Abstract
Pirellula-like planctomycetes are ubiquitous aquatic bacteria, which are often detected in anoxic or micro-oxic habitats. By contrast, the taxonomically described representatives of these bacteria, with very few exceptions, are strict aerobes. Here, we report the isolation and characterization of the facultatively anaerobic planctomycete, strain PX69T, which was isolated from a boreal lake. Its 16S rRNA gene sequence is affiliated with the Pirellula-related Pir4 clade, which is dominated by environmental sequences retrieved from a variety of low-oxygen habitats. Strain PX69T was represented by ellipsoidal cells that multiplied by budding and grew on sugars, some polysaccharides and glycerol. Anaerobic growth occurred by means of fermentation. Strain PX69T grew at pH 5.5–7.5 and at temperatures between 10 and 30 °C. The major fatty acids were C18:1ω9c, C16:0 and C16:1ω7c; the major intact polar lipid was dimethylphosphatidylethanolamine. The complete genome of strain PX69T was 6.92 Mb in size; DNA G + C content was 61.7 mol%. Among characterized planctomycetes, the highest 16S rRNA gene similarity (90.4%) was observed with ‘Bythopirellula goksoyri’ Pr1d, a planctomycete from deep-sea sediments. We propose to classify PX69T as a novel genus and species, Lacipirellula parvula gen. nov., sp. nov.; the type strain is strain PX69T (=KCTC 72398T = CECT 9826T = VKM B-3335T). This genus is placed in a novel family, Lacipirellulaceae fam. nov., which belongs to the order Pirellulales ord. nov. Based on the results of comparative genome analysis, we also suggest establishment of the orders Gemmatales ord. nov. and Isosphaerales ord. nov. as well as an emendation of the order Planctomycetales.
Keywords: Planctomycetes, Anoxic habitats, Pir4 clade, Lacipirellula parvula, Lacipirellulaceae, Pirellulales
Introduction
Pirellula-related planctomycetes represent one of the widespread groups of aquatic bacteria, which are also found in wetlands and soils [4], [7], [10], [15], [27], [32], [45]. The first cultured and described organism in this group, Pirellula staleyi, was isolated from Lake Lansing, Michigan, USA [34], [35], [42]. This aerobic freshwater planctomycete has a low salinity tolerance, of up to 50% artificial salt water (ASW) [34]. Several Pirellula-like isolates were later obtained from various freshwater habitats in northern Germany [32] but were not characterized taxonomically. Members of all other described genera in the phylogenetic clade defined by the genus Pirellula, such as Blastopirellula [22], [33], [36], Rhodopirellula [2], [36], Rubripirellula [3], Roseimaritima [3] and Mariniblastus [21], were isolated from brackish and marine habitats. Growth of these bacteria occurs in media containing increased concentrations (12–200%) of ASW. With the only exception of Blastopirellula marina, which is capable of fermenting glucose and reducing nitrate to nitrite [33], these planctomycetes were characterized as strict aerobes that compose microbial biofilms of macro-algae and participate in the breakdown of sulfated polysaccharides [4], [13], [46].
The first evidence for the occurrence of as-yet-uncultivated Pirellula-related planctomycetes with anaerobic metabolism was obtained in the course of microbial diversity analysis in an anaerobic digestor of a municipal wastewater treatment plant [8]. This study showed that activated sludge contains a highly diverse community of planctomycetes, which grow under both aerobic and anaerobic conditions. Two groups of Pirellula-related microorganisms, clades Pir3 and Pir4, were defined in that study. Since then, 16S rRNA gene sequences affiliated with these groups of planctomycetes were recovered from various environments, such as marine sediments [44], oxic/anoxic interfaces of wetlands [16], swamp meadows [38] and other habitats, suggesting the occurrence of as-yet-uncultured anaerobic representatives in this group.
Two recently described, phylogenetically divergent clades of Pirellula-related planctomycetes are represented by ‘Bythopirellula goksoyri’ Pr1d, which was isolated from deep sea iron-hydroxide deposits [44], and two genera of thermophilic planctomycetes from terrestrial and marine thermal habitats, Thermogutta [40] and Thermostilla [41]. ‘Bythopirellula goksoyri’ Pr1d was described as an aerobic planctomycete, while Thermogutta and Thermostilla are facultative anaerobes.
The vast majority of Pirellula-related planctomycetes in the environment, however, remains uncharacterized. One particular example is the ‘Pir4’ clade. According to the SILVA database (release 132, SSU Ref) [29], this clade currently includes 4015 environmental clone sequences, which were retrieved from various habitats. Here, we describe strain PX69T, a member of this phylogenetic clade, which was isolated from a boreal lake in Northern Russia, and analyze its genome-encoded traits and the metabolic potential. We also define the taxonomic position of this novel planctomycete based on the results of a comparative genome analysis, and propose a number of re-arrangements within the order Planctomycetales.
Materials and methods
Sampling site
Strain PX69T was isolated from water collected from the upper oxic layer (0−10 cm) of the boreal eutrophic lake Morotskoye (Vologda region, European North Russia, 58°43′28.5"N, 37°39′07ʺE) in August 2017. The lake area is 6.24 km2 and its maximum depth is 2.1 m. Site-specific parameters are as follows (range given in parenthesis): water conductivity (30–50 μS cm−1), dissolved organic carbon (25.0–37.5 mg L−1), total nitrogen (2.0 and 3.7 mg L−1), and total phosphorus (45−77 μg L−1). The pH is 7.5–7.8.
Isolation and cultivation
The enrichment culture was obtained using an agar (Agar-Bacto B/D, Difco) medium prepared with original lake water and containing 0.05 g L−1 carbenicillin (sodium salt). An aliquot of lake water (10 ml) was spread onto this medium, the plates were placed in bags to prevent drying, and incubated at at 22 °C. Cell suspensions that developed on plates after 4 weeks of incubation were enriched with microorganisms having a planctomycete-like morphology. Aliquots (20 μl) of these enrichment cultures were spread plated onto the MPYVG medium (modification of medium 621 DSMZ), solidified with 10 g phytagel (Sigma - Aldrich) and containing (per litre distilled water): 0.1 g peptone (Fluka), 0.25 g yeast extract (Fluka), 0.1 g NH4NO3, 20 ml Hutner’s basal salts [43]. After sterilization, the medium was complemented with 5 ml L−1 5% glucose solution, 1 ml L−1 Staley’s vitamin solution [43], 0.05 g carbenicillin (sodium salt), pH 6.5. The plates were subsequently incubated at 22 °C for 4 weeks. Colonies that developed on plates were screened microscopically for the presence of budding cells with a planctomycete-like morphology. The selected cell material was re-streaked onto the same medium MPYVG, supplemented with 0.05% (w/v) glucose. Once obtained in pure culture, the isolate was maintained on MPYVG medium and was sub-cultured at 2 month intervals.
Microscopic studies
Morphological observations and cell size measurements were made with a Zeiss Axioplan 2 microscope and Axiovision 4.2 software (Zeiss, Germany). For negative staining, cells were dried onto grids and treated with 1% (w/v) phosphotungstic acid. The specimen samples were examined with JEM-1011 (JEOL, Japan) transmission electron microscope.
Physiological tests
Physiological tests were performed in liquid MPYVG medium. Growth of strain PX69T was monitored by nephelometry at 600 nm in an Eppendorf BioPhotometer for 2–3 weeks under a variety of conditions, including temperatures of 4−37 °C, pH 3.8–8.0 and NaCl concentrations of 0–3.0% (w/v). Incubations at various temperatures were made under static conditions in triplicate; OD600 was determined after 2 weeks of incubation. Variations in the pH were achieved by mixing 0.1 M solutions of H2SO4 and KOH. Carbon source utilization was determined using mineral medium M1 [19], supplemented with 0.0.5% yeast extract and the individual carbon sources given in the species description in a concentration 0.05 % (w/v). Cultivation was done in 120 ml flasks containing 20 ml medium. Cultures were incubated at 22 °C for 2–3 weeks on a shaker (100 r.p.m.). All experiments were performed in triplicate.
The ability to grow under micro-oxic conditions was tested in 120-ml flasks filled with 100 ml of liquid medium MPYVG. The flasks were incubated under static conditions for 3 weeks. Strain PX69T was also examined for growth under anaerobic conditions using tightly closed 120 ml serum flasks containing 80 ml of liquid medium MPYVG. Before autoclaving, these flasks were flushed for 5 min with a mixture of CO2 and N2 (7: 93). Growth was assessed by measuring OD600 after 3 weeks of incubation. The presence of fermentation products (organic acids, alcohols) in culture liquids was verified by using a Stayer HPLC chromatograph (Aquilon, Russia) equipped with a refractometric detector (Knauer, Germany) and an Aminex HPX-87H column (Bio-Rad, USA), operated isocratically with 5 mM H2SO4 as eluent at 0.6 ml/min.
Oxidative and fermentative utilization of carbohydrates was determined as described for the Hugh-Leifson test [12]. Analyzes of enzymatic profiles, oxidase test, gelatin and urease hydrolysis, indole production were made with API ZYM and API 20NE kits (bioMérieux). Catalase test was carried out by standard method [12]. Susceptibility to antibiotics was determined on solid medium MPYVG using discs (Oxoid) containing the following antibiotics: ampicillin (10 μg), gentamycin (10 μg), kanamycin (30 μg), neomycin (10 μg), novobiocin (30 μg), streptomycin (10 μg), chloramphenicol (30 μg), rifampicin (10 μg), tetracycline (10 μg), and lincomycin (10 μg). Growth of strain PX69T and occurrence of growth inhibition zones around these discs was assessed after 4 weeks of incubation at 22 °C. Only the inhibition zones exceeding 2 mm were taken into account.
Lipid and quinone analyses
For lipid analysis, cells of strain PX69T were grown in liquid MPVYG medium and harvested in the late exponential growth phase. Fatty acids were analyzed after acid hydrolysis of whole cells following the procedure described elsewhere [39]. The main intact polar lipids (IPLs) in strain PX69T were analyzed following procedures reported previously [25].
Genome sequencing, annotation and analysis
Genomic DNA was isolated from strain PX69T using the PowerSoil DNA isolation kit (Mo Bio Laboratories Inc., Carlsbad, CA). The sequencing library for Illumina sequencing was prepared using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, USA) following the manufacturer's instructions. The sequencing of this library on the Illumina HiSeq2500 platform using HiSeq Rapid Run v2 sequencing reagents generated 19,701,367 single-end reads with a length of 250 nt (4.9 Gbp in total). Primer sequences were removed from the Illumina reads using Cutadapt v.1.17 [24] with the default settings, and low quality read ends were trimmed using Sickle v.1.33 (option q = 30) (https://github.com/najoshi/sickle). For Nanopore sequencing the library was prepared using the 1D ligation sequencing kit (SQK-LSK108, Oxford Nanopore, UK). Sequencing of this library in an R9.4 flow cell (FLO-MIN106) using MinION device yielded 1,703,953 reads with a total length of 1.23 Gbp. Hybrid assembly of Illumina and Nanopore reads was performed using Unicycler v. 0.4.8 [47]. A single circular contig of 6,922,258 bp was obtained.
Gene search and annotation were performed using the RAST server [6], followed by manual correction. Signal peptides were predicted using Signal P v. 5.0 for Gram-negative bacteria (http://www.cbs.dtu.dk/services/SignalP/). The average amino acid identity (AAI) between the selected genomes was calculated using the aai.rb script from the enveomics collection [31].
The annotated genome sequence of strain PX69T has been deposited at DDBJ/ENA/GenBank under the accession number AP021861.
Phylogenetic analyses
16S rRNA gene-based phylogenetic analysis was carried out using the MEGAX [20]. The phylogenetic tree was built applying the maximum-likelihood statistical method. Visualization of the tree was performed on iTOL platform [23].
In addition, the GTDB-Tk v. 0.3.2 toolkit [26] was used to identify the 120 single-copy, phylogenetically informative bacterial marker genes used in the Genome Taxonomy Database (GTDB) classification system in the genome of strain PX69T. These were used to construct a multiple alignment of concatenated single-copy gene sequences, comprising those from PX69T and all species from the GTDB. The recently sequenced genomes of Planctomycetes bacterium I41 (GenBank CP36339) and ‘Bythopirellula goksoyri’ Pr1d (GenBank CP042913), as well as the single-cell genome assembly of Planctomyces bekefii (SRHE00000000) were additionally included in this analysis. A selected part of the multiple alignment built in GTDB-Tk was used to construct a phylogenetic tree in PhyML v. 3.3 [14] with default parameters. The level of support for internal branches was assessed using the Bayesian test in PhyML.
Results
Cell morphology and physiology
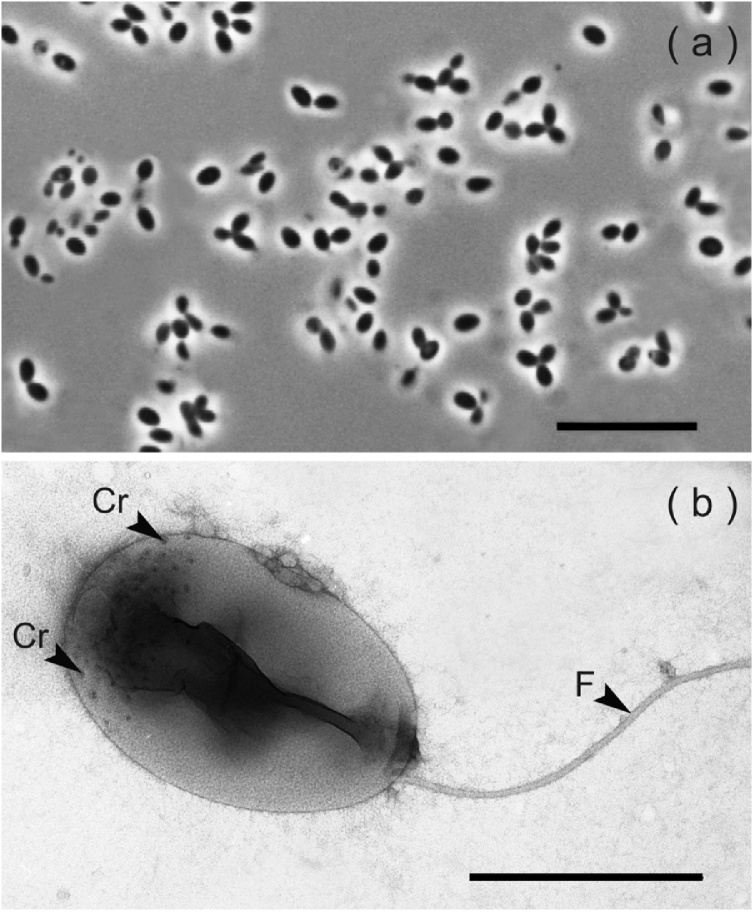
On phytagel-solidified medium MPYVG, strain PX69T formed small (0.5−1 mm), circular, unpigmented colonies with an entire edge and a smooth surface. These colonies were composed of bacteria with ellipsoidal cells, 0.5–0.9 μm wide and 0.9–1.4 μm long, which multiplied by budding. The cells occurred singly, in pairs, in rosettes composed of 3 cells or in aggregates (Fig. 1a). Large cell rosettes characteristic for Pirellula-like planctomycetes were not observed. Daughter cells were highly motile by means of one polar flagellum (Fig. 1b). Examination of negatively stained cells of strain PX69T using electron microscopy showed the presence of crateriform-like structures, which were distributed on a reproductive cell pole only (Fig. 1b).
Fig. 1.
(a) Phase-contrast image of cells of strain PX59T grown for 7 days on solid medium MPVGY; bar, 10 μm. (b) Electron micrographs of a negatively stained daughter cell with flagellum (F) and crateriform pits (Cr) distributed over one cell pole; bar, 0.5 μm.
Strain PX69T was capable of growth at pH values between 5.0 and 7.5 (with an optimum at pH 6.5), and at temperatures between 10 and 30 °C (with an optimum at 20−25 °C). NaCl inhibited growth at concentrations above 0.5% (w/v). Strain PX69T grew on various sugars, including N-acetylglucosamine, and some polymeric compounds, such as aesculin, arabinogalactan, dextrin, laminarin, locust bean gum, gelatin, starch, xanthan gum, pectin and xylan, some organic acids (glucuronic acid, lactate, pyruvate) and polyalcohols (glycerol, sorbitol and dulcitol) (see the species description). Chondroitin sulfate was used by strain PX69T not only as a source of carbon but also as a source of nitrogen (given that glucose was provided as a carbon source). Pullulan, cellulose, carboxymethylcellulose, casein, chitin and chitosan were not hydrolyzed. Growth also occurred under microaerobic conditions by fermentation of glucose, lactose, trehalose, sucrose, and xanthan gum. The only detectable end product of glucose fermentation was lactate. Weak growth was observed under strictly anaerobic conditions.
Strain PX69T was catalase- and cytochrome oxidase- positive but urease-negative. Dissimilatory nitrate reduction was also positive (API 20NE strip). In the API ZYM strip, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase were detected but trypsin, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-galactosidase, α-fucosidase, lipase (C4), cystine arylamidase, α-chymotrypsin, β-glucuronidase and α-mannosidase were absent. Strain PX69T was resistant to ampicillin, carbenicillin, chloramphenicol, gentamicin, neomycin, kanamycin, streptomycin, but sensitive to rifampicin and tetracycline.
Lipid composition
The major fatty acids of strain PX69T were C18:1ω9c, C16:0, C16:1ω7c (Table 1). Cells also contained a wide variety of hydroxy and dihydroxy fatty acids and a C31 polyunsaturated alkene, which is often present in planctomycetes. The major intact polar lipid was dimethylphosphatidylethanolamine. Minor amounts of phosphatidylcholine, monomethylphosphatidylethanolamine, and hexyl phosphatidylglycerol were also detected.
Table 1.
Relative abundance (%; normalized on their sum) of fatty acids and hydrocarbons present in the acid hydrolysate of cell material of strain PX69T. Only fatty acids comprising ≥0.4% of the total are shown. Major components (>5%) are given in bold type face.
| Lipid type | Relative abundance (%) |
|---|---|
| Fatty acids | |
| C14:0 | 0.5 |
| C15:0 | 0.2 |
| isoC16:0 | 1.1 |
| C16:1ω7c | 6.2 |
| C16:1ω7t | 1.0 |
| C16:0 | 18.6 |
| C17:1ω8 | 2.9 |
| C17:0 | 0.5 |
| C18:1ω9 | 34.7 |
| C18:1ω7 | 5.2 |
| C18:0 | 4.6 |
| C20:1ω11+ω9 | 0.7 |
| C20:1ω7 | 0.5 |
| C20:0 | 0.9 |
| C22:1ω9 | 0.8 |
| C22:1ω7 | 0.8 |
| C22:0 | 0.4 |
| Hydroxy fatty acids | |
| α+β-OH-C18:0 | 0.6 |
| (ω-1)OH-C28:1ω11 | 0.6 |
| (ω-1)OH-C32:1ω11 | 2.6 |
| (ω-1)OH-C32:0 | 0.5 |
| (ω-1)OH-C33 cyclopropyl | 3.6 |
| diOH-C32:1ω11a | 1.9 |
| (ω-1)OH-C34:1ω11 | 2.2 |
| diOH-C33 cyclopropyla | 2.8 |
| (ω-1)OH-C35 cyclopropyl | 1.1 |
| diOH-C34:1ω11a | 1.1 |
| diOH-C35 cyclopropyla | 0.9 |
| Hydrocarbons | |
| n-C31:9 | 2.8 |
For these FAs, one of the OH group is at the (ω-1) position, the other is unknown.
Genome characteristics
The genome of strain PX69T was sequenced using a combination of Illumina and Nanopore technique. Combined assembly yielded a single circular contig with a length of 6.92 Mbp. The DNA G + C content was 61.7%. Separately located 16S and 23S-5S rRNA genes, and 61 tRNA genes were identified. Annotation of the genome sequence revealed 5275 potential protein-coding genes of which 2876 (55%) could be functionally assigned.
The examination of the strain PX69T genome revealed a set of genes encoding a bacterial flagellar machinery and chemotaxis functions, consistently with observed morphological characteristics.
Analysis of metabolic pathways revealed that strain PX69T is metabolically versatile and has the genetic potential for fermentation, aerobic and anaerobic respiration. The genome encodes a complete Embden-Meyerhof glucolytic pathway, both oxidatative and non-oxidatative stages of pentose phosphate pathway, and the tricarboxylic acid cycle. The pyruvate generated in the glycolysis could be oxidized by pyruvate dehydrogenase, followed by conversion of acetyl-CoA to acetate. The presence of pyruvate decarboxylase and alcohol dehydrogenase is consistent with the ability of fermentative production of ethanol. Activity of lactate dehydrogenase could result in the production of lactate among the fermentation products. Interestingly, genome analysis revealed no hydrogenases, frequently found in heterotrophic bacteria.
Consistently with the ability of strain PX69T to grow under microaerobic conditions, all components of an aerobic respiratory chain were found, namely NADH ubiquinone oxidoreductase, succinate dehydrogenase, cytochrome bc1 complex, cytochrome c oxidases and an F1F0-type ATP synthase. Interestingly, in addition to three gene clusters, encoding cytochrome bc1 complexes, the genome also contained five-gene cluster coding for alternative complex III (ACIII). The genes encoding ACIII are clustered in the genome with cytochrome c oxidase genes, indicating that the ACIII complex in strain PX69T is likely involved in aerobic rather than anaerobic respiratory processes [30]. Terminal oxygen reductases in strain PX69T are represented by three proton translocating cytochrome c oxidases, two of which could be classified as caa3-type. Cytochrome c oxidases of this type have low affinity to oxygen and are usually used by aerobic microorganisms for respiration (reviewed in [11], [28]). The presence of multiple oxidases could be an important factor for adaptation of strain PX69T to changes in oxygen concentration in the environment. Interestingly, the cytochrome bd ubiquinol oxidase, an enzyme with high affinity to oxygen often found in anaerobes and microaerophiles [5], was absent in the genome of strain PX69T.
The capacities of strain PX69T to grow by anaerobic respiration seems to be limited. The genome contained an operon with genes for the catalytic molybdopterin-binding NapA subunit of nitrate reductase and an electron-transfer iron-sulfur subunit fused to NrfD-like membrane subunit. Genes for nitrate/nitrite transporter NarK and assimilatory NAD(P)H-dependent cytoplasmic nitrite reductase are located in the same region. Together, these reductases convert nitrate through nitrite to ammonium. In addition, NrfAH-type cytochrome c nitrite reductase could enable dissimilatory reduction of nitrite to ammonium.
Analysis of the strain PX69T genome revealed >60 genes encoding carbohydrate-active enzymes predicted to have N-terminal signal peptides and thus possibly involved in extracellular hydrolysis of substrates (Table S1). The most notable is the presence of 39 genes encoding proteins with significant similarity to sulfatases. For 37 of sulfatases in strain PX69T a signal peptide is predicted with high probability, suggesting an extracytosolic localization of these proteins. Bacterial sulfatases are supposed to be primarily used in sulfur scavenging [17]. Alternatively, strain PX69T could use sulfatases for a more efficient access to the carbon skeleton of sulfated compounds used as an energy and carbon sources, as it was proposed for marine Pirellula sp. strain 1, whose genome contained ca. 110 sulfatase genes [13]. Remarkably, several sulfatases were classified as N-acetylglucosamine-6-sulfatase and N-sulfoglucosamine sulfohydrolase. They could be involved in the degradation of sulfated glycosaminoglycans and cleaning of N-acetylglucosamine, shown to support the growth of strain PX69T. In addition, three sulfatases contained additional conserved domains such as DUF1080 (LamG) and Dockerin_II that could facilitate their interaction with the substrates, and one enzyme was predicted to contain the GH43 beta-xylosidase domain. Two iduronate-2-sulfatases could catalyze the hydrolysis of sulfate ester bonds from a wide variety of substrates, including steroids, carbohydrates and proteins. Therefore, the presence of sulfatases in strain PX69T could play an important role in the degradation of sulfated glycopolymers, which are commonly produced by macroalgae and other aquatic macrophytes.
According to the growth experiments, strain PX69T was capable of growth on xylan, laminarin and pectin, and the corresponding pathways were found in the genome. The presence of an extracellular endo-1,4-beta-xylanase and beta-xylosidase, and intracellular enzymes of the isomerase pathway of xylose metabolism, xylose isomerase and xylulokinase is consistent with the ability to grow on xylans. Extracellular pectate lyases and endo-1,3-beta-d-glucosidase could be involved in utilization of pectin and laminarin, respectively. Despite the reported inability of strain PX69T to grow on cellulose, its genome encodes several signal peptide-containing endoglucanases and beta-glucosidase. Several other signal peptide-containing glycosyl hydrolases could be responsible for extracellular hydrolysis of polysaccharides, namely, alpha-galactosidases, alpha-glucosidases, alpha-l-arabinofuranosidases, beta-l-arabinofuranosidases, alpha-l-rhamnosidase, alpha-l-fucosidase, and glycoside hydrolases of GH28 and GH43 families (Table S1). An intracellular metabolism of imported sugars is probably linked to central glycolytic pathways. For example, N-acetylglucosamine, imported via the NagX transporters, in the cytoplasm may be phosphorylated by N-acetylglucosamine kinase. Upon deamination and deacetylation N-acetylglucosamine-6-phosphate is converted into fructose-6- phosphate that enters the Embden-Meyerhof pathway [1], while ammonium could be used as a nitrogen source. Consistently with the inability of strain PX69T to grow on chitin, the polymer of N-acetylglucosamine, the genome encoded no N-acetyl-beta-hexosaminidases and chitinases, and the bacterium probably relies of monomers produced by chitinolytic community members. The ability of strain PX69T to grow on glycerol is consistent with the presence of glycerol uptake facilitator, glycerol kinase and downstream enzymes of glycerol metabolism.
Phylogenetic placement of strain PX69T
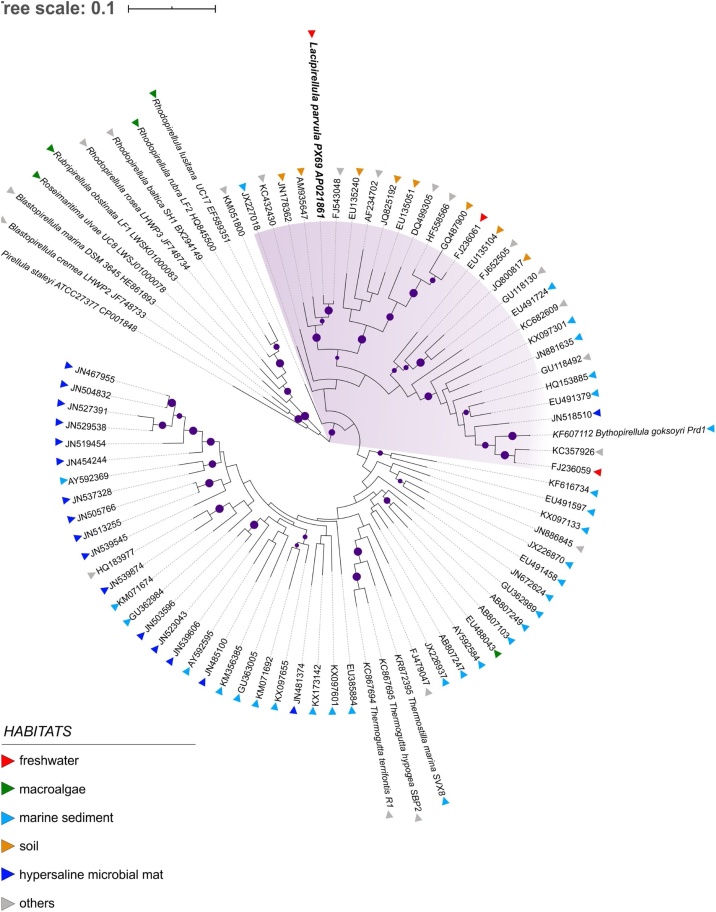
According to the SILVA database (release 132), the 16S rRNA gene sequence of strain PX69T was affiliated with the Pirellula-related Pir4 lineage and grouped with a large number of environmental clone sequences obtained from various low-oxygen habitats, such as wastewater sludge, earthworm gut, lake moss pillars, soils, marine sediments, and seafloor lava (Fig. 2). Among all taxonomically characterized planctomycetes, the highest 16S rRNA gene similarity (90.4%) was observed with ‘Bythopirellula goksoyri’ Pr1d, a planctomycete from deep-sea sediments [44].
Fig. 2.
16S rRNA gene-based maximum-likelihood tree showing the phylogenetic relationship of strain PX69T to other described Pirellula-related planctomycetes and some environmental clone sequences. Habitats are coded by colored triangles. The clade defined by strain PX69T and ‘Bythopirellula goksoyri’ Pr1dT is depicted with a lilac background. Bootstrap values are depicted as circles (60–100%). The root was composed of nine 16S rRNA gene sequences from 9 described members of the family Pirellulaceae (LWSK01000083, LWSJ01000078, JF748734,JF748733, HQ845500, HE861893, EF589351, CP001848, BX294149). Tree scale, 0.1 substitutions per nucleotide position.
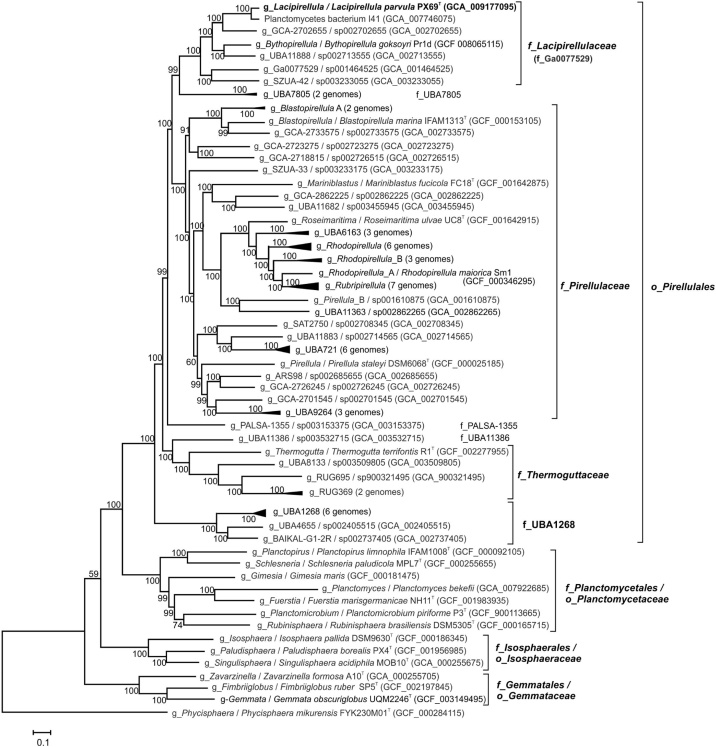
To determine the genome-based phylogenetic position of strain PX69T within the Planctomycetes, a phylogenetic tree based on concatenated sequences of conservative marker genes was constructed. This placed strain PX69T and ‘Bythopirellula goksoyri’ Pr1d within the candidate family Ga0077529 of the order ‘Pirellulales’, as defined by the GTDB taxonomy (Fig. 3). This candidate family also included the taxonomically uncharacterized Planctomycetes bacterium I41, which was isolated from a freshwater pond in Northern Germany, and four other uncultured bacteria. The AAI value between strain PX69T and its closest relative with determined genome sequence, Planctomycetes bacterium I41, was 77.3%. According to AAI thresholds recently proposed for taxonomic delineation (i.e. 45–65% for the same family, 65–95% for the same genus and 95–100% for the same species, [18]), these two bacteria represented different species in one genus. The differences in AAI values between strain PX69T and five other members of the candidate family Ga0077529 were in the range of 54–59% (Table 3). We, therefore, propose to place strain PX69T in a novel genus and species, Lacipirellula parvula, which belongs to a novel family, Lacipirellulaceae fam. nov. According to the pairwise comparison of AAI values (Table 3) and genome-based phylogeny (Fig. 3), five other genera in addition to Lacipirellula, those represented by ‘Bythopirellula’, UBA11888, GCA-2702655, Ga0077529 and SZUA-42, likely comprise the novel Lacipirellulaceae family. The characteristics that distinguish strain PX69T and ‘Bythopirellula goksoyri’ Pr1d are displayed in Table 2.
Fig. 3.
Genome-based phylogeny of the class Planctomycetacia defined in the GTDB taxonomy. A selected part of the GTDB-Tk multiple alignment was used for tree construction in PhyML v. 3.3 with default parameters. The tree was inferred from the concatenation of 120 conserved bacterial marker genes. The support values for the internal nodes were estimated by approximate Bayes tests in PhyML. Taxonomy is shown according to the GTDB (o_, order; f_, family; g_, genus). Phycisphaera mikurensis, a member of the class Phycisphaerae, was used to root the tree.
Table 3.
Pairwise AAI determined for members of the family Lacipirellulaceae.
| Genus / species | Lacipirellula parvula P X 69 | Planctomycetes bacterium I41 | g_GCA-2702655/sp002702655 | Bythopirellula goksoyri Pr1d | g_UBA11888/sp002713555 | g_Ga0077529/sp001464525 | g_SZUA-42/sp003233055 |
|---|---|---|---|---|---|---|---|
| Lacipirellula parvula PX69 | 100% | 77.3 | 58.6 | 56.4 | 56.4 | 54.0 | 54.4 |
| Planctomycetes bacterium I41 | 77.3 | 100% | 59.1 | 56.7 | 56.2 | 54.5 | 54.5 |
| g_GCA-2702655/sp002702655 | 58.6 | 59.1 | 100% | 57.4 | 56.5 | 53.9 | 55.2 |
| Bythopirellula goksoyri Pr1d | 56.4 | 56.7 | 57.4 | 100% | 60.1 | 53.2 | 55.1 |
| g_UBA11888/sp002713555 | 56.4 | 56.2 | 56.5 | 60.1 | 100% | 52.2 | 54.7 |
| g_Ga0077529/sp001464525 | 54.0 | 54.5 | 53.9 | 53.2 | 52.2 | 100% | 56.0 |
| g_SZUA-42/sp003233055 | 54.4 | 54.5 | 55.2 | 55.1 | 54.7 | 56.0 | 100% |
Table 2.
Major characteristics that distinguish strain PX69T and ‘Bythopirellula goksoyri’ Pr1dT[44].
| Characteristic | Strain PX69T | ‘Bythopirellula goksoyri’ |
|---|---|---|
| Habitat | Freshwater | Seawater |
| Cell size (μm) | 0.5–1.4 | 0.5–1.5 |
| Distribution of crateriform structures | On reproductive cell pole only | Around both cell poles |
| Anaerobic metabolism | + | n.d. |
| pH growth range (optimum) | 5.0–7.5 (6.5) | 4.0–6.8 (5.5–6.0) |
| Major fatty acids | C18:1ω9c, C16:0, C16:1ω7c | C16:1ω7, C16:0, C18:1ω9, C18:0 |
| Carbon sources: | ||
| Glucose | + | – |
| Cellobiose | + | – |
| Gelatin | + | – |
| Starch | – | + |
| DNA G + C content, mol% | 61.7 | 52.8 |
n.d., not determined.
Although the phylogenetic boundaries of the putative order ‘Pirellulales’ have been suggested by the GTDB taxonomy, this order has never been formally proposed and, till now, Pirellula-like planctomycetes belong to the order Planctomycetales. The major reason that precluded taxonomic re-arrangements within the order Planctomycetales was the absence of information regarding the phylogenetic position of Planctomyces bekefii, the type genus and species of the family Planctomycetaceae and the order Planctomycetales [37]. The recent identification of Planctomyces bekefii [9] now opens the way to a taxonomic rearrangement of the Planctomycetales. According to genome-based phylogeny, the latter should be divided into four orders, i.e. Planctomycetales, Pirellulales, Gemmatales and Isosphaerales (Fig. 3). The order Planctomycetales includes a single family, Planctomycetaceae, which accommodates genera Planctomyces, Gimesia, Planctopirus, Planctomicrobium, Rubinisphaera, Schlesneria and ‘Fuerstella’. The newly proposed order Pirellulales includes the families Pirellulaceae, Thermoguttaceae, Lacipirellulaceae, and four other family-level lineages still lacking cultivated members (Fig. 3). Finally, the orders Gemmatales and Isosphaerales include one family each, i.e. Gemmataceae and Isosphaeraceae. Descriptions of Lacipirellula gen. nov. and Lacipirellula parvula sp. nov. are given in Table 4. Descriptions of the newly proposed families and orders are given below.
Table 4.
Descriptions of Lacipirellula gen. nov. and Lacipirellula parvula sp. nov.
| Genus name | Lacipirellula | |
| Species name | Lacipirellula parvula | |
| Genus status | gen. nov | |
| Genus etymology | La.ci.pi.rel’lu.la. L. masc. n. lacus lake; N.L. fem. n. Pirellula a bacterial genus; N.L. fem. n. Lacipirellula a lake-dwelling Pirellula | |
| Type species of the genus | Lacipirellula parvula | |
| Specific epithet | – | parvula |
| Species status | – | sp. nov |
| Species etymology | – | par'vu.la. L. fem. adj. parvula small |
| Description of the new taxon and diagnostic traits | Gram-stain negative, ellipsoidal cells that occur singly or in aggregates. Reproduce by budding; daughter cells are motile by means of a polar flagellum. Mature cells are non-motile. Free cell poles are covered with crateriform pits. Cells occurred singly, in pairs, in rosettes composed of 3 cells or in aggregates. Stalks are not formed. Non-pigmented. Oxidase- and catalase-positive. Mesophilic. Chemoorganotrophic. Sugars are the preferred growth substrates. Capable of hydrolyzing several polysaccharides but not cellulose or chitin. Micro-aerobic and facultatively anaerobic. Anaerobic growth occurs by means of fermentation. The major fatty acids are C16:0, C18:1ɷ9c, and C16:1ɷ7c. The major polar lipid is dimethylphosphatidylethanolamine. The genus belongs to the family Lacipirellulaceae of the order Pirellulales. | Colonies are small (0.5–1 mm in diameter), circular and unpigmented. The cells are 0.5-0.9 μm wide and 0.9–1.4 μm long. The temperature range for growth is 10–30 °C with the optimum at 20–25 °C. The pH range for growth is 5.0–7.5 with the optimum at 6.5. Growth does not occur at NaCl concentrations above 0.5%. Do not require sea salt (ASW). Facultatively anaerobic, capable of microaerobic growth. Growth under oxic and micro-oxic conditions occurs on glucose, fructose, galactose, lactose, cellobiose, maltose, mannose, melibiose, melezitose, rhamnose, ribose, trehalose, sucrose, xylose, raffinose, N-acetylglucosamine, pyruvate, salicin, arbutin, glycerol, tween 80, glucuronic acid, lactate, pyruvate, glycerin, sorbitol and dulcitol. Capable of hydrolyzing aesculin, arabinogalactan, chondroitin sulfate, dextrin, laminarin, locust bean gum, gelatin, starch, xanthan gum, pectin and xylan. Cannot utilize fructose, raffinose, sorbose, ribose, methanol, acetate, benzoate, caproate, citrate, fumarate, glutarate, malate, propionate, mannitol, tartrate, alanine, arginine, asparagine, aspartate, cysteine, cystine, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, norleucine, ornithine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine or valine. Cannot hydrolyze casein, chitosan, chitin, starch, carboxymethylcellulose and cellulose. Grow anaerobically by fermentation of glucose, lactose, trehalose, sucrose, and xanthan gum. The end product of glucose fermentation is lactate. Urease is negative. Dissimilatory nitrate reduction is positive (API 20NE strip). In the API ZYM strip, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase are present but trypsin, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-galactosidase, α-fucosidase, lipase (C4), cystine arylamidase, α-chymotrypsin, β-glucuronidase and α-mannosidase are absent. The major fatty acids are C18:1ω9c, C16:0 and 16:1ω7c. The major polar lipid is dimethylphosphatidylethanolamine. Resistant to ampicillin, carbenicillin, chloramphenicol, gentamicin, neomycin, kanamycin, streptomycin, but sensitive to rifampicin and tetracycline. |
| Country of origin | – | Russian Federation |
| Region of origin | – | Vologda region |
| Date of isolation | – | 25/10/2017 |
| Source of isolation | – | Boreal lake water |
| Sampling date | – | 08/2017 |
| Latitude | – | 58°43′28.5ʺN |
| Longitude | – | 37°39′07ʺE |
| Genome accession number | – | GenBank: AP021861 |
| Genome status | – | complete |
| Genome size | – | 6.92 |
| GC mol% | – | 61.17 |
| Number of strains in study | – | 1 |
| Information related to the Nagoya Protocol | – | Not applicable |
| Designation of the Type Strain | – | PX69T |
| Strain Collection Numbers | – | KCTC 72398T = CECT 9826T = VKM B-3335T |
Description of Lacipirellulaceae fam. nov
Lacipirellulaceae (La.ci.pi.rel.lu.la.ce´ae. N.L. fem. n. Lacipirellula type genus of the family; -aceae ending to denote a family; N.L. fem. pl. n. Lacipirellulaceae the Lacipirellula family). Gram-stain negative, budding bacteria with oval or ellipsoidal cells, which occur singly, in pairs, in rosettes composed of 3 cells or in aggregates. Stalks are not formed. Crateriform pits are located around free cell poles. Daughter cells are motile, while mother cells are non-motile. Aerobes and facultative anaerobes. Neutrophilic and acidotolerant mesophiles. Chemoorganotrophs. DNA G + C content is in the range 52–62 mol%. Inhabit a wide variety of low-oxygen aquatic and terrestrial habitats. The type genus is Lacipirellula. Another described genus in this family is ‘Bythopirellula’. The family belongs to the order Pirellulales of the class Planctomycetacia.
Description of Pirellulaceae fam. nov
Pirellulaceae (Pi.rel.lu.la.ce´ae. N.L. fem. n. Pirellula type genus of the family; -aceae ending to denote a family; N.L. fem. pl. n. Pirellulaceae the Pirellula family). Gram-stain negative, budding bacteria with ovoid, ellipsoidal, pear- or teardrop-shaped cells. Cells do not possess a stalk, but may form a fibrillar holdfast. Frequently form rosettes by attachment at the narrow cell pole. Crateriform pits are located around free cell poles. Daughter cells are motile in most species, while mother cells are non-motile. Chemoorganotrophs. Aerobes and facultative anaerobes. Main habitats are marine and freshwater environments. The type genus is Pirellula. Other described genera in this family are Blastopirellula, Rhodopirellula, Rubripirellula, Roseimaritima and Mariniblastus. The family belongs to the order Pirellulales of the class Planctomycetacia.
Description of Thermoguttaceae fam. nov
Thermoguttaceae (Ther.mo.gut.ta.ce´ae. N.L. fem. n. Thermogutta type genus of the family; -aceae ending to denote a family; N.L. fem. pl. n. Thermoguttaceae the Thermogutta family). Gram-stain negative, budding bacteria with oval cells that occur singly or in aggregates. Daughter cells are motile; mature cells are non-motile. Thermophilic and neutrophilic. Facultatively anaerobic. Chemoorganotrophic. Oxidize organic substrates with external electron acceptors. Grow by fermentation of mono-, di- and polysaccharides. DNA G + C content is in the range 57−67 mol%. The type genus is Thermogutta. Another described genus in this family is Thermostilla. The family belongs to the order Pirellulales of the class Planctomycetacia.
Emended description of the family Planctomycetaceae Schlesner and Stackebrandt 1987, 179VP, emend. Ward 2010
Gram-stain-negative, budding bacteria. Cell shape may be tear-drop to pear-shaped, spherical to ovoid, or bulbiform. Mature cells may have multifibrillar appendages described as stalks, spikes, spines, fimbria, or bristles. Holdfasts may be present at the distal end of the stalk. Formation of rosettes may occur. Crateriform structures are present on a reproductive pole of mature cells. Daughter cells may be motile by means of one or two flagella. Most axenic cultures are aerobic or facultatively anaerobic chemo-organotrophs, with carbohydrates serving as preferred carbon sources. The type genus is Planctomyces. Other genera in the family are Gimesia, Planctopirus, Planctomicrobium, Rubinisphaera, Schlesneria and ‘Fuerstella’. The family belongs to the order Planctomycetales of the class Planctomycetacia.
Description of Pirellulales ord. nov
Pirellulales (Pi.rel.lu.la´les. N.L. fem. n. Pirellula type genus of the order; -ales ending to denote an order; N.L. fem. pl. n. Pirellulales the Pirellula order). Gram-stain-negative, budding, chemoheterotrophic bacteria with ovoid, ellipsoidal, pear- or teardrop-shaped cells. The type genus is Pirellula. The order contains the families Pirellulaceae, Lacipirellulaceae and Thermoguttaceae, and belongs to the class Planctomycetacia.
Description of Gemmatales ord. nov
Gemmatales (Gem.ma.ta´les. N.L. fem. n. Gemmata type genus of the order; -ales ending to denote an order; N.L. fem. pl. n. Gemmatales the Gemmata order). Gram-stain negative, budding, chemoheterotrophic bacteria with spherical or ellipsoidal cells. The type genus is Gemmata. The order contains the single family Gemmataceae and belongs to the class Planctomycetacia.
Description of Isosphaerales ord. nov
Isosphaerales (I.so.sphae.ra´les. N.L. fem. n. Isosphaera type genus of the order; -ales ending to denote an order; N.L. fem. pl. n. Isosphaerales the Isosphaera order). Gram-stain negative, budding, chemoheterotrophic bacteria with spherical cells. Aerobes or micro-aerobes. The type genus is Isosphaera. The order contains the single family Isosphaeraceae and belongs to the class Planctomycetacia.
Emended description of the order Planctomycetales Schlesner and Stackebrandt 1987, 179VP, emend. Ward 2010
The description is the same as for the family Planctomycetaceae, which is the single family in this order. The type genus is Planctomyces. The order belongs to the class Planctomycetacia.
Funding information
This study was supported by the Russian Science Foundation (project No 16-14-10210). WICR and JSSD received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 694569).
Acknowledgements
The authors thank N.E. Suzina for electron microscopy analysis.
Footnotes
The annotated genome sequence of strain PX69T has been deposited in NCBI GenBank under the accession number AP021861.
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.syapm.2019.126050.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Beier S., Bertilsson S. Bacterial chitin degradation — mechanisms and ecophysiological strategies. Front. Microbiol. 2013;4:149. doi: 10.3389/fmicb.2013.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bondoso J., Albuquerque L., Lobo-da-Cunha A., da Costa M.S., Harder J., Lage O.M. Rhodopirellula lusitana sp. nov. and Rhodopirellula rubra sp. nov., isolated from the surface of macroalgae. Syst. Appl. Microbiol. 2014;37:157–164. doi: 10.1016/j.syapm.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Bondoso J., Albuquerque L., Nobre M.F., Lobo-da-Cunha A., da Costa M.S., Lage O.M. Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst. Appl. Microbiol. 2015;38(1):8–15. doi: 10.1016/j.syapm.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Bondoso J., Godoy-vitorino F., Gasol J.M., Harder J., Lage O.M., Juan S., Lage O.M. Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol. Ecol. 2017;93(3) doi: 10.1093/femsec/fiw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borisov V.B., Gennis R.B., Hemp J., Verkhovsky M.I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta Bioenerg. 2011;1807(11):1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., Shukla M., Thomason J.A., Stevens R., Vonstein V., Wattam A.R., Xia F. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brümmer I.H.M., Felske A.D.M., Wagner-Döbler I. Diversity and seasonal changes of uncultured Planctomycetales in river biofilms. Appl. Environ. Microbiol. 2004;70(9):5094–5101. doi: 10.1128/AEM.70.9.5094-5101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chouari R., Le Paslier D., Daegelen P., Ginestet P., Weissenbach J., Sghir A. Molecular evidence for novel planctomycete diversity in a municipal wastewater treatment plant. CEUR Workshop Proc. 2015;1542(12):33–36. doi: 10.1128/AEM.69.12.7354-7363.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedysh S.N., Henke P., Ivanova A.A., Kulichevskaya I.S., Philippov D.A., Meier-Kolthoff J.P., Göker M., Huang S., Overmann J. 100-year-old enigma solved: identification, genomic characterization and biogeography of the yet uncultured Planctomyces bekefii. Environ. Microbiol. 2020 doi: 10.1111/1462-2920.14838. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst J.A. The Planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology. 1995;141(7):1493–1506. doi: 10.1099/13500872-141-7-1493. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Horsman J.A., Barquera B., Rumbley J., Ma J., Gennis R.B. The superfamily of heme-copper respiratory oxidases. J. Bacteriol. 1994;176(18):5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerhardt P. American Society for Microbiology; 1981. Manual of Methods for General Bacteriology. [Google Scholar]
- 13.Glöckner F.O., Kube M., Bauer M., Teeling H., Lombardot T., Ludwig W., Gade D., Beck A., Borzym K., Heitmann K., Rabus R., Schlesner H., Amann R., Reinhardt R. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. U. S. A. 2003;100(14):8298–8303. doi: 10.1073/pnas.1431443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 15.Ivanova A.A., Philippov D.A., Kulichevskaya I.S., Dedysh S.N. Distinct diversity patterns of Planctomycetes associated with the freshwater macrophyte Nuphar lutea (L.) Smith. Antonie Van Leeuwenhoek. 2018;111:811–823. doi: 10.1007/s10482-017-0986-4. [DOI] [PubMed] [Google Scholar]
- 16.Ivanova A.O., Dedysh S.N. Abundance, diversity, and depth distribution of Planctomycetes in acidic northern wetlands. Front. Microbiol. 2012;3:5. doi: 10.3389/fmicb.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kertesz M.A. Riding the sulfur cycle — metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 2000;24(2):135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 18.Konstantinidis K.T., Rosselló-Móra R., Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulichevskaya I.S., Ivanova A.A., Detkova E.N., Rijpstra W.I.C., Sinninghe Damsté J.S., Dedysh S.N. Tundrisphaera lichenicola gen. nov., sp. nov., a psychrotolerant representative of the family Isosphaeraceae from lichen-dominated tundra soils. Int. J. Syst. Evol. Microbiol. 2017;67(9):3583–3589. doi: 10.1099/ijsem.0.002172. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(May):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lage O.M., Albuquerque L., Cunha A.L., Costa M.S. da. Mariniblastus fucicola gen. nov., sp. nov. a novel planctomycete associated with macroalgae. Int. J. Syst. Evol. Microbiol. 2017;67(5):1571–1576. doi: 10.1099/ijsem.0.001760. [DOI] [PubMed] [Google Scholar]
- 22.Lee H.W., Roh S.W., Shin N.R., Lee J., Whon T.W., Jung M.J., Yun J.H., Kim M.S., Hyun D.W., Kim D., Bae J.W. Blastopirellula cremea sp. nov., isolated from a dead ark clam. Int. J. Syst. Evol. Microbiol. 2013;63:2314–2319. doi: 10.1099/ijs.0.044099-0. [DOI] [PubMed] [Google Scholar]
- 23.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17.1:10–12. [Google Scholar]
- 25.Moore E.K., Hopmans E.C., Rijpstra W.I.C., Villanueva L., Dedysh S.N., Kulichevskaya I.S., Wienk H., Schoutsen F., Sinninghe Damsté J.S. Novel mono-, di-, and trimethylornithine membrane lipids in northern wetland planctomycetes. Appl. Environ. Microbiol. 2013;79(22):6874–6884. doi: 10.1128/AEM.02169-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.A., Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 27.Pollet T., Tadonléké R.D., Humbert J.-F. Comparison of primer sets for the study of Planctomycetes communities in lentic freshwater ecosystems. Environ. Microbiol. Rep. 2011;3(2):254–261. doi: 10.1111/j.1758-2229.2010.00219.x. [DOI] [PubMed] [Google Scholar]
- 28.Poole R.K., Cook G.M. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv. Microb. Physiol. 2000;43:165–224. doi: 10.1016/s0065-2911(00)43005-5. [DOI] [PubMed] [Google Scholar]
- 29.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Refojo P.N., Sousa F.L., Teixeira M., Pereira M.M. The alternative complex III: a different architecture using known building modules. Biochim. Biophys. Acta Bioenerg. 2010;1797(12):1869–1876. doi: 10.1016/j.bbabio.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-R L., Konstantinidis K. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016;4 e1900v1. [Google Scholar]
- 32.Schlesner H. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst. Appl. Microbiol. 1994;17:135–145. [Google Scholar]
- 33.Schlesner H. Pirella marina sp. nov., a budding, peptidoglycan-less bacterium from brackish water. Syst. Appl. Microbiol. 1986;8:177–180. [Google Scholar]
- 34.Schlesner H., Hirsch P. Assignment of ATCC 27377 to Pirella gen. nov. as Pirella staleyi comb. nov. Int. J. Syst. Bacteriol. 1984;34:492–495. [Google Scholar]
- 35.Schlesner H., Hirsch P. Rejection of the genus name Pirella for pear-shaped budding bacteria and proposal to create the genus Pirellula gen. nov. Int. J. Syst. Bacteriol. 1987;37:441. [Google Scholar]
- 36.Schlesner H., Rensmann C., Tindall B.J., Gade D., Rabus R., Pfeiffer S., Hirsch P. Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int. J. Syst. Evol. Microbiol. 2004;54(5):1567–1580. doi: 10.1099/ijs.0.63113-0. [DOI] [PubMed] [Google Scholar]
- 37.Schlesner H., Stackebrandt E. Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov. Syst. Appl. Microbiol. 1986;8(3):174–176. [Google Scholar]
- 38.Shao K., Gao G. Soil microbial communities of three grassland ecosystems in the bayinbuluke, China. Can. J. Microbiol. 2018;64(3):209–213. doi: 10.1139/cjm-2017-0585. [DOI] [PubMed] [Google Scholar]
- 39.Sinninghe Damsté J.S., Rijpstra W.I.C., Hopmans E.C., Weijers J.W.H., Foesel B.U., Overmann J., Dedysh S.N. 13,16-Dimethyl octacosanedioic acid (iso-diabolic acid), a common membrane-spanning lipid of Acidobacteria subdivisions 1 and 3. Appl. Environ. Microbiol. 2011;77(12):4147–4154. doi: 10.1128/AEM.00466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slobodkina G.B., Kovaleva O.L., Miroshnichenko M.L., Slobodkin A.I., Kolganova T.V., Novikov Aa., van Heerden E., Bonch-Osmolovskaya Ea. Thermogutta terrifontis gen. nov., sp. nov. and Thermogutta hypogea sp. nov., novel thermophilic anaerobic representatives of the phylum Planctomycetes. Int. J. Syst. Evol. Microbiol. 2015;65(3):760–765. doi: 10.1099/ijs.0.000009. [DOI] [PubMed] [Google Scholar]
- 41.Slobodkina G.B., Panteleeva A.N., Beskorovaynaya D.A., Bonch-Osmolovskaya E.A., Slobodkin A.I. Thermostilla marina gen. nov., sp. nov., a thermophilic, facultatively anaerobic planctomycete isolated from a shallow submarine hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2016;66(2):633–638. doi: 10.1099/ijsem.0.000767. [DOI] [PubMed] [Google Scholar]
- 42.Staley J.T. Budding bacteria of the Pasteuria – Blastobacter group. Can. J. Microbiol. 1973;19(5):609–614. doi: 10.1139/m73-100. [DOI] [PubMed] [Google Scholar]
- 43.Staley J.T., Fuerst J.A., Giovannoni S., Schlesner H. The order Planctomycetales and the genera Planctomyces, Pirellula, Gemmata, and Isosphaera. In: Balows A., Trüper H.G., Dworkin M., Harder W., Schleifer K.-H., editors. The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications. Springer; New York, New York, NY: 1992. pp. 3710–3731. [Google Scholar]
- 44.Storesund J.E., Øvreås L. Diversity of planctomycetes in iron-hydroxide deposits from the arctic mid ocean ridge (AMOR) and description of Bythopirellula goksoyri gen. nov., sp. nov., a novel Planctomycete from deep sea iron-hydroxide deposits. Antonie Van Leeuwenhoek. 2013;104(4):569–584. doi: 10.1007/s10482-013-0019-x. [DOI] [PubMed] [Google Scholar]
- 45.Ward N.L. Phylum XXV. Planctomycetes Garrity and Holt 2001, 137 emend. Ward. In: Krieg N.R., Staley J.T., Brown D.R., Hedlund B.P., Paster B.J., Ward N.L., Ludwig W., Whitman W.B., editors. Bergey’s Manual® of Systematic Bacteriology: Volume Four The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes. Springer; New York, New York, NY: 2010. pp. 879–925. [Google Scholar]
- 46.Wegner C.E., Richter-Heitmann T., Klindworth A., Klockow C., Richter M., Achstetter T., Glöckner F.O., Harder J. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar. Genomics. 2013;9:51–61. doi: 10.1016/j.margen.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13(6) doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.