Abstract
There is no clearly established association between the gene NUP188 and human pathology. Only a few reports of patients with different clinical presentation and different heterozygous or compound heterozygous missense or splice region variants have been identified in several sequencing projects; however, a causative association between the clinical features and the identified variants has not been established. For the first time, we report 2 unrelated patients with 2 different homozygous nonsense gene variants of NUP188, p.Tyr96* and p.Gln113*, respectively. Although having different supposedly truncating mutations, the patients presented with strikingly comparable phenotypes including pre- and postnatal microcephaly, trigonocephaly, congenital bilateral cataract, microphthalmia, cleft lip and palate or high-arched palate, camptodactyly, rocker-bottom feet, heart anomalies, specific brain changes (such as loss of periventricular white matter), thin corpus callosum, and delayed myelinization. Both patients showed very similar facial features such as laterally extended arched eyebrows, wide convex nose with a wide prominent nasal bridge, and prominent angulated antihelix. They were both born small for gestational age and died shortly after birth at the age of 67 and 140 days, respectively, as a result of central respiratory failure. Our findings strongly suggest a correlation between the homozygous nonsense gene variants of NUP188 and a severe phenotype of a new developmental syndrome with poor prognosis resulting from nucleoporin 188 homolog protein insufficiency.
Keywords: Exome sequencing, Homozygous truncating mutation, New syndrome, Nucleoporin 188 insufficiency, NUP188
Established Facts
There is no clearly established association between the gene NUP188 and human pathology.
The gene NUP188 has a lower than expected proportion of loss-of-function variants (observed/expected = 0.2), suggesting that biallelic loss of function of the gene is poorly tolerated.
RNA knockdown of nup188 resulted in abnormal left/right patterning in Xenopus embryos.
Novel Insights
This study indicates a new autosomal recessive developmental syndrome resulting from nonsense (truncating) homozygous variants of the NUP188 gene.
The suggested novel syndrome was well-characterized in 2 unrelated patients with a different homozygous nonsense variant of the NUP188 gene. Both patients showed close similarity and specificity of clinical features including the course of the disease and a poor prognosis.
This study gives a new insight into the role of the NUP188 gene in human pathology.
The NUP188 (nucleoporin, 188-kDa, OMIM 615587) gene is located at 9q34.11 and encodes the nucleoporin NUP188 homolog (hNup188), which is a part of the large nuclear pore complexes (NPCs). NPCs are aqueous channels generated from a complex network of evolutionarily conserved proteins known as nucleoporins that function at the gate controlling all trafficking between the nucleus and cytoplasm [Jühlen and Fahrenkrog, 2018]. hNup188 is a part of the hNup93 subcomplex, the second major structural unit of NPC, which has been shown to control the passage of membrane or transmembrane proteins through NPC. The nuclei of cells lacking the hNup188-hNup93 complex have been indicated to considerably increase in size, likely as a result of dysregulation of nuclear import of these membranous proteins [Theerthagiri et al., 2010]. Knockdown of hNup188 or its binding partner hNup93 leads to a loss of cilia during embryonic development while leaving the NPC function largely intact [Del Viso et al., 2016]. hNup188 has also been shown to have additional functionality during mitosis, where it appears to be important for K-fiber and spindle formation [Itoh et al., 2013]. It has been shown to specifically bind to flexible gate domains and to be able to transverse NPC, characteristics typically associated with nuclear transport receptors [Andersen et al., 2013; Kabachinski and Schwartz, 2015]. RNA knockdown of nup188 resulted in abnormal left/right patterning (heterotaxy) in Xenopus embryos [Fakhro et al., 2011]. There is, however, no clearly established association between the gene NUP188 and human pathology. Nevertheless, the gene has a lower than expected proportion of loss-of-function variants (observed/expected = 0.2), suggesting that loss of function of the gene is poorly tolerated (https://gnomad.broadinstitute.org/gene/ENSG00000095319). Strauss et al. [2018] reported 2 variants of the NUP188 gene, c.313 C>T; p.(Arg105Trp) and c.3429+5 G>A, in a 3-year-old boy with motor delay, C1 dysplasia, hearing loss, tracheomalacia, and cryptorchidism. The gene variants were a missense and an intronic variant, respectively. No functional characterization was performed, and no other patients with a similar phenotype-genotype combination were reported. Thus, the causative association between the clinical features of this patient and the shown gene variants was not established. Yates et al. [2017] found a heterozygous missense variant of the NUP188 gene, p.R202H, in a fetus with diaphragmatic hernia. Haskell et al. [2017] reported another heterozygous variant of the NUP188 gene, c.4737+1G>T, in an adult patient with mitral valve prolapse. None of these publications could display a clear association between the NUP188 gene variant and the respective phenotype.
Missense variants in other nucleoporin genes have recently been reported to result in various disorders, including autosomal recessive Galloway-Mowat syndrome characterized by renal symptoms in conjunction with developmental delay, edema, dysmorphic features, and microcephaly (NUP107) [Rosti et al., 2017]; nephrotic syndrome with variable extrarenal manifestation (NUP107, NUP85, NUP133, and NUP160) [Braun et al., 2018], and a lethal fetal akinesia with dysmorphic features (NUP88) [Bonnin et al., 2018]. Interestingly, Braun et al. [2018] noted that complete loss of the nup107 homolog caused early lethality in Zebrafish. None of the NUP107, NUP85, NUP133, or NUP160 variants found in human patients were biallelic truncating variants.
Here, for the first time, we report 2 different homozygous nonsense gene variants of NUP188, namely NM_015354.2:c.287dupA; p.(Tyr96*) and c.337C>T; p.(Gln113*), in 2 unrelated newborn female patients showing strikingly similar phenotypes. They both presented with pre- and postnatal microcephaly, trigonocephaly, congenital bilateral cataract, either cleft lip and palate or high-arched palate, camptodactyly, rocker-bottom feet, heart anomalies, brain changes (such as loss of periventricular white matter), thin corpus callosum, and delayed myelinization as well as very similar facial gestalt. Both patients were born small for gestational age, both developed respiratory insufficiency, and both died in central respiratory failure at the age of 67 and 140 days, respectively. Supposedly, the homozygous truncating NUP188 mutations observed in both children underlie all of these abnormalities, which collectively indicate the existence of a “Nucleoporin 188 insufficiency syndrome.”
Clinical Reports
Patient 1
The patient, a newborn girl, was the fifth child to healthy consanguineous parents (first cousins) of Syrian origin. She had 4 healthy brothers. There were no miscarriages in the family, and no other affected family members were reported. Reduced fetal movements and −3 SD intrauterine growth restriction, calculated on the basis of 3 measurements (biparietal diameter, femur length, and median abdominal circumference), were noted at gestational week 32. The girl was born small for gestational age with acute cesarean section at 36 + 4 weeks of gestation due to progression of intrauterine growth restriction and abnormal cardiotocography traces. Apgar score was 9/10/10. The birth measurements were weight 1,915 g (−3 SD), length 42.5 cm (−3 SD), and head circumference 29.5 cm (−3 SD). She presented with a microcephaly, trigonocephaly, small fontanelle, microphthalmia, bilateral congenital cataracts, unilateral cleft lip and palate, camptodactyly, rocker-bottom feet, muscle hypotonia, and a muscular ventricular septal defect (VSD) (Table 1). Dysmorphic features included narrow lateral forehead, sparse medial and laterally extended arched eyebrows, epicanthus, a wide prominent nasal bridge, wide convex nasal ridge, retrognathia, low-set ears with hypoplastic tragus and prominent angulated antihelix, short neck, laterally placed nipples, long gracile fingers, bilateral single palmar crease, hammer toe, and facial hypertrichosis (Fig. 1). Head circumference progressively decreased from −3 SD at birth to −4 SD at 2 months of age. Her length increased from −3 SD at birth to approximately −1 SD at 2 months of age, and her weight improved slightly from −3 SD to −2 SD. All growth parameters were age adjusted for prematurity.
Table 1.
Clinical manifestations of 2 independent patients first shown to have NUP188 nonsense homozygous gene variants
| Features | Patient 1 | Patient 2 |
|---|---|---|
| NUP188 variants | c. 287dupA; p.(Tyr96*) | c.337C>T; p.(Gln113*) |
| Gender/age of death | Female/67 days | Female/140 days |
| Ethnic origin | Syrian | Indian |
| Consanguinity | + | − |
| Abnormal CTG | + | + |
| Preterm/SGA | + | + |
| Bilateral congenital cataract | + | + |
| Microphthalmia | + | + |
| Microcephaly | ||
| Prenatal | + | + |
| Postnatal | + | + |
| Metopic ridge | − | + |
| Trigonocephaly | + | + |
| Cleft lip and palate | + | − |
| Arched palate | − | + |
| Respiratory failure | + | + |
| Heart anomalies | Muscular VSD | Perimembranous VSD |
| Brain MRI | ||
| Wide ventricles | + | + |
| Loss of periventricular white matter | + | + |
| Thin corpus callosum | + | + |
| Delayed myelination | − | + |
| Epilepsy/EEG changes | −/– | −/+ |
| Narrow lateral forehead | + | + |
| Sparse medial eyebrows | + | − |
| Laterally extended arched eyebrows | + | + |
| Epicanthus | + | + |
| Wide prominent nasal bridge | + | + |
| Wide convex nasal ridge | + | + |
| Retrognathia | + | + |
| Low-set ears | + | + |
| Hypoplastic tragus | + | + |
| Prominent angulated antihelix | + | + |
| Short neck | + | + |
| Laterally placed nipples | + | + |
| Bilateral single palmar crease | + | + |
| Camptodactyly | + | + |
| Clinodactyly | + | + |
| Rocker-bottom feet | + | + |
Note the large number of features shared between the 2 patients. CTG, cardiotocography; SGA, small for gestational age; VSD, ventricular septal defect.
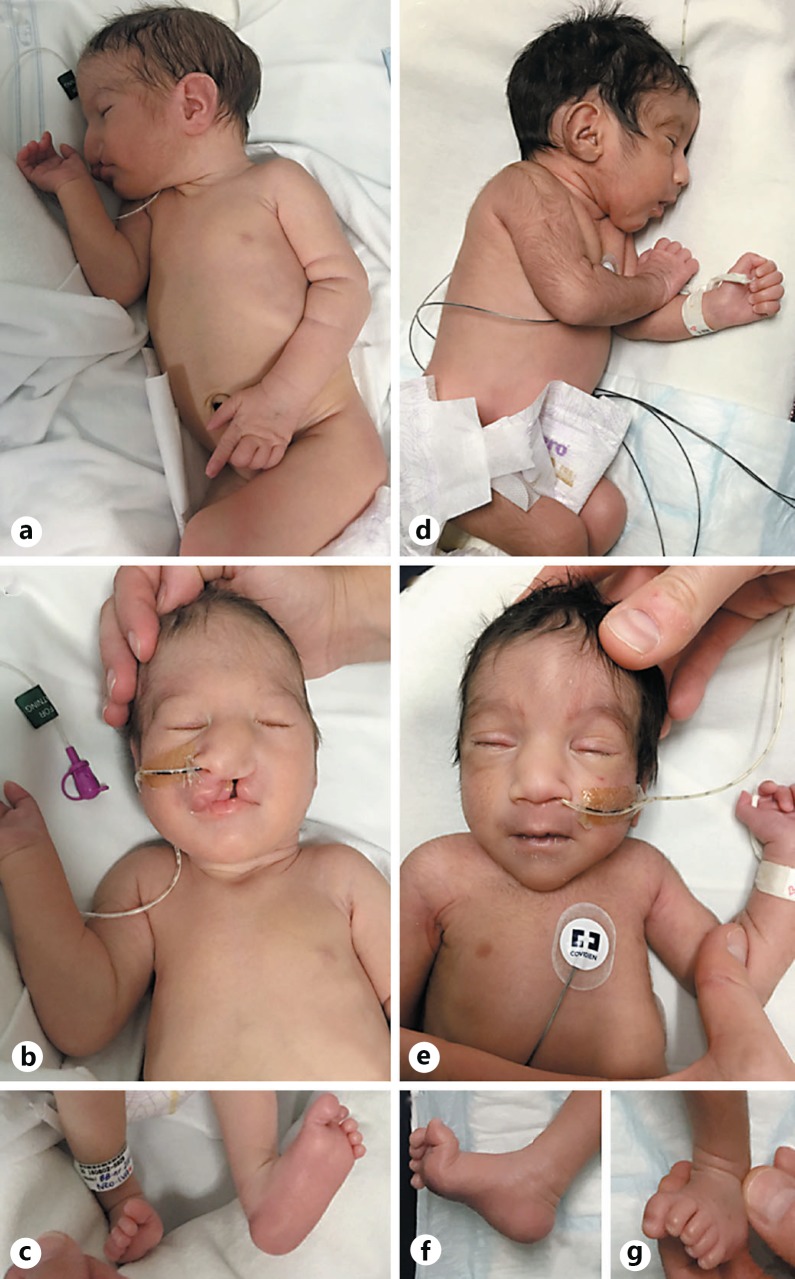
Fig. 1.
a-c Patient 1 at 6 days of age showing microcephaly, trigonocephaly, facial hypertrichosis, retrognathia, low-set ears with hypoplastic tragus and prominent angulated antihelix, short neck, long gracile fingers, camptodactyly (a), narrow lateral forehead, microphthalmia, epicanthus, unilateral cleft lip and palate, sparse medial and laterally extended arched eyebrows, a wide prominent nasal bridge, wide convex nasal ridge, laterally placed nipples (b), rocker-bottom feet, and hammer toe (c). d-g Patient 2 at 10 days of age showing microcephaly, trigonocephaly, retrognathia, low-set ears with hypoplastic tragus and prominent angulated antihelix, generalized hypertrichosis, short neck, gracile fingers, camptodactyly (d), narrow lateral forehead, metopic ridge, microphthalmia, laterally extended arched eyebrows, epicanthus, a wide prominent nasal bridge, wide convex nasal ridge, laterally placed nipples (e), rocker-bottom feet, and hammer toe (f, g).
Ultrasound of the heart showed a muscular VSD without circulatory changes. Ultrasound of the abdomen was normal. Brain MRI showed wide ventricles, loss of periventricular white matter (mostly in the occipital area), and a thin corpus callosum. EEG was performed because of occasional jerking movements and chewing in association with apnea and showed no epileptiform activity. Hearing screening with the otoacoustic emission test was not performed. Metabolic screening showed a high level of galactitol (251.3 mmol/mol, reference value <95) in urine, but galactose, galactose-1-phophate, and galactose-1P-uridyl transferase GALT levels were screened normal in blood, and no pathogenic variants of the genes associated with galactosemia were found by exome sequencing. The high level of galactitol was therefore interpreted as probably secondary to immature liver function or liver dysfunction and not as a sign of galactosemia. A slight increase in tyrosine was seen, which was also interpreted to be due to immature liver function.
During her lifetime, the girl was lethargic, hypotonic and hypoactive, and almost constantly in need of extra oxygen supply and a feeding tube. Her general health status progressively worsened, and after a period of recurrent episodes of decreased oxygen saturation, she died of respiratory failure at 67 days of age. Consent for autopsy was not given.
Patient 2
The patient, a newborn girl, was the second child to healthy reportedly unrelated parents coming from different regions in India. She had 1 healthy sister. No miscarriages were reported in the family, but one extrauterine pregnancy was observed. No other similarly affected family members were noted. The pregnancy was uneventful apart from a breech presentation and abnormal cardiotocography observed at a routine visit at 35 weeks of gestation. The girl was born small for gestational age by acute cesarean delivery at 35 + 3 weeks of gestation because of the abnormal cardiotocography. Apgar score was 7/8/9. The birth measurements were weight 1,960 g (−2 SD) and length 45 cm (−1 SD). The head circumference measurement at birth (30.5 cm) cannot be relied on; at 2 weeks of age, it was 30 cm (−3 SD). She presented with microcephaly, trigonocephaly, small fontanelle (10/10 mm), metopic ridge, microphthalmia, bilateral congenital cataracts, camptodactyly, rocker-bottom feet, muscle hypotonia, and a perimembranous VSD (Table 1). Dysmorphic features included narrow lateral forehead, laterally extended arched eyebrows, epicanthus, a wide prominent nasal bridge, wide convex nasal ridge, very high-arched palate, retrognathia, low-set ears with hypoplastic tragus and prominent angulated antihelix, short neck, laterally placed nipples, long gracile fingers, bilateral single palmar crease, camptodactyly, rocker-bottom feet, hammer toe, and generalized hypertrichosis (Fig. 1).
The girl developed a postnatal microcephaly. Her head circumference decreased from −3 SD at 2 weeks of age to −5 SD at 4 months of age. Length was steady at approximately −1 SD and her weight increased from −3 SD at birth to ±0 SD at 4 months of age. All growth parameters are age adjusted for prematurity. Ultrasound of the heart showed a perimembranous VSD without circulatory changes. Ultrasound of the abdomen was normal. Brain MRI showed distinct ventriculomegaly with loss of periventricular white matter (pronounced in the occipital area), a corpus callosum hypoplasia, and apparent delayed myelinization. EEG showed multifocal epileptiform activity; however the girl did not present with seizures. Otoacoustic emission test at approximately 5 weeks of age showed bilateral abnormal results. Metabolic screening showed a mildly increased level of galactitol (122.8 mmol/mol creatinin, reference value <90), but galactose, galactose-1-phophate, and galactose-1P-uridyl transferase (GALT) was screened normal in blood, and no genetic variants of the genes causing galactosemia were found by exome sequencing. The elevated level of galactitol was therefore interpreted as due to immature liver function or liver dysfunction and not as a sign of galactosemia. Elevated levels of some amino acids were also seen in urine and also interpreted to be due to immature liver function. Biochemical mitochondrial examination of a muscle biopsy showed normal results, no signs of mitochondrial defects or mitochondrial myopathy.
During her lifetime, the girl was lethargic and hypotonic, and often in need of extra oxygen supply as well as a feeding tube. She had some episodes of acute circulatory and respiratory failure, requiring cardiopulmonary resuscitation, and she was briefly intubated after one of those episodes. She died following respiratory failure at 140 days of age. Permission for autopsy was not given.
Warburg (MICRO) syndrome and cerebro-oculo-facio-skeletal (COFS) syndrome were considered as differential diagnoses in both patients, but the clinical features did not match sufficiently with the abovementioned entities. Furthermore, trio-exome sequencing analyses did not show any pathogenic mutations in genes known to be associated with these conditions.
Materials and Methods
Previous genetic testing included SNP array on both patients, with normal results. Analysis of patient 1 showed higher than usual number of loss of heterozygosity (LOH) regions, corresponding to the known parental consanguinity.
Trio-exome sequencing analysis was performed in the settings of a research project in order to screen for disease-causing gene variants and was performed by CeGaT GmbH, Tübingen, Germany.
Sequencing was performed on DNA extracted from blood samples from both patients and their parents. The sequencing library was prepared using the SureSelectXT workflow (Agilent, Santa Clara, CA, USA) and the Human All Exon enrichment kit (version 7). Library preparation and capture was performed according to the manufacturer's instructions and paired-end sequencing was done on a NovaSeq6000 instrument (Illumina, San Diego, CA, USA) with 2×100-bp read length. After demultiplexing (Illumina bcl2fastq 2.20), adapters were trimmed with Skewer (0.2.2). Trimmed raw reads were aligned to the human genome (hg19) with the Burrows-Wheeler Aligner (BWA-mem version 0.7.17). Average coverage on target was >120× (96% >30×). Copy number variations (CNVs) were computed using an internally developed method based on expected sequencing coverage depth versus observed sequencing coverage depth. Sequence variants were called (CeGaT stratacall) with a minimum variant allele frequency of 5%. Resulting variants were annotated with population frequencies from dbSNP (release 151), GnomAD (2.1), and an in-house database, with functional predictions from dbNSFP (3.4c), with publications from HGMD (19.1), and with transcript information from Ensembl, RefSeq, and CCDS (release 22). SNVs and small indels in the coding regions and the flanking intronic regions (±8 bp) with a minor allele frequency (MAF) <1.5% were evaluated. MAFs were taken from public databases (gnomAD, dbSNP) and an in-house database. Known disease-causing variants (according to HGMD) were evaluated in up to ±30 bp of flanking regions and up to 5% MAF. Trio-exome filtering was performed in order to obtain variants with expected mendelian patterns of inheritance: autosomal recessive (compound heterozygous or homozygous variants) and autosomal dominant (de novo variants, potential parental germline mosaicism, and LOH).
Evaluation was based on the ACMG guidelines for the interpretation of sequence variants.
Sanger sequencing was performed on the relevant region of the NUP188 gene, based on the NM_015354.2 transcript. Sanger sequencing analysis for the NUP188 gene variant found in patient 1 was performed on her 3 healthy brothers.
Results
Trio-exome sequence analyses of patient 1 revealed a novel homozygous nonsense variant of the NUP188 gene (NM_015354.2, CCDS35156.1, UniProt accession: Q5SRE5): c. 287dupA; p.(Tyr96*), which introduces a premature termination codon in exon 5 of 44 of the NUP188 gene, likely resulting in nonsense-mediated decay of the mRNA product. The parents were heterozygous carriers of the same variant. None of the patient's 3 healthy brothers were homozygous carriers of the variant. No de novo or other rare biallelic variants fulfilling the inclusion criteria were found in patient 1.
Trio-exome sequence analyses of patient 2 revealed a novel homozygous nonsense variant of the NUP188 gene (NM_015354.2, CCDS35156.1, UniProt accession: Q5SRE5): c.337C>T; p.(Gln113*), which introduces a premature termination codon in exon 6 of 44 of the NUP188 gene, likely resulting in nonsense-mediated decay of the mRNA product. The parents were heterozygous carriers of the same variant. The healthy sister of the patient has not been tested for this variant since the parents did not agree.
Both gene variants create a premature stop codon, and they are both absent from the gnomAD global population dataset and from the ClinVar dataset.
No de novo or other rare biallelic variants fulfilling the inclusion criteria were found in patient 2.
Discussion
Until now, the association between NUP188 gene variants and human pathology has remained elusive. Only a few patients, all with different pathology and different gene variants of NUP188 have been reported, but with no common denominator [Haskell et al., 2017; Yates et al., 2017; Strauss et al, 2018].
Here, we studied 2 unrelated preterm newborn patients and, for the first time, report a new syndrome, which we tentatively named “Nucleoporin 188 insufficiency syndrome” because the apparent disease-causing mechanism was identified as a homozygous nonsense gene variant of NUP188. The 2 patients, having different gene variants and coming from 2 different ethnic groups - Syrian and Indian, respectively - presented with strikingly similar phenotypes, including their facial gestalt, congenital anomalies, the course of the disease, a poor prognosis, and the cause of death. The common clinical features included pre- and postnatal microcephaly, trigonocephaly, congenital bilateral cataract, microphthalmia, camptodactyly, rocker-bottom feet, heart anomalies, brain changes including loss of periventricular white matter and thin corpus callosum, very similar facial gestalt with laterally extended arched eyebrows, wide convex nasal ridge with a wide prominent nasal bridge, ear dysmorphology (low-set ears, hypoplastic tragus, and prominent angulated antihelix) as well as bilateral single palmar crease, and clinodactyly (Table 1). We observed very few clinical features not shared by the 2 patients (Table 1). These could be referred to either minor dysmorphic features (sparse medial eyebrows) or to incomplete overlap because of a large variation of the same feature. For example, a metopic ridge was only found in patient 2 (Fig. 1), but both patients had trigonocephaly; patient 2 did not have a cleft lip and palate, but she presented with a very high-arched palate; patient 1 showed the same brain changes on MRI as patient 2 without delayed myelinization, but the MRIs were performed at the age of 2 weeks for patient 1 versus 4 months of age for patient 2. The variants identified in NUP188 in both patients are very likely loss of function caused by the presence of a premature termination codon within a small exon close to the 5′ end of the gene. They are expected most likely to result in nonsense-mediated decay of the mRNA product and considerably reduced expression of an aberrant gene product.
To the best of our knowledge, homozygous nonsense gene variants of NUP188 have not been described previously. Our search outside published literature found only 1 short communication about 2 sisters with compound heterozygous nonsense gene variants of NUP188 gene who presented with similar phenotypes as the ones described here (abstract presented at the ASHG Meeting 2012). There may be 6 more patients in USA with a similar clinical condition and a biallelic truncating gene variant of NUP188 (pers. communication A. Holz).
Our findings suggest a strong correlation between the homozygous nonsense gene variants of NUP188 and a severe phenotype of a novel autosomal recessive developmental syndrome with pre- and postnatal microcephaly, trigonocephaly, congenital cataract, microphthalmia, particular facial gestalt, camptodactyly, loss of periventricular white matter, thin corpus callosum, delayed myelinization, and poor prognosis as main features.
We suggest the name Nucleoporin 188 insufficiency syndrome. One possible reason that this condition has remained unknown so far could be its severity and high lethality at early age as well as the fact that whole-exome sequencing has, until quite recently, been rarely used as a diagnostic tool for severely affected infants. One could also speculate about the possibility of a skewed distribution of the disease-causing heterozygous gene variants among the different ethnic groups. Future reports of patients with the same condition and homozygous, or compound, nonsense NUP188 gene variants will contribute to our knowledge of this new syndrome.
Statement of Ethics
This study was performed in the settings of a research project and was approved by the Linköping University Ethics Committee. Both parents and one brother gave their written informed consent for the analyses and publication, including the photographs of the patients.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was funded by the Forskningsrådet Sydöstra Sverige (FORSS) and the Styrgruppen “Sjukvård och Välfärd,” Region Östergotland, Linköping, Sverige.
Acknowledgments
We gratefully acknowledge the families for participating in this study. Special thanks go to our colleague Dr. Jon Jonasson for his valuable advice and for the revision of the manuscript.
References
- Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, et al. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. Elife. 2013;2:e00745. doi: 10.7554/eLife.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin E, Cabochette P, Filosa A, Jühlen R, Komatsuzaki S, et al. Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence. PLoS Genet. 2018;14:e1007845. doi: 10.1371/journal.pgen.1007845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Lovric S, Schapiro D, Schneider R, Marquez J, et al. Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J Clin Invest. 2018;128:4313–4328. doi: 10.1172/JCI98688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Viso F, Huang F, Myers J, Chalfant M, Zhang Y, et al. Congenital heart disease genetics uncovers context-dependent organization and function of nucleoporins at cilia. Dev Cell. 2016;38:478–492. doi: 10.1016/j.devcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, et al. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskell GT, Jensen BC, Samsa LA, Marchuk D, Huang W, et al. Whole exome sequencing identifies truncating variants in nuclear envelope genes in patients with cardiovascular disease. Circ Cardiovasc Genet. 2017;10:e001443. doi: 10.1161/CIRCGENETICS.116.001443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh G, Sugino S, Ikeda M, Mizuguchi M, Kanno S, et al. Nucleoporin Nup188 is required for chromosome alignment in mitosis. Cancer Sci. 2013;104:871–879. doi: 10.1111/cas.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühlen R, Fahrenkrog B. Moonlighting nuclear pore proteins: tissue-specific nucleoporin function in health and disease. Histochem Cell Biol. 2018;150:593–605. doi: 10.1007/s00418-018-1748-8. [DOI] [PubMed] [Google Scholar]
- Kabachinski G, Schwartz TU. The nuclear pore complex - structure and function at a glance. J Cell Sci. 2015;128:423–429. doi: 10.1242/jcs.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti RO, Sotak BN, Bielas SL, Bhat G, Silhavy JL, et al. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway-Mowat syndrome. J Med Genet. 2017;54:399–403. doi: 10.1136/jmedgenet-2016-104237. [DOI] [PubMed] [Google Scholar]
- Strauss KA, Gonzaga-Jauregui C, Brigatti KW, Williams KB, King AK, et al. Genomic diagnostics within a medically underserved population: efficacy and implications. Genet Med. 2018;20:31–41. doi: 10.1038/gim.2017.76. [DOI] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates CL, Monaghan KG, Copenheaver D, Retterer K, Scuffins J, et al. Whole-exome sequencing on deceased fetuses with ultrasound anomalies: expanding our knowledge of genetic disease during fetal development. Genet Med. 2017;19:1171–1178. doi: 10.1038/gim.2017.31. [DOI] [PubMed] [Google Scholar]