Abstract
Purpose
NUSAP1 has been reported to be involved in the progression of several types of cancer. However, its expression and exact role in bladder cancer (BLCA) remains elusive. The aim of this study was to determine the expression and role of NUSAP1 in BLCA.
Methods
Tissue microarray, real-time PCR, Western blot and immunohistochemistry assays were carried out to determine NUSAP1 expression in BLCA tissues and cells. The biological roles of NUSAP1 were investigated using CCK-8, EdU labeling, flow cytometry, Transwell, and wound healing assays. Additionally, the effect of NUSAP1 on epithelial-mesenchymal transition (EMT) was investigated by Western blotting and real-time PCR.
Results
We found that NUSAP1 was upregulated in BLCA, and its expression was closely related to the poor prognosis of patients. Subsequently, we transfected 5637 and T24 cell lines with NUSAP1 siRNA and an NUSAP1 overexpression plasmid, respectively. NUSAP1 downregulation in 5637 cells inhibited cell proliferation, migration, and invasiveness and enhanced chemosensitivity to gemcitabine, while NUSAP1 overexpression in T24 cells resulted in the inverse effects. Moreover, NUSAP1 regulated EMT via the TGF-β signaling pathway, and when TGF-beta receptor 1 (TGFBR1) was inhibited with the inhibitor SB525334, the invasion and metastasis ability of BLCA cells was significantly suppressed, as well as p-Smad2/3 and vimentin expression.
Conclusion
Our above data demonstrate that NUSAP1 contributes to BLCA progression via the TGF-β signaling pathway.
Keywords: NUSAP1, bladder cancer, tumor progression, epithelial-mesenchymal transition, TGF-β signaling pathway
Introduction
Bladder cancer (BLCA) is the tenth most common malignancy worldwide, with an estimated 80,470 new cancer cases in the United States in 2019.1 Such cancers may be caused by certain known risk factors, such as smoking, urinary tract infections and occupational hazards.2 Despite the advances in surgical methods, radiotherapy and chemotherapy, the 5-year survival rate and prognosis of BLCA have only slightly improved in recent years. Therefore, finding a more effective therapeutic strategy to prevent BLCA progression and recurrence is urgently needed.
NUSAP1 is a protein with a molecular weight of 55 kDa that plays key roles in mitotic progression, spindle formation and stability, which makes it an important regulator of mitosis and cell proliferation.3–5 Increased expression of NUSAP1 has been observed in many tumors, such as prostate cancer, acute myeloid leukemia, cervical cancer, oral squamous cell carcinoma, and astrocytoma, and NUSAP1 has been found to be closely associated with tumor progression.6–10 However, the potential relationship between NUSAP1 expression in BLCA and clinical outcomes remains unknown and requires further investigation.
In the present study, we investigated NUSAP1 expression in BLCA tissues and cell lines and found that NUSAP1 was highly expressed in BLCA. NUSAP1 plays an important role in cell proliferation, cell cycle regulation, apoptosis, migration, invasion and chemoresistance. Moreover, we found that NUSAP1 could regulate EMT through the TGF-β signaling pathway. Therefore, these results demonstrated that NUSAP1 might be a potential target for the diagnosis and treatment of BLCA.
Materials and Methods
Data Collection
The mRNA expression profile matrix files for GSE3167 were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). GSE3167 data, which comprised 14 normal bladder tissues and 46 bladder cancer tissues, were collected on the Affymetrix Human Genome U133A Array platform.11
The mRNA sequencing expression profile and clinical data from patients with BLCA were obtained from the Cancer Genome Atlas (TCGA) data portal (https://gdc-portal.nci.nih.gov/). Only 19 matched cases of BLCA were included in our study. Additionally, our research followed TCGA access rules and publication guidelines.
Patients and Tissue Specimens
Fresh BLCA tissues and tumor-adjacent tissues were obtained from patients who underwent radical cystectomy at the Department of Urology of the First Affiliated Hospital of Chongqing Medical University. This study was approved by the Human Research Ethics Committee of the First Affiliated Hospital of Chongqing Medical University, the written informed consent was obtained from all patients which was conducted in accordance with the Declaration of Helsinki.
Cell Culture and Treatment
The human BLCA cell lines 5637, T24, BIU87, UM-UC-3, and RT4 and the human ureteric epithelial cell line SV-HUC-1 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The 5637, T24 and BIU87 cells were grown in RPMI 1640 medium (Corning Incorporated, Corning, NY, USA) containing 10% fetal bovine serum (FBS, Gibco, Thermo Scientific, Waltham, MA, USA). UM-UC-3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Corning Incorporated, Corning, NY, USA) containing 10% FBS, RT4 cells were grown in McCoy’s 5A (modified) medium (Boster Biological Technology, Wuhan, China) containing 10% FBS, SV-HUC-1 cells were grown in F-12K medium (Thermo Scientific, Waltham, MA, USA) containing 10% FBS, and all cells were maintained in medium supplemented with 1% penicillin/streptomycin. Furthermore, all cells were maintained at 37 °C in a humidified incubator with 5% CO2. Cell transfection was performed with Lipofectamine 2000 (Invitrogen, Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s protocol; 5637 and T24 cells were seeded into six-well plates (5–8×104 cells per well), and after the cells reached 60% to 80% confluency, the medium was exchanged with 1.5 mL of basal medium containing 500 μL of Lipofectamine 2000 mix and 0.2 nmol of siRNA or 5 μg of DNA. Small interfering RNA (siNUSAP1; siRNA1, 5′-CCACUGAAUUCCAGAAUCATTUGAUUCUGGAAUUCAGUGGTT-3′; siRNA2, 5′-CUGCUACUAAAGAUAAUGATTUCAUUAUCUUUAGUAGCAGTT-3′; siRNA3, 5′-CAACAUGUCAACAGAAUUATTUAAUUCUGUUGACAUGUUGTT-3′), a negative control (NC) sequence (5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′) and pcDNA3.1-NUSAP1 and pcDNA3.1 vectors were purchased from GenePharma (Shanghai, China). An inhibitor of TGF‐β receptor 1, sb525334 (10 μmol/L, Selleck Chemicals, Houston, TX, USA), was used to treat transfected T24 cell lines (NC/NUSAP1) to inhibit the TGF-β signaling pathway. A recombinant human TGF-β1 (20 ng/mL; R&D System, Minneapolis, MN) was used to treat transfected 5637 cell lines (NC/siNUSAP1) to activate the TGF-β signaling pathway.
Gene Set Enrichment Analysis (GSEA)
To identify NUSAP1-related biological processes and pathways, GSEA was performed using a TCGA dataset that included 414 patients with BLCA divided into high and low expression groups according to the median value; the analysis was executed by GSEA software 3.0 from the Broad Institute.12 The gene sets of canonical pathways (c2.all.v6.0.symbols.gmt) were obtained from the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). Gene set permutations were performed 1000 times for each analysis to obtain a normalized enrichment score (NES), which was used for sorting pathways enriched in each phenotype. A result was regarded as significant when the nominal P-value was <0.05 and the false discovery rate (FDR) was <0.2.
Cell Proliferation Assay
Cell proliferation was analyzed via Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Inc.) assays. Forty-eight hours after transfection, 4 × 103 T24 cells or 4.5 × 103 5637 cells were seeded into 96-well flat-bottomed microplates. CCK-8 reagent was added after 24, 48, and 72 h of culture, and absorbance was measured at 450 nm. Similarly, a Cell-Light EdU Apollo 567 in vitro kit (cat. no. C10310-1; Guangzhou RiboBio Co., Ltd.) was used to assess cell proliferation ability according to the manufacturer’s protocol. Images were captured with a fluorescence microscope (Olympus Corporation).
RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR
Total RNA was extracted from cell lines and tissues with TRIzol® reagent (Takara Biotechnology Co., Ltd., China), and cDNA samples were synthesized using random primers and a Reverse Transcriptase PCR kit (Takara Biotechnology Co., Ltd., China) according to the manufacturer’s protocols. NUSAP1 mRNA expression and its correlation with TGF-beta receptor 1 expression were assessed by real-time PCR. β-actin expression was used as the internal control. The following primers were used: human NUSAP1, forward, 5ʹ-CGTCCCCTCAACTATGAACCAC-3ʹ, reverse, 5ʹ-GCGTTTCTTCCGTTGCTCTT-3ʹ; β-actin, forward, 5ʹ-CCACGAAACTACCTTCAACTCC-3ʹ, reverse, 5ʹ-GTGATCTCCTTCTGCATCCTGT-3ʹ; TGFBR1, forward, 5ʹ-ACATGATTCAGCCACAGATACC-3ʹ, reverse, 5ʹ-GCATAGATGTCAGCACGTTTG-3ʹ.
Western Blotting Analysis
Total proteins were extracted from BLCA tissue samples or cells using RIPA lysis buffer (Beyotime Institute of Biotechnology, Jiangsu, China) containing protease inhibitors. The protein concentrations were determined using a BCA Protein Assay Kit (Beyotime Institute of Biotechnology, Jiangsu, China). Proteins (30 μg) were separated on 10% SDS-PAGE gels and then transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% skim milk for 1 h and then incubated overnight at 4 °C with primary antibodies against NUSAP1 and TGFBR1 (cat. nos. ab137230 and ab31013, respectively; 1:1000 dilution for each; Abcam); GAPDH (cat. no. AB0037, 1:5000 dilution; Abways); E-cadherin, vimentin, N-cadherin, CDK1, cyclin B1, Smad2/3, P-Smad2/3, Bax, Bcl2 and cleaved-caspase3 (cat. nos. 3195T, 5741T, 13116T, 9116T, 12231T, 8685T, 8828s, 5023T, 3498T and 9964T, respectively; 1: 1000 dilution for each; Cell Signaling Technology). After washing with TBST three times, the membranes were incubated with secondary antibodies conjugated to HRP (Cell Signaling Technology, USA) for 1 h at room temperature. After washing with TBST three times, the bands were visualized using an ECL system (Bio-Rad Laboratories). The relative protein expression was analyzed using Quantity One software (Bio-Rad).
Immunohistochemistry (IHC)
Tissue sections were dewaxed and rehydrated in 100, 95, 85, 75 and 50% ethanol, boiled at 100 °C in 0.01 mol/L sodium citrate buffer (pH 6.0) for 15 min and incubated with 3% hydrogen peroxide for 20 min. Subsequently, the samples were blocked with normal goat serum at 37 °C for 30 min and then incubated with an anti-NUSAP1 antibody (cat no. 12024-1-AP, Proteintech, Wuhan, China) at 4 °C overnight; next, the samples were incubated for 30 min at 37 °C with a horseradish peroxidase (HRP)-conjugated secondary antibody (Zhonghshan Golden Bridge Biotechnology, Beijing, China). Immunostaining was performed using a DAB Detection Kit (Polymer) (Zhonghshan Golden Bridge Biotechnology, Beijing, China), and counterstaining was performed with hematoxylin. Scoring criteria were based on the intensity and extent of section staining (0, negative; 1, weak; 2, moderate; and 3, strong) and the percentage of stained tumor cells (<10% = 0, 10–25% = 1, 26–50% = 2, 51–75% = 3, 76–100% = 4). The sections with a score of 1–4 were defined as having low NUSAP1 expression, while the sections with a score of 5–12 were defined as having high NUSAP1 expression. The scores were confirmed independently by two pathologists.
Flow Cytometry
Cell cycle and apoptosis assays were performed 48 h after cells were transfected. For cell cycle analysis, the transfected cells were trypsinized and fixed in 75% ethanol at 4 °C overnight and then stained with propidium iodide (PI) at room temperature for 30 min. For apoptosis analysis, the transfected cells were trypsinized and resuspended in PBS, and the cells were then stained using an Annexin V-fluorescein isothiocyanate/PI kit (cat. no. 556570, FITC Annexin V Apoptosis Detection Kit II, BD Biosciences) according to the manufacturer’s protocol. The stained cells were analyzed by flow cytometry.
Wound Healing Assay
At 48 h post-transfection, cells were grown to confluence in 6-well plates. Six wounds were made in each well with a sterile pipette tip. The wells were washed with PBS and then cultured in RPMI-1640. After 0 or 24 h of incubation, the migration status was assessed by photographing the same location in the wells. Then, the area of the healed wound was compared with the area of the initial wound.
Cell Migration and Invasion Assays
Cell migration and invasion ability were evaluated with Transwell chambers (Corning Incorporated, Corning, NY, USA). For the migration assay, cells were harvested at 48 h post-transfection and then resuspended in serum-free RPMI 1640; cells (5 × 104/100 µL) were loaded into the upper chamber. The lower chambers contained 600 μL of medium with 10% FBS. The incubation time was 24 h for the T24 cell line and 48 h for the 5637 cell line. After washing with PBS, the cells on the upper surface of the chamber were removed with a cotton swab. The cells on the lower surface of the membrane were fixed and stained with a 0.5% crystal violet solution. Similarly, the invasion assay was performed almost identically to the migration assay except that a Transwell chamber with Matrigel was used instead.
Statistical Analysis
The data are expressed as the mean ± standard deviation (SD). GraphPad Prism 7 software (GraphPad Software, Inc., San Diego, CA) and SPSS 22.0 (IBM Corporation, Armonk, NY) were used for statistical analyses. T-tests or analysis of variance was used for statistical comparisons. *p <0.05 was considered statistically significant.
Results
Expression of NUSAP1 in BLCA
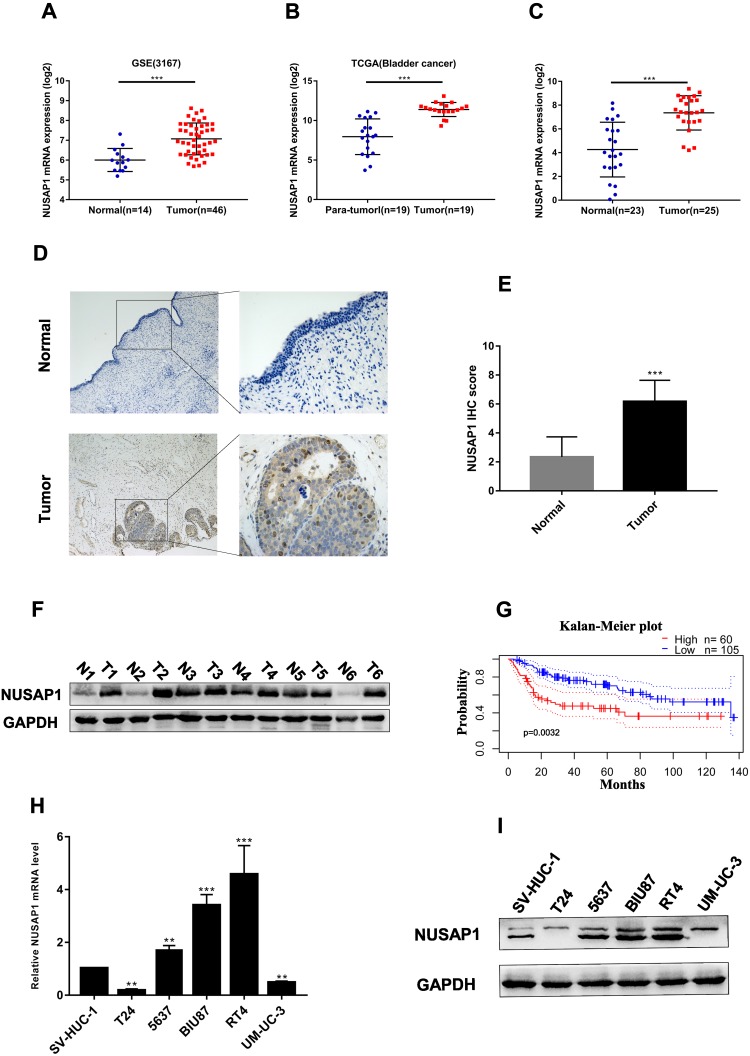
To explore the expression levels of NUSAP1 in BLCA, we analyzed several BLCA RNA sequencing datasets from TCGA and GEO and found that NUSAP1 mRNA levels were dramatically higher in BLCA tissues than in adjacent normal bladder tissues (Figure 1A and B). We further measured NUSAP1 expression levels in the BLCA tissues and normal bladder tissues in this study through real-time PCR, IHC and Western blot analysis, as shown in Figure 1C–E. NUSAP1 was significantly upregulated in BLCA tissues compared with normal bladder tissues (the clinical characteristics of the patients are listed in Table 1). Western blotting showed that NUSAP1 was upregulated in 6 paired BLCA tissues (Figure 1F). Subsequently, a PrognoScan online analysis demonstrated that patients with high NUSAP1 expression had shorter 5-year metastasis-free survival rates than those with low NUSAP1 expression (Figure 1G). Consistently, we also found through real-time PCR and Western blotting that NUSAP1 expression levels were higher in most BLCA cells than in normal ureteric epithelial SV-HUC-1 cells (Figure 1H and I). Therefore, these results indicate that NUSAP1 expression is significantly upregulated in BLCA and that high NUSAP1 expression predicts a poor prognosis in BLCA patients.
Figure 1.
NUSAP1 expression in BLCA tissues and cell lines and survival analysis. (A). NUSAP1 mRNA expression levels in 46 BLCA tumors and 14 normal tissues extracted from GSE3167. ***P<0.001 vs Normal. (B). Nineteen paired BLCA tumors and normal tissues extracted from the TCGA data portal. ***P<0.001 vs Normal. (C). NUSAP1 mRNA expression levels in 25 BLCA tumors and 23 normal tissues obtained from the Department of Urology of the First Affiliated Hospital of Chongqing Medical University. ***P<0.001 vs Normal. (D and E). NUSAP1 protein expression in BLCA and normal tissues as detected by immunohistochemistry (100× and 400× magnification). ***P<0.001 vs Normal. (F). NUSAP1 protein expression in 6 paired BLCA tissues. (G). Survival analysis of patients with BLCA obtained from PrognoScan (GSE13507). (H and I). The expression of NUSAP1 in BLCA cell lines was detected by real-time PCR and Western blotting. **P<0.01, ***P<0.001 vs SV-HUC-1 cells.
Table 1.
Clinicopathological Characteristics of Patients with BLCA
| Case No. | Age, Years | Sex | Diagnosis | TNM Stage |
|---|---|---|---|---|
| 1 | 76 | Male | BLCA | T2bN0M0 |
| 2 | 57 | Male | BLCA | T2bN0M0 |
| 3 | 71 | Male | BLCA | T3bN0M0 |
| 4 | 60 | Female | BLCA | T2N0M0 |
| 5 | 71 | Male | BLCA | T1N0M0 |
| 6 | 83 | Male | BLCA | T2bN0M0 |
| 7 | 65 | Male | BLCA | T2bNxM0 |
| 8 | 71 | Male | BLCA | T1N0M0 |
| 9 | 59 | Male | BLCA | T1N0M0 |
| 10 | 69 | Female | BLCA | T2bN0M0 |
| 11 | 33 | Male | BLCA | T2NxM0 |
| 12 | 67 | Male | BLCA | T2bN0M0 |
| 13 | 57 | Male | BLCA | T2N0M0 |
| 14 | 71 | Male | BLCA | T1N0M0 |
| 15 | 69 | Female | BLCA | T2bN0M0 |
| 16 | 68 | Male | BLCA | T4aNxMx |
| 17 | 72 | Female | BLCA | T2N0M0 |
| 18 | 48 | Male | BLCA | T2N0M0 |
| 19 | 58 | Male | BLCA | T2aN0M0 |
| 20 | 61 | Female | BLCA | T2N0M0 |
| 21 | 62 | Male | BLCA | T2bN0M0 |
| 22 | 63 | Male | BLCA | T2bN0M0 |
| 23 | 81 | Male | BLCA | T2bN0M0 |
| 24 | 57 | Male | BLCA | T1N0M0 |
| 25 | 79 | Female | BLCA | T2N0M0 |
| 26 | 51 | Male | BLCA | T2N0M0 |
| 27 | 69 | Male | BLCA | T2N0M0 |
| 28 | 68 | Female | BLCA | T1N0M0 |
| 29 | 48 | Male | BLCA | T1N0M0 |
| 30 | 65 | Male | BLCA | T2bN0M0 |
| 31 | 72 | Male | BLCA | T2bN0M0 |
| 32 | 80 | Male | BLCA | T2N0M0 |
| 33 | 76 | Female | BLCA | T2bN0M0 |
| 34 | 55 | Female | BLCA | T1N0M0 |
| 35 | 75 | Male | BLCA | T2aN0M0 |
Detection of NUSAP1 Expression in Transfected BLCA Cells
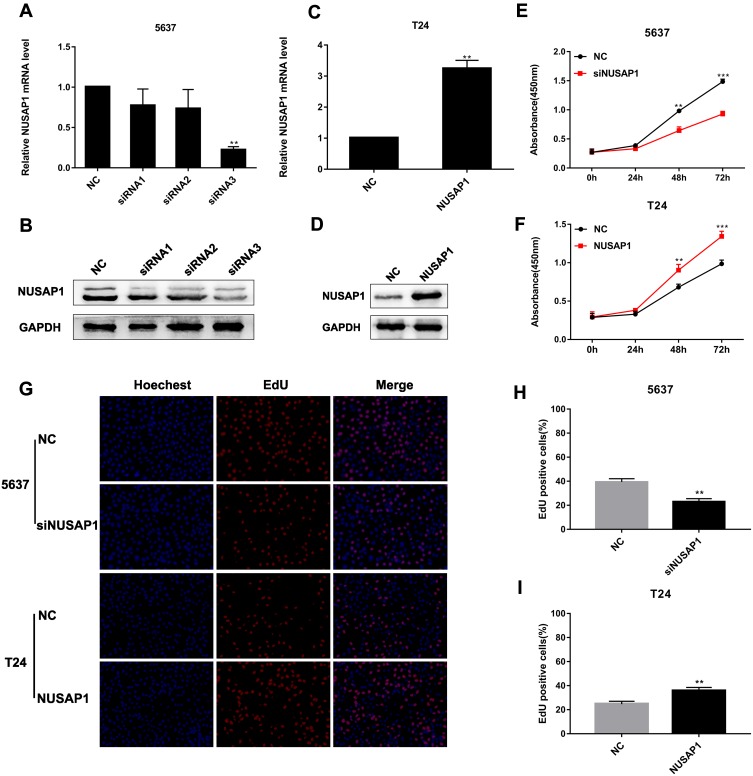
To further validate the role of NUSAP1 in BLCA cells, 5637 cells were transfected with NC or siNUSAP1, and T24 cells were transfected with a NUSAP1 expression plasmid or an empty vector (NC); then, NUSAP1 expression was measured via real-time PCR and Western blotting. The results revealed that NUSAP1 expression was significantly lower in 5637 cells in the siRNA3 group than in the normal control and other siRNA groups (Figure 2A and B). In T24 cells, NUSAP1 expression was increased compared with that in the vector (NC) group (Figure 2C and D). These results indicate that NUSAP1 was successfully knocked down in 5637 cells and overexpressed in T24 cells.
Figure 2.
NUSAP1 enhanced the proliferation of BLCA cells. (A and B). NUSAP1 expression in transfected 5637 cells determined by PCR and Western blotting. **P<0.01 vs NC. (C and D). NUSAP1 expression in transfected T24 cells by real-time PCR and Western blotting. **P<0.01 vs NC. (E and F). Cell proliferation was determined by a Cell Counting Kit-8 assay. **P<0.01, ***P<0.001 vs NC (5637) or NC (T24). (G, H and I). Cell proliferation was determined by an EdU assay (100× magnification). **P<0.01 vs NC (5637) or NC (T24).
NUSAP1 Enhanced BLCA Cell Proliferation
The effect of NUSAP1 on BLCA cell proliferation was evaluated with CCK-8 and EdU assays. The CCK-8 assay demonstrated that 5637-siNUSAP1 cells grew slower than 5637-NC cells, whereas the growth rate of T24-NUSAP1 cells was faster than that of T24-NC cells (Figure 2E and F). In addition, the EdU assay demonstrated that NUSAP1 overexpression promoted cell proliferation, whereas NUSAP1 downregulation suppressed cell proliferation (Figure 2G–I). These results indicated that high NUSAP1 expression significantly enhanced the proliferative capacity of BLCA cells.
NUSAP1 Downregulation Induced G2/M Phase Cell Arrest and Apoptosis in BLCA Cells
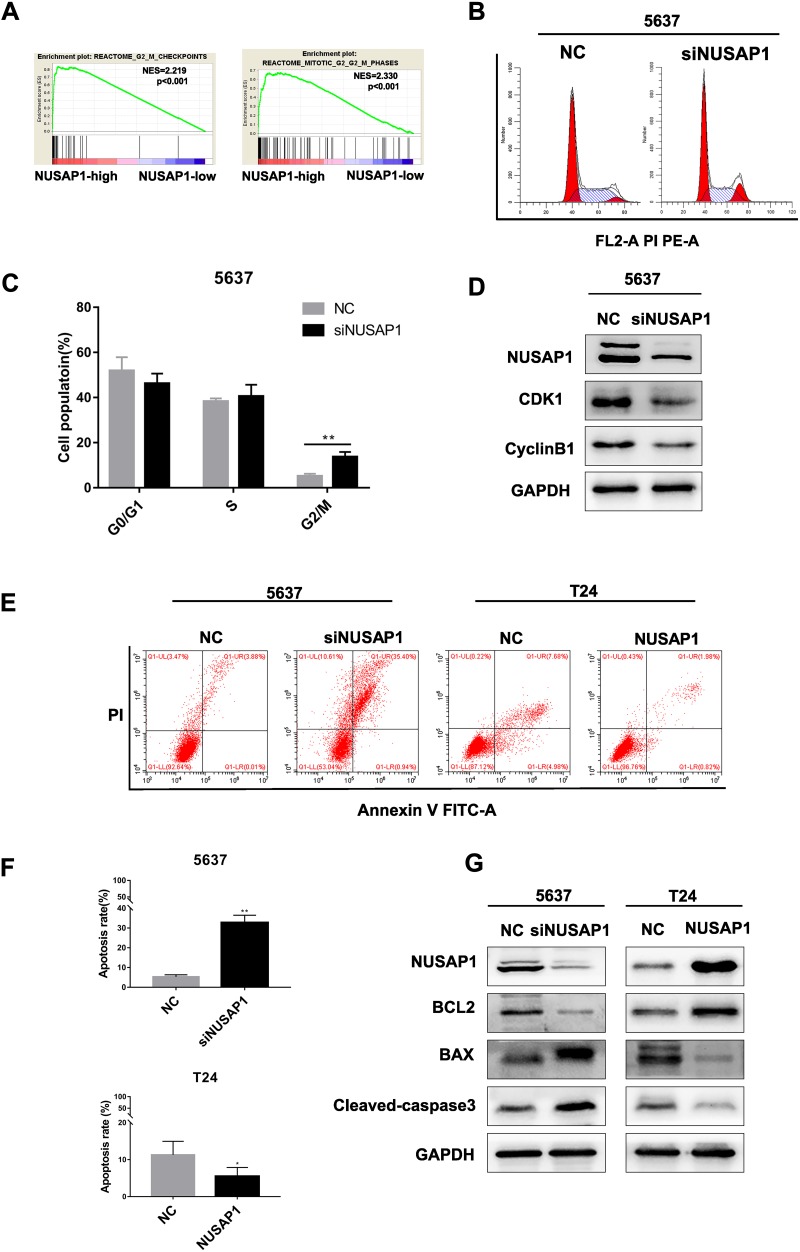
GSEA suggested that NUSAP1 is closely related to cell cycle regulation, especially at the G2/M phase (Figure 3A). Thus, flow cytometry cell cycle distribution and Western blotting assays were performed to evaluate the role of NUSAP1 in cell cycle regulation, as shown in Figure 3B and C. The percentage of cells in the G2/M phase was greater in the 5637-siNUSAP1 group than in the 5637-NC group. Western blotting showed that CDK1 and cyclin B1 expression was inhibited in the 5637-siNUSAP1 group compared with the 5637-NC group (Figure 3D). In addition, the effect of NUSAP1 on BLCA cell apoptosis was evaluated via flow cytometry. The results revealed that compared with the 5637-NC group, the 5637-siNUSAP1 group had significantly higher levels of apoptosis. In contrast, the apoptosis rate was lower in the T24-NUSAP1 group than in the T24-NC group (Figure 3E and F). Furthermore, Western blotting analysis was carried out to detect the protein expression levels of Bcl-2, Bax and cleaved caspase-3. Bax and cleaved-caspase3 protein expression levels were increased in the 5637-siNUSAP1 group and decreased in the T24-NUSAP1 group, whereas Bcl-2 expression was suppressed in the 5637-siNUSAP1 group and increased in the T24-NUSAP1 group compared with the NC groups (Figure 3G). These results indicated that NUSAP1 downregulation induced cell cycle arrest at the G2/M phase and induced apoptosis in BLCA cells.
Figure 3.
NUSAP1 downregulation induced cell cycle arrest and apoptosis in BLCA cells. (A). GSEA suggests that NUSAP1 is closely related to the G2/M cell cycle phase. (B and C). Cell cycle analysis via flow cytometry. **P<0.01 vs NC. (D). Western blotting analysis of CDK1 and cyclin B1 protein expression. (E and F). Apoptosis was analyzed by flow cytometry. **P<0.01 vs NC (5637), *P<0.05 vs NC (T24). (G). Western blotting analysis detected the protein expression levels of Bax, Bcl-2 and cleaved-caspase3.
NUSAP1 Silencing in BLCA Cells Enhanced Their Chemosensitivity to Gemcitabine
Because of the effect of NUSAP1 on tumor cell proliferation, cycle regulation and apoptosis, we hypothesized that NUSAP1 was involved in tumor chemoresistance.
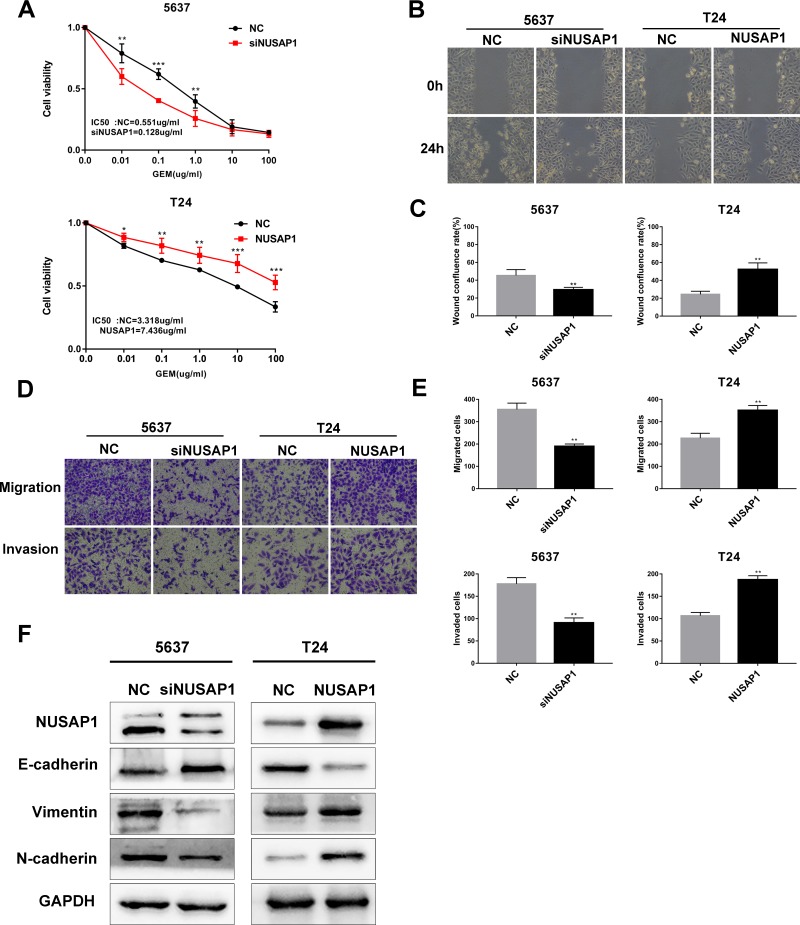
To validate our hypothesis, a CCK-8 assay was performed to confirm the effect of NUSAP1 on gemcitabine (GEM) chemosensitivity in BLCA. 5637 and T24 cells were treated with GEM at various concentrations, from 0.01 to 100 μg/mL, for 48 h, and cell viability was measured by CCK-8 assay. As expected, our data revealed that the 5637-siNUSAP1 group had higher chemosensitivity than the 5637-NC group, whereas the T24-NUSAP1 group exhibited reduced GEM chemosensitivity compared with the T24-NC group (Figure 4A). These data demonstrate that NUSAP1 downregulation in BLCA enhanced GEM chemosensitivity.
Figure 4.
NUSAP1 downregulation in BLCA cells enhanced GEM chemosensitivity and suppressed cell migration, invasion, and EMT. (A). The chemosensitivity of BLCA cells to GEM was determined by a Cell Counting Kit-8 assay. *P<0.05, **P<0.01, ***P<0.001 vs NC (5637) or NC (T24). (B and C). A wound healing assay was performed to detect the migration of cells (100× magnification). **P<0.01, vs NC (5637) or NC (T24). (D and E). Transwell assays were performed to evaluate the migration and invasion of cells (100× magnification). **P<0.01, vs NC (5637) or NC (T24). (F). Western blotting analysis detected the protein expression levels of E-cadherin, vimentin and N-cadherin.
NUSAP1 Downregulation in BLCA Cells Suppressed Cell Migration, Invasion, and Epithelial-Mesenchymal Transition
To investigate the biological role of NUSAP1 in metastasis, Transwell and wound healing assays were performed. We found that the cell migration and invasion abilities were markedly increased in the 5637-NC and T24-NUSAP1 groups, while those abilities were apparently decreased in the 5637-siNUSAP1 and T24-NC groups (Figure 4B–E). Previous studies indicated that EMT is a critical step for tumor metastasis and relapse; consequently, Western blotting was performed to confirm the effect of NUSAP1 on EMT-associated proteins. Our results revealed that E-cadherin levels were markedly increased, but vimentin and N-cadherin levels were obviously decreased in the 5637-siNUSAP1 group compared with the 5637-NC group; in contrast, the opposite pattern was observed in the T24-NUSAP1 group (Figure 4F). These results revealed that NUSAP1 could regulate cell migration, invasion, and EMT in BLCA.
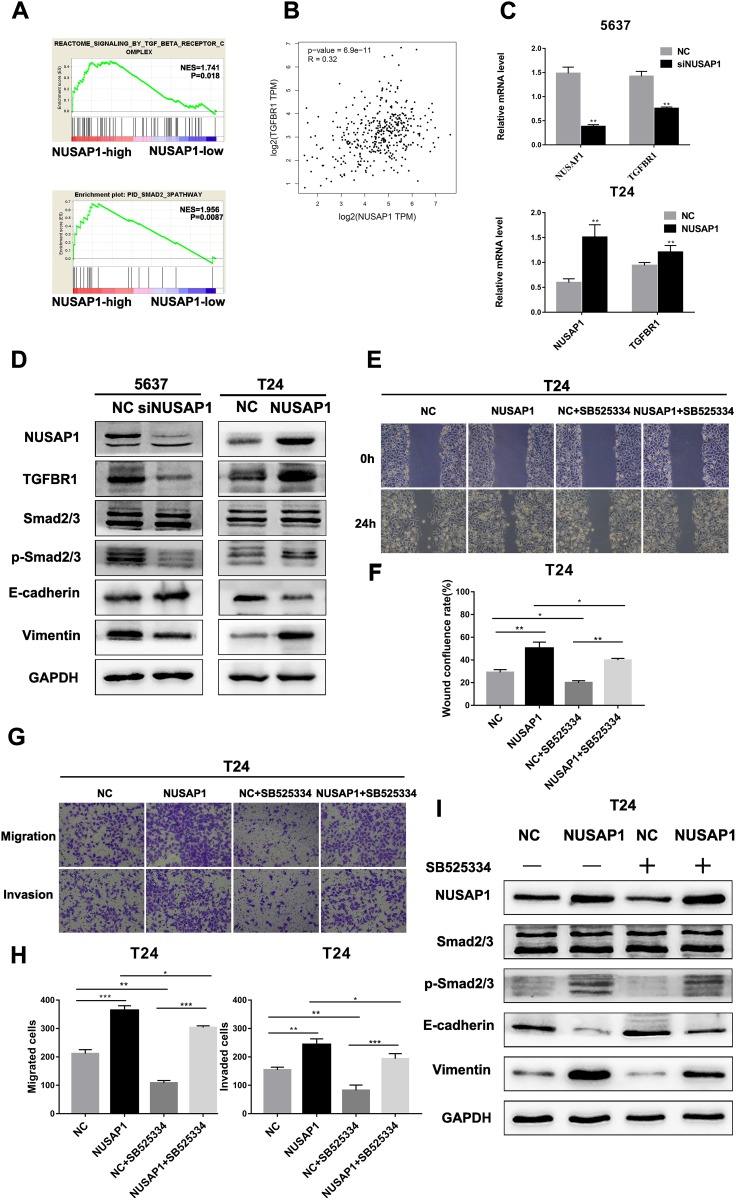
The TGF-β Signaling Pathway Is Involved in NUSAP1-Regulated EMT in BLCA
The TGF-β signaling pathway has been reported to induce EMT, which is closely related to tumor progression and metastasis. GSEA revealed that the expression level of NUSAP1 in the TCGA BLCA dataset was positively correlated with the expression of the TGFBR complex and Smad2/3 in BLCA (Figure 5A). Moreover, NUSAP1 expression was found to be positively correlated with TGFBR1 expression, according to Gene Expression Profiling Interactive Analysis (GEPIA) (Figure 5B) (http://gepia.cancer-pku.cn/). Thus, we suspect that the TGF-β signaling pathway is involved in the NUSAP1-regulated epithelial-mesenchymal transition in BLCA. To validate our hypothesis, real-time PCR and Western blotting assays were performed to confirm the mechanism. As shown in Figure 5C, we found that TGFBR1 and NUSAP1 maintained consistent mRNA levels in both T24 and 5637 cells. In addition, silencing NUSAP1 in 5637 cells resulted in decreases in TGFBR1, p-Smad2/3, vimentin, and N-cadherin but an increase in E-cadherin; however, NUSAP1 overexpression in T24 cells resulted in increases in TGFBR1, p-Smad2/3, vimentin and N-cadherin and a decrease in E-cadherin (Figure 5D). SB525334 inhibited TGFBR1 in NUSAP1-overexpressing T24 cells, and cell invasion and metastasis abilities were also significantly suppressed (Figure 5E–H); p-Smad2/3 and vimentin expression was also reduced following treatment (Figure 5I). On the other hand, after treating with exogenous recombinant TGF-β1 in NUSAP1-knockedown 5637 cells, the expression of p-smad2/3 and Vimentin was increased, compared with the control group (Figure S1). These data confirmed that the TGF-β signaling pathway was involved in the NUSAP1-regulated invasion, migration and EMT of BLCA.
Figure 5.
NUSAP1 regulated the epithelial-mesenchymal transition of BLCA via the TGF-β signaling pathway. (A). GSEA revealed that NUSAP1 expression was positively correlated with the TGFBR complex and the Smad2/3 pathway in BLCA. (B). Gene correlation analysis of NUSAP1 and TGFBR1 obtained from Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/). ***P<0.001. (C). Real-time PCR was performed to detect the relationship between NUSAP1 and TGFBR1. **P<0.01, vs NC (5637) or NC (T24). (D). Western blotting analysis of the protein expression levels of TGFBR1, Smad2/3, p- Smad2/3, E-cadherin, and vimentin. (E and F). Wound healing assays were performed to detect the migration of cells (100× magnification) *P<0.05, **P<0.01. (G and H). Transwell assays were performed to evaluate the migration and invasion of cells (100× magnification). *P<0.05, **P<0.01, ***P<0.001. (I). Western blotting analysis of the protein expression levels of Smad2/3, p- Smad2/3, E-cadherin, and vimentin.
Discussion
NUSAP1 is a protein with a molecular weight of 55 kDa that plays key roles in mitotic progression, spindle formation and stability.3 The role of NUSAP1 in tumorigenesis has been revealed in various types of tumors. For instance, in cervical cancer, NUSAP1 induced the sumoylation of TCF4 by interacting with the SUMO-E3 ligase Ran binding protein 2 (RanBP2), thereby inducing cervical cancer cell metastasis and enhancing Wnt/β-catenin signaling.8 Similarly, high NUSAP1 expression levels promoted astrocytoma progression through the Hedgehog signaling pathway.10 In addition, NUSAP1 is highly expressed in colon cancer tissues and cells, and NUSAP1 upregulation indicates a poor prognosis.13 In colorectal cancer, NUSAP1 knockdown induced apoptosis and inhibited tumor cell migration, invasion, cell proliferation and EMT by inhibiting the expression of DNA methyltransferase 1 (DNMT1), indicating that NUSAP1 can be used as an independent prognostic biomarker.14 In prostate cancer, high NUSAP1 expression promoted prostate cancer cell proliferation and invasion with the loss of RB1 via the RB1/E2F1 axis.15 However, the functional roles of NUSAP1 in BLCA are not clear.
In the Oncomine database, the data showed that NUSAP1 had remarkably higher expression in both infiltrating urothelial BLCA and superficial BLCA. In the present study, we found that NUSAP1 was upregulated in BLCA tissues and cell lines. Moreover, the GSE13507 database revealed that patients with high NUSAP1 expression had shorter 5-year metastasis-free survival rates. Hence, we explored the biological function of NUSAP1 in BLCA. In addition, NUSAP1 overexpression promoted the proliferation, invasion and metastasis of BLCA cells, whereas NUSAP1 downregulation induced an increase in apoptosis and enhanced the antitumor effect of GEM. Furthermore, we observed that E-cadherin levels were markedly increased, but vimentin and N-cadherin levels were obviously decreased in the 5637-siNUSAP1 group compared with the 5637-NC group; however, the opposite pattern was observed in the T24-NUSAP1 group, which means that NUSAP1 was involved in regulating EMT in BLCA.
EMT is regarded as an important step in cancer invasion and metastasis and can be induced by various signaling pathways (TGF-β, Wnt/β-catenin, Notch, EGF, and HGF). It is well known that the TGF-β signaling pathway is widely involved in the EMT of various tumors, and TGFBR1 is an important participant in this pathway.16 For instance, Zhang revealed that circCACTIN promoted proliferation, migration, invasion and EMT in gastric cancer by sponging miRNA-331-3p and regulating TGFBR1 mRNA expression.17 Similarly, a previous report illustrated that SHOX2 acts as a transcription factor to upregulate TGFBR1 expression, and the TGFBR1 inhibitor LY364497 abolishes EMT induction by ectopic SHOX2 expression, suggesting that TGF-β signaling is essential for SHOX2-induced EMT.18 A recent study reported that linc00462 may function as an effective invasiveness marker for pancreatic cancer patients, and linc00462 upregulation may promote pancreatic cancer cell invasiveness through the miR-665/TGFBR1-TGFBR2/Smad2/3 pathway.19 In our study, we first found that NUSAP1 expression was positively correlated with the TGFBR1 and Smad2/3 pathways, as shown by GSEA. Furthermore, NUSAP1 expression was positively correlated with TGFBR1 expression, as shown by real-time PCR. Therefore, we speculated that NUSAP1 regulates EMT in BLCA by affecting TGFBR1. As expected, we found that silencing NUSAP1 in cells caused TGFBR1, Smad2/3, vimentin and N-cadherin downregulation and E-cadherin upregulation. However, when NUSAP1 was overexpressed, the opposite expression phenomenon was observed. Next, SB525334 inhibited TGFBR1 in NUSAP1-overexpressing T24 cells, and cell invasion and metastasis were significantly suppressed; p-Smad2/3 and EMT-associated protein expression levels were also decreased. These results indicate that NUSAP1 is involved in the regulation of EMT in BLCA, probably via the TGF-β signaling pathway.
In previous reports, NUSAP1 was found to be involved in the regulation of EMT in tumors via the HH signaling pathway10 or the Wnt/β-catenin signaling pathway.8 In this study, we reported a novel approach to the regulation of EMT by NUSAP1. However, we do not have any direct evidence to demonstrate how NUSAP1 affects TGFBR1 and thus participates in EMT regulation in BLCA through the TGF-β signaling pathway. A recent study indicated that SENP2 overexpression suppresses TGF-β signaling activation in BLCA cells owing to TGFBR1 deSUMOylation.20 Moreover, Li et al reported that NUSAP1 induced cervical cancer cell metastasis by inducing the SUMOylation of TCF4.8 Considering the above two reports, we speculate that NUSAP1 may be involved in the TGF-β signaling pathway by regulating the SUMOylation of TGFBR1, but we have not performed further verification experiments.
EMT has also been recognized as an important mechanism of tumor resistance. Tumor cells tend to be interstitial while they are in the process of producing acquired drug resistance, and tumor cells with mesenchymal differentiation are usually characterized by primary drug resistance.21 This phenomenon was observed in adriamycin-resistant breast cancer, cisplatin-resistant ovarian cancer cell lines/pancreatic cancer cell lines, and OXA-resistant CRC cell lines/colorectal cancer cells.22–25 Atsushi Okamoto et al revealed that NUSAP1 in oral squamous cell carcinoma enhanced the antitumor effect of paclitaxel.9 Consistent with this report, we also found that NUSAP1 downregulation enhanced the chemosensitivity of BLCA cells to GEM. However, we did not verified that the change in chemosensitivity caused by NUSAP1 was achieved by EMT regulation. Thus, these topics require further research.
Conclusion
In conclusion, our study revealed that NUSAP1 plays an important role in BLCA progression by promoting tumor cell proliferation, cell migration, cell invasion, and chemoresistance. Therefore, NUSAP1 may be a potential prognostic biomarker and a therapeutic target for BLCA.
Acknowledgments
This study was supported by funds from the Natural Science Foundation of China (Grant Nos. 81372758 and 81874092).
Disclosure
The authors report no conflicts of interest regarding this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.v69.1 [DOI] [PubMed] [Google Scholar]
- 2.Hartge P, Harvey EB, Linehan WM, et al. Unexplained excess risk of bladder cancer in men. J Natl Cancer Inst. 1990;82(20):1636–1640. doi: 10.1093/jnci/82.20.1636 [DOI] [PubMed] [Google Scholar]
- 3.Raemaekers T, Ribbeck K, Beaudouin J, et al. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol. 2003;162(6):1017–1029. doi: 10.1083/jcb.200302129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribbeck K, Groen AC, Santarella R, et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol Biol Cell. 2006;17(6):2646–2660. doi: 10.1091/mbc.e05-12-1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanden Bosch A, Raemaekers T, Denayer S, et al. NuSAP is essential for chromatin-induced spindle formation during early embryogenesis. J Cell Sci. 2010;123(Pt 19):3244–3255. doi: 10.1242/jcs.063875 [DOI] [PubMed] [Google Scholar]
- 6.Gulzar ZG, McKenney JK, Brooks JD. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene. 2013;32(1):70–77. doi: 10.1038/onc.2012.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadia PP, Coram M, Armstrong RJ, Mindrinos M, Butte AJ, Miklos DB. Antibodies specifically target AML antigen NuSAP1 after allogeneic bone marrow transplantation. Blood. 2010;115(10):2077–2087. doi: 10.1182/blood-2009-03-211375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Zhang W, Yan M, et al. Nucleolar and spindle associated protein 1 promotes metastasis of cervical carcinoma cells by activating Wnt/β-catenin signaling. J Exp Clin Cancer Res. 2019;38(1):33. doi: 10.1186/s13046-019-1037-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamoto A, Higo M, Shiiba M, et al. Down-regulation of nucleolar and spindle-associated protein 1 (NUSAP1) expression suppresses tumor and cell proliferation and enhances anti-tumor effect of paclitaxel in oral squamous cell carcinoma. PLoS One. 2015;10(11):e0142252. doi: 10.1371/journal.pone.0142252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Xu B, Yang C, et al. Nucleolar and spindle associated protein 1 promotes the aggressiveness of astrocytoma by activating the Hedgehog signaling pathway. J Exp Clin Cancer Res. 2017;36(1):127. doi: 10.1186/s13046-017-0597-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyrskjøt L, Kruhøffer M, Thykjaer T, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64(11):4040–4048. doi: 10.1158/0008-5472.CAN-03-3620 [DOI] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Guan C, Lu C, et al. High NUSAP1 expression predicts poor prognosis in colon cancer. Pathol Res Pract. 2018;214(7):968–973. doi: 10.1016/j.prp.2018.05.017 [DOI] [PubMed] [Google Scholar]
- 14.Han G, Wei Z, Cui H, et al. NUSAP1 gene silencing inhibits cell proliferation, migration and invasion through inhibiting DNMT1 gene expression in human colorectal cancer. Exp Cell Res. 2018;367(2):216–221. doi: 10.1016/j.yexcr.2018.03.039 [DOI] [PubMed] [Google Scholar]
- 15.Gordon CA, Gulzar ZG, Brooks JD. NUSAP1 expression is upregulated by loss of RB1 in prostate cancer cells. Prostate. 2015;75(5):517–526. doi: 10.1002/pros.v75.5 [DOI] [PubMed] [Google Scholar]
- 16.Katsuno Y, Lamouille S, Derynck R. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol. 2013;25(1):76–84. doi: 10.1097/CCO.0b013e32835b6371 [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Song X, Chen X, et al. Circular RNA CircCACTIN promotes gastric cancer progression by sponging MiR-331-3p and regulating TGFBR1 expression. Int J Biol Sci. 2019;15(5):1091–1103. doi: 10.7150/ijbs.31533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong S, Noh H, Teng Y, et al. SHOX2 is a direct miR-375 target and a novel epithelial-to-mesenchymal transition inducer in breast cancer cells. Neoplasia. 2014;16(4):279–290.e1–5. doi: 10.1016/j.neo.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis. 2018;9(6):706. doi: 10.1038/s41419-018-0724-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan M, Zhang D, Zhang E, et al. SENP2 suppresses epithelial-mesenchymal transition of bladder cancer cells through deSUMOylation of TGF-βRI. Mol Carcinog. 2017;56(10):2332–2341. doi: 10.1002/mc.v56.10 [DOI] [PubMed] [Google Scholar]
- 21.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67(5):1979–1987. doi: 10.1158/0008-5472.CAN-06-1479 [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Sun J, Cai B, et al. NANOG regulates epithelial-mesenchymal transition and chemoresistance through activation of the STAT3 pathway in epithelial ovarian cancer. Tumour Biol. 2016;37(7):9671–9680. doi: 10.1007/s13277-016-4848-x [DOI] [PubMed] [Google Scholar]
- 24.Yang AD, Fan F, Camp ER, et al. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12(14 Pt 1):4147–4153. doi: 10.1158/1078-0432.CCR-06-0038 [DOI] [PubMed] [Google Scholar]
- 25.Yin J, Wang L, Wang Y, Shen H, Wang X, Wu L. Curcumin reverses oxaliplatin resistance in human colorectal cancer via regulation of TGF-β/Smad2/3 signaling pathway. Onco Targets Ther. 2019;12:3893–3903. doi: 10.2147/OTT.S199601 [DOI] [PMC free article] [PubMed] [Google Scholar]