Abstract
Background: We previously revealed that the expression of galectin-1 (LGALS1) was significantly reduced after neoadjuvant chemotherapy treatment in cervical cancer patients. The objective of this study is to investigate the effects of LGALS1 expression on biological behaviors of cervical cancer cells.
Methods: Immunohistochemistry and immunocytochemistry were performed to detect the expression of LGALS1 in cervical cancer tissues and cells (SiHa and C33A). Western blot analysis was performed to evaluate the efficacy of lentivirus-mediated upregulation or downregulation of LGALS1 in cervical cancer cells. Cell viability and proliferation were detected by CCK-8 and BrdU assays, respectively. Annexin V-FITC/PI apoptosis detection kit was employed to measure the apoptosis of cervical cancer cells. Transwell invasion and migration assays were also conducted to explore the invasive and migratory capabilities of cervical cancer cells. The expression of apoptosis- (Bcl-2 and Bax), invasion- (MMP-2 and MMP-9), and migration-related (Fascin and Ezrin) proteins, were detected by Western blot analysis. Xenograft mouse model of cervical cancer was generated to explore whether LGALS1 overexpression could promote tumor growth in vivo.
Results: LGALS1 was overexpressed in cervical cancer tissues and cell lines compared to that in normal cervical tissues and epithelium cells. Upregulation of LGALS1 significantly promoted the cell proliferation, inhibited cell apoptosis, and enhanced the migratory and invasive abilities of both SiHa and C33A cells, whereas downregulation of LGALS1 led to the opposite results. The level of Bcl-2, MMP-2, MMP-9, Fascin, and Erzin expression was significantly upregulated in cervical cancer cells with LGALS1 overexpression, while converse results were obtained in LGALS1 knockdown cancer cells. In vivo study also showed that LGALS1 overexpression facilitated tumor growth of cervical cancer cells.
Conclusion: Overexpression of LGALS1 significantly promoted and enhanced the aggressive features of cervical cancer both in vitro and in vivo, which may be associated with high expression of Bcl-2, MMP-2, MMP-9, Fascin, and Erzin proteins.
Keywords: Galectin-1, cervical cancer, proliferation, apoptosis, migration, invasion.
Introduction
Cervical cancer, originating from epithelial neoplastic transformation of uterine cervix, is the fourth cause of cancer-related death among women worldwide 1, 2. However, due to inadequate screening protocols for early detection of lesion in cervix, cervical cancer is still considered as the most common malignancy and poses a vital public health problem among Chinese women 3. According to statistics, the number of new cases of cervical cancer in China is estimated to reach 93,500 in 2030 and 186,600 in 2050 4, respectively. Thus, it is required to find the potential diagnostic markers as well as therapeutic targets, aiming to reduce the mortality and improve overall survival in patients with cervical cancer.
Galectin-1 (LGALS1) is a member of the carbohydrate-binding protein family, which shares a conserved amino acid sequence in the carbohydrate recognition domain and shows high affinity for β-galactoside-containing glycol-conjugates 5, 6. Several studies have demonstrated that LGALS1 plays an important role in tumorigenesis, progression and metastasis of human cancers 7, serving as a reliable diagnostic and prognostic biomarker in cancerous patients 8, 9. For instance, Schulkens et al. 10 have observed that LGALS1 expression in non-small cell lung cancer tissues was elevated compared with adjacent normal tissues. Additionally, overexpression of LGALS1 has also been reported in hepatocellular carcinoma 11, 12 and bladder urothelial carcinoma 13. Furthermore, upregulation of LGALS1 is also implicated in gastric tumor growth and metastasis 14. However, the role of LGALS1 in cervical cancer is still not fully elucidated. Notably, our previous proteomic study demonstrated that the expression of LGALS1 is significantly reduced after neoadjuvant chemotherapy treatment in cervical cancer patients, which may be associated with neoadjuvant chemotherapy exposure and response 15. Nevertheless, the function of LGALS1 on biological behavior in cervical cancer has not been extensively investigated yet.
In the current study, we explored the expression of LGALS1 protein in cervical cancer tissues and cells compared to adjacent non-tumor tissues and epithelial cells of cervix, respectively. Furthermore, we also investigated the effect of LGALS1 overexpression and knockdown on the proliferation, apoptosis, and migratory and invasive capabilities of cervical cancer cells and the related proteins in vitro. More importantly, the influence of LGALS1 on the tumor growth of cervical cancer in vivo was further studied.
Materials and Methods
Ethics statement
This study was approved by the ethical committee of the Second Affiliated Hospital of Wenzhou Medical University. Informed consent was obtained from each subject for the sample collection and analysis. All animal experiments were carried out according to the Guide for the Care and Use of Laboratory Animals published by the United States National Institutes of Health. They were approved by the Animal Care and Use Committee of Wenzhou Medical University.
Patients and tissue samples
Women with stage IB-IIA cervical cancers were recruited for this study, who underwent radical hysterectomy at the Second Affiliated Hospital of Wenzhou Medical University between January 2013 and August 2015. All these patients were retrospectively reviewed using electronic medical records. After exclusion of patients without complete clinicopathological data, 20 patients were enrolled in our study with a median age of 43 years (range, 24-59 years). All patients were pathologically diagnosed with squamous cell carcinoma of cervix after surgery (Differentiation: 13 moderate and 7 well; stage: 12 IB and 8 IIA). None of the patients received chemotherapy or radiotherapy prior to surgery. None of the patients had other synchronous malignancies or serious systemic diseases. Formalin-fixed cervical cancer tissues and matching adjacent non-tumor tissues from these patients were used for immunohistochemistry (IHC) staining.
Cell lines and culture
The human cervical squamous cancer cell lines (SiHa and C33A) and normal cervical epithelial cell (Ect1/E6E7) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in Dulbecco's Modifed Eagle medium (DMEM) (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and 1% antibiotics (penicillin-streptomycin). All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. Cells were cultured to a confluence of 80% and passaged by using 1× trypsin with 0.2% Ethylene Diamine Tetraacetic Acid (EDTA).
Immunohistochemistry (IHC) and immunocytochemistry (ICC)
IHC staining was performed using the SPlink Detection Kits (Biotin-Streptavidin HRP Detection Systems, ZSGB-BIO, SP-9000) in accordance with the manufacturer's instruction. Paraffin-embeded sections were cut at 4 μm thickness and deparaffinized in xylene and rehydrated in a gradient of ethanol solutions. After that, the tissue slides were washed with phosphate-buffered saline (PBS), and then placed in 80 mL plastic jars containing citrate buffer (pH 6.0) and repeatedly heated for 20 min at 95°C in a microwave oven for antigen retrieval. Endogenous peroxidase activity was suppressed with 3% hydrogen peroxide in methanol for 15 min and nonspecific binding was prevented through incubation with non-immune serum for 15 min. Tissue sections were then incubated with primary mouse anti-human LGALS1 monoclonal antibody (Santa Cruze, USA, 166618; 1:200) overnight at 4°C, followed by further incubation with biotin-conjugated secondary antibodies for 30 min at room temperature. Subsequently, the samples were exposed to streptavidin peroxidase as a label for 20 min. The sections were stained with diaminobenzidine for 10 min and counterstained with hematoxylin to enhance the nuclear detection. Finally, the slides were mounted, dehydrated through xylene and cover slipped. Appropriate positive and negative controls were stained in parallel. The results were assessed by two independent observers, who were blinded to the study. LGALS1 immunoreactivity was observed in the cytoplasm and cells that showed yellowish brown were recognized as positive. The percentage of positive cells was scored as following: 0 (0-5%), 1 point (6%-24%), 2 points (25%-49%), 3 points (50%-74%), and 4 points (75%-100%). Staining intensity was graded semiquantitatively into four levels as following: 0 (negative), 1 point (weak), 2 points (moderate), and 3 points (strong). The immunoreactive score was derived by the fraction of positive cell scores multiplied by staining intensity score.
Additional ICC analyses of LGALS1 expression were performed in SiHa, C33A and Ect1/E6E7 cells, which were grown on Chamber Slides System (Lab-Tek, USA) in a humidified incubator at 37°C with 5% CO2. After 24 h, the cells were fixed with acetic acid and methanol solution (ratio 1:3) at room temperature for 10 min. ICC was conducted using mouse anti-human LGALS1 (overnight incubation at 4°C and 1:200 dilution). The cells were then stained with avidin-biotin-peroxidase complex (UltramarqueTM-HRP-Detection kit, Greenwood, USA). Negative controls were treated identically by using PBS instead of primary antibodies. The ICC staining results were quantified based on the system identical to IHC and evaluated by two individual observers blindly.
Lentivirus construction and transfection of target cells
The LGALS1 sequences were amplified by polymerase chain reaction and then inserted into the lentiviral expression vector pLVX-IRES-ZsGreen1 (Origene, USA). Four different LGALS1 small hairpin RNAs (shRNAs) and the negative control plasmid were inserted into pGFP-C-shLenti vectors (Origene, USA). After confirmation using gene sequencing, LGALS1-pLVX-IRES-ZsGreen1, LGALS1-pGFP-C-shLenti and the corresponding control vectors, together with two packaging plasmids psPAX2 and pMD2.G (donated by Dr Luzhe Sun, The University of Texas Health Science Center at San Antonio) were transfected into 293T cells for 48 h, using lipofectamine 2,000 (Gibco, USA). Cells were seeded in 60-mm dish and cultured in DMEM medium without antibiotics for 24 h. Then, recombinant lentiviral particles were harvested and filtered to infect cell lines with the help of polybrene (final concentration 8 μg/mL). The expression of LGALS1 in all transfected cells were confirmed by Western blot. The primers and shRNA used for gene upregulation or downregulation were listed in Table 1.
Table 1.
Description of the selected genes and specific primers used in this study
| Gene | Primer name | Primer sequence (5'-3') |
|---|---|---|
| LGALS1 | LGALS1 (XhoI). F | AATCTCGAGATGGCTTGTGGTCTGGTCGC |
| LGALS1 (BamHI). R | TTGGATTCTCAGTCAAAGGCCACACATT | |
| LGALS1-shA | LGALS1-shRNA-A | GATCCGGGTCGCCAGCAACCTGAATCTTCAAGAGAGATTCAGGTTGCGGCGACCTTTTTTG |
| LGALS1-shB | LGALS1-shRNA-B | GATCCGACCTGTGCCTGCACTTCAACCTCAAGAGGGTTGAAGTGCAGGCACAGGTTTTTTTG |
| LGALS1-shC | LGALS1-shRNA-C | GATCCGGCCAGATGGATACGAATTCAATCAAGAGTTGAATTCGTATCCATCTGGCTTTTTTG |
| LGALS1-shD | LGALS1-shRNA-D | GATCCGGCCATCAACTACATGGCAGCTTCAAGAGAGCTGCCATGTAGTTGATGGCTTTTTTG |
Abbreviations: LGALS1, galectin-1; shRNA, small hairpin RNA.
Western blot analysis
The cells were washed with PBS and then lysed in a buffer (40 mM Tris-Cl, 10 mM EDTA, 120 mM NaCl, 0.1% Nonidet P-40) containing protease inhibitor cocktail (Beyotime, China). The lysates were incubated on ice for 30 min, followed by centrifugation at 12,000 g at 4°C for 20 min. The protein concentration was determined by a Bicinchoninic Acid Protein Assay Kit (Beyotime, China). Equal amounts of protein (30 μg) were separated by 12% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, USA). The membranes were blocked for 2 h with 5% skimmed milk in Tris-buffered saline with 0.1% Tween-20 (TBS-T, pH 7.4) at room temperature and then incubated overnight at 4°C with one of the appropriate primary antibodies at 1:2000 dilutions, unless specified otherwise. The primary antibodies used in this study were as following: mouse anti-human LGALS1 antibody (Santa Cruze, USA, 166618), rabbit anti-human Bcl-2 antibody (Abcam, USA, ab59348), rabbit anti-human Bax antibody (Abcam, USA, ab199677), mouse anti-human MMP-2 antibody (Abcam, USA, ab2462), rabbit anti-human MMP-9 antibody (Abcam, USA, ab38898), rabbit anti-human Fascin antibody (Abcam, USA, ab126772), rabbit anti-human Erzin antibody (Abcam, USA, ab47293), mouse anti-human α-Tubulin antibody (Abcam, USA, ab7291), and rabbit anti-human GAPDH antibody (Abcam, USA, ab181602). The membranes were rinsed and incubated with a goat anti-rabbit (Abcam, USA, ab205718) or anti-mouse (Abcam, USA, ab205719) horseradish peroxidase-conjugated secondary antibody at a concentration of 1:4000 for 2 h at room temperature. Finally, immunoreactive bands were visualized with an enhanced chemiluminescence detection reagent (Beyotime, China). The films were scanned using a GS-800 scanner and Quantity One software (Bio-Rad, USA) was employed to analyze the visualized protein bands. The expression levels of above proteins were quantified with densitometry and normalized by corresponding levels of α-Tubulin or GAPDH, respectively. All experiments were performed in triplicate.
Cell viability assay
Viability of cervical cancer cells was assessed using Cell Counting Kit-8 (CCK-8) assay (Dojindo, Japan). Briefly, cells (2000 cells/well) from the control, overexpression and knockdown groups were dispensed into 96-well plates with a volume of 100 μl/well and incubated at 37°C in 5% CO2 for 48 h. Subsequently, 10 μl CCK-8 solution in 100 μl complete DMEM was added to each well, followed by incubation for 4 h. After that, the absorbance of each well at 450 nm was detected using Micro-plate Reader (Bio-Tek Instruments, USA). Cell viability (%) was calculated by average absorbance of transfected group/average absorbance of control group × 100%. The experiment was repeated three times.
Cell proliferation assay
BrdU incorporation assay (Calbiochem, USA) was employed to determine cell proliferation. Briefly, transfected or non-transfected cells were seeded into 6-well plate with a density of 1 × 105 cells/well for 48 h. Then, the cells were incubated with BrdU (1 mg/ml) for 4 h and stained with anti-BrdU antibody (Cell Signaling Technology, USA) according to the manufacturer's instruction. After that, the number of BrdU positive (+) cells in each well was counted under microscope (Nikon, Japan), which was proportional to cell proliferation.
Cell apoptosis assay
Apoptosis was analyzed using Annexin V-FITC/PI apoptosis detection kit (BD Pharmingen, USA) according to the manufacturer's instructions. Briefly, the cells in 80% confluency at 48 h were collected with 0.25% trypsin, washed with pre-cooled PBS for twice, and stained using kit solution for 15 min at room temperature in the dark. The rate of apoptotic cells was recorded using flow cytometry (Beckman Coulter, USA). Data were quantified by FlowJo sofeware (Tree Star Incorporation, USA).
Transwell migration and invasion assays
The abilities of cell migration and invasion were assessed in 24-well plates with 8 μm pore transwell polycarbonate filters (Corning, USA). For the migration assay, cells with or without transfection were resuspended in serum-free DMEM and seeded to the upper chambers without Matrigel pre-coating at 1 × 104 cells/well, while DMEM containing 2.5% FBS was added to the lower chamber. Following incubation at 37°C in 5% CO2 for 48 h, the filters were fixed in 4% paraformaldehyde at room temperature for 20 min and stained with 0.1% crystal violet (Beyotime, China) for 10 min. Then, cells on the upper surface of the filter were removed using a cotton swab gently. Images of the cells migrated to the lower side of the filter were captured (magnification, × 40) and counted (magnification, × 100) under microscope (Nikon, Japan) by an independent investigator.
For the invasion assay, the upper transwell inserts were pre-coated with 100 μl 1:5 diluted Matrigel (BD Biosciences, USA), where cells from each group were seeded next. The medium in the lower chamber was complete DMEM with 10% FBS. After incubation at 37°C in 5% CO2 for 48 h, a cotton swab was used to gently remove the cells and Matrigel on the upper surface of the membrane. The number of invaded cells was determined in the same way as stated for the migration assay. All experiments were repeated in triplicate.
Animal studies
Four-week-old female nude mice were purchased from Beijing Vital River Laboratory Animal Technology of China, which were housed under specific pathogen free (SPF) condition in Wenzhou Medical University and used for in vivo animal experiments. Exponentially growing SiHa and C33A cells with LGALS1 overexpression (LGALS1-Lt), LGALS1 knockdown (LGALS1-shRNA), or corresponding empty vectors (Control-Lt and Control-shRNA) were suspended in 50% Matrigel in cold PBS and injected subcutaneously into the right flank of the mice (4 × 105 cells/mouse, 7 mice/group). One week after inoculation, the length (a) and width (b) of tumor tissue were monitored using a caliper once a week. The tumor volumes were calculated by the following formula: volume (mm3) = a*b2/2. Mice were sacrificed after tumor inoculation for a total of six weeks. Then the tumor samples were dissected, weighed and imaged with the camera.
Statistical analyses
All data analyses were performed using SPSS software (version 18.0; Chicago, USA). Continuous variables were expressed as the means ± SD. Two-tailed Student t test was applied for comparison of continuous variables. When group numbers were more than two, One-way ANOVA analysis was performed to determine the difference. P <0.05 was defined as statistical significance.
Results
LGALS1 is overexpressed in cervical cancer tissues and cell lines
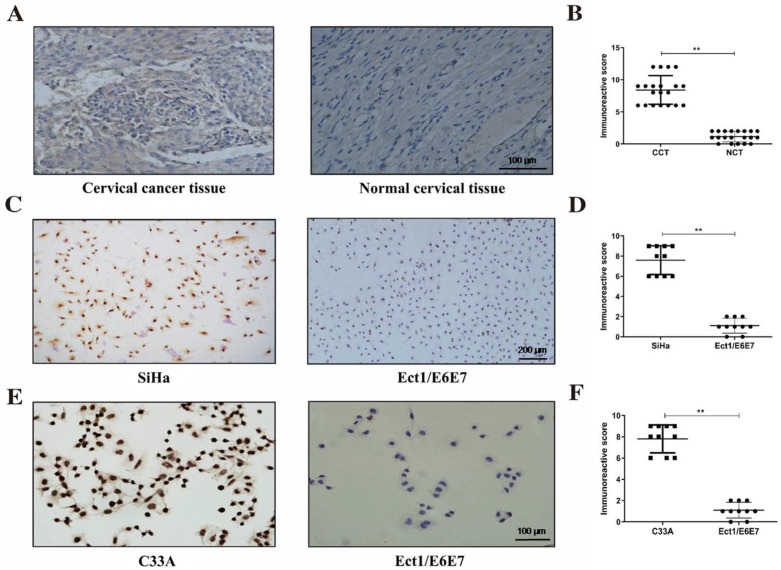
To understand the expression of LGALS1 in cervical cancer, we conducted IHC analyses of 20 cervical cancer specimens and matched non-tumor tissues in this study. Significantly higher expression of LGALS1 was exhibited in cervical cancer tissues (mainly in the cytoplasm) than normal cervical samples, while the latter showed only weak expression of LGALS1 (Figure 1A). The mean IHC scores of LGALS1 in cervical cancer and normal cervical tissues were 8.40 ± 2.23 and 1.15 ± 0.81, respectively. A significant difference in LGALS1 expression was observed between cervical cancer and adjacent non-tumor cervical tissues (P <0.01; Figure 1B).
Figure 1.
LGALS1 is overexpressed in cervical cancer tissues and cells. (A and B) Representative images and scores of IHC for LGALS1 in human cervical cancer tissue and matched normal cervical tissue (×200). (C, D, E and F) Representative images and scores of ICC for LGALS1 in SiHa cells, C33A cells and Ect1/E6E7 cells (×100). Values are expressed as the mean ± SD. *P <0.05, **P <0.01. LGALS1, galectin-1; IHC, immunohistochemistry; ICC, immunocytochemistry; CCT, cervical cancer tissue; NCT, normal cervical tissue.
To further validate the results of IHC, additional ICC analyses of LGALS1 expression were also performed in SiHa and C33A cervical cancer cells compared to normal cervical Ect1/E6E7 cells. As shown in Figure 1C and 1E, LGALS1 protein expression in the SiHa and C33A cells was detected mainly in the cytoplasm and nucleus, whereas the Ect1/E6E7 cells reflected nearly negative staining of LGALS1. The immunoreactive score of ICC for LGALS1 expression was significantly increased in the SiHa (7.60 ± 1.43) and C33A (7.80 ± 1.32) cell lines compared with Ect1/E6E7 cells (1.10 ± 0.74) (P <0.01; Figures 1D-1F).
Lentivirus-mediated upregulation and downregulation of LGALS1 in SiHa and C33A cells
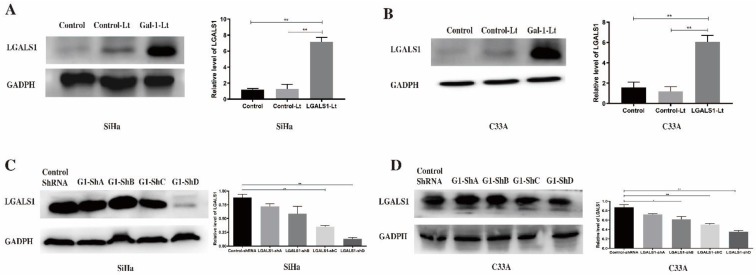
As shown in Figure 2, the efficacy of upregulation and downregulation of LGALS1 mediated by lentivirus was confirmed using Western blot analysis. Transfection of both SiHa and C33A cells with LGALS1-pLVX-IRES-ZsGreen1 lentivirus markedly increased the expression of LGALS1 by approximately 4-fold (P <0.01; Figures 2A-2B). In comparison, approximately 20%-85% reduction of LGALS1 expression was shown in both SiHa and C33A cells transfected with LGALS1-shRNA (P <0.01; Figures 2C-2D). The levels of LGALS1 expression in empty-vector group showed no significant difference compared to the blank control group (P >0.05). Therefore, LGALS1-shRNA-D was used in the following experiments.
Figure 2.
LGALS1 is upregulated or downregulated by LGALS1-Lt or LGALS1-shRNA in both SiHa and C33A cell lines. (A and B) Protein expression of LGALS1 in SiHa and C33A cells transfected with LGALS1-Lt was determined by Western blot analysis at 48 h post-transfection. (C and D) LGALS1 protein expression in SiHa and C33A cells transfected with four various LGALS1 shRNAs was detected by Western blot analysis 48 h post-transfection. Values are expressed as the mean ± SD. *P <0.05, **P <0.01. LGALS1, galectin-1; Control-Lt, control lentivirus; LGALS1-Lt, galectin-1 lentivirus; shRNA, small hairpin RNA.
Overexpression of LGALS1 enhances the viability and promotes the proliferation of SiHa and C33A cells
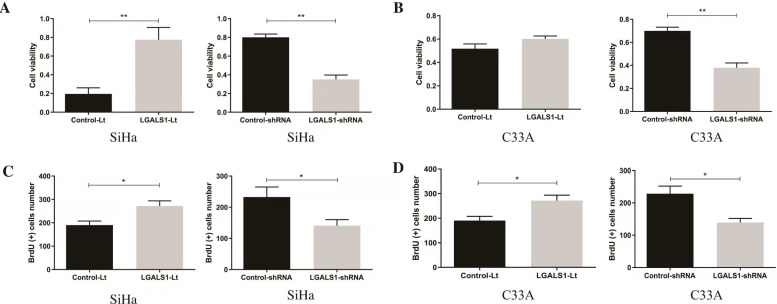
At 48 h after transfection, the effects of LGALS1 on the viability and proliferation of SiHa and C33A cells were assessed using CCK-8 and BrdU assay, respectively. A significant decrease of cell viability and proliferation were detected in the LGALS1-shRNA group compared with that in the control-shRNA group (P <0.01, Figures 3A-3B; P <0.05, Figures 3C-3D). While transfection with LGALS1-Lt did not dramatically influence the viability of C33A cells (P >0.05; Figure 3B), the viability was remarkably increased in SiHa cells following transfection (P <0.01, Figure 3A). Moreover, BrdU positive rates were significantly enhanced in both SiHa and C33A cells with LGALS1 overexpression (P <0.05, Figures 3C-3D).
Figure 3.
LGALS1 promotes proliferation in both SiHa and C33A cell lines. (A and B) The viability of SiHa and C33A cells at 48 h after transfection with LGALS1-Lt or LGALS1-shRNA were measured using CCK-8 assay. (C and D) The proliferation of SiHa cells at 48 h after transfection with LGALS1-Lt or LGALS1-shRNA-D were assessed using BrdU assay. Values are expressed as the mean ± SD. *P <0.05, **P <0.01. LGALS1, galectin-1; LGALS1-Lt, galectin-1-lentivirus; shRNA, small hairpin RNA; CCK-8, cell counting Kit-8.
Overexpression of LGALS1 inhibits the apoptosis of SiHa and C33A cells
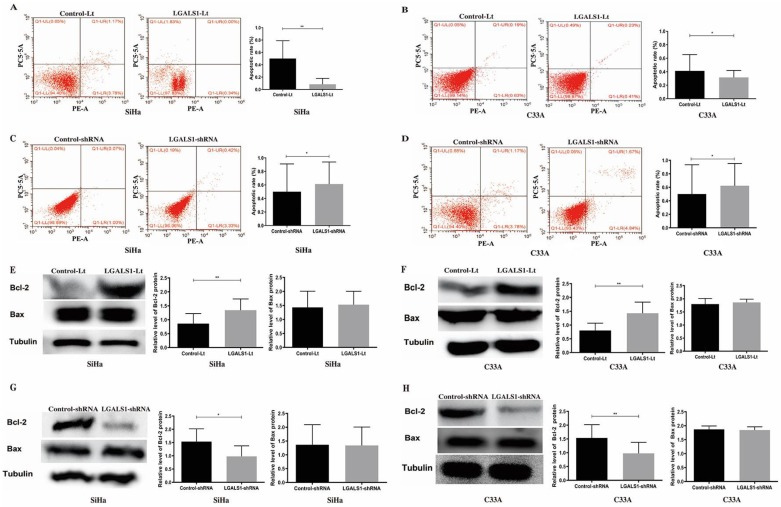
The role of LGALS1 on cell apoptosis was further studied and flow cytometry was utilized to measure the apoptotic cells. As shown in Figure 4, SiHa and C33A cells with LGALS1 overexpression had significantly lower apoptotic rate (P <0.01, Figure 4A; P <0.05, Figure 4B) whereas LGALS1 knockdown weakly enhanced the apoptotic rate in both SiHa and C33A cells (P <0.05; Figures 4C-4D). Moreover, the level of apoptosis-related protein Bcl-2 was significantly elevated in both SiHa and C33A cells after upregulation of LGALS1 (P <0.01; Figures 4E-4F), and reduced after LGALS1 knockdown (P <0.05, Figure 4G; P <0.01, Figure 4H). However, the expression level of Bax protein was not changed in SiHa and C33A cells after LGALS1 overexpression or knockdown (P >0.05; Figures 4E-4H).
Figure 4.
LGALS1 inhibits apoptosis in both SiHa and C33A cell lines. (A, B, C and D) The apoptosis of SiHa and C33A cells with LGALS1 overexpression or knockdown were analyzed by flow cytometric analysis. (E, F, G and H) Levels of apoptosis-related proteins Bcl-2 and Bax in SiHa and C33A cells with LGALS1 overexpression or knockdown were detected by Western blot analysis. Values are expressed as the mean ± SD. *P <0.05, **P <0.01. LGALS1, galectin-1; LGALS1-Lt, galectin-1-lentivirus; shRNA, small hairpin RNA.
Overexpression of LGALS1 promotes the migratory and invasive capabilities of SiHa and C33A cells
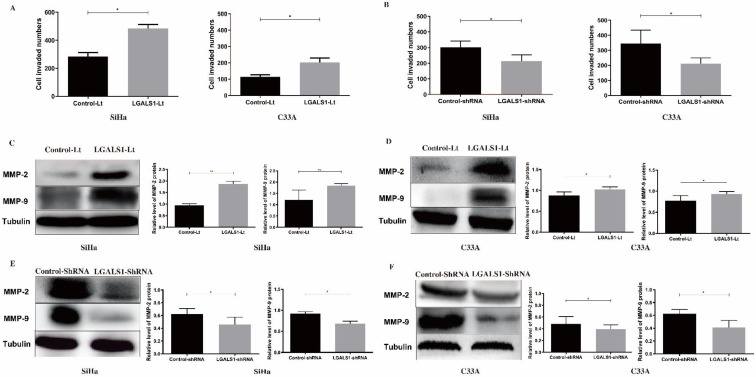
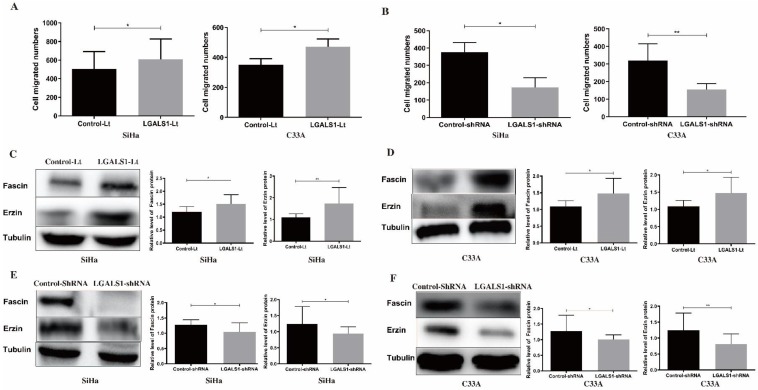
Subsequently, the effect of LGALS1 on the invasive (Figure 5) and migratory (Figure 6) capabilities of SiHa and C33A cell lines was examined using transwell assay. For both SiHa and C33A cells, a significant increase of cell invasion (P <0.05; Figure 5A) and migration (P <0.05; Figure 6A) were detected in the LGALS1-Lt group compared with that in the control-Lt group. Conversely, LGALS1 knockdown markedly decreased the invasive (P <0.05; Figure 5B) and migratory (P <0.05 for SiHa, P <0.01 for C33A; Figure 6B) abilities of both SiHa and C33A cells compared with control-shRNA cells.
Figure 5.
LGALS1 enhances invasive abilities in both SiHa and C33A cell lines. (A and B) Quantification of invaded SiHa and C33A cells with LGALS1 overexpression or knockdown were counted under microscope. (C, D, E and F) Levels of invasion-related proteins MMP-2 and MMP-9 in LGALS1 overexpression or knockdown SiHa and C33A cells were detected by Western blot analysis. Values are expressed as the mean ± SD. *P <0.05, **P <0.01. LGALS1, galectin-1; LGALS1-Lt, galectin-1-lentivirus; shRNA, small hairpin RNA; MMP, matrix metalloproteinases.
Figure 6.
LGALS1 enhances migration abilities in both SiHa and C33A cell lines. (A and B) Quantification of migrated SiHa and C33A cells with LGALS1 overexpression or knockdown were counted under microscope. (C, D, E and F) Levels of migration-related proteins Fascin and Erzin in LGALS1 overexpression or knockdown SiHa and C33A cells were detected by Western blot analysis. Values are expressed as the mean ± SD. *P <0.05, **P <0.01. LGALS1, galectin-1; LGALS1-Lt, galectin-1-lentivirus; shRNA, small hairpin RNA.
Next, the expression of invasion and migration related proteins were evaluated by Western blot analyses, including MMP-2, MMP-9, Fascin and Erzin. The expression of MMP-2, MMP-9, Fascin, Erzin were dramatically increased in LGALS1 overexpressing SiHa and C33A cells compared to the control-Lt cells (P <0.01, Figures 5C; P <0.01, Figure 5D; P <0.05 for Fascin, P <0.01 for Erzin, Figure 6C; P <0.05, Figure 6D). On the contrary, downregulation of LGALS1 significantly suppressed the expression of MMP-2, MMP-9, Fascin and Erzin protein expressions in both SiHa and C33A cells (P <0.05, Figure 5E; P <0.05 for MMP-2, P <0.01 for MMP-9, Figure 5F; P <0.05, Figure 6E; P <0.05 for MMP-2, P <0.01 for MMP-9, Figure 6F).
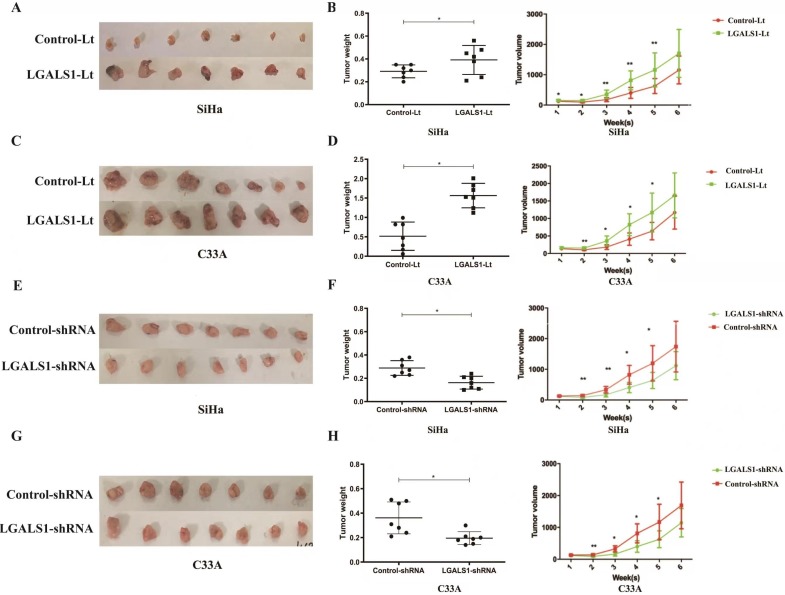
Overexpression of LGALS1 promotes tumor growth of cervical cancer in vivo
To further investigate whether LGALS1 promotes tumor growth in vivo, subcutaneous cervical cancer xenografts in nude mice were generated by injection with SiHa and C33A cells transfected with LGALS1-Lt, LGALS1-shRNA and related empty control vectors respectively. The diameter of the implanted tumors was measured every week. Six weeks after subcutaneous implantation (both for SiHa and C33A), tumors in the LGALS1-Lt group were larger and heavier than that in the control-Lt group (P <0.05; Figures 7A-7D), while tumors in LGALS1-shRNA group were remarkably smaller and lighter than that in the control-shRNA group (P <0.05; Figure 7E-7H). Tumor volumes (both for SiHa and C33A) were significantly higher in the LGALS1-Lt group than that in the control-Lt group from week 2 onwards (P <0.01; Figures 7B and 7D), while apparently smaller in the LGALS1-shRNA group compared with the control-shRNA group from the 2nd weeks after inoculation (P <0.05; Figures 7F and 7H).
Figure 7.
LGALS1 promotes cervical cancer growth in vivo. (A and C) Overexpression of LGALS1 promoted SiHa and C33A xenografts growth. (B and D) Tumor volumes and weights were measured in LGALS1 overexpression SiHa and C33A xenografts nude mice. (E and G) Knockdown of LGALS1 inhibited SiHa and C33A xenografts growth. (F and H) Tumor volumes and weights were measured in LGALS1 knockdown SiHa and C33A xenografts nude mice. Values are expressed as the mean ± SD. *P <0.05, **P <0.01. LGALS1, galectin-1; LGALS1-Lt, galectin-1-lentivirus; shRNA, small hairpin RNA.
Discussion
In this study, we observed that LGALS1 was upregulated in cervical cancer tissues and cell lines, and it was significantly associated with aggressive behavior of cervical squamous carcinoma. One study has shown that LGALS1 expression was increased remarkably according to the pathologic grade of cervical tissues 16. In our study, overexpression of LGALS1 was also observed in human cervical cancer tissues as compared with matched non-tumor samples using IHC staining, corresponding with previous reports 17-19. Furthermore, similar results were obtained in ICC studies that level of LGALS1 expression was significantly elevated in SiHa and C33A cell lines comparing to normal epithelial cells of cervix. Therefore, these findings together with previous reports strongly suggest that LGALS1 is overexpressed in cervical cancer cells 20, 21.
To further investigate the effects of LGALS1 on multiple biological functions of human cervical cancer cells, we employed lentivirus-mediated vectors to successfully overexpress and knockdown endogenous LGALS1 in SiHa and C33A cell lines. Cell viability is defined as the amount of living cells in a certain population and apoptosis of cells is an important indicator for action of cell survival or death when exposing to various stimulations 22. Recently, Kim et al. 23 has reported that LGALS1 promoted the proliferation of human keratinocytes and fibroblasts thereby accelerating wound healing, which may be achieved via triggering the accumulation of intracellular reactive oxygen species (ROS) 24. Similar to neuroblastoma 25, oral squamous cell carcinoma, lung cancer 26, and breast cancer 27, downregulation of LGALS1 had been shown to reduce the viability and proliferation of both SiHa and C33A cervical cancer cells, whereas LGALS1 overexpression significantly enhanced the viable and proliferative rates of SiHa cells in the present study. However, although the number of BrdU positive C33A cells was significantly elevated after upregulation of LGALS1, the viability of C33A cells with LGALS1 overexpression did not change. Moreover, LGALS1 overexpression promoted cervical cancer growth in mice by in vivo study. Interestingly, one group has previously indicated that LGALS1 inhibited cell proliferation and induced apoptosis in human colorectal 28 and prostate cancer cells 29. Moreover, decrease of LGALS1 expression has been demonstrated to suppress the proliferation of A172 and U343 but had no effect on U118 and U87 glioma cells 30, 31. Hence, LGALS1 may exhibit dual functions on proliferation in a tissue and cellular context dependent manner in different human malignant tumors or cell lines.
Numerous signaling molecules have been reported to be involved in cell apoptotic pathways, of which the Bcl-2 and Bax family proteins are the best characterized 32. It is widely accepted that Bcl-2 protein-mediated apoptosis blockade contributes to malignant transformation in multiple human cancers 33. Conversely, Bax protein enhances the permeability of the mitochondrial outer membrane, induces release of cytochrome c and consequently promotes apoptosis, which can be counteracted by Bcl-2 33. In this study, overexpression of LGALS1 inhibited the apoptosis of both SiHa and C33A cells while opposite results were obtained after LGALS1 downregulation. Furthermore, LGALS1 knockdown resulted in a sharp decline in the Bcl-2 protein expression in both SiHa and C33A cell lines accompanied with no variation in the level of Bax protein, suggesting that LGALS1 influenced the apoptosis of cervical cancer cells mainly through regulating the expression of Bcl-2 but not Bax. However, the underlying mechanism involved in LGALS1-mediated regulation of Bcl-2 remains to be explored in the future. Intriguingly, accumulated evidences have revealed that the sensitivity to cisplatin is conferred by suppression of Bcl-2 expression in various types of human cancers 34-37. Indeed, it has been documented that elevated levels of LGALS1 significantly induced cisplatin resistance in ovarian cancer partly via upregulation of Bcl-2 38. Moreover, our previous study using proteomics profiling has also confirmed that expression of LGALS1 was significantly decreased in cervical cancer patients after neoadjuvant chemotherapy administration 15. These data collectively indicate that the sensitivity to chemotherapies may be regulated by LGALS1 expression in cervical cancer patients, which also needs to be further validated.
Cell migration and invasion is a complex process initiating from proteolytic degradation of extracellular matrix (ECM) 39. It is well-known that matrix metalloproteinases (MMPs) have the ability to degrade almost all components of ECM and basement membrane, thereby facilitating the invasion and migration of cancer cells 40. Moreover, augmented levels of MMPs are functionally correlated with cancer cell metastasis 41. MMP-2 and MMP-9 belong to gelatinase-A and gelatinase-B, respectively, and are activated in several human cancers as the key enzymes controlling the rate of cell invasion and metastasis 42, 43. Subsequently, Ezrin and Fascin are also found to play important roles in the tumorigenesis and metastasis of several malignancies. Ezrin is closely associated to p85 subunit, which activates the PI3K/Akt pathway and plays an important role in modulating Ewing's sarcoma cell survival, invasion, and metastasis 44. Fascin-1, a member of cytoskeleton proteins, induces the formation of filopodia, lamellipodia, and microspikes of the cell membrane after cross-linking with F-actin, by facilitating the movement, metastasis, and invasion of tumor cells 45. Multiple investigations have speculated that overexpression of Fascin-1 was associated with a variety of human malignancies, such as pancreatic 46, esophageal 47, breast 48 and colorectal cancers 49. In line with these notions, our study provided further evidence to satisfy an unmet clinical need. Our study also found that overexpression of LGALS1 significantly promoted the migration and invasion of both SiHa and HeLa cells, whereas LGALS1 knockdown led to opposing results. Biochemically, we also observed that LGALS1 overexpression was significantly correlated with higher expression of MMP-2, MMP-9, Fascin, Ezrin protein expression in SiHa and C33A cells, while treating with LGALS1-shRNA markedly decreased the expression of these proteins. These results suggested that the oncogenic effects of LGALS1 on cervical cancer migration and invasion could be regulated by the downregulation of MMP-2, MMP-9, Fascin and Ezrin. Several signaling pathways in which LGALS1 is involved have been shown to promote the progression of cancer cells, such as H-Ras-dependent pathway 50, MAPK JNK/p38 pathway 51, and Akt/ERK1/2 signaling pathway 52. However, more studies should be carried out to verify whether these pathways regulate the invasion and migration through altering the level of MMP-2, MMP-9, Fascin and Erzin proteins.
Up to date, several clinicopathological studies have revealed an independent role of LGALS1 for predicting tumor progression and poor survival of cervical cancer patients 17-19. However, the research focusing on the role of LGALS1 in cervical cancer cells has not been extensively conducted. One previous study by Kim et al. 18 first showed that knockdown of LGALS1 using small interfering RNA (siRNA) significantly inhibited the proliferation and invasion of SiHa and HeLa cells, which were further confirmed in the current study. Strikingly, our study used shRNA to downregulate LGALS1 more stably and explored several signaling molecules related to the biological behavior of cervical cancer cells, which strengthened the results of this study. Unfortunately, due to the loss of several relevant information of subjects, the association between LGALS1 expression and these clinicopathological parameters could not be analyzed. Moreover, we did not explore how LGALS1 promoted the tumorigenesis and progression of cervical cancer and the potential signaling pathways, which need to be elucidated in the future. Further studies are also warranted to elucidate whether LGALS1 could regulate cisplatin chemosensitivity of squamous cervical cancer cells and the underlying mechanisms.
Taken together, this study revealed that LGALS1 is overexpressed in cervical cancer patients and cervical cancer cells, while LGALS1 downregulation had the opposed results in cervical cancer cells, indicating that LGALS1 may play an oncogenic role in the cervical carcinogenesis. LGALS1 overexpression in cervical cancer cell lines resulted in the promotion of cell proliferation, migration and invasion, as well as suppression of cell apoptosis. Moreover, LGALS1 overexpression also promoted the tumor growth of cervical cancer in vivo. Furthermore, LGALS1 downregulation had the opposed results in cervical cancer cells. Due to that downregulation of LGALS1 might contribute to the interference with tumor progression, LGALS1 could be evolved as a promising therapeutic target in the treatment of cervical cancer.
Acknowledgments
We thank Dr. Luzhe Sun and his group for donating the packaging plasmids psPAX2 and pMD2.G used in this study. This work was supported by Science and Technology Innovation Team of Wenzhou city-Gynecological Oncology (No. C20170004). The study sponsors had no involvement in the collection, analysis, or interpretation of data or in the writing of the manuscript.
ORCID: 0000-0002-8389-928X.
Abbreviations
- LGALS1
galectin-1
- IHC
immunohistochemistry
- DMEM
Dulbecco's Modifed Eagle medium
- FBS
fetal bovine serum
- EDTA
Ethylene Diamine Tetraacetic Acid
- ICC
immunocytochemistry
- PBS
phosphate-buffered saline
- DAB
phosphate-buffered saline
- shRNA
small hairpin RNA
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel
- PVDF
polyvinylidene difluoride
- ECL
enhanced chemiluminescence
- CCK-8
Cell Counting Kit-8
- SPF
specific pathogen free
- ROS
reactive oxygen species
- ECM
extracellular matrix
- MMPs
matrix metalloproteinases
- EMT
epithelial-mesenchymal transition
- CSC
cancer stem cell
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Koh WJ, Abu-Rustum NR, Bean S, Bradley K, Campos SM, Cho KR. et al. Cervical Cancer, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. Journal of the National Comprehensive Cancer Network: JNCCN. 2019;17:64–84. doi: 10.6004/jnccn.2019.0001. [DOI] [PubMed] [Google Scholar]
- 3.Di J, Rutherford S, Chu C. Review of the Cervical Cancer Burden and Population-Based Cervical Cancer Screening in China. Asian Pacific journal of cancer prevention: APJCP. 2015;16:7401–7. doi: 10.7314/apjcp.2015.16.17.7401. [DOI] [PubMed] [Google Scholar]
- 4.Shi JF, Canfell K, Lew JB, Qiao YL. The burden of cervical cancer in China: synthesis of the evidence. International journal of cancer. 2012;130:641–52. doi: 10.1002/ijc.26042. [DOI] [PubMed] [Google Scholar]
- 5.Barondes SH, Castronovo V, Cooper DN, Cummings RD, Drickamer K, Feizi T. et al. Galectins: a family of animal beta-galactoside-binding lectins. Cell. 1994;76:597–8. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 6.Thiemann S, Baum LG. Galectins and Immune Responses-Just How Do They Do Those Things They Do? Annual review of immunology. 2016;34:243–64. doi: 10.1146/annurev-immunol-041015-055402. [DOI] [PubMed] [Google Scholar]
- 7.Chou FC, Chen HY, Kuo CC, Sytwu HK. Role of Galectins in Tumors and in Clinical Immunotherapy. International journal of molecular sciences; 2018. p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu R, Wu T, Wang K, Luo S, Chen Z, Fan M. et al. Prognostic significance of galectin-1 expression in patients with cancer: a meta-analysis. Cancer cell international. 2018;18:108. doi: 10.1186/s12935-018-0607-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang MY, He JP, Zhang WQ, Liu JL. Pooling analysis reveals that galectin-1 is a reliable prognostic biomarker in various cancers. Journal of cellular physiology; 2019. [DOI] [PubMed] [Google Scholar]
- 10.Schulkens IA, Heusschen R, van den Boogaart V, van Suylen RJ, Dingemans AM, Griffioen AW. et al. Galectin expression profiling identifies galectin-1 and Galectin-9Delta5 as prognostic factors in stage I/II non-small cell lung cancer. PloS one. 2014;9:e107988. doi: 10.1371/journal.pone.0107988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Chen P, Liao R, Li YW, Yi Y, Wang JX. et al. Overexpression of galectin-1 is associated with poor prognosis in human hepatocellular carcinoma following resection. Journal of gastroenterology and hepatology. 2012;27:1312–9. doi: 10.1111/j.1440-1746.2012.07130.x. [DOI] [PubMed] [Google Scholar]
- 12.Yeh CC, Hsu CH, Shao YY, Ho WC, Tsai MH, Feng WC. et al. Integrated Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) Quantitative Proteomic Analysis Identifies Galectin-1 as a Potential Biomarker for Predicting Sorafenib Resistance in Liver Cancer. Molecular & cellular proteomics: MCP. 2015;14:1527–45. doi: 10.1074/mcp.M114.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu TF, Li CF, Chien LH, Shen KH, Huang HY, Su CC. et al. Galectin-1 dysregulation independently predicts disease specific survival in bladder urothelial carcinoma. The Journal of urology. 2015;193:1002–8. doi: 10.1016/j.juro.2014.09.107. [DOI] [PubMed] [Google Scholar]
- 14.You X, Wang Y, Wu J, Liu Q, Chen D, Tang D. et al. Galectin-1 Promotes Metastasis in Gastric Cancer Through a Sphingosine-1-Phosphate Receptor 1-Dependent Mechanism. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;51:11–30. doi: 10.1159/000495157. [DOI] [PubMed] [Google Scholar]
- 15.Zou S, Shen Q, Hua Y, Jiang W, Zhang W, Zhu X. Proteomic identification of neoadjuvant chemotherapy-related proteins in bulky stage IB-IIA squamous cervical cancer. Reproductive sciences. 2013;20:1356–64. doi: 10.1177/1933719113485291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohrenhagen N, Volker HU, Kapp M, Dietl J, Kammerer U. Increased expression of galectin-1 during the progression of cervical neoplasia. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2006;16:2018–22. doi: 10.1111/j.1525-1438.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 17.Huang EY, Chanchien CC, Lin H, Wang CC, Wang CJ, Huang CC. Galectin-1 is an independent prognostic factor for local recurrence and survival after definitive radiation therapy for patients with squamous cell carcinoma of the uterine cervix. International journal of radiation oncology, biology, physics. 2013;87:975–82. doi: 10.1016/j.ijrobp.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 18.Kim HJ, Do IG, Jeon HK, Cho YJ, Park YA, Choi JJ. et al. Galectin 1 expression is associated with tumor invasion and metastasis in stage IB to IIA cervical cancer. Human pathology. 2013;44:62–8. doi: 10.1016/j.humpath.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Punt S, Thijssen VL, Vrolijk J, de Kroon CD, Gorter A, Jordanova ES. Galectin-1, -3 and -9 Expression and Clinical Significance in Squamous Cervical Cancer. PloS one. 2015;10:e0129119. doi: 10.1371/journal.pone.0129119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Zhao Y, Wang Y, Wu X. The Role of Galectins in Cervical Cancer Biology and Progression. BioMed research international. 2018;2018:2175927. doi: 10.1155/2018/2175927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chetry M, Thapa S, Hu X, Song Y, Zhang J, Zhu H. et al. The Role of Galectins in Tumor Progression, Treatment and Prognosis of Gynecological Cancers. Journal of Cancer. 2018;9:4742–55. doi: 10.7150/jca.23628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adan A, Kiraz Y, Baran Y. Cell Proliferation and Cytotoxicity Assays. Current pharmaceutical biotechnology. 2016;17:1213–21. doi: 10.2174/1389201017666160808160513. [DOI] [PubMed] [Google Scholar]
- 23.Kim MH, Wu WH, Choi JH, Kim J, Jun JH, Ko Y. et al. Galectin-1 from conditioned medium of three-dimensional culture of adipose-derived stem cells accelerates migration and proliferation of human keratinocytes and fibroblasts. Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2018;26(Suppl 1):S9–S18. doi: 10.1111/wrr.12579. [DOI] [PubMed] [Google Scholar]
- 24.Lin YT, Chen JS, Wu MH, Hsieh IS, Liang CH, Hsu CL. et al. Galectin-1 accelerates wound healing by regulating the neuropilin-1/Smad3/NOX4 pathway and ROS production in myofibroblasts. The Journal of investigative dermatology. 2015;135:258–68. doi: 10.1038/jid.2014.288. [DOI] [PubMed] [Google Scholar]
- 25.Cimmino F, Schulte JH, Zollo M, Koster J, Versteeg R, Iolascon A. et al. Galectin-1 is a major effector of TrkB-mediated neuroblastoma aggressiveness. Oncogene. 2009;28:2015–23. doi: 10.1038/onc.2009.70. [DOI] [PubMed] [Google Scholar]
- 26.Wu MH, Hong TM, Cheng HW, Pan SH, Liang YR, Hong HC. et al. Galectin-1-mediated tumor invasion and metastasis, up-regulated matrix metalloproteinase expression, and reorganized actin cytoskeletons. Molecular cancer research: MCR. 2009;7:311–8. doi: 10.1158/1541-7786.MCR-08-0297. [DOI] [PubMed] [Google Scholar]
- 27.Jung EJ, Moon HG, Cho BI, Jeong CY, Joo YT, Lee YJ. et al. Galectin-1 expression in cancer-associated stromal cells correlates tumor invasiveness and tumor progression in breast cancer. International journal of cancer. 2007;120:2331–8. doi: 10.1002/ijc.22434. [DOI] [PubMed] [Google Scholar]
- 28.Satelli A, Rao US. Galectin-1 is silenced by promoter hypermethylation and its re-expression induces apoptosis in human colorectal cancer cells. Cancer letters. 2011;301:38–46. doi: 10.1016/j.canlet.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valenzuela HF, Pace KE, Cabrera PV, White R, Porvari K, Kaija H. et al. O-glycosylation regulates LNCaP prostate cancer cell susceptibility to apoptosis induced by galectin-1. Cancer research. 2007;67:6155–62. doi: 10.1158/0008-5472.CAN-05-4431. [DOI] [PubMed] [Google Scholar]
- 30.Strik HM, Schmidt K, Lingor P, Tonges L, Kugler W, Nitsche M. et al. Galectin-1 expression in human glioma cells: modulation by ionizing radiation and effects on tumor cell proliferation and migration. Oncology reports. 2007;18:483–8. [PubMed] [Google Scholar]
- 31.Jung TY, Jung S, Ryu HH, Jeong YI, Jin YH, Jin SG. et al. Role of galectin-1 in migration and invasion of human glioblastoma multiforme cell lines. Journal of neurosurgery. 2008;109:273–84. doi: 10.3171/JNS/2008/109/8/0273. [DOI] [PubMed] [Google Scholar]
- 32.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nature reviews Molecular cell biology. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 33.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J. et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tung MC, Lin PL, Cheng YW, Wu DW, Yeh SD, Chen CY. et al. Reduction of microRNA-184 by E6 oncoprotein confers cisplatin resistance in lung cancer via increasing Bcl-2. Oncotarget. 2016;7:32362–74. doi: 10.18632/oncotarget.8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Zhang Y, Shi Y, Lian H, Tu H, Han S. et al. MiR-873 acts as a novel sensitizer of glioma cells to cisplatin by targeting Bcl-2. International journal of oncology. 2015;47:1603–11. doi: 10.3892/ijo.2015.3143. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Yang L, Zhang J, Zhang J, Chen Y, Li K. et al. Knock-down of NDRG2 sensitizes cervical cancer Hela cells to cisplatin through suppressing Bcl-2 expression. BMC cancer. 2012;12:370. doi: 10.1186/1471-2407-12-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jing Z, Heng W, Xia L, Ning W, Yafei Q, Yao Z. et al. Downregulation of phosphoglycerate dehydrogenase inhibits proliferation and enhances cisplatin sensitivity in cervical adenocarcinoma cells by regulating Bcl-2 and caspase-3. Cancer biology & therapy. 2015;16:541–8. doi: 10.1080/15384047.2015.1017690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Zhang P, Shi B, Zhou M, Jiang H, Zhang H. et al. Galectin-1 overexpression promotes progression and chemoresistance to cisplatin in epithelial ovarian cancer. Cell death & disease. 2014;5:e991. doi: 10.1038/cddis.2013.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albini A. Extracellular Matrix Invasion in Metastases and Angiogenesis: Commentary on the Matrigel "Chemoinvasion Assay". Cancer research. 2016;76:4595–7. doi: 10.1158/0008-5472.CAN-16-1971. [DOI] [PubMed] [Google Scholar]
- 40.Jablonska-Trypuc A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. Journal of enzyme inhibition and medicinal chemistry. 2016;31:177–83. doi: 10.3109/14756366.2016.1161620. [DOI] [PubMed] [Google Scholar]
- 41.Yadav L, Puri N, Rastogi V, Satpute P, Ahmad R, Kaur G. Matrix metalloproteinases and cancer - roles in threat and therapy. Asian Pacific journal of cancer prevention: APJCP. 2014;15:1085–91. doi: 10.7314/apjcp.2014.15.3.1085. [DOI] [PubMed] [Google Scholar]
- 42.Xu W, Xu H, Fang M, Wu X, Xu Y. MKL1 links epigenetic activation of MMP2 to ovarian cancer cell migration and invasion. Biochemical and biophysical research communications. 2017;487:500–8. doi: 10.1016/j.bbrc.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Hung CY, Lee CH, Chiou HL, Lin CL, Chen PN, Lin MT. et al. Praeruptorin-B Inhibits 12-O-Tetradecanoylphorbol-13-Acetate-Induced Cell Invasion by Targeting AKT/NF-kappaB via Matrix Metalloproteinase-2/-9 Expression in Human Cervical Cancer Cells. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2019;52:1255–66. doi: 10.33594/000000088. [DOI] [PubMed] [Google Scholar]
- 44.Krishnan K, Bruce B, Hewitt S, Thomas D, Khanna C, Helman LJ. Ezrin mediates growth and survival in Ewing's sarcoma through the AKT/mTOR, but not the MAPK, signaling pathway. Clinical & experimental metastasis. 2006;23:227–36. doi: 10.1007/s10585-006-9033-y. [DOI] [PubMed] [Google Scholar]
- 45.Ma Y, Reynolds LE, Li A, Stevenson RP, Hodivala-Dilke KM, Yamashiro S. et al. Fascin 1 is dispensable for developmental and tumour angiogenesis. Biology open. 2013;2:1187–91. doi: 10.1242/bio.20136031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai WC, Lin CK, Lee HS, Gao HW, Nieh S, Chan DC. et al. The correlation of cortactin and fascin-1 expression with clinicopathological parameters in pancreatic and ampulla of Vater adenocarcinoma. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2013;121:171–81. doi: 10.1111/j.1600-0463.2012.02952.x. [DOI] [PubMed] [Google Scholar]
- 47.Cao HH, Zheng CP, Wang SH, Wu JY, Shen JH, Xu XE. et al. A molecular prognostic model predicts esophageal squamous cell carcinoma prognosis. PloS one. 2014;9:e106007. doi: 10.1371/journal.pone.0106007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esnakula AK, Ricks-Santi L, Kwagyan J, Kanaan YM, DeWitty RL, Wilson LL. et al. Strong association of fascin expression with triple negative breast cancer and basal-like phenotype in African-American women. Journal of clinical pathology. 2014;67:153–60. doi: 10.1136/jclinpath-2013-201698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams JC. Fascin-1 as a biomarker and prospective therapeutic target in colorectal cancer. Expert review of molecular diagnostics. 2015;15:41–8. doi: 10.1586/14737159.2015.976557. [DOI] [PubMed] [Google Scholar]
- 50.Huang EY, Chen YF, Chen YM, Lin IH, Wang CC, Su WH. et al. A novel radioresistant mechanism of galectin-1 mediated by H-Ras-dependent pathways in cervical cancer cells. Cell death & disease. 2012;3:e251. doi: 10.1038/cddis.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J, Zheng Y, Zhang H, Liu Y, Sun H, Zhang P. Galectin-1 induces metastasis and epithelial-mesenchymal transition (EMT) in human ovarian cancer cells via activation of the MAPK JNK/p38 signalling pathway. Am J Transl Res. 2019;11:3862–78. [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu BQ, Zhang PF, Xiong D, Xu JJ, Long X, Zhu SQ. et al. CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non-small cell lung cancer by regulating Galectin-1-AKT/ERK1/2 signaling. Journal of cellular physiology. 2019;234:11256–64. doi: 10.1002/jcp.27783. [DOI] [PubMed] [Google Scholar]