mehta KR, Fridlyand J, et al. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis 2004;25: 1345–57. [DOI] [PubMed] [Google Scholar]
- 37.Lassmann S, Weis R, Makowiec F, et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med (Berl) 2007;85: 293–304. [DOI] [PubMed] [Google Scholar]
- 38.Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nature Rev Mol Cell Biol 2002;3: 919–31. [DOI] [PubMed] [Google Scholar]
- 39.Gordan JD, Bertout JA, Hu CJ, et al. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 2007;11: 335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue X, Shah YM. Hypoxia-inducible factor-2alpha is essential in activating the COX2/mPGES-1/PGE2 signaling axis in colon cancer. Carcinogenesis 2013;34: 163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciampa J, Yeager M, Amundadottir L, et al. Large-scale exploration of gene-gene interactions in prostate cancer using a multistage genome-wide association study. Cancer Res 2011;71: 3287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purdue MP, Johansson M, Zelenika D, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet 2011;43: 60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W, Yuan L, Cai Y, et al. Identification of chromosomal copy number variations and novel candidate loci in hereditary nonpolyposis colorectal cancer with mismatch repair proficiency. Genomics 2013;102: 27–34. [DOI] [PubMed] [Google Scholar]
- 44.Miyoshi N, Ishii H, Mimori K, et al. SCRN1 is a novel marker for prognosis in colorectal cancer. JSurg Oncol 2010;101: 156–9. [DOI] [PubMed] [Google Scholar]
- 45.Edwards SL, Beesley J, French JD, et al. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 2013;93: 779–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quan L, Stassen AP, Ruivenkamp CA, et al. Most lung and colon cancer susceptibility genes are pair-wise linked in mice, humans and rats. PLoS One 2011;6: e14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castellvi-Bel S, Ruiz-Ponte C, Fernandez-Rozadilla C, et al. Seeking genetic susceptibility variants for colorectal cancer: the EPICOLON consortium experience. Mutagenesis 2012;27: 153–9. [DOI] [PubMed] [Google Scholar]
- 48.Lesueur F, Pharoah PD, Laing S, et al. Allelic association of the human homologue of the mouse modifier Ptprj with breast cancer. Human Mol Genet 2005;14: 2349–56. [DOI] [PubMed] [Google Scholar]
- 49.Mita Y, Yasuda Y, Sakai A, et al. Missense polymorphisms of PTPRJ and PTPN13 genes affect susceptibility to a variety of human cancers. J Cancer Res Clin Oncol 2010;136: 249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Primers for Sequenom MassARRAY quantitative SNP genotyping
Supplemental Table 2: Genes showing differential expression between Balb/cHeA and STS/A by RNA-seq
Supplemental Table 3: Coding SNPs identified in Balb/cHeA and STS/A by RNA-seq
Supplemental Table 4: Allele-specific imbalance data for 525 SNPs in discovery sample set
Supplemental Table 5: Allele-specific imbalance data for 103 SNPs in validation sample set
Supplemental Table 6: Allele-specific imbalance data for 103 SNPs in discovery sample set and validation sample set combined