Abstract
Purpose
A lack of early diagnostic biomarkers and therapeutic targets has led to poor prognosis for gastric cancer patients. However, the analysis of cancer-associated genomic data has been shown to be effective in identifying potential markers. Recently, the long non-coding RNA LINC00365 and SCGB2A1 gene (as known as mammaglobin B) were predicted to be co-expressed in gastric cancer based on the Gene Expression Omnibus database. However, their precise role in gastric cancer tumors is still not clear.
Methods
The expressions of LINC00365 and SCGB2A1 in gastric cancer tissues were investigated using qPCR and their expressions were detected in a gastric cancer tissue microarray by in situ hybridization and immunohistochemical staining. The functions of LINC00365 in BGC-823 and MGC-803 gastric cancer cells were tested using the MTT assay, flow cytometry, colony formation assay, EDU staining, immunofluorescence and luciferase assay.
Results
We found that LINC00365 and SCGB2A1 mRNA were both expressed at low levels in 30 cases of gastric cancer. Gastric cancer tissue microarray analysis indicated that LINC00365 and SCGB2A1 were expressed at low levels in tumor tissue, and low expression of both factors correlated with shorter survival time. Functional studies showed that LINC00365 overexpression significantly inhibited gastric cancer cell viability through the impairment of proliferation rather than the promotion of apoptosis. Furthermore, overexpressed LINC00365 upregulated SCGB2A1 in gastric cancer cell lines. Immuno-fluorescence and luciferase assay analysis indicated that LINC00365 overexpression inhibited the NF-κB pro-survival signaling pathway. Consistent with the effects of LINC00365, SCGB2A1 upregulation also reduced cell survival and inactivated NF-κB.
Conclusion
Collectively, our findings revealed that SCGB2A1 may be the target coding protein regulated by LINC00365 in gastric cancer. LINC00365 and SCGB2A1 may function as tumor suppressors and may serve as potential prognostic and therapeutic markers in gastric cancer treatment.
Keywords: LncRNA, NF-κB, proliferation, gastric cancer
Introduction
Gastric cancer (GC) is one of the most common gastrointestinal tumors. Although chemotherapy, radiotherapy, and surgical treatments have achieved success, the survival time of GC patients has not increased substantially and the overall 5-year survival rate is still only 25%.1,2 Recently, microarray technology in association with high-throughput gene expression has been used as an efficient method for potential GC biomarker identification.3–5 After analyzing GC datasets GSE30727, GSE13195, and GSE33335 Li et al reported the co-expression of LINC00365, located on chromosome 13q12.3, and secretoglobin family 2A member 1 (SCGB2A1), which indicated that these molecules have the potential to be biomarkers for GC treatment.6 However, their regulatory mechanism is unclear and further confirmation is needed.
SCGB2A1 (also known as mammaglobin B) is a secreted protein that belongs to the secretoglobin superfamily.7 Several studies have indicated a role for SCGB2A1 in ovarian and breast cancers; the expression of SCGB2A1 was higher in ovarian cancer tissues compared with normal tissues, and SCGB2A1 may function as an immunotherapy target to overcome drug resistance.8,9 However, the role of SCGB2A1 in gastric cancer is poorly understood.
Increasing evidence has shown that long non-coding (lnc)RNAs play important roles in cellular events such as differentiation, cell cycle progression, and tumorigenesis through regulating coding proteins and miRNA.10,11 Yang et al demonstrated that the lncRNA H19 led to partial inactivation of p53, which caused GC cells to escape apoptosis by inhibiting the expression of the p53 downstream protein Bax.12 Identifying potential lncRNAs that may be involved in regulating SCGB2A1 may be helpful to understand the role of SCGB2A1 in GC.
Abnormal activation of nuclear factor (NF)-κB signaling has been detected in a variety of tumors.13–15 Zhang et al found that LINC01410 activated NF-κB signaling through targeting mir-532-5p, resulting in increased angiogenesis and metastasis in GC.16 Activated NF-κB triggers a series of molecular reactions, including the upregulation of protein-encoding genes involved in cell proliferation.17,18 Claerhout et al found that the histone deacetylase inhibitor vorinostat up-regulated SCGB2A1 mRNA expression in GC cell lines AGS and KATO-III while inhibiting cell proliferation, suggesting that SCGB2A1 may be involved in the regulation of pro-survival signals in GC.19
lncRNA may function through regulating the expression of encoding genes and this “lncRNA-encoding proteins pattern” has been demonstrated in a variety of tumors including GC.20,21 Based on the findings by Li et al,66 we explored the relationship and in vitro regulatory mechanism of LINC00365 and SCGB2A1. This study provide theoretical support for the clinical application of LINC00365 and SCGB2A1 as prognostic biomarkers and targets in GC treatment.
Materials and Methods
Cell Lines and Cell Culture
Human GC cell lines (SGC-7901, BGC-823, MGC-803) and the human fetal gastric epithelial cell line GES-1 were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM or RPMI-1640 medium (Gibco Life Technologies, Carlsbad, CA, USA) supplemented with penicillin (100 IU/mL), streptomycin (100 µg/mL), and 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37°C in a 5% CO2 incubator.
Antibodies and Reagents
Antibodies used in this study were anti-p65 and anti-β-actin (Proteintech, Chicago, IL, USA), anti-SCGB2A1 (ABclonal Biotechnology Co., Ltd.), anti-inhibitor of κB (IκB) and anti-pκB (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Reagents used in this study included 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Sigma-Aldrich, St. Louis, MO, USA) and ViaFect™ transfection reagent (Promega, Madison, MI, USA). The FITC Annexin V Apoptosis Detection Kit was purchased from BD Biosciences (Franklin Lakes, NJ, USA).
Plasmids and Transfection
pcDNA3.1-LINC00365, and pcDNA3.1-SCGB2A1 were constructed by Sangon Biotech (Shanghai, China); pcDNA3.1 vector was used as negative control. Cells were transfected using ViaFect™ transfection reagent (Promega, Madison, MI, USA) according to the manufacturer’s instructions.
GC Tissue Microarray
A total of 90 pairs of GC tissues collected between 2010 and 2016 were purchased from Shanghai Outdo Biotech Co., Ltd. All participants in this study provided informed consent for the use of their tissue samples for research. The use of tissue samples was approved by the local ethics committee (YB M-05-01). Clinical characteristics are listed in Table 1. Immunohistochemistry (IHC) was carried out using primary antibodies against SCGB2A1 and in situ hybridization (ISH) was performed using oligonucleotides targeting LINC00365 (Exiqon, Vedbæk, Denmark). Sections were fixed in 4% (v/v) formaldehyde in phosphate-buffered saline (PBS) and then dehydrated and embedded in paraffin. Tissues were sliced into 5 µm sections and antigen retrieval was performed by heating for 15 min in a microwave. Horse serum albumin (5%) was used to block nonspecific interactions. Sections were incubated with primary antibodies and then horseradish peroxidase (HRP)-conjugated secondary antibodies; nuclei were stained with 4′,6-diamidino-2-phenylindole. Images were acquired using an Olympus microscope.
Table 1.
Clinical Pathological Characteristics of Gastric Cancer Cases
| Characteristic | GC, N=90 | % |
|---|---|---|
| Sex | ||
| Male | 68 | 75.6 |
| Female | 21 | 23.3 |
| Miss | 1 | 1.1 |
| Age (years) | ||
| < 65 | 53 | 58.9 |
| ≥ 65 | 37 | 41.1 |
| Miss | 0 | 0 |
| Tumor Size (cm) | ||
| < 5 | 34 | 37.8 |
| ≥ 5 | 54 | 60 |
| Miss | 2 | 2.2 |
| Histological grade | ||
| I–III | 48 | 53.3 |
| III | 42 | 46.7 |
| Miss | 0 | 0 |
| T stage | ||
| T1–T2 | 16 | 17.8 |
| T3–T4 | 60 | 66.7 |
| Miss | 14 | 15.6 |
| Lymph nodes metastasis | ||
| Absence | 24 | 26.7 |
| Presence | 65 | 72.2 |
| Miss | 1 | 1.1 |
| Distant metastasis | ||
| Absence | 88 | 97.8 |
| Presence | 2 | 2.2 |
| Miss | 0 | 0 |
| Follow-ups | ||
| Dead | 60 | 66.7 |
| Survival | 30 | 33.3 |
| Miss | 0 | 0 |
Cell Viability Assay
Cell viability was estimated by the MTT assay. Cells transfected with pcDNA3.1-LINC00365, pcDNA3.1-SCGB2A1 or empty vector were plated in 96-well plates at a density of 1.2 × 104 cells/well. MTT reagents were added and cells were incubated for 4 h. Absorbance values were then measured at 490 nm using a Vmax Microplate Reader (Molecular Devices, Sunnyvale, CA, USA).
RNA Extraction and Quantitative Real-Time PCR
GC tissues were obtained from The China-Japan Union Hospital of Jilin University. Written informed consent was obtained from all patients. Experiments followed the Helsinki Declaration (as revised in 2013) and were approved by The China-Japan Union Hospital of Jilin University ethics committee (No.2018-NSFC-051). Total RNA was extracted and first-strand cDNA was synthesized as previously described.22 Quantitative real-time PCR was performed using the MX300P instrument (Agilent Technologies, Santa Clara, CA, USA) followed by a two-step PCR protocol. Relative expression was calculated by ΔCt among different experimental groups normalized to ACTB mRNA expression. Primer sequences are listed in Table 2.
Table 2.
Primer Sequences
| PRIMERS | SEQUENCES (5ʹ-3ʹ) |
|---|---|
| LINCOO365 | Forward:TGCTATTCTATGGCTGGGCTTC |
| Reverse:ACTGTTGAATTTCCTGGATGTGTC | |
| SCGB2A1 | Forward:AAACTCCTGGAGGACATGGTT |
| Reverse:ACTGCTTGAATTTCCCCATAGC | |
| CXCL8 | Forward:ATGACTTCCAAGCTGGCCGTGGCT |
| Reverse:TCTCAGCCCTCTTCAAAAACTTCTC | |
| TGFΒ1 | Forward:GGATAACACACTGCAAGTGG |
| Reverse:GAGCTGAAGCAATAGTTGGTG | |
| ACTIN | Forward:ATATCGCGTCGCTGGTCGTC |
| Reverse:AGGATGGCGTGAGGGAGAGC |
Abbreviations: EdU, 5-Ethynyl-2′-deoxyuridine; GC, gastric cancer; lncRNA, long non-coding RNA; NF-κB, nuclear factor-κB; p65, RELA proto-oncogene; IHC, Immunohistochemistry; ISH, in situ hybridization; PI, propidium iodide; SCGB2A1, secretoglobin family 2A member 1.
Colony Formation Assay
Cells transfected with the LINC00365 overexpression plasmid or empty vector were seeded in 6-well culture plates (500 cells each well). After incubation in fresh medium for 1 week, cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min, and stained with crystal violet (0.5% crystal violet, 1% paraformaldehyde and 20% methanol in PBS) for 30 min. Colonies were counted under a microscope.
Luciferase Assay
Cells were transfected with the NF-κB-Luc reporter plasmid (Beyotime Biotechnology) using ViaFect™ transfection reagent. Luciferase activity was then measured using a Dual-Luciferase assay kit (Promega) and normalized to Renilla luciferase activity.
5-Ethynyl-2′-Deoxyuridine (EdU) Staining
Transfected cells were seeded in 96-well plates and exposed to 50 µM of EdU (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. DNA contents were stained with Hoechst 33342 for 30 min and visualized under a microscope (Olympus, Tokyo, Japan).
Flow Cytometry Analysis
Cell death was analyzed by Annexin-V FITC/propidium iodide (PI) staining (BD Biosciences). Cells were seeded in 6-well plates and transfected with the LINC00365 overexpression plasmid or empty vector. The attached and detached cells were harvested and subjected to Annexin-V FITC/PI staining according to the manufacturer’s instructions. Samples were analyzed using an Accuri C6 Flow Cytometer (BD Biosciences).
Western Blotting
Cells were lysed in radioimmunoprecipitation assay buffer with protease inhibitors. Lysates were cleared by centrifugation at 1000 × g for 15 min at 4°C, boiled in loading buffer, and resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Proteins were transferred to polyvinylidene difluoride membranes, which were then blocked with 5% milk, followed by incubation with primary antibodies overnight at 4°C. Membranes were incubated with HRP-conjugated secondary antibodies (Proteintech), and ECL reagent (Thermo Fisher Scientific, Rockford, IL, USA) was used for immunodetection with visualization by Syngene Bio Imaging (Synoptics, Cambridge, UK).
Immunofluorescence and Confocal Microscopy
Cells were fixed in 4% (w/v) paraformaldehyde/PBS for 20 min and then permeabilized with 0.1% Triton X-100 for 15 min. After blocking with bovine serum albumin for 30 min, cells were incubated with primary antibody overnight at 4°C. Cells were then incubated with FITC/Texas Red-conjugated secondary antibodies (Proteintech) at room temperature for 1 h. Images were acquired using an Olympus FV1000 confocal laser microscope.
Statistical Analysis
Experimental data were presented as means ± standard deviation (SD) and compared using the Student’s t-test. P < 0.05 was considered statistically significant. Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA).
Results
Low Expression of LINC00365 and SCGB2A1 Is Correlated with Poor Prognosis in Gastric Cancer
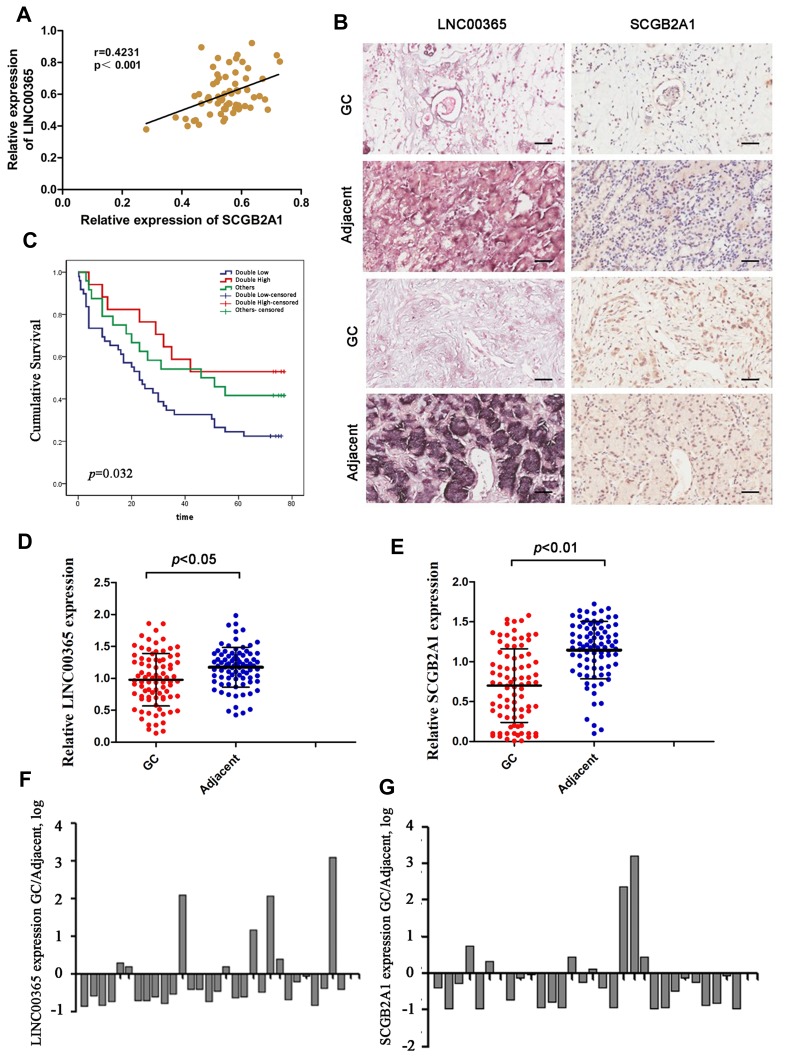
We first analyzed LINC00365 and SCGB2A1 expression by in situ hybridization and IHC staining, respectively, in tissue samples from 90 GC patients. Pearson’s correlation coefficient analysis showed a positive correlation between LINC00365 and SCGB2A1 in GC tissues (r=0.4231, P<0.001) (Figure 1A and B). Kaplan–Meier analysis indicated that patients with low expression of both factors had shorter overall survival (Figure 1C). Furthermore, compared with adjacent tissues, tumor tissues showed lower expression of LINC00365 and SCGB2A1 (Figure 1D and E). To further confirm these findings, we performed quantitative PCR to evaluate the expression of LINC00365 and SCGB2A1 in 30 pairs of GC and adjacent tissues. Consistent with the tissue microarray results, the expressions of LINC00365 and SCGB2A1 were decreased in tumor tissues compared with adjacent tissues (Figure 1F and G). These results indicated that LINC00365 and SCGB2A1 may function as tumor suppressors in GC.
Figure 1.
The expression of LINC003365 and SCGB2A1 in gastric cancer tissues. (A) Pearson’s correlation coefficient analysis for LINC00365 and SCGB2A1 expression in gastric cancer tissues. (B) Representative images of LINC00365 and SCGB2A1 expression in human gastric cancer tissues and adjacent tissues (scale bar=20µm). SCGB2A1 was analyzed by IHC and LINC00365 was detected by ISH. (C) Kaplan–Meier survival curve showing a significant association between LINC00365 and SCGB2A1 expression and survival in gastric cancer patients (P=0.032). Double high: higher expression of both LINC00365 and SCGB2A1 in gastric cancer tissues compared with adjacent tissues (n=17); Double low: low expression of both LINC00365 and SCGB2A1 in gastric cancer tissues compared with adjacent tissues (n=49); other: higher expression of either LINC00365 or SCGB2A1 in gastric cancer tissues compared with the adjacent tissues (n=24). (D, E) The expression of SCGB2A1 and LINC00365 in gastric cancer tissues and adjacent tissues. Statistical significance was determined using a two-tailed, unpaired Student’s t-test. (F, G) Relative mRNA expression of SCGB2A1 and LINC00365 in tissues from 30 gastric cancer patients.
LINC00365 Positively Regulates the Expression of SCGB2A in Gastric Cancer
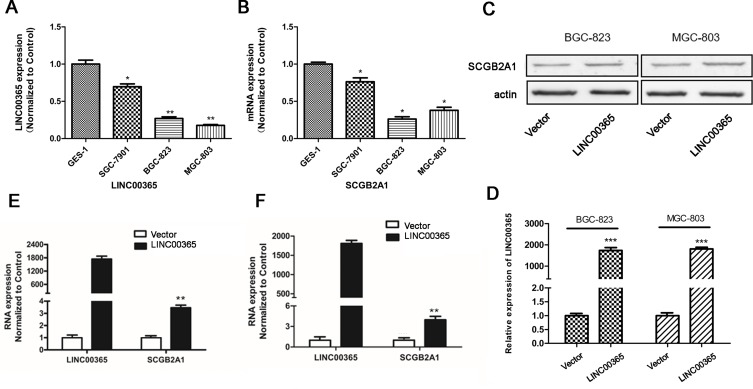
To explore the regulatory relationship between LINC00365 and SCGB2A1, we examined their expression in human GC cell lines SGC-7901, BGC-823, and MGC-803 and the human fetal gastric epithelial cell line GES-1. Consistent with the database findings, our results showed that LINC00365 and SCGB2A1 were expressed at lower levels in GC cell lines compared with normal gastric epithelial cells (Figure 2A and B). Upon overexpression of LINC00365 in BGC-823 and MGC-803 cells, which had low levels of both LINC00365 and SCGB2A1, up-regulation of SCGB2A1 was detected at both the mRNA and protein level (Figure 2C–F). This indicated that SCGB2A1 may be a target gene of LINC00365.
Figure 2.
LINC00365 regulates the expression of SCGB2A1 in gastric cancer cell lines. (A, B) Relative mRNA expression of LINC00365 and SCGB2A1 in three gastric cancer cell lines; data were normalized to levels detected in the human fetal gastric mucosa epithelium cell line GES-1. Data are presented as means ± SD; *P < 0.05; **P < 0.01. (C, D) BGC-823 and MGC-803 cells were transfected with LINC00365 overexpression plasmid or empty vector. The expression of SCGB2A1 was analyzed using Western blotting, and LINC000365 expression was determined using quantitative PCR. Data are presented as means ± SD; ***P < 0.001. (E, F) BGC-823 and MGC-803 cells were treated as in C, and the mRNA expressions of LINC00365 and SCGB2A1 were examined. Data are presented as means ± SD; **P < 0.01.
LINC00365 Inhibits Proliferation of Gastric Cancer Cells
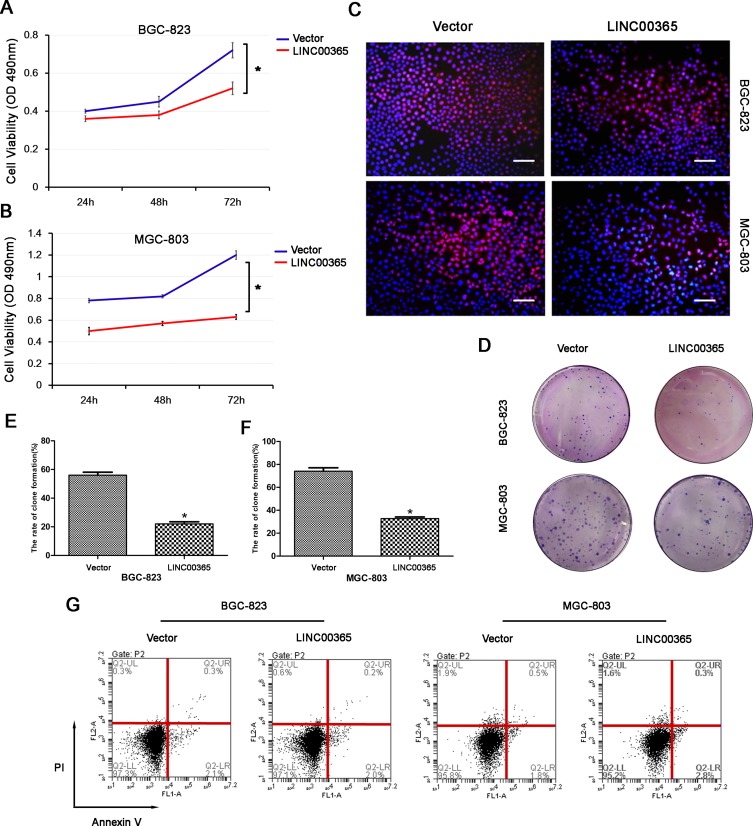
Given that low LINC00365 and SCGB2A1 expression were correlated with poor prognosis in GC, we next investigated the effect of LINC00365 overexpression on the viability of BGC-823 and MGC-803 cells using the MTT assay. High LINC00365 expression significantly suppressed cell survival in both GC cell lines (Figure 3A and B). EdU staining and colony formation analysis further confirmed that overexpression of LINC00365 inhibited proliferation of GC cell lines compared with empty vector transfection (Figure 3C–F). However, when we evaluated the effect of LINC00365 in cell death, we found that LINC00365 overexpression had no significant impact on apoptosis (Figure 3G). These findings suggested inhibiting the GC cells proliferation may be one of the anti-tumor mechanism of LINC00365.
Figure 3.
LINC00365 inhibits proliferation through regulating SCGB2A1 in gastric cancer cells. (A, B) BGC-823 and MGC-803 cells were transfected with LINC00365 overexpression plasmid or empty vector. Cell viability was analyzed by the MTT assay. Data are presented as means ± SD; *P < 0.05. (C) BGC-823 and MGC-803 cells were treated as in A. Cells were stained after 48 h with EdU and Hoechst 33342 and observed using a fluorescence microscope (scale bar, 50 µm). (D–F) Clone formation assays in BGC-823 and MGC-803 cells transfected with SCGB2A1 overexpression plasmid or empty vector. Data are presented as means ± SD; *P < 0.05. (G) Transfected BGC-823 and MGC-803 cells were stained with PI and Annexin V-FITC. Positively stained cells were detected by flow cytometry.
LINC00365 Inhibits Activation of the NF- κB Signaling Pathway Through Regulating SCGB2A1
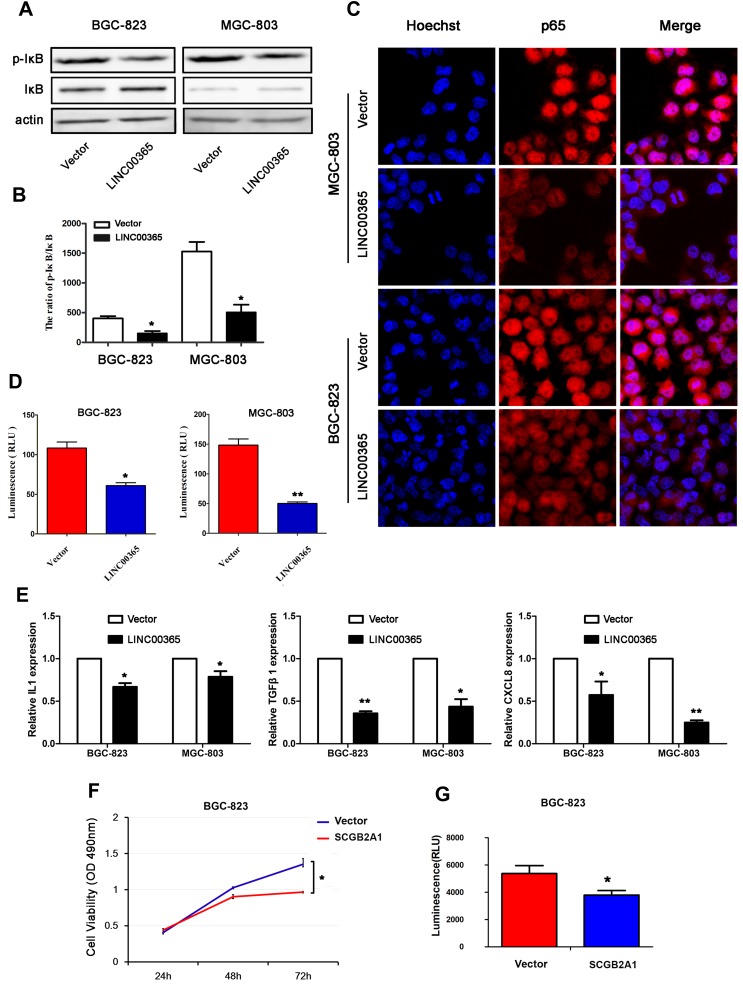
Aberrant activation of NF-κB signaling is known to be critical for GC survival.18,23,24 To verify whether LINC00365 is involved in NF-κB pathway regulation, we examined the phosphorylation of IκB by Western blotting and found that up-regulation of LINC00365 was accompanied by a decrease in IκB phosphorylation (Figure 4A and B). Immunofluorescence revealed that LINC00365 overexpression inhibited translocation of the p65 subunit of NF-κB from the cytoplasm to the nucleus in GC cells (Figure 4C). Luciferase assays also showed that LINC00365 overexpression down-regulated the transcriptional activity of NF-κB (Figure 4D).
Figure 4.
LINC00365 promotes the activation of NF-κB signaling in gastric cancer cells. (A, B) BGC-823 and MGC-803 cells were transfected with empty vector or LINC00365. The expression of p-IκB was analyzed by Western blotting. Data are presented as means ± SD; *P < 0.05. (C) BGC-823 and MGC-803 cells were transfected with the LINC00365 overexpression plasmid or empty vector. After 24h, cells were fixed in 4% (w/v) paraformaldehyde/PBS and incubated with p65 antibody overnight at 4°C. Nuclei were stained with Hoechst 33342. Images were acquired by an Olympus FV1000 confocal laser microscope (scale bar=25 µm). (D) BGC-823 and MGC-803 cells were transfected with LINC00365 overexpression plasmid or empty vector. The transcriptional activity of NF-κB was determined by the luciferase reporter assy. Data are presented as means ± SD; *P < 0.05; **P < 0.01. (E) The mRNA expressions of IL-1, TGFB1, and CXCL8 were determined by quantitative PCR. Data are presented as means ± SD; *P < 0.05, **P < 0.01. (F) BGC-823 cells were transfected with SCGB2A1 overexpression plasmid or empty vector. Cell viability was analyzed by the MTT assay. Data are presented as means ± SD (n=3); *P < 0.05. (G) BGC-823 cells were transfected with SCGB2A1 overexpression plasmid or empty vector. The transcriptional activity of NF-κB was determined by the luciferase reporter assay. Data are presented as means ± SD (n=3); *P < 0.05.
We next examined the expression of NF-κB target genes IL-1, TGFB1, and CXCL8 using quantitative PCR and found that LINC00365 overexpression resulted in downregulation of all three genes (Figure 4E). Because our results indicated that SCGB2A1 is regulated by LINC00365, we hypothesized that LINC00365 may inhibit NF-κB signaling by targeting SCGB2A1. To test this possibility, we overexpressed SCGB2A1 in BGC-823 cells and found that overexpressed SCGB2A1 inhibited NF-κB, similar to LINC00365 overexpression (Figure 4F). Moreover, the MTT assay confirmed that SCGB2A1 overexpression decreased cell viability (Figure 4G). Taken together, our results indicated that the upregulation of LINC00365 in GC cells inhibited activation of the pro-tumorigenesis NF-κB pathway, which was at least partially dependent on SCGB2A1.
Discussion
The identification of early diagnostic and treatment biomarkers is key to improve prognosis in GC.25–27 In this study, we verified that the expression of LINC00365 and SCGB2A1 were both lower in tumor tissues compared with adjacent tissues and low expression of both LINC00365 and SCGB2A1 correlated with poor prognosis of GC. Furthermore, up-regulation of LINC00365 resulted in decreased SCGB2A1 expression at both mRNA and protein levels. Overexpression of LINC00365 suppressed the proliferation of GC cells by inhibiting NF- κB signals.
During the past few decades, the main therapeutic strategy for GC has been surgery combined with perioperative chemotherapy.28 Finding effective targets has become a promising approach for developing novel treatments.29 LINC00365 is a newly identified lncRNA and how it functions in cancer is still unknown. Fan et al showed that esophageal cancer patients with high LINC00365 expression have shorter survival times than those with low LINC00365 expression.30 As Li et al found SCGB2A1 may be co-expressed with LINC00365 in GC cancer, we kept this coding gene in focus.6 SCGB2A1 is a secreted protein that was first isolated by Becker in 1988 and has been detected in the breast, secretory mucosal epithelium, uterus, and lacrimal, prostate, and pituitary glands.31 Munakata et al evaluated 222 colorectal cancer patients and found that patients with low SCGB2A1 mRNA expression had longer disease-free survival than those with high expression.32 Different from these findings in esophageal and colorectal cancer, our study showed that low expression of both LINC00365 and SCGB2A1 were related to poor prognosis in GC. These results indicate that LINC00365 and SCGB2A1 may have tissue specificity and they function as tumor suppressors in GC. Detection of both may increase the accuracy of determining the prognosis of GC patients.
To further explore the possible regulatory mechanism between LINC00365 and SCGB2A1, we overexpressed LINC00365 in BGC-823 and MGC-803 GC cell lines and found that overexpression of LINC00365 up-regulated the transcriptional and protein levels of SCGB2A1. Furthermore, we evaluated the role of LINC00365 in proliferation and apoptosis of GC cells and found that LINC00365 overexpression had no significant impact on cell death but it impaired cell survival through proliferation inhibition.
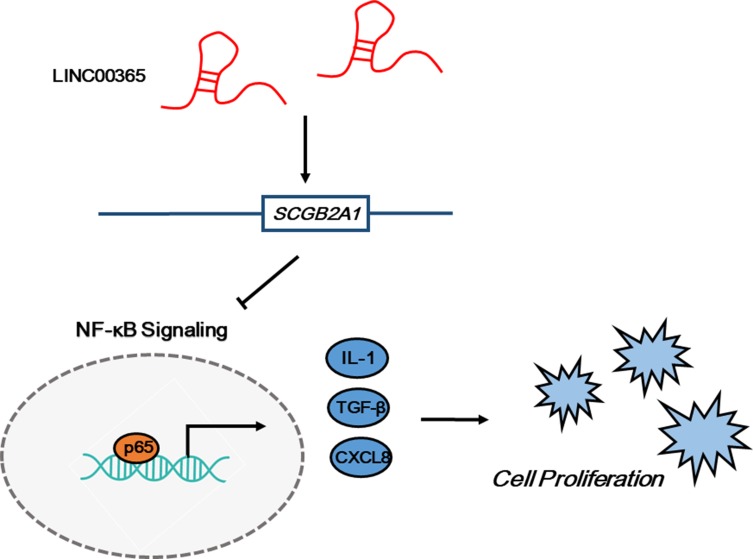
Previous studies reported the involvement of lncRNAs in NF-κB pathway regulation. For example, Zhang et al found that overexpression of the lncRNA BANCR promoted the transcriptional activity of NF-κB and the translocation of p65 in BGC-823 and MGC-803 GC cells, while Yo et al showed that the lncRNA FLJ46906 enhanced the transcription of NF-κB downstream genes ILB, IL6, CXCL8, and TGF-β1.33,34 Our data showed that overexpression of LINC00365 in BGC-823 and MGC-803 cells decreased IκB phosphorylation, which may stabilize the IκB-p50-p65 complex. Moreover, immunofluorescence results confirmed that LINC00365 overexpression inhibited translocation of the NF-κB key subunit p65 from the cytoplasm to nucleus. Additionally, the transcriptional activity of NF-κB was suppressed upon LINC00365 overexpression, which caused reduced gene expression of NF-κB downstream genes IL1, CXCL8, and TGF-β1. Similar to the effects of LINC00365 overexpression, up-regulated SCGB2A1 also inhibited cell viability and NF-κB signaling. These findings suggest that the low expression of LINC00365 in GC promotes cell survival through relieving the inhibition of NF-κB signaling in a process partially dependent on SCGB2A1 regulation (Figure 5).
Figure 5.
The LINC00365/SCGB2A1 axis suppresses cell proliferation through inhibiting NF-κB signaling in gastric cancer.
Conclusion
Here we investigated the role and regulatory mechanism of LINC00365 and SCGB2A1 in GC. We found that low expression of LINC00365 and SCGB2A1 was associated with poor prognosis of GC. Furthermore, our results suggest that LINC00365 may target SCGB2A1 to suppress GC cell proliferation through NF-κB signaling inhibition. These data indicate that LINC00365 and SCGB2A1 could be potential biomarkers to improve the accuracy of GC diagnosis and treatment, and the secreted protein properties of SCGB2A1 may be advantageous for subsequent clinical application.
Acknowledgments
We are grateful to State Key Laboratory on Integrated Optoelectronics College of Electronic Science and Engineering Jilin University for providing confocal laser microscope. We also thank Sarah Williams, PhD, from Liwen Bianji, Edanz Group China, for editing the English text of a draft of this manuscript.
Funding Statement
This work was supported by National Natural Science Foundation of China (81672948, 81772794, 81501982); Jilin Provincial Health Special Project (2018SCZ021); Jilin Provincial Industrial Innovation Project (2018C052-7); Jilin University Bethune Plan B Projects (2015222); and the Fundamental Research Funds for the Central Universities, Jilin University.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zhao J, Liu Y, Huang G, Cui P, Zhang W, Zhang Y. Long non-coding RNAs in gastric cancer: versatile mechanisms and potential for clinical translation. Am J Cancer Res. 2015;5(3):907. [PMC free article] [PubMed] [Google Scholar]
- 2.Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42(2):211. doi: 10.1016/j.gtc.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;141893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F, Huang C, Li Q, Wu X. Construction and comprehensive analysis for dysregulated long non-coding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) network in gastric cancer. Med Sci Monit. 2018;24:37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guoxing HU, Xiaoru YU. Analysis of gastric cancer drug resistance gene expression profile and its effect on prognosis based on GEO database. Pract Radiat Oncol. 2018. [Google Scholar]
- 6.Li Y, He Y, Han S, Liang Y. Identification and functional inference for tumor-associated long non-coding RNA. IEEE/ACM Trans Comput Biol Bioinform. 2017;PP(99):1. [DOI] [PubMed] [Google Scholar]
- 7.Pandey M, Sunil Kumar BV, Gupta K, Sethi RS, Kumar A, Verma R. Over-expression of mammaglobin-B in canine mammary tumors. BMC Vet Res. 2018;14(1):184. doi: 10.1186/s12917-018-1507-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellone S, Tassi R, Betti M, et al. Mammaglobin B (SCGB2A1) is a novel tumour antigen highly differentially expressed in all major histological types of ovarian cancer: implications for ovarian cancer immunotherapy. Br J Cancer. 2013;109(2):462–471. doi: 10.1038/bjc.2013.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nissan A, Jager D, Roystacher M, et al. Multimarker RT|[ndash]|PCR assay for the detection of minimal residual disease in sentinel lymph nodes of breast cancer patients. Cuadernos Value. 2006;94:681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 2018;41(6):1–19. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Tang Y, Xiong F, et al. LncRNAs regulate cancer metastasis via binding to functional proteins. Oncotarget. 2018;9(1):1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F, Bi J, Xue X, et al. Up‐regulated long non‐coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279(17):3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x [DOI] [PubMed] [Google Scholar]
- 13.Xiang Z, Zhou ZJ, Xia GK, et al. A positive crosstalk between CXCR4 and CXCR2 promotes gastric cancer metastasis. Oncogene. 2017;36(36):5122. doi: 10.1038/onc.2017.108 [DOI] [PubMed] [Google Scholar]
- 14.Fusella F, Secli L, Busso E, et al. The IKK/NF-kappaB signaling pathway requires morgana to drive breast cancer metastasis. Nat Commun. 2017;8(1):1636. doi: 10.1038/s41467-017-01829-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J, Li Y, Chen C, et al. NF-κB p65 overexpression promotes bladder cancer cell migration via FBW7-mediated degradation of RhoGDIα protein. Neoplasia. 2017;19(9):672–683. doi: 10.1016/j.neo.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang JX, Chen ZH, Chen DL, et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37(20):2660–2675. doi: 10.1038/s41388-018-0162-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Hoesel B, Schmid AJ. The complexity of NF-kappa B signaling in inflammation and cancer. Mol Cancer. 2013;12(1):86. doi: 10.1186/1476-4598-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokolova O, Naumann M. NF-κB signaling in gastric cancer. Toxins. 2017;9(4):119. doi: 10.3390/toxins9040119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claerhout S, Lim JY, Choi W, et al. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One. 2011;6(9):e24662. doi: 10.1371/journal.pone.0024662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song X, Cao G, Jing L, et al. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med. 2014;18(6):991–1003. doi: 10.1111/jcmm.2014.18.issue-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batista P, Chang H. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan XY, Zhang Y, Zhang JJ, et al. p62/SQSTM1 as an oncotarget mediates cisplatin resistance through activating RIP1-NF-kappaB pathway in human ovarian cancer cells. Cancer Sci. 2017;108(7):1405–1413. doi: 10.1111/cas.13276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang T, Wei K, Zhang B, et al. miR-508-3p concordantly silences NFKB1 and RELA to inactivate canonical NF-κB signaling in gastric carcinogenesis. Mol Cancer. 2016;15(1):1–15. doi: 10.1186/s12943-016-0493-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki N, Morisaki T, Hashizume K, et al. Nuclear factor-kappaB p65 (RelA) transcription factor is constitutively activated in human gastric carcinoma tissue. Clin Cancer Res. 2001;7(12):4136–4142. [PubMed] [Google Scholar]
- 25.Hudler P. Challenges of deciphering gastric cancer heterogeneity. World J Gastroenterol. 2015;21(37):10510. doi: 10.3748/wjg.v21.i37.10510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lordick F, Janjigian YY. Clinical impact of tumour biology in the management of gastroesophageal cancer. Nat Rev Clin Oncol. 2016;13(6):348–360. doi: 10.1038/nrclinonc.2016.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10(11):643–655. doi: 10.1038/nrclinonc.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasako M, Sakuramoto S, Katai H, et al. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J Clin Oncol. 2011;29(33):4387. doi: 10.1200/JCO.2011.36.5908 [DOI] [PubMed] [Google Scholar]
- 29.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde VDV. Gastric cancer. Lancet. 2009;374(9688):477–490. doi: 10.1016/S0140-6736(09)60617-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan Q, Liu B. Identification of a RNA-seq based 8-long non-coding RNA signature predicting survival in esophageal cancer. Med Sci Monit. 2016;22:5163–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker RM, Darrow C, Zimonjic DB, Popescu NC, Watson MA, Fleming TP. Identification of mammaglobin B, a novel member of the uteroglobin gene family. Genomics. 1998;54(1):70–78. doi: 10.1006/geno.1998.5539 [DOI] [PubMed] [Google Scholar]
- 32.Munakata K, Uemura M, Takemasa I, et al. SCGB2A1 is a novel prognostic marker for colorectal cancer associated with chemoresistance and radioresistance. Int J Oncol. 2014;44(5):1521–1528. doi: 10.3892/ijo.2014.2316 [DOI] [PubMed] [Google Scholar]
- 33.Zhang ZX, Liu ZQ, Jiang B, et al. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-κB1. Biochem Biophys Res Commun. 2015;465(2):225–231. doi: 10.1016/j.bbrc.2015.07.158 [DOI] [PubMed] [Google Scholar]
- 34.Yo K, Rünger TM. The long non-coding RNA FLJ46906 binds to the transcription factors NF-κB and AP-1 and regulates expression of aging-associated genes. Aging. 2018;10(8):2037–2050. doi: 10.18632/aging.v10i8 [DOI] [PMC free article] [PubMed] [Google Scholar]