To the Editor
Juvenile myelomonocytic leukemia (JMML) is an aggressive myeloproliferative neoplasm of childhood with nearly half of all patients relapsing within 3 years [1, 2]. More than 90% of JMML patients present with initiating mutations in NF1, KRAS, NRAS, RRAS, RRAS2, CBL, and PTPN11, all leading to hyperactivation of the Ras pathway [3]. Patients with additional molecular alterations including cytogenetic abnormalities, and in particular, point mutations in SETBP1 have inferior outcomes [4]. At present, hematopoietic stem cell transplantation (HSCT) is the only curative therapy, with the primary cause of death being relapse or progression to acute myeloid leukemia (AML) [5]. Recently, JMML patients who did not display Ras pathway mutations were found to have ALK and ROS1 fusions that were sensitive to ALK inhibition [6]. Given the poor overall survival of JMML patients even after intensive treatments such as HSCT, opportunities to implement precision medicine should be explored in this disease. Here, we present a JMML patient who failed to respond to conventional cytotoxic chemotherapy and was found to have a novel CCDC88C–FLT3 fusion driving the patient’s leukemia that responded to FLT3 inhibition with sorafenib. Of note, FLT3 fusions and internal tandem duplication (ITD)s give rise to myeloproliferative disorders in mouse models but have yet to be reported in pediatric patients [7].
DNA and RNA were extracted using standard methods from bone marrow, spleen, and peripheral blood mononuclear cells obtained at various time points through the clinical course. Approvals for these studies were obtained from the University of California San Francisco (UCSF) Committee on Human Research. The guardians provided informed consent in accordance with the Declaration of Helsinki.
For additional details, see Supplemental Methods, available on the Leukemia website. All relevant data used in this study are available for download from Synapse (https://doi.org/10.7303/syn18913127).
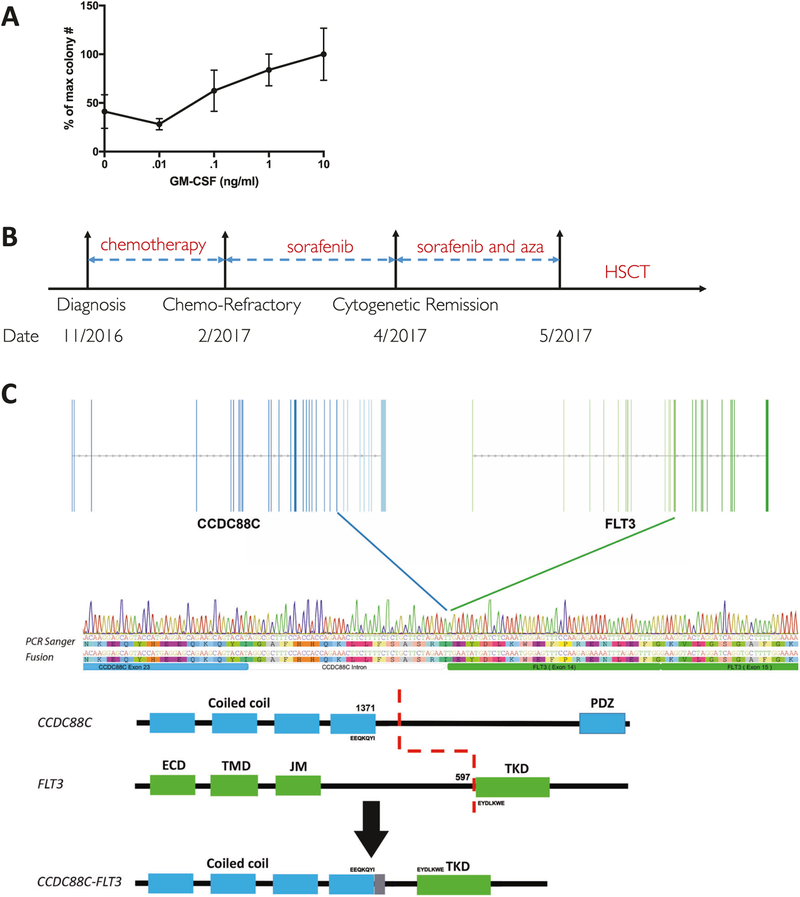
A 20-week-old boy presented with hepatosplenomegaly, an elevated white blood count (WBC) of 92 K/μl with 60% monocytes, and leukoerythroblastosis on peripheral blood smear. A diagnostic bone marrow aspirate revealed <5% blasts, absence of BCR/ABL1, and no known JMML mutations. The patient’s peripheral blood mononuclear cells were hypersensitive to the cytokine granulocyte-macrophage colony-stimulating factor (Fig. 1a). The patient was treated with fludarabine and high-dose cytarabine per our institutional standard of care in an attempt to reduce the disease burden prior to HSCT [1]. However, the patient was refractory to chemotherapy and experienced worsening hepatosplenomegaly, eventually requiring splenectomy. During these therapies, the patient’s clinical status continued to deteriorate and he was not an eligible candidate for HSCT (Fig. 1b).
Fig. 1.
Clinical course related to CCDC88C–FLT3 fusion. a Colony forming assay of patient PBMCs with increasing concentrations of GM-CSF (mean ± SEM is displayed, n = 4, N = 1). b Schematic timeline of JMML patient course (5-azacitidine is abbreviated as Aza). c Schematic illustration of CCDC88C–FLT3 fusion: the top panel illustrates exon structure indicated by blue (CCDC88C) or green (FLT3) vertical lines. The middle panel shows a Sanger sequence trace from patient cDNA aligned with an annotated sequence of the fusion junction. The lower panel shows an illustration of protein domains from both partners retained in the fusion
An institutional DNASeq panel assaying 480 cancer relevant genes and RNA-Seq both revealed an inframe CCDC88C–FLT3 fusion (Fig. 1c). Although FLT3 mutations are found in both adult and pediatric AML, FLT3 mutations have previously been reported as rare in JMML, and no FLT3 fusions have ever been reported in a pediatric malignancy. Given the patient’s lack of response to cytotoxic chemotherapy and because he was deemed ineligible for HSCT, sorafenib, an oral FLT3 inhibitor was identified as a potential tolerable and targeted therapy. Sorafenib is FDA approved for adults with hepatocellular and renal cell carcinoma and has been shown to effectively treat a FLT3–ETV6 fusion in an adult with a myeloproliferative fusion [8]. Prior to initiating sorafenib, we performed a bone marrow aspirate which revealed a t(13;14)(q12;q32) translocation consistent with the CCDC88C–FLT3 fusion, as well as monosomy 7.
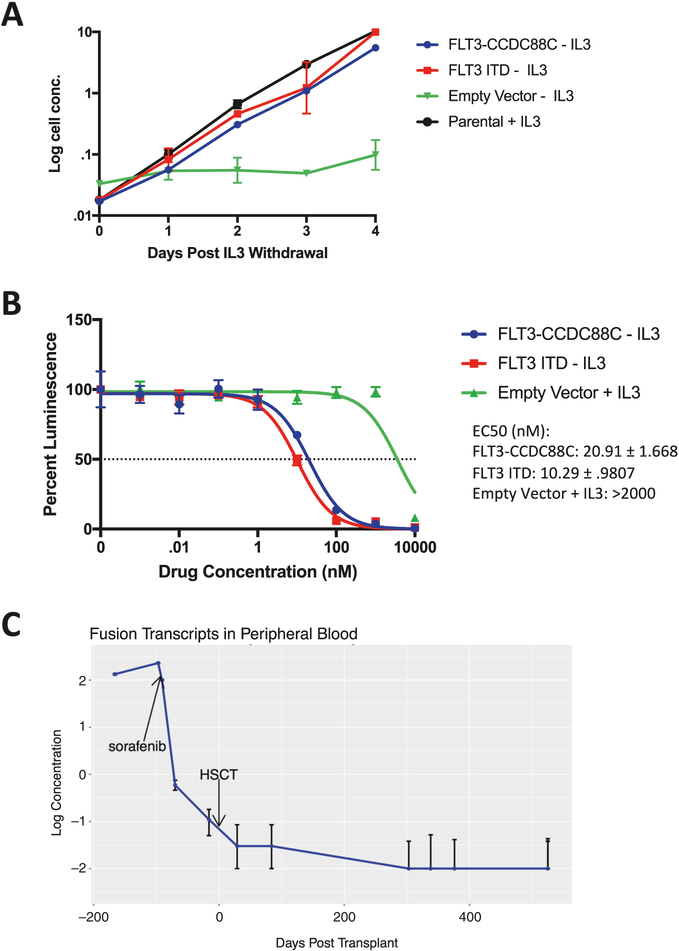
To determine whether the CCDC88C–FLT3 fusion protein was oncogenic, we transduced the murine pro-B cell line Ba/F3, which is well established for studying FLT3 alterations in myeloid malignancies with viral constructs containing either CCDC88C–FLT3, a FLT3 ITD mutant, or an empty vector. Ba/F3 cells transduced with a FLT3 fusion construct showed IL3-independent growth at a comparable rate to cells expressing a FLT3 ITD mutant (Fig. 2a). Furthermore, in response to sorafenib drug exposure, CCDC88C–FLT3-transduced cells were more sensitive compared with mock cells grown in the presence of IL3 (Fig. 2b). These data suggest that this CCDC88C–FLT3 fusion drives cytokine-independent growth and is sensitive to inhibition by sorafenib in vitro. Of note, the fusion junction in FLT3 in this patient occurs in exon 14, the same site where ITDs occur in AML [9]. FLT3 ITD-driven AML is associated with resistance to cytotoxic chemotherapy and may explain our patients lack of response to AML-like chemotherapy [10].
Fig. 2.
FLT3 fusion causes cytokine-independent growth and is sensitive to FLT3 inhibition. a Growth rates of Ba/F3 cells transduced with CCDC88C–FLT3, FLT3 ITD, or mock vector after mouse interleukin-3 (mIL3) withdrawal (mean ± SEM is displayed, n = 3, representative of N = 3 independent experiments). b Drug sensitivity of Ba/F3 cells to a 48 h sorafenib exposure. EC50 ± SEM values are reported as the drug concentration that achieves a 50% decrease in luminescence by Cell Titer Glo assay (mean ± SEM is displayed, n = 3, representative of N = 3 independent experiments). c FLT3 fusion transcript levels in patient peripheral blood over time as assayed by RT-ddPCR (mean ± SEM is displayed, n = 3, N = 1)
Following sorafenib monotherapy treatment for 2 weeks, the patient’s WBC decreased to a normal range. A repeat bone marrow was performed and the patient was noted to be in a cytogenetic remission. Given the novelty of this CCDC88C–FLT3 fusion, no commercial assays were available to identify fusion transcripts below the sensitivity of cytogenetics. We therefore designed a droplet digital PCR assay to detect levels of the FLT3 fusion and FLT3 wild-type transcripts using banked patient samples (Supplemental Fig. 1). Fusion transcript levels in peripheral blood dropped following sorafenib monotherapy and continued to decrease on a combination of azacitidine and sorafenib. This clinical response rendered the patient eligible for HSCT 10 weeks after initiating sorafenib and he received a haploidentical transplant from his father using an alpha/beta T-cell-depleted approach following conditioning with busulfan, cyclophosphamide, thiotepa, and rabbit anti-thymocyte globulin (Fig. 1b). Fusion transcripts reached undetectable levels following HSCT (Fig. 2c) by day +30 and the patient remains on sorafenib monotherapy post-transplant with fusion transcripts remaining undetectable > 300 days after transplant.
There are now several reports of fusions in JMML patients lacking Ras mutations. Interestingly, there is an additional case of a fusion involving t(13;14)(q12;q32) in a pediatric patient with a myeloproliferative neoplasm with hepatosplenomegaly and an elevated monocyte count [11]. Although the fusion genes were not identified in that case, we hypothesize this could also have reflected a CCDC88C–FLT3 fusion. Recently, several ALK and ROS1 fusions have been reported in JMML patients lacking canonical Ras mutations but did also have monosomy 7 similar to our patient [6]. Fusions involving FIP1L1-RARA, HCMOGT-1-PDGFRB, NDEL1-PDGFRB, and NUP98-HOXA11 have all been previously reported in JMML patients, several of which would be suspected to respond to targeted therapies, including tyrosine kinase inhibitors [12–15].
In this study, we report and characterize a novel CCDC88C–FLT3 fusion in a child with JMML that was refractory to cytotoxic chemotherapy. We demonstrated that this FLT3 fusion transforms Ba/F3 cells and is sensitive to FLT3 inhibition with sorafenib in vitro. We administered sorafenib, enabling the patient to achieve a molecular remission and proceed safely to HSCT. We also developed a custom ddRT-PCR assay to follow FLT3 fusion transcript levels throughout his treatment course. In summary, all JMML patients who lack canonical Ras pathway mutations should have RNA-Seq performed to identify potentially targetable fusions including FLT3.
Supplementary Material
Acknowledgements
We thank the family for their willingness to share their story. This work was supported by the National Institutes of Health, National Cancer Institute grant 1U54CA196519 (M.L.L., E.S.); National Institutes of Health, National Heart, Lung, and Blood Institute grant K08HL135434 (E.S.); Alex’s Lemonade Stand, Center of Excellence (M.L.L., E.S.) and the Frank A. Campini Foundation (M.L.L. and E.S.). C.S. is the Damon Runyon-Richard Lumsden Foundation Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation (CI-99–18).
Footnotes
Conflict of interest C.S. received research funding from FujiFilm and Astellas Pharma. A.K.C. received an honoraria from Bio-Rad for speaking at a symposium. No other authors declare conflicts of interest.
Supplementary information The online version of this article (https://doi.org/10.1038/s41375-019-0549-y) contains supplementary material, which is available to authorized users.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stieglitz E, Ward AF, Gerbing RB, Alonzo TA, Arceci RJ, Liu YL, et al. Phase II/III trial of a pre-transplant farnesyl transferase inhibitor in juvenile myelomonocytic leukemia: a report from the Children’s Oncology Group. Pediatr blood cancer. 2015;62: 629–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dvorak CC, Satwani P, Stieglitz E, Cairo MS, Dang H, Pei Q, et al. Disease burden and conditioning regimens in ASCT1221, a randomized phase II trial in children with juvenile myelomonocytic leukemia: sa Children’s Oncology Group study. Pediatr blood cancer. 2018;65:e27034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stieglitz E, Taylor-Weiner AN, Chang TY, Gelston LC, Wang YD, Mazor T, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nat Genet. 2015;47:1326–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stieglitz E, Troup CB, Gelston LC, Haliburton J, Chow ED, Yu KB, et al. Subclonal mutations in SETBP1 confer a poor prognosis in juvenile myelomonocytic leukemia. Blood. 2015;125: 516–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locatelli F, Nollke P, Zecca M, Korthof E, Lanino E, Peters C, et al. Hematopoietic stem cell transplantation (HSCT) in children with juvenile myelomonocytic leukemia (JMML): results of the EWOG-MDS/EBMT trial. Blood. 2005;105:410–9. [DOI] [PubMed] [Google Scholar]
- 6.Murakami N, Okuno Y, Yoshida K, Shiraishi Y, Nagae G, Suzuki K, et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018;131:1576–86. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin BR, Li L, Tse KF, Small S, Collector M, Whartenby KA, et al. Transgenic mice expressing Tel-FLT3, a constitutively activated form of FLT3, develop myeloproliferative disease. Leukemia. 2007;21:764–71. [DOI] [PubMed] [Google Scholar]
- 8.Falchi L, Mehrotra M, Newberry KJ, Lyle LM, Lu G, Patel KP, et al. ETV6-FLT3 fusion gene-positive, eosinophilia-associated myeloproliferative neoplasm successfully treated with sorafenib and allogeneic stem cell transplant. Leukemia. 2014;28:2090–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stirewalt DL, Kopecky KJ, Meshinchi S, Engel JH, Pogosova-Agadjanyan EL, Linsley J, et al. Size of FLT3 internal tandem duplication has prognostic significance in patients with acute myeloid leukemia. Blood. 2006;107:3724–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–9. [DOI] [PubMed] [Google Scholar]
- 11.Bown N, Y SM, Evans J, Kernahan J, Reid MM. Chronic myelomonocytic leukemia with t(13;14) in a child. Cancer Genet Cytogenet. 1992;60:190–2. 1992 [DOI] [PubMed] [Google Scholar]
- 12.Buijs A, Bruin M. Fusion of FIP1L1 and RARA as a result of a novel t(4;17)(q12; q21) in a case of juvenile myelomonocytic leukemia. Leukemia. 2007;21:1104–8. [DOI] [PubMed] [Google Scholar]
- 13.Morerio CA M, Rosanda C, Rapella A, Dufour C, Locatelli F, Maserati E, et al. HCMOGT-1 is a novel fusion partner to PDGFRB in juvenile myelomonocytic leukemia with t(5;17)(q33; p11.2). Cancer Res. 2014;64:2649–51. [DOI] [PubMed] [Google Scholar]
- 14.Byrgazov K, Kastner R, Dworzak M, Hoermann G, Haas OA, Ulreich R, et al. A novel fusion gene NDEL1-Pdgfrb in a patient with JMML with a new variant of TKI-resistant mutation in the kinase domain of PDGFR beta. Blood. 2014;124:613. [Google Scholar]
- 15.Mizoguchi Y, Fujita N, Taki T, Hayashi Y, Hamamoto K. Juvenile myelomonocytic leukemia with t(7;11)(p15; p15) and NUP98-HOXA11 fusion. Am J Hematol. 2009;84:295–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.