Abstract
PURPOSE:
Ki67 is a proliferation marker commonly assessed by immunohistochemistry in breast cancer, and it has been proposed as a clinical marker for subtype classification, prognosis and prediction of therapeutic response. However, the clinical utility of Ki67 is limited by the lack of consensus on the optimal cut point for each application.
METHODS:
We assessed Ki67 by immunohistochemistry using Definiens digital image analysis (DIA) in 2,653 cases of incident invasive breast cancer diagnosed in the Nurses’ Health Study from 1976–2006. Ki67 was scored as continuous percentage of positive tumor cells, and dichotomized at various cut points. Multivariate hazard ratios (HR) and 95% confidence intervals (CI) were calculated using Cox regression models for distant recurrence, breast cancer-specific mortality and overall mortality in relation to luminal subtypes defined with various Ki67 cut points, adjusting for breast cancer prognostic factors, clinico-pathologic features and treatment.
RESULTS:
DIA was highly correlated with manual scoring of Ki67 (Spearman correlation ρ=0.86). Mean Ki67 score was higher in grade-defined luminal B (12.6%), HER2-enriched (17.9%) and basal-like (20.6%) subtypes compared to luminal A (8.9%). In multivariable-adjusted models, luminal B tumors had higher breast cancer-specific mortality compared to luminal A cancer classified using various cut points for Ki67 positivity including the 14% cut point routinely reported in the literature (HR=1.38, 95% CI 1.11–1.72, p=0.004). There was no significant difference in clinical outcomes for ER- tumors according to Ki67 positivity defined at various cut points.
CONCLUSIONS:
Assessment of Ki67 in breast tumors by DIA was a robust and quantitative method. Results from this large prospective cohort study provide support for the clinical relevance of using Ki67 at the 14% cut point for luminal subtype classification and breast cancer prognosis.
Keywords: Ki67, Breast cancer, Subtype, Survival, Risk factor
Introduction
Breast cancer is a heterogeneous disease with four major molecular subtypes: luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-enriched, and basal-like (1, 2). These subtypes vary in their genomic, clinical, and pathologic features, has and have important implications for treatment (3, 4) and clinical outcome (5). In particular, subtype classification is clinically relevant for predicting recurrence risk and survival. Patients with basal-like tumors have a poorer prognosis than patients with luminal subtypes, although patients with luminal B subtype have significantly worse clinical outcome than those with luminal A subtype (1, 5–7).
Breast cancer subtype classification based on immunohistochemical (IHC) surrogate methods is widely used in clinical practice in accordance with St. Gallen International Breast Cancer Consensus recommendations (8, 9). Molecular subtyping derived using tumor grade and IHC is highly correlated with intrinsic subtypes (6, 10, 11), and is a practical and cost-effective alternative to gene expression profiling (8). While tumor grade is a valuable prognostic factor for breast cancer prognosis (12), it may not be optimal for distinguishing luminal A versus B subtypes due to heterogeneity among moderately differentiated (grade 2) tumors (13, 14).
Ki67 – also known as Ki67 antigen or MKI67 (marker of proliferation Ki67) – is a marker of proliferation expressed exclusively during active phases of the cell cycle (15, 16). Ki67 is commonly assessed by IHC in clinical settings and has been correlated with clinical outcome (17). However, use of Ki67 in the clinical management of breast cancer patients is limited by the lack of analytic validity in its assessment (18). Ki67 scoring reproducibility is only moderate when manual scoring methods are used (19), and thus there is currently no consensus on the optimal Ki67 cut point for molecular subtyping and prediction of breast cancer prognosis (18).
Using prospective data from the Nurses’ Health Study cohort, we systematically evaluated the robustness of Ki67 staining by Definiens digital image analysis (DIA). In addition, we examined the prognostic value of using Ki67 at various cut points to distinguish Luminal tumors for distant recurrence, breast cancer-specific and overall mortality, adjusting for established prognostic clinico-pathologic and lifestyle factors.
Materials and Methods
Study Population
The Nurses’ Health Study (NHS), established in 1976, is an ongoing prospective cohort study of 121,701 female registered nurses aged 30–55 at enrollment. Biennial questionnaires are used to collect data on lifestyle factors and health outcomes, including breast cancer, with a follow-up rate of over 90% (20). Return of questionnaires was considered implied consent. Incident breast cancer cases were ascertained by biennial questionnaire and the National Death Index, and confirmed by medical record review (21). Informed consent was obtained from all participants to collect and use tissue specimens for research. This study was approved by the Human Subjects Committee at Brigham and Women’s Hospital (Boston, Massachusetts).
Breast Cancer Tissue Block Collection and Selection
The collection of archived formalin-fixed paraffin-embedded (FFPE) breast cancer blocks from participants diagnosed with primary incident breast cancer began in 1993 and currently includes 30 years of follow-up (1976–2006). Tissue microarray (TMA) construction was performed as previously described (21, 22). Participants were eligible for this study if they were diagnosed with non-metastatic primary invasive breast cancer between 1976 and 2006 with no previous history of cancer and had FFPE breast cancer tissue available with pathologist-confirmed tumor on the TMA. We identified 3,284 tumors and excluded cases with in situ breast cancer (n=339), stage IV disease (n=57), diagnosis before 1976 (n=1), and previous non-skin cancer diagnosis (n=234). Our final sample included 2,653 breast tumors.
Immunohistochemical Analysis
We previously performed IHC staining and scoring for ER-α, PR, HER2, cytokeratin 5/6 (CK5/6), and epidermal growth factor receptor (EGFR) on 5 μm paraffin sections from TMA blocks (22, 23). Ki67 immunostaining was optimized in the BWH Specialized Histopathology Core and performed on a Dako Autostainer (Dako Corporation, Carpinteria, CA, USA). Briefly, tissue sections were deparaffinized in xylene and rehydrated in a series of ethanol. After heat-induced inactivation of endogenous peroxidase activity and antigen retrieval in citrate buffer (pH 6.1), tissue sections were incubated with Ki67 antibody (1:250 dilution of clone SP6 antibody from VP-RM04, Vector, Burlingame, CA, USA). SP6 clone from VP-RM04 has been used previously in large studies of FFPE TMA breast tumor tissue (10). In addition, SP6 performs better than MIB1 in image analysis on FFPE TMA breast tumor tissue due to its reduced background (24).
Scoring of Ki67
Nuclear staining of Ki67 was assessed in up to three cores per breast tumor. The percentage of Ki67 positive tumor cells and the intensity of Ki67 staining were measured using DIA with the Definiens Tissue Studio package (Definiens Tissue Studio software, Munich, Germany; Scanner: Pannoramic SCAN by 3DHISTECH; Scanner software: Pannoramic Scanner by 3DHISTECH (Version 1.17); Scanner viewer: Pannoramic Viewer by 3DHISTECH (Version 1.15.4). DIA was trained to distinguish malignant breast epithelial cells from non-malignant cells (e.g., stroma, lymphocytes and normal breast cells) based on nuclear size, contour and other presets. For each tumor, the sum of Ki67 positive tumors cells in all cores was divided by the total number of detectable tumor cell nuclei in all cores to create a continuous Ki67 score. We dichotomized Ki67 score at various cut points – 6.7% (median), 10%, 14%, 20%, 25% and 30% – to generate different definitions of Ki67 positivity. Ki67 histological score, which sums the weighted proportion of Ki67 positive tumor cells in three levels of staining intensity (low/medium/high), correlated nearly perfectly with Ki67 score (Spearman ρ=0.99). In a representative subset of tumors (n=159), we validated DIA Ki67 continuous score with manual (visual estimate of the percent positive tumor cells) Ki67 continuous score ascertained by an expert pathologist (LCC) and found strong agreement between methods (Spearman ρ=0.86).
Classification of Breast Cancer Molecular Phenotype
Five breast cancer molecular subtypes were defined by immunostaining for ER-α, PR, HER2, cytokeratin 5/6 (CK5/6) and epidermal growth factor receptor (EGFR), and histologic grade in the primary definition (23) or Ki67 in the secondary definition, for this study. Luminal A cases were ER+ and/or PR+ and HER2- and grades 1 or 2 (low or intermediate grade). Luminal B cases were ER+ and/or PR+ and HER2- and grade 3 (high grade), or ER+ and/or PR+ and HER2+ with any grade. HER2-enriched cases were ER- and PR- and HER2+. Basal-like cases were ER- and PR- and HER2-, and CK5/6+ and/or EGFR+. Unclassified cases were negative for all five markers. Separately, we defined triple-negative breast cancer (TNBC) as ER-, PR- and HER2- in subanalyses.
Clinical Outcomes
Distant recurrence, breast-cancer specific mortality and overall mortality were the primary outcomes. Women with incident invasive breast cancer who reported subsequent cancer of the lung, liver, bone or brain were considered to have breast cancer recurrence. Women who died from breast cancer and did not report recurrence were considered to have recurred two years prior to death (25).
Exposures
Ki67 percent positivity (continuous score), Ki67 high and Ki67 low (dichotomous) and luminal breast cancer subtypes defined with Ki67 at various cut points were the primary exposure variables. Luminal subtype classification based on Ki67 cut points was compared to classification using tumor grade.
Covariates
Information was collected on age at diagnosis (continuous), and on several risk factors prior to diagnosis including birth index (continuous) (26), oral contraceptive (OC) use (categorical), menopausal status and menopausal hormone (MH) use (categorical), BMI (categorical), and smoking status (categorical). Weight change (categorical) and physical activity (categorical) were assessed >12 months after diagnosis (25, 27). Clinico-pathological features and treatment factors included tumor stage (categorical), ER/PR status (categorical), chemotherapy (yes/no), radiotherapy (yes/no) and hormone therapy (yes/no).
Statistical Analysis
Spearman correlations and Wilcoxon two-sample tests were used to assess the significance of staining agreement among tumor cores. Associations of Ki67 with tumor features, breast cancer risk factors, and molecular subtypes defined using tumor grade were evaluated using chi square (χ2) tests and Kruskal-Wallis tests to assess significance.
We used multivariate Cox regression models to estimate hazard ratios (HR) and 95% confidence intervals (CI) for the association between luminal subtypes defined with various Ki67 cut points and clinical outcomes, and for the relationship between Ki67 score (and Ki67 at the 14% cut point) and clinical outcomes in all breast cancer and ER+ breast cancer.
All analyses were conducted with SAS version 9.3 (Cary, NC, USA). All statistical tests were two-sided and p-values <0.05 were considered statistically significant.
Results
Table 1 shows the distribution of clinico-pathologic features in 2,653 breast tumors according to Ki67 positivity at the 14% cut point. Women with Ki67 high (≥14% positive nuclei), tumors tended to be older at diagnosis (p<0.0001).. Compared with Ki67 low (<14% positive nuclei), Ki67 high tumors were larger size, higher grade, higher stage, and more likely to be ER-, HER2+, CK5/6+ and EGFR+ (p<0.0001). EGFR+ tumors had a mean Ki67 score of 19.0% compared to 9.4% for EGFR- tumors. ER+ and PR+ tumors had a lower Ki67 score (p<0.0001). ER+ tumors had a mean Ki67 score of 9.9% compared to 17.5% for ER- tumors. Significant associations were observed at all cut points for Ki67 positivity (data not shown), suggesting that the relationship between Ki67 and these breast tumor features is quite robust.
Table 1.
Distribution of clinico-pathological features of 2,653 incident invasive breast tumors by Ki67 positivity, Nurses’ Health Study, 1976–2006
| Ki67 <14% n=1954 (%) | Ki67 ≥14% n=699 (%) | p valuea | |
|---|---|---|---|
| Age at diagnosis | 58.9 (8.9) | 60.7 (9.4) | <0.0001 |
| Tumor size (cm) | 0.0007 | ||
| < 2.0 | 1250 (67) | 384 (58) | |
| 2.1–4.0 | 459 (25) | 222 (33) | |
| 4.0+ | 160 (9) | 61 (9) | |
| Tumor grade | <0.0001 | ||
| Well differentiated | 403 (22) | 86 (13) | |
| Moderately differentiated | 963 (51) | 282 (42) | |
| Poorly differentiated | 508 (27) | 310 (46) | |
| Lymph node involvement | 0.21 | ||
| No nodes involved | 1218 (67) | 405 (64) | |
| 1–3 nodes | 361 (20) | 146 (23) | |
| 4–9 nodes | 146 (8) | 47 (7) | |
| 10+ nodes | 89 (5) | 38 (6) | |
| Tumor stage at diagnosis | 0.004 | ||
| Stage 1 | 1073 (55) | 324 (46) | |
| Stage 2 | 607 (31) | 272 (39) | |
| Stage 3 | 274 (14) | 103 (15) | |
| ER status | <0.0001 | ||
| ER+ | 1603 (82) | 449 (64) | |
| ER− | 342 (18) | 249 (36) | |
| PR status | <0.0001 | ||
| PR+ | 1405 (72) | 380 (55) | |
| PR− | 541 (28) | 316 (45) | |
| Joint ER/PR status | <0.0001 | ||
| ER+/PR+ | 1376 (71) | 363 (52) | |
| ER+/PR− | 225 (12) | 85 (12) | |
| ER−/PR+ | 27 (1) | 17 (2) | |
| ER−/PR− | 315 (16) | 231 (33) | |
| HER2 status | <0.0001 | ||
| HER2+ | 360 (19) | 196 (28) | |
| HER2− | 1565 (81) | 493 (72) | |
| EGFR status | <0.0001 | ||
| EGFR+ | 288 (15) | 250 (37) | |
| EGFR− | 1624 (85) | 427 (63) | |
| CK5/6 status | <0.0001 | ||
| CK5/6+ | 172 (9) | 145 (21) | |
| CK5/6− | 1745 (91) | 545 (79) | |
| Surgery | 0.12 | ||
| No | 3 (0) | 2 (0) | |
| Lumpectomy | 642 (34) | 247 (37) | |
| Mastectomy | 1235 (66) | 412 (62) | |
| Unknown type of surgery | 1 (0) | 1 (0) | |
| Radiation | 0.24 | ||
| Yes | 833 (55) | 321 (58) | |
| No | 674 (45) | 231 (42) | |
| Chemotherapy | <0.0001 | ||
| Yes | 681 (45) | 324 (58) | |
| No | 845 (55) | 234 (42) | |
| Tamoxifen | 0.01 | ||
| Yes | 1139 (76) | 371 (70) | |
| No | 360 (24) | 157 (30) |
Numbers may not add to column totals due to missing data and percentages may not add to 100 due to rounding
χ2 tests for all variables except Kruskal-Wallis test for age at diagnosis; p-trend for categorical variables with more than two categories
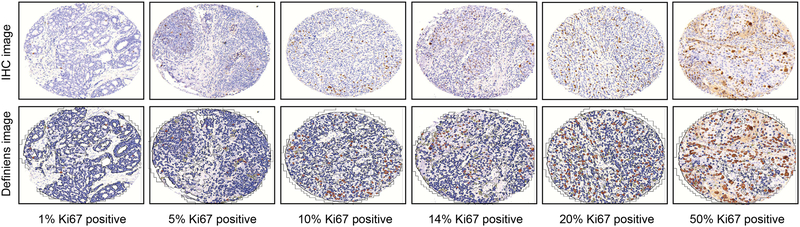
Figure 1 shows representative images for IHC staining used for manual scoring (top row) and DIA (bottom row) for Ki67 in breast tumor tissue specimens at Ki67 scores of 1%, 5%, 10%, 14%, 20%, and 50%.
Figure 1.
Ki67 staining in breast tumor tissue specimens using immunohistochemistry (IHC)
Representative Ki67 staining images at various percentages of tumor positivity. Top panel is IHC staining image used for manual scoring, bottom panel is Definiens digital analysis image at 20X magnification. Ki67 staining was scored continuously as the percentage of Ki67+ tumor cells relative to the total number of detected nuclei
Molecular subtypes were defined for 2,555 cases. Mean Ki67 score varied significantly across breast cancer subtypes (p<0.0001) (Table 2). Mean Ki67 score was higher in grade-defined luminal B (12.6%), HER2-enriched (17.9%) and basal-like (20.6%) subtypes compared to luminal A (8.9%).
Table 2.
Mean Ki67 score in 2,555 incident invasive breast tumors by molecular subtype defined with tumor grade, Nurses’ Health Study, 1976–2006
| Breast Cancer Molecular Subtypea | ||||||
|---|---|---|---|---|---|---|
| Luminal A | Luminal B | HER2-enriched | Basal-like | Unclassified | ||
| n=1287 (50%) | n=738 (29%) | n=177 (7%) | n=268 (11%) | n=85 (3%) | p-valueb | |
| Mean Ki67 score, % (SD) | 8.9 (10.6) | 12.6 (13.9) | 17.9 (19.7) | 20.6 (18.5) | 9.5 (10.3) | <0.0001 |
Molecular breast cancer subtypes defined using ER, PR, HER2, EGFR, CK5/6 and tumor grade
Kruskal-Wallis test
Next, luminal subtype classification based on Ki67 cut points was compared to classification using tumor grade. Reclassification occurred when a case defined as luminal A by grade was classified as luminal B using Ki67, or vice versa (Table 3). The extent of reclassification varied by Ki67 cut point, ranging from 18.8% to 34.7%. At the Ki67 14% cut point, 24.5% of luminal cases (n=496) were reclassified, with 47.0% of these being reclassified from luminal A to luminal B. The vast majority of reclassified luminal B cases (n=233), 72.0% were moderately differentiated (grade 2).
Table 3.
Luminal subtype reclassification comparing subtypes defined with tumor grade to subtypes defined with Ki67 at various cut points in 2,025 incident invasive luminal breast tumors, Nurses’ Health Study, 1976–2006
| Reclassified, n (%) | ||||
|---|---|---|---|---|
| Ki67 Cut Point | No | Yes | Luminal A to B | Luminal B to A |
| 6.7% (median) | 1322 (65.3) | 703 (34.7) | 523 (25.8) | 180 (8.9) |
| 10% | 1434 (70.8) | 591 (29.2) | 364 (18.0) | 227 (11.2) |
| 14% | 1529 (75.5) | 496 (24.5) | 233 (11.5) | 263 (13.0) |
| 20% | 1600 (79.0) | 425 (21.0) | 128 (6.3) | 297 (14.7) |
| 25% | 1636 (80.8) | 389 (19.2) | 76 (3.8) | 313 (15.5) |
| 30% | 1645 (81.2) | 380 (18.8) | 51 (2.5) | 329 (16.3) |
Percentages may not add to 100 due to rounding
After adjusting for clinico-pathologic features, lifestyle prognostic factors and treatment, there was a modest increased risk of breast cancer-specific death comparing luminal B to luminal A breast cancer consistent across Ki67 cut points (Table 4). The association appeared to be strongest for Ki67 cut points ≤ 20% (6.7% cut point: HR=1.38, 95% CI (1.13–1.70), p=0.002; 10% cut point: HR=1.32, 95% CI 1.07–1.63, p=0.009; 14% cut point: HR=1.38, 95% CI 1.11–1.72, 0.004; 20% cut point: HR=1.28, 95% CI 1.01–1.62, p=0.04). We observed several suggested increased risks of distant recurrence comparing luminal B to luminal A breast cancer (6.7% cut point: HR=1.23, 95% CI 1.01–1.50, p=0.04; 10% cut point: HR=1.19, 95% CI 0.97–1.45, p=0.09; 14% cut point: HR=1.22, 95% CI 0.99–1.51, p=0.06; 20% cut point: HR=1.17, 95% CI 0.93–1.46, p=0.19; 25% cut point: HR=1.17, 95% CI 0.92–1.49, p=0.19; 30% cut point: HR=1.24, 95% CI 0.96–1.59, p=0.10). We also observed a slight increased risk of all-cause death comparing luminal B to luminal A breast cancer at lower Ki67 cut points (6.7% cut point: HR=1.18, 95% CI 1.02–1.36 p=0.03; 10% cut point: HR=1.13, 95% 0.97–1.31, p=0.12; 14% cut point: HR=1.17, 95% CI 1.00–1.37, p=0.05; 20% cut point: HR=1.08, 95% CI 0.91–1.28, p=0.36; 25% cut point: HR=1.07, 95% CI 0.89–1.28, p=0.46; 30% cut point: HR=1.11, 95% CI 0.92–1.33, p=0.28). Strikingly, there were no significant associations of luminal B (compared to luminal A) defined using tumor grade and risk of distant recurrence (HR=1.18, 95% CI 0.96–1.44, p=0.11), breast cancer-specific mortality (HR=1.16, 95% CI 0.94–1.43, p=0.16), and risk of overall mortality (HR=1.05, 95% CI 0.91–1.22, p=0.49).
Table 4.
Age-adjusted and multivariable-adjusted relative hazards of clinical outcomes for luminal B (versus luminal A) breast cancer molecular subtypes defined with Ki67 at various cut points or tumor grade, Nurses’ Health Study, 1976–2006
| Distant recurrence | Breast cancer-specific mortality | Overall mortality | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Luminal breast cancer | Person-years | Recurrences | Person-years | Deaths | Person-years | Deaths | |||||
| 32,530 | 439 | 31,309 | 410 | 31,309 | 830 | ||||||
| Model | HR | p-valuea | Model | HR | p-valuea | Model | HR | p-valuea | |||
| Luminal B (6.7%)d | Age-adjusted | 1.19 (0.99–1.44) | 0.07 | Age-adjusted | 1.37 (1.13–1.67) | 0.002 | Age-adjusted | 1.15 (1.00–1.32) | 0.05 | ||
| Multivariableb | 1.23 (1.01–1.50) | 0.04 | Multivariableb | 1.38 (1.13–1.70) | 0.002 | Multivariableb | 1.18 (1.02–1.36) | 0.03 | |||
| Luminal B (10%)d | Age-adjusted | 1.16 (0.96–1.41) | 0.13 | Age-adjusted | 1.33 (1.09–1.63) | 0.005 | Age-adjusted | 1.13 (0.98–1.30) | 0.09 | ||
| Multivariableb | 1.19 (0.97–1.45) | 0.09 | Multivariableb | 1.32 (1.07–1.63) | 0.009 | Multivariableb | 1.13 (0.97–1.31) | 0.12 | |||
| Luminal B (14%)d | Age-adjusted | 1.16 (0.94–1.42) | 0.16 | Age-adjusted | 1.33 (1.08–1.64) | 0.007 | Age-adjusted | 1.13 (0.98–1.32) | 0.10 | ||
| Multivariableb | 1.22 (0.99–1.51) | 0.06 | Multivariableb | 1.38 (1.11–1.72) | 0.004 | Multivariableb | 1.17 (1.00–1.37) | 0.05 | |||
| Multivariablec | 1.22 (0.99–1.51) | 0.07 | Multivariablec | 1.38 (1.10–1.72) | 0.005 | Multivariablec | 1.20 (1.02–1.40) | 0.03 | |||
| Luminal B (20%)d | Age-adjusted | 1.11 (0.89–1.38) | 0.36 | Age-adjusted | 1.25 (1.00–1.57) | 0.05 | Age-adjusted | 1.05 (0.89–1.24) | 0.54 | ||
| Multivariableb | 1.17 (0.93–1.46) | 0.19 | Multivariableb | 1.28 (1.01–1.62) | 0.04 | Multivariableb | 1.08 (0.91–1.28) | 0.36 | |||
| Luminal B (25%)d | Age-adjusted | 1.05 (0.84–1.33) | 0.66 | Age-adjusted | 1.17 (0.92–1.49) | 0.20 | Age-adjusted | 1.03 (0.86–1.22) | 0.78 | ||
| Multivariableb | 1.17 (0.92–1.49) | 0.19 | Multivariableb | 1.26 (0.98–1.62) | 0.08 | Multivariableb | 1.07 (0.89–1.28) | 0.46 | |||
| Luminal B (30%)d | Age-adjusted | 1.04 (0.82–1.33) | 0.74 | Age-adjusted | 1.15 (0.89–1.47) | 0.29 | Age-adjusted | 1.03 (0.86–1.24) | 0.71 | ||
| Multivariableb | 1.24 (0.96–1.59) | 0.10 | Multivariableb | 1.27 (0.97–1.65) | 0.08 | Multivariableb | 1.11 (0.92–1.33) | 0.28 | |||
| Luminal B (grade)d | Age-adjusted | 1.39 (1.14–1.68) | <0.001 | Age-adjusted | 1.47 (1.21–1.80) | <0.001 | Age-adjusted | 1.18 (1.02–1.36) | 0.03 | ||
| Multivariableb | 1.18 (0.96–1.44) | 0.11 | Multivariableb | 1.16 (0.94–1.43) | 0.16 | Multivariableb | 1.05 (0.91–1.22) | 0.49 | |||
HR hazard ratio
χ2 tests
Adjusted for age and breast cancer survival risk factors including menopausal status and MH use (premenopausal, never user, past user, current estrogen user, current estrogen and progesterone user, current other user, unknown status), BMI (<20, 20–21.9, 22–23.9, 24–26.9, 27+ kg/m2), smoking status (never, current and past smoker), weight change at least 12 months after diagnosis (>10% loss, 5–10% loss, <5% loss to <5% gain, 5–10% gain, >10% gain), physical activity at least 12 months after diagnosis (<3, 3–8.9, 9–14.9, 15–23.9, 24+ metabolic equivalents/week), stages I-III, chemotherapy (yes/no), radiation therapy (yes/no), tamoxifen use (yes/no), other hormone therapy (yes/no) and date of diagnosis and time since diagnosis.
Same as multivariable model excluding cases with no information on tumor grade (for comparison with multivariable model defined with tumor grade)
Reference category is luminal A breast cancer
We also examined the prognostic value of Ki67 at various cut points according to ER status. In multivariable models, there was no difference in risk of distant recurrence comparing ER+/Ki67 low tumors to other ER/Ki67 subtypes (ER+/Ki67 high, ER-/Ki67low and ER-/Ki67 high; data not shown). We observed a modest increased risk of breast-cancer specific mortality comparing ER+/Ki67 low to ER-/Ki67 high tumors defined at Ki67 6.7%, 10% and 14% cut points (14% cut point: HR=1.54, 95% CI 1.16–2.02, p=0.002). We also observed a modest increased risk of overall mortality comparing ER+/Ki67 low to ER-/Ki67 high tumors defined with the Ki67 6.7% and 10% cut points (10% cut point: HR=1.26, 95% CI 1.04–1.53, p=0.02). There was no difference in risk of distant recurrence, breast cancer-specific mortality or overall mortality comparing ER-/Ki67 low to ER-/Ki67 high tumors, or comparing TNBC/Ki67 high to TNBC/Ki67 low tumors.
Finally, we explored the relationship of Ki67 score (continuous) with clinical outcomes (Supplementary Table 1). In multivariable models, Ki67 score was not associated with clinical outcomes in all tumors but was associated with breast cancer-specific mortality in ER+ tumors (HR=2.94, 95% CI 1.32–6.54, p=0.008). Further adjustment for tumor grade slightly attenuated this association (HR=2.75, 95% CI 1.22–6.21, p=0.02).
Discussion
Breast cancer subtype classification is a valuable clinical tool for prognosis and clinical management of breast cancer patients, and this study establishes the Ki67 14% cut point as a predictor of breast cancer-specific mortality in luminal subtypes, independent of risk factors for breast cancer survival, clinico-pathological features, and treatment.
Median Ki67 in all breast cancer (6.7%) was lower than the cut points of 14% and 20% often cited in the literature to distinguish luminal subtypes. This difference is likely due to two factors. First, 50% of our cases were luminal A tumors, which we demonstrated have the lowest mean Ki67 among the subtypes. The distribution of subtypes in this large population of women was not enriched for luminal subtypes, and is similar to other population-based cohorts (28–30) although classification methodologies vary. Second, manual reading tended to overestimate Ki67 staining (62% of cases), which could explain higher mean Ki67 scores and a higher cut off value in studies that use manual Ki67 scoring. The significantly higher mean Ki67 scores in HER2+ tumors, EGFR+ tumors and CK5/6+ tumors supports the theory that increased proliferative capacity may explain in part their aggressive behavior and poor prognosis (6). Mean Ki67 was lower in luminal breast cancers than in HER2-enriched and basal-like breast cancers, consistent with previous studies (10).
Smaller studies have found that mean Ki67 varied significantly between HER2+ luminal B and HER2- luminal B breast cancer (31). There was no apparent difference in mean Ki67 score between these two subsets of luminal B in our well-powered study, suggesting that the extent of proliferative activity in these subsets is similar.
Breast cancer may be classified into molecular subtypes using a panel of immunohistochemical markers with tumor grade in clinical settings, but gene expression profiling is the gold standard. Because we do not have gene expression profiling on our breast tumors, we did not aim to validate cut points for subtyping. Instead, we used molecular subtypes defined with tumor grade to assess whether various cut points of Ki67 reasonably well classified luminal breast cancers, and to identify features associated with reclassified tumors. Our results are consistent with previous findings that Ki67 at 14% is a good marker for luminal subtype classification. Importantly, the vast majority of reclassified tumors were luminal A tumors of intermediate grade. In ER+ breast cancer, low grade (grade 1) and high grade (grade 3) tumors have been found to be strongly associated with a gene expression grade index based mostly on cell cycle regulation and proliferation. In contrast, intermediate grade (grade 2) tumors are highly variable in their gene expression grade index (14). In this study, Ki67 appears to distinguish different groups of luminal tumors that are moderately differentiated (grade 2) based on their variation in proliferative activity. Thus, Ki67 staining may provide a relatively simple and clinically applicable method to refine the classification of ER+ tumors with intermediate grade.
This study is among the first to evaluate Ki67 by DIA, and the first large-scale study to examine the relationship between Ki67 and clinical breast outcomes, adjusting for breast cancer prognostic factors. We observed a small but consistent increased risk of distant recurrence in luminal B compared to luminal A tumors at the Ki67 6.7%, 10% and 14% cut points, consistent with previous studies demonstrating that Ki67 may be a valuable clinical marker for predicting breast cancer recurrence in luminal breast cancer (10, 32). Ki67 predicts recurrence in subgroups of TNBC (33, 34), and it could plausibly predict worse breast cancer-specific mortality in ER- breast cancer. However, there was no difference in breast cancer-specific mortality between ER- tumors or TNBC tumors according to Ki67 positivity at any cut point in our study (data not shown). Our data support the clinical utility of Ki67 in predicting recurrence in luminal breast cancer, but suggest that it may not be as informative in ER- breast cancer, which is consistent with other studies (35).
Although our data suggest that there may be a small increased risk of overall mortality comparing luminal B to luminal A, these results were inconsistent across Ki67 cut points. There was no association between Ki67 positivity (>14%) and overall mortality in ER+ tumors in multivariate models; further investigation with time-varying treatment data may be warranted.
Importantly, there was a significant increased risk of breast cancer-specific mortality comparing luminal B to luminal A defined with the Ki67 14% cut point, but not with tumor grade. These data suggest that the Ki67 14% cut point better distinguishes luminal subtypes that differ in breast cancer prognosis. Although higher Ki67 cut points have recently been suggested (9, 36), we have shown that manual IHC scoring tends to overestimate Ki67 positivity in breast tumor specimens in this study. Identifying distinct luminal breast cancers based on proliferative activity may lead to improved clinical management of breast cancer patients, including enhanced prediction of prognosis. Although the Ki67 14% cut point was not data-derived in our study, we have shown that this cut point may have independent prognostic value for breast cancer-specific mortality. Whether the Ki67 14% cut point is a marker for two distinct luminal subtypes with different underlying prognoses, or a surrogate for luminal tumor aggressiveness and response to chemotherapy, cannot be determined within the scope of this study. A recent study found that Ki67 positivity in normal mammary epithelial cells predicts breast cancer risk among premenopausal women, which argues that Ki67 may play an early role in the etiology of luminal breast cancer (37).
Although we did not have gene expression profiling to benchmark our results, Ki67 in combination with ER, PR and HER2 has previously been shown to be a cost-effective and robust biomarker panel for classifying luminal tumors. More recently, PR≥ 20% has been proposed to distinguish luminal A versus luminal B tumors (38), but we do not currently have manual scoring or DIA at the 20% cut point. Another limitation is that information on neoadjuvant endocrine treatment, type of chemotherapy and duration of regimen is not known. Therefore, we were not able to explore the potential predictive value of Ki67 for treatment response and there is the possibility of some residual confounding by treatment. This study has several strengths, including the use and validation of DIA to assess Ki67, which is an important step towards standardizing Ki67 assessment for clinical use. In addition, our study includes a large sample size of breast tumors, which provides sufficient statistical power, particularly for luminal subtype analyses. Another strength is that we were able to assess the independent prognostic value of Ki67 in breast cancer with adjustment for breast cancer prognostic factors.
In one of the largest prospective cohort study to date examining the utility of Ki67 for luminal breast cancer classification and prognosis, we have demonstrated that DIA is a robust method for accurately quantitating Ki67 in breast tumors. Further, our data suggest that the previously established Ki67 14% cut point has prognostic value for luminal tumors independent of clinico-pathological features and breast cancer prognosis factors. Overall, our study provides additional support for the clinical relevance of using Ki67 in a molecular marker panel for luminal subtype classification and breast cancer prognosis.
Supplementary Material
Acknowledgements
We would like to thank the participants and staff of the Nurses’ Health Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Acknowledgement of Research Support
This study was supported by the National Cancer Institute (UM1 CA186107 and Dietary and Hormonal Determinants of Cancer in Women NIH P01 CA87969). MAH and KAH were supported by the National Institutes of Health Cancer Epidemiology Training Grant (NIH T32 CA09001).
Abbreviations
- BMI
body mass index
- CI
confidence interval
- CK5/6
cytokeratin 5/6
- DAB
diaminobenzidine
- DIA
digital image analysis
- EGFR
epidermal growth factor receptor
- ER
estrogen receptor
- FFPE
formalin-fixed paraffin-embedded
- HER2
human epidermal growth factor receptor 2
- NHS
Nurses’ Health Study
- IHC
immunohistochemistry
- HR
hazard ratio
- PMH
post-menopausal hormone
- PR
progesterone receptor
- TMA
tissue microarray
- TNBC
triple-negative breast cancer
Footnotes
Ethical Standards
All data collection was conducted with approval of appropriate institutional review boards to protect human subjects with consent and data protection systems in place. Data analysis for this manuscript was conducted on de-identified data sets.
Conflict of interest
The authors declare that they have no conflict of interest. AHB has an equity interest in PathAI,Inc.
References
- 1.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98(19):10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A 2003;100(18):10393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer 2011;11(143):1471–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Lluch A, Albanell J, Barry WT, Fan C, Chacon JI, et al. Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br J Cancer 2014;111(8):1532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comprehensive molecular portraits of human breast tumours. Nature 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004;10(16):5367–74. [DOI] [PubMed] [Google Scholar]
- 7.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res 2009;7(1–2):4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013;24(9):2206–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, et al. Tailoring therapies--improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol 2015;26(8):1533–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 2009;101(10):736–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama 2006;295(21):2492–502. [DOI] [PubMed] [Google Scholar]
- 12.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991;19(5):403–10. [DOI] [PubMed] [Google Scholar]
- 13.Simpson JF, Gray R, Dressler LG, Cobau CD, Falkson CI, Gilchrist KW, et al. Prognostic value of histologic grade and proliferative activity in axillary node-positive breast cancer: results from the Eastern Cooperative Oncology Group Companion Study, EST 4189. J Clin Oncol 2000;18(10):2059–69. [DOI] [PubMed] [Google Scholar]
- 14.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 2006;98(4):262–72. [DOI] [PubMed] [Google Scholar]
- 15.Schonk DM, Kuijpers HJ, van Drunen E, van Dalen CH, Geurts van Kessel AH, Verheijen R, et al. Assignment of the gene(s) involved in the expression of the proliferation-related Ki-67 antigen to human chromosome 10. Hum Genet 1989;83(3):297–9. [DOI] [PubMed] [Google Scholar]
- 16.Bullwinkel J, Baron-Luhr B, Ludemann A, Wohlenberg C, Gerdes J, Scholzen T. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J Cell Physiol 2006;206(3):624–35. [DOI] [PubMed] [Google Scholar]
- 17.Urruticoechea A, Smith IE, Dowsett M. Proliferation marker Ki-67 in early breast cancer. J Clin Oncol 2005;23(28):7212–20. [DOI] [PubMed] [Google Scholar]
- 18.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst 2011;103(22):1656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, et al. An international Ki67 reproducibility study. J Natl Cancer Inst 2013;105(24):1897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Giovannucci EL, Wu K, Smith-Warner SA, Fuchs CS, Pollak M, et al. Magnesium intake, plasma C-peptide, and colorectal cancer incidence in US women: a 28-year follow-up study. Br J Cancer 2012;106(7):1335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamimi RM, Baer HJ, Marotti J, Galan M, Galaburda L, Fu Y, et al. Comparison of molecular phenotypes of ductal carcinoma in situ and invasive breast cancer. Breast Cancer Res 2008;10(4):R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Healey MA, Hu R, Beck AH, Collins LC, Schnitt SJ, Tamimi RM, et al. Association of H3K9me3 and H3K27me3 repressive histone marks with breast cancer subtypes in the Nurses’ Health Study. Breast Cancer Res Treat 2014;147(3):639–51. [DOI] [PubMed] [Google Scholar]
- 23.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2011;24(7):924–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zabaglo L, Salter J, Anderson H, Quinn E, Hills M, Detre S, et al. Comparative validation of the SP6 antibody to Ki67 in breast cancer. J Clin Pathol 2010;63(9):800–4. [DOI] [PubMed] [Google Scholar]
- 25.Holmes MD, Chen WY, Feskanich D, Kroenke CH, Colditz GA. Physical activity and survival after breast cancer diagnosis. Jama 2005;293(20):2479–86. [DOI] [PubMed] [Google Scholar]
- 26.Colditz GA, Rosner B. Cumulative risk of breast cancer to age 70 years according to risk factor status: data from the Nurses’ Health Study. Am J Epidemiol 2000;152(10):950–64. [DOI] [PubMed] [Google Scholar]
- 27.Kroenke CH, Chen WY, Rosner B, Holmes MD. Weight, weight gain, and survival after breast cancer diagnosis. J Clin Oncol 2005;23(7):1370–8. [DOI] [PubMed] [Google Scholar]
- 28.Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol 2010;28(10):1684–91. [DOI] [PubMed] [Google Scholar]
- 29.Engstrom MJ, Opdahl S, Hagen AI, Romundstad PR, Akslen LA, Haugen OA, et al. Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients. Breast Cancer Res Treat 2013;140(3):463–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parise CA, Caggiano V. Breast Cancer Survival Defined by the ER/PR/HER2 Subtypes and a Surrogate Classification according to Tumor Grade and Immunohistochemical Biomarkers. J Cancer Epidemiol 2014;2014:469251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanagawa M, Ikemot K, Kawauchi S, Furuya T, Yamamoto S, Oka M, et al. Luminal A and luminal B (HER2 negative) subtypes of breast cancer consist of a mixture of tumors with different genotype. BMC Res Notes 2012;5(376):1756–0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams DJ, Cohen C, Darrow M, Page AJ, Chastain B, Adams AL. Proliferation (Ki-67 and phosphohistone H3) and oncotype DX recurrence score in estrogen receptor-positive breast cancer. Appl Immunohistochem Mol Morphol 2011;19(5):431–6. [DOI] [PubMed] [Google Scholar]
- 33.Keam B, Im SA, Lee KH, Han SW, Oh DY, Kim JH, et al. Ki-67 can be used for further classification of triple negative breast cancer into two subtypes with different response and prognosis. Breast Cancer Res 2011;13(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang L, Liu Z, Chen S, Liu Y, Shao Z. A prognostic model for triple-negative breast cancer patients based on node status, cathepsin-D and Ki-67 index. PLoS One 2013;8(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abubakar M, Orr N, Daley F, Coulson P, Ali HR, Blows F, et al. Prognostic value of automated KI67 scoring in breast cancer: a centralised evaluation of 8088 patients from 10 study groups. Breast Cancer Res 2016;18(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bustreo S, Osella-Abate S, Cassoni P, Donadio M, Airoldi M, Pedani F, et al. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: a large case series study with a long-term follow-up. Breast Cancer Res Treat 2016;157(2):363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huh SJ, Oh H, Peterson MA, Almendro V, Hu R, Bowden M, et al. The Proliferative Activity of Mammary Epithelial Cells in Normal Tissue Predicts Breast Cancer Risk in Premenopausal Women. Cancer Res 2016;76(7):1926–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maisonneuve P, Disalvatore D, Rotmensz N, Curigliano G, Colleoni M, Dellapasqua S, et al. Proposed new clinicopathological surrogate definitions of luminal A and luminal B (HER2-negative) intrinsic breast cancer subtypes. Breast Cancer Res 2014;16(3):R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.