Abstract
Autophagy is the major cellular pathway to degrade dysfunctional organelles and protein aggregates. Autophagy is particularly important in neurons, terminally differentiated cells that must last the lifetime of the organism. There are both constitutive and stress-induced pathways for autophagy in neurons, which catalyze the turnover of aged or damaged mitochondria, endoplasmic reticulum, other cellular organelles, and aggregated proteins. These pathways are required in neurodevelopment as well as in the maintenance of neuronal homeostasis. Here we review the core components of the pathway for autophagosome biogenesis, as well as the cell biology of bulk and selective autophagy in neurons. Finally, we discuss the role of autophagy in neuronal development, homeostasis, and aging, and the links between deficits in autophagy and neurodegeneration.
Keywords: Autophagy, mitophagy, ERphagy, aggrephagy, neuronal homeostasis, neurodegeneration
INTRODUCTION: DEGRADATIVE PATHWAYS IN THE NEURON
Neurons are post-mitotic and long-lived cells. Both neurodevelopment and the long-term maintenance of neuronal health require effective removal of aggregated proteins and aged or defective organelles. Genetic and cellular studies indicate that defects in protein and organelle turnover closely correlate with cellular stress, degeneration, and death.
Multiple degradative pathways are at work in neurons, such as the degradation of misfolded proteins via the ubiquitin-proteasome pathway (Bingol & Sheng 2011). Protein degradation also occurs via the endolysosomal trafficking pathway (Lie & Nixon 2018, Winckler et al. 2018), as well as by chaperone-mediated autophagy and endosomal microautophagy (Tekirdag & Cuervo 2018). Recently identified pathways may also contribute to the clearance of misfolded or aggregated proteins in neurons, such as the MAGIC (mitochondria as guardian in cytosol) pathway involving import of misfolded proteins into mitochondria (Ruan et al. 2017).
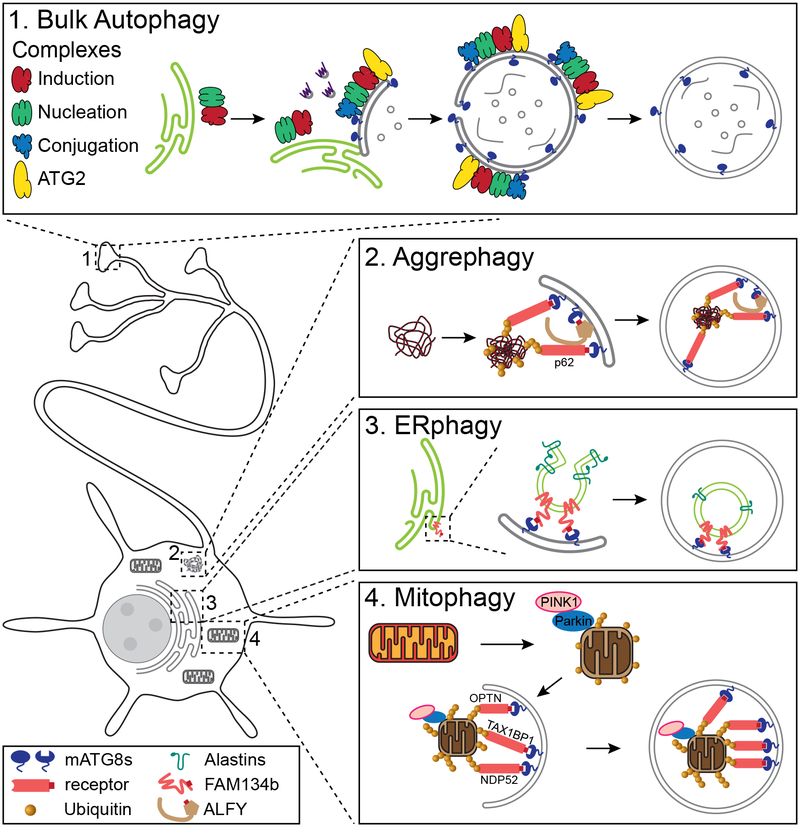
Macroautophagy, reviewed here, is the major pathway to clear larger targets for degradation, such as protein aggregates and dysfunctional organelles. The molecular players in this pathway have been well-defined from decades of elegant work in yeast (Ohsumi 2014). Many of the proteins initially identified in yeast screens have clear homologs required for autophagy in mammalian cells, and function in an analogous manner, giving us an overall understanding of the canonical pathway. However, we are just beginning to understand how this pathway is specialized for the turnover of specific cargos in selective autophagy, and how it has been adapted to fit the distinct needs of terminally differentiated and highly polarized cells such as neurons (Figure 1).
Figure 1. The spatial organization of autophagy in neurons.
A cartoon neuron is depicted with the different forms of autophagy in spatially distinct neuronal compartments: (1) nonselective autophagy in the distal axon and (2) aggrephagy, (3) ERphagy, and (4) mitophagy in the soma. For the types of selective autophagy (2–4), relevant LIRs are depicted by red boxes on the autophagy receptors. In aggrephagy (2), WDR81 performs a synonymous function to ALFY (depicted). In ERphagy (3), RTN3 acts in a similar manner to FAM134b (depicted).
Here we focus on the dynamics of autophagy in neurons, where issues of cellular compartmentalization, long-distance trafficking, activity-dependent plasticity and high levels of metabolic stress have led to cell-type specific adaptation of the autophagy pathway. For example, neuronal axons can extend up to 1m from the soma, the principle site of protein biosynthesis and degradation. How proteins that take hours to days to arrive at cellular destinations such as synapses are effectively turned over is a topic of great interest. Another question is how autophagy contributes to synaptic plasticity, which is required for learning and memory. Significant interest is focused on the role of autophagy in neuronal homeostasis. Neurons often operate under conditions of high metabolic demand, induced for example by repetitive stimulation. This can lead to oxidative stress and organelle damage, and so damaged organelles such as mitochondria must be subject to vigilant quality control mechanisms including targeted removal by selective autophagy. While many questions remain, the importance of autophagy in neurons is clear, as the neuron-specific ablation of autophagy leads to neurodegeneration (Hara et al. 2006, Komatsu et al. 2006).
CANONICAL PATHWAY FOR MACROAUTOPHAGY
Bulk and selective macroautophagy (hereafter autophagy) pathways share most of the molecular machinery required to execute these related processes. Autophagy begins with autophagosome biogenesis. In non-neuronal cells, autophagosome biogenesis is activated by cellular stressors such as starvation via the suppression of mTOR (mammalian target of rapamycin 1) kinase activity. In turn, suppression of mTOR removes repression of the induction complex to activate autophagosome biogenesis. Neurons are more dependent on constitutive autophagy than other cell types, and do not exhibit a robust starvation-induced upregulation of autophagy (Mizushima et al., 2004), so the specific role of mTOR in neurons requires further study.
The induction complex is composed of the kinase ULK1, along with ATG101, ATG13, and FIP200. When activated, ULK1 phosphorylates other autophagy pathway components, including Beclin1 (BECN1) and ATG9 (Feng et al. 2014, Kamada et al. 2000, Russell et al. 2013, Zhou et al. 2017). Beclin1 is a component of the nucleation complex, along with ATG14, lipid kinase VPS34/PI3KIII, and VPS15/PIK3R4 (Itakura et al. 2008, Matsunaga et al. 2009, Sun et al. 2008, Zhong et al. 2009). The nucleation complex generates PI3P, an important component of the autophagosome membrane during biogenesis (Kihara et al. 2001, Obara et al. 2006). Several components of the nucleation complex also function in cellular processes other than autophagy (Liang et al. 2008, Lindmo & Stenmark 2006, Matsunaga et al. 2009, Yan et al. 2009, Zhong et al. 2009). In contrast, ATG14 is specifically required for the proper localization of the nucleation complex to the forming autophagosome (Matsunaga et al. 2010); ATG14 also recruits other autophagosome biogenesis components to the initial autophagosome membrane, termed the isolation membrane in mammals.
Elongation of the autophagosome membrane is mediated by the aptly named elongation complex, composed of two ubiquitin-like conjugation complexes (Ichimura et al. 2000, Mizushima et al. 1998). In the first complex, E1-like ATG7 and E2-like ATG10 conjugate ATG12 to ATG5. In the second complex, ATG7 is again the E1-like activating enzyme and ATG3 is the E2-like conjugating enzyme. The ATG12-ATG5 conjugate, along with ATG16L1, acts as the E3-like enzyme to conjugate mATG8s to phosphatidylethanolamine (PE) (Mizushima et al. 2001, Suzuki et al. 2001). While yeast has one Atg8 that is conjugated to PE, there are five or six Atg8 homologs in mammals (mATG8s): LC3A, LC3B, GABARAP, GABARAPL1, GABARAPL2, and LC3C (expressed in humans, but not mice or rats). All mATG8s must be processed before conjugation to PE; ATG4 cleaves the C-terminal amino acids to expose Glycine. ATG4 also acts at the end of autophagy to cleave mATG8s from the cytosolic face of the completed autophagosome (Kauffman et al. 2018, Tanida et al. 2004).
ATG9 is a six-pass transmembrane protein, the only transmembrane protein in the core machinery for autophagy. Several accessory transmembrane proteins have recently been discovered, such as VMP1 and TMEM41B (Moretti et al. 2018, Morita et al. 2018), which primarily localize to the ER. ATG9 is thought to shuttle between the membrane source and the growing autophagosome, possibly bringing lipids to the autophagosome. ATG9 interacts with ATG2, a large protein with regions of homology to VPS13. VPS13 is a lipid transporter between ER and other membranes, raising the possibility that ATG2 also functions as a lipid transporter that supplies lipids to the growing autophagosomal membrane (Kumar et al. 2018). Yeast Atg2 also interacts with Atg18, which has four homologs in mammals, WIPI1-WIPI4 (WIPI: WD-repeat protein interacting with phosphoinositides). WIPI1 and WIPI2 are thought to act early in autophagosome biogenesis, linking the nucleation and elongation steps. As WIPIs interact with membranes and can oligomerize, it is intriguing to hypothesize that WIPI assembly may bend the membrane of the growing autophagosome.
Since autophagy requires de novo membrane formation and elongation around cargo in the cytoplasm, the source of the lipids that make up the autophagosome membrane has been a pressing question. Virtually every possible membrane source, including plasma membrane (Hollenbeck 1993; Ravikumar 2010), mitochondria (Hailey et al. 2010), Golgi (van der Vaart et al. 2010), ER (Hamasaki et al. 2013, Hayashi-Nishino et al. 2009, Ylä-Anttila et al. 2009), and recycling endosomes (Puri et al. 2018), has been implicated in autophagy at some point. However, the endoplasmic reticulum has been identified repeatedly as crucial for autophagosome formation, both as a source of donor membrane and as a platform for initial biogenesis of the organelle at the omegasome. The importance of the ER has been confirmed in primary hippocampal neurons, in which autophagosomes colocalized with ER marker Sec61β, but not plasma membrane or mitochondrial markers (Maday & Holzbaur 2014). In neurons, ATG9 appears to colocalize and be transported with synaptic vesicles (Stavoe et al. 2016), but there is currently no evidence suggesting that synaptic vesicles directly donate lipids to autophagosomes.
In bulk, or nonselective, autophagy cytoplasmic contents are nonspecifically engulfed within the isolation membrane during autophagosome formation. In contrast, during selective autophagy, specific cargoes are recognized and/or clustered by receptors that directly link the cargo to the forming autophagosome membrane via mATG8s, discussed in more detail below. Once the contents of the autophagosome are engulfed, the isolation membrane must close by fusing with itself, yielding a double layer membrane surrounding the contents. Around the time the autophagosome membrane fuses with itself, the biogenesis machinery dissociates from the fully formed autophagosome, with the exception of the mATG8s, which remain tightly associated with the limiting membrane (Mizushima et al. 2003). The protein machinery necessary for membrane fission at the extremities of the isolation membrane is not yet known. However, the mATG8s have been implicated in this step (Tsuboyama et al. 2016).
After autophagosome closure, PI3P is removed from the outer membrane by PI3P phosphatases (Cebollero et al. 2012) and mATG8s are removed from the outer membrane by ATG4 (Kauffman et al. 2018, Nair et al. 2012, Yu et al. 2012), possibly destabilizing the association of other autophagy components with the cytosolic face of the autophagosomal membrane. Autophagosomes then fuse with late endosomes or lysosomes. Following fusion to form an autophagolysosome, the internal pH decreases, activating lysosomal enzymes that digest the engulfed cargos for eventual recycling of components.
SELECTIVE AUTOPHAGY
Macroautophagy can occur as either a non-selective or a selective process. Starvation-induced autophagy is the classic example of non-selective cargo uptake, as the rapid turnover of cytosolic components upon nutrient deprivation is not mediated by molecular selectivity. While starvation does not appear to induce significant upregulation of autophagy in neurons as compared to other cell types (Mizushima et al., 2004; Maday and Holzbaur, 2016), there is a conserved pathway for constitutive and non-selective turnover of cytosolic proteins, aggregates and organelles in the axon, as discussed below. In addition to this non-selective pathway, there is a growing list of cargos in the neuron that are turned over by selective autophagy.
In a selective mechanism, there is a specific cargo recognition step mediated by one or more receptors, which then enhances the engulfment and degradation of targeted substrates. Selective autophagy uses the same core components of the non-selective macroautophagy pathway, but generates specialized autophagosomes that selectively engulf specific cargoes (Figure 1). The specificity is mediated by receptors which connect the selected cargo to the autophagy machinery via the mATG8 proteins, stimulating autophagosome formation around the specified cargo. p62/SQSTM1 (sequestome 1) was the first autophagy receptor to be identified; p62 interacts with ubiquitin chains through its UBAN (ubiquitin binding in ABIN and NEMO) domain and with mATG8s via its LIR (LC3-interacting region), also known as AIM (Atg8 family-interacting motif). Other receptors involved in selective autophagy such as OPTN (Optineurin) are also characterized by the presence of both ubiquitin-binding and LIR motifs. These receptors can be either selective for a single cargo or promiscuous, implicated in the removal of multiple types of cargo such as damaged mitochondria or aggregated proteins. Further, there is evidence of functional redundancy, as some receptors appear capable of substituting for other receptors within a specific type of selective autophagy.
The best understood example of selective autophagy, at least within the context of the neuron, is mitophagy, the clearance of defective mitochondria. However, there is growing interest in parallel pathways mediating the selective turnover of ER by ERphagy, protein aggregates by aggrephagy, RNA granules by granulophagy, lysosomes by lysophagy, peroxisomes by pexophagy, etc. Only a few of these pathways have been examined in detail in neurons, so here we focus on progress in the understanding of mitophagy, ERphagy, and aggrephagy.
Mitophagy
Interest in the mechanisms driving mitophagy was sparked by the discovery that two genes causal for familial forms of Parkinson’s disease (PD), PINK1 and Parkin, are part of a conserved mitophagy pathway. Work has shown that PINK1 is a mitochondrially-targeted kinase involved in the recruitment of the E3 ubiquitin ligase Parkin to impaired mitochondria (Narendra et al. 2008, 2010a). PINK1 is usually turned over very rapidly at the outer mitochondrial membrane (OMM) leading to a low steady state level. Conditions such as mitochondrial depolarization or a block in the mitochondrial import machinery lead to the accumulation of PINK1 on the OMM (Narendra et al. 2010a). This allows PINK1 to phosphorylate Parkin, leading to Parkin recruitment and activation (Kondapalli et al. 2012, Shiba-Fukushima et al. 2012). PINK1 also phosphorylates ubiquitin, which further enhances Parkin activity (Kane et al. 2014). This feedforward mechanism leads to the rapid ubiquitination of OMM proteins by Parkin (Narendra et al. 2010b) that serve as a binding platform for ubiquitin-binding mitophagy receptors.
Mitophagy can be rapidly induced by either global or focal mitochondrial damage. Either mitochondrial uncouplers such as CCCP or the localized production of reactive oxygen species (ROS) can effectively induce mitophagy in cells expressing Parkin (Wong & Holzbaur 2014a). The ubiquitin-binding domains of mitophagy receptors such as OPTN, NBR1 (neighbor of BRCA1 gene1), NDP52/CALCOCO2 (nuclear dot protein 52 kDa or Calcium binding and coiled-coil 2), and TAX1BP1 (Tax1 binding protein 1) mediate their recruitment to damaged, ubiquitinated mitochondria (Lazarou et al. 2015, Moore & Holzbaur 2016). These receptors then bind LC3 via their LIR motifs to enhance the formation of LC3-positive autophagosomes that surround and engulf damaged mitochondria. Knockout studies in non-neuronal cells indicate that these receptors are partially redundant. Depletion of a single receptor such as OPTN may induce a delay in mitochondrial engulfment (Moore & Holzbaur 2016) but does not block mitophagy completely (Lazarou et al. 2015), indicating some redundancy in the system. One reason there may be multiple mitophagy receptors is to allow for differential regulation. For example, OPTN binds to and is activated by the kinase TBK1 (Wild et al., 2011; Richter et al., 2016) and is not recruited to damaged mitochondria when TBK1 activity is inhibited, whereas NDP52 is not affected by TBK1 inhibition (Moore & Holzbaur 2016). Of note, p62 has both a UBAN domain and an LIR motif, but does not promote autophagosome formation around damaged mitochondria; rather, p62 appears to cluster damaged mitochondria (Lazarou et al. 2015, Narendra et al. 2010b, Wong & Holzbaur 2014a).
The observation that OPTN and TBK1 are involved in mitophagy downstream from PINK1 and Parkin, is particularly interesting because mutations in both OPTN and TBK1 are causative for rare forms of familial amyotrophic lateral sclerosis (ALS) (Cirulli et al. 2015, Freischmidt et al. 2015, Maruyama et al. 2010). Thus, proteins involved in mitophagy are implicated in two major neurodegenerative diseases, both PD and ALS, making it particularly important to understand how this pathway contributes to neuronal homeostasis and neurodegeneration.
In mice, knockout of neither PINK1 nor Parkin is lethal. Knockout mice exhibit relatively subtle phenotypes, including altered synaptic excitability and decreased dopamine release, which do not clearly model human PD (Goldberg et al. 2003, Kitada et al. 2007, Perez & Palmiter 2005). Analysis of mitochondrial turnover in PINK1 knockout mice suggests that there are additional, PINK1-independent pathways leading to mitophagy in neurons under basal conditions (McWilliams et al. 2018), which remain to be more fully characterized. For example, mitochondrial ubiquitin ligase 1 (MUL1 or Mulan) has been proposed to function in a parallel pathway to Parkin in the clearance of damaged mitochondria (Yun et al. 2014). One possibility is that MUL1 or another ubiquitin ligase pathway may compensate for loss of PINK1/Parkin activity in some contexts such as neurodevelopment, but not others, for example in aging dopaminergic neurons. The importance of more fully understanding mitochondrial quality control is emphasized by the observation that both PINK1 and Parkin knockout mice are susceptible to stress, and that this in turn leads to inflammation (Sliter et al. 2018). Further work exploring the links between mitophagy and inflammation, and the possible contribution of this crosstalk to neurodegenerative disease in humans is now required.
ERphagy
The ER forms an extensive and dynamic network of sheets, tubules and cisternae that extends throughout the cell. Three-dimensional reconstructions of the ER in mouse brain show the dramatic extension of ER tubules throughout the soma, dendrites and axon of neurons in vivo (Wu et al. 2017). The ER must be remodeled and renewed in neurons, especially under conditions of stress. One mechanism for turnover is ERphagy (also referred to as reticulophagy), the selective removal of ER segments by autophagy (Grumati et al. 2018).
Recent progress has identified multiple ER-associated proteins with LIR motifs that mediate binding to LC3-family proteins, including FAM134B/RETREG1 (reticulophagy regulator 1), reticulon 3L (RTN3), and CCPG1 (cell cycle progression 1) (Fumagalli et al. 2016, Grumati et al. 2018, Khaminets et al. 2015, Smith et al. 2018). FAM134B and RTN3 are proposed to function in the basal remodeling of ER membranes by autophagy, while CCPG1 is induced by ER stress (Grumati et al. 2018, Smith et al. 2018). As resident ER transmembrane proteins, Atlastins are proposed to remodel the ER to sequester FAM134B-marked membrane for delivery to the autophagosome (Liang et al. 2018). Future studies are likely to further implicate the dynamics of ER remodeling and ERphagy in the maintenance of neuronal homeostasis and link further defects in this pathway to neurodegenerative disease.
Aggrephagy
Misfolded or damaged proteins can be cleared by the ubiquitin-proteasome system, but once misfolded proteins aggregate, these accumulations are cleared by autophagy. The selective removal of aggregated proteins is termed aggrephagy and is mediated in neurons by several receptors that may work either independently or in combination.
The best characterized autophagy receptor known to function in aggrephagy is p62/SQSTM1 (Bjørkøy et al. 2005, Komatsu et al. 2007a, Pankiv et al. 2007). Like other receptors involved in selective autophagy, p62 is recruited to cargo via its ubiquitin binding domain and recruits autophagy machinery through its LIR motif, which mediates binding to the ATG8 proteins. In vitro studies indicate that p62 is sufficient to induce the clustering of ubiquitinated proteins, known as cargo nucleation, into aggregates that can be targeted by autophagy (Zaffagnini et al. 2018). In cells, p62 also induces cargo clustering (Komatsu et al. 2007a), but is also known to interact with other proteins to facilitate aggrephagy. Supporting the importance of p62 in neurons, mutations in p62/SQSTM1 are implicated in ALS/FTD (frontotemporal dementia) and ataxia (Fecto et al. 2011, Haack et al. 2016).
ALFY (autophagy-linked FYVE protein, also known as WDFY3 for WD repeat and FYVE domain-containing protein) is a large scaffolding molecule that binds to p62, the ATG8 proteins LC3C and the GABARAPs, and ATG5, a component of the core autophagy machinery (Lystad et al. 2014). In vivo studies support a role for ALFY in aggrephagy, as the Drosophila mutant blue cheese demonstrates accumulation of ubiquitin-positive inclusions, neurodegeneration, and shortened lifespan (Finley et al. 2003). In mice, deletion of the gene encoding Alfy leads to pronounced defects in axon outgrowth, due to a failure to respond correctly to axon guidance cues. Baseline defects in autophagy were not observed, but an exogenous aggregation-prone substrate accumulated more rapidly in Alfy knockout MEFs (Dragich et al. 2016).
Another, similar, protein that can function along with p62 in aggrephagy is WDR81, a BEACH (beige and Chediak-Higashi) and WD40 repeat protein. WDR81 (WD repeat domain 81) can bind directly to LC3, but also interacts with p62, and has been proposed to facilitate the ability of p62 to bind to aggregated and ubiqutinated proteins (Liu et al. 2017). In cells, depletion of WDR81 leads to accumulation of both p62 and ubiquitinated proteins. p62 appears to sit at a crucial juncture between the ubiquitin proteasome and selective autophagy by detecting the buildup of ubiquitinated substrates (Danieli & Martens 2018), suggesting a key role in regulation of protein turnover.
SPATIAL ORGANIZATION OF AUTOPHAGY IN NEURONS
Compartmentalization of Neurons
Neurons are highly specialized cells that are polarized, long-lived, very metabolically active, and post-mitotic. Elucidated by Santiago Ramón y Cajal using Golgi stain in the 19th century, neuronal structure is generally divided into three compartments: the soma, the axon, and the dendrites. Axons can grow from a few micrometers up to many feet to reach their targets, while dendrites are typically much shorter but can form elaborate highly branched networks. Neurons perform the tasks of processing and transmitting information, placing high metabolic demands on these cells. Furthermore, these demands are primarily placed at the isolated regions of the neurons, far away from the cell body, in the axon and dendrites. Thus, the most vulnerable regions of neurons bear the brunt of the stress.
In addition, neurons are post-mitotic cells, with no ability to dilute damaged or accumulated proteins and organelles by cell division, a strategy commonly employed by mitotic cells. Furthermore, neurons typically terminally differentiate very early during development, almost always during embryogenesis, and must survive the lifetime of the organism. While there is some ability to regenerate axons and replace neurons, depending on species, age, neuronal type, and other factors, most organisms do not appear capable of replacing every neuron throughout the organism’s lifetime. Therefore, neurons need to robustly manage the stresses placed on remote compartments throughout the lifetime of the organism by removing aggregated and/or damaged proteins and organelles. Autophagy can, and does, perform this function in each neuronal compartment.
Axonal Autophagy
Landmark work in cultured embryonic peripheral neurons determined that large (~1 μm) acidified vesicles were transported retrogradely in axons toward the soma, suggesting that autophagosomes formed in the distal axon (Hollenbeck 1993). This work was consistent with electron microscopy studies of primary rat sympathetic motor neurons identifying autophagic vesicles in distal growth cones (Bunge 1973). Subsequently, several labs used a variety of neuronal culture paradigms, including mouse and rat dorsal root ganglion (DRG) neurons and mouse embryonic cortical and hippocampal neurons expressing autophagosomal marker GFP-LC3 (Mizushima et al. 2004) to observe autophagosome biogenesis and dynamics in axons. These studies consistently observed large, LC3-positive organelles forming constitutively at the axonal tip. These autophagosomes were then rapidly transported back to the cell body (Cheng et al. 2015, Lee et al. 2011, Maday & Holzbaur 2014, Maday et al. 2012).
Furthermore, these findings have been corroborated in vivo in model organisms. In Caenorhabditis elegans, autophagosome biogenesis was observed at synaptic sites in the distal axon of interneuron AIY at a consistent rate (Stavoe et al. 2016). In Drosophila, autophagosome biogenesis also occurred at the neuromuscular junction (NMJ) in the distal axon of motor neurons (Neisch et al. 2017, Soukup et al. 2016). While autophagosome biogenesis was observed consistently at the NMJ, newly formed autophagosomes were only rarely detected in the soma (Soukup et al. 2016). At the Drosophila NMJ presynapse, autophagy is dependent on LRRK2, a protein implicated in Parkinson’s disease, and EndophilinA, which recruits Atg3 to membranes (Soukup et al. 2016, Vanhauwaert et al. 2017). Similarly, in the Zebrafish photoreceptor, synaptojanin, a lipid phosphatase and EndophilinA interactor, is necessary for autophagy (George et al. 2016).
The step-wise autophagosome biogenesis pathway elucidated in yeast and mammalian cell culture (Itakura & Mizushima 2010, Koyama-Honda et al. 2013) appears to be consistent with the pathway for biogenesis in the distal axon (Maday & Holzbaur 2014). One significant difference is that axonal autophagosomes form constitutively under fed conditions (Cheng et al. 2015, Lee et al. 2011, Maday & Holzbaur 2014, 2016; Maday et al. 2012, Stavoe et al. 2016), in contrast, to the induction of autophagy in nutrient-limiting conditions observed in many non-neuronal cell types.
One intriguing aspect of axonal autophagy is the temporal regularity of biogenesis in the distal axon. While the rates of autophagosome biogenesis vary with species and neuronal type, rates of formation are highly consistent across individual neurons within a type, even from different individual animals (Maday & Holzbaur 2014, Maday et al. 2012, Stavoe et al. 2016). Another striking aspect of this pathway is the spatial specificity, with the majority of autophagosome biogenesis events, as labeled with ATG13 or ATG5 and LC3B, occurring in the distal axon. Autophagosome formation is only rarely observed in the dendrites or soma in primary mouse adult DRG or embryonic hippocampal neurons under resting conditions. (Maday & Holzbaur 2014).
Active zone proteins Bassoon and Piccolo may contribute to modulation of presynaptic autophagy. In primary hippocampal embryonic rat neurons, knockdown of Bassoon caused an accumulation of LC3-positive structures at presynaptic boutons. Bassoon negatively regulated autophagy by interacting, sequestering, and preventing ATG5 from acting in autophagosome biogenesis (Okerlund et al. 2017). Engulfed cargos include mitochondrial fragments as well as cytosolic proteins and protein aggregates, indicating that uptake is via non-selective, bulk autophagy (Maday et al. 2012, Wong & Holzbaur 2014b)
Once formed, axonal autophagosomes undergo bidirectional movement along microtubules in the distal axon driven by kinesin and dynein motors that localize to neuronal autophagosomes (Maday et al. 2012). Dynein is recruited to autophagosomes by RAB7 and RILP (Cheng et al. 2015, Jordens et al. 2001, Wijdeven et al. 2016). However, the mechanisms regulating kinesin recruitment have yet to be identified. After an initial phase of bidirectional movement, autophagosomes transition to highly processive retrograde transport toward the cell body driven by dynein and regulated by its scaffolding proteins JIP1, huntingtin, and HAP1 (huntingtin-associated protein 1) (Fu et al. 2014, Wong & Holzbaur 2014b). This robust retrograde trafficking of autophagosomes has been confirmed in vivo in C. elegans (Stavoe et al. 2016) and Drosophila (Neisch et al. 2017), where the STRIPAK complex was implicated as a key regulatory factor.
Following initial formation and as they transit through the axon toward the soma, autophagosomes encounter and fuse with late endosomes and lysosomes, leading to increasingly acidic and eventually degradation-competent autolysosomes (Maday et al. 2012). While there are lysosomes present in the axon, they lack the full complement of degradative enzymes found in somal lysosomes (Cheng et al. 2018, Gowrishankar et al. 2015). It is likely that as an autophagosome is transported retrogradely along the axon, it can fuse with additional, and more degradatively competent, lysosomes. In support of this possibility, blocking the retrograde trafficking of autophagosomes is sufficient to block their acidification and their degradation of engulfed cargos (Fu et al. 2014, Wong & Holzbaur 2014b).
Dendritic autophagy
The GABARAP subfamily of mATG8s, as its name suggests, was initially identified to interact with GABAA receptors by yeast-two-hybrid screening (Wang et al. 1999). GABARAP was then subsequently found to be important for GABAA receptor intracellular trafficking (Leil 2004), but the cellular mechanism of GABARAP regulation of GABAA receptor trafficking remains unclear. In PC12 cells, primary hippocampal neurons, and Xenopus oocytes, GABARAP lipidation was required for increased surface expression of GABAA receptors (Chen et al. 2007). In contrast, in C. elegans, GABAA receptors colocalized with autophagosome markers and autophagy reduced GABAA receptor surface expression in nonenervated muscle. In contrast, acetylcholine (ACh) receptor subunits did not colocalize with autophagosome markers, and disruption of autophagy did not increase ACh currents. These data indicate that autophagy selectively regulates surface expression of GABAA receptors, suggesting that autophagy could modulate neuronal excitation and inhibition (Rowland 2006).
Autophagy has also been implicated in AMPA receptor degradation in primary rat embryonic hippocampal neurons. Upon stimulation with low-dose NMDA, dendritic spines displayed an increase in GFP-LC3 puncta. When treated with bafilomycin A to inhibit lysosomal acidification, LC3-positive puncta were also detectable in the dendritic shaft with or without stimulation by NMDA (Shehata et al. 2012).
Autophagy can also modulate dendritic branching. In Drosophila, knockdown of autophagy genes reduced dendritic arbor growth and terminal branching in multidendritic sensory neurons in vivo (Clark et al. 2018). Surprisingly, overexpression of Atg1 also decreased dendritic arbor growth and terminal branching in the same neurons. These results suggest that constitutive autophagy must be tightly regulated during dendrite outgrowth and branching.
Additionally, autophagy can regulate dendritic degeneration. Specifically deleting Atg7 in DA neurons using the tyrosine hydroxylase (TH) promoter in conditional knock-out mice resulted in dopaminergic dendrites with many large swellings and dendritic dystrophy in TH+ neurons. These swellings contained ubiquitinated filamentous inclusions that were also observed in cell bodies (Friedman et al. 2012). When Atg7 was specifically deleted from forebrain excitatory neurons in vivo, dendritic spines failed to undergo pruning, leading to social behavioral defects. Similar results were observed in primary hippocampal neurons subjected to Atg7 shRNA, with fewer dendritic spines being eliminated compared to control neurons. These results indicated that basal autophagy is necessary for postnatal spine pruning (Tang et al. 2014).
Somal autophagy
Little is known about bulk autophagy in the neuronal cell body. In mouse primary DRGs, autophagosome biogenesis primarily occurs in the distal axon, with very few autophagosome formation events, as marked by ATG13 or ATG5, observed in the soma (Maday & Holzbaur 2014). In mouse primary hippocampal neurons, basal autophagosome biogenesis was observed in the cell body, but autophagosomes generated in the soma were characteristically distinct from axonally-generated autophagosomes; soma-derived autophagosomes were less dynamic and mature (Maday & Holzbaur 2016).
There is some debate about whether neuronal mitophagy occurs in the axon or in the soma. In vitro experiments with primary neurons have shown that mitochondrial fragments are engulfed within autophagosomes formed constitutively in the distal axon; however there is limited evidence that the engulfment of these fragments involves the machinery for selective mitophagy such as PINK1, Parkin and OPTN. Studies looking more specifically at the mitophagy pathway provide conflicting views. One group reported localized mitophagy along the axons of hippocampal neurons upon induction of cellular stress via mitochondrial depolarization or ROS generation (Ashrafi et al. 2014). In contrast, mitochondrial depolarization in cortical neurons resulted in Parkin-labeled mitochondria accumulation in somatodendritic regions that were subsequently degraded locally by the autophagosomal-lysosomal pathway. Conversely, GFP-LC3 was rarely observed associated with axonal mitochondria in these experiments (Cai et al. 2012).
In vivo studies provide more convincing evidence that mitophagy occurs predominately in the cell body. Detailed characterization of neurons from PINK1 or Parkin mutant flies indicate that loss of either protein decreases the flux of mitochondria along the axon, but does not lead to the accumulation of senescent mitochondria either in the axons of motor neurons or at neuromuscular junctions. Instead, there is a loss of integrity of somal mitochondria, indicating that active turnover of mitochondria by PINK1/Parkin-dependent mitophagy may be part of a mitochondrial quality control mechanism that is specific to the soma (Devireddy et al. 2015, Sung et al. 2016). Furthermore, initial observations from a mouse model expressing a mitophagy reporter (mito-QC) also support a soma-specific degradation pathway (McWilliams et al. 2016). This pathway may become more important in the maintenance of neuronal function through aging (Cornelissen et al. 2018).
An intriguing possibility is that neurons deploy alternative mechanisms to maintain mitochondrial health in the distal axon. Compared to mitochondria in cell lines, neuronal mitochondria are more resistant to initiation of mitophagy. Independent of Parkin, DRP1 (dynamin-related protein 1), and autophagy, mitochondrial fragments from stressed mitochondria are transported out of the axon in primary rodent neurons. Additionally, mitochondrial anchoring protein syntaphilin (SNPH) is released from axonal mitochondria upon stress, allowing their retrograde transport out of the axon (Lin et al. 2017). Thus, damaged axonal mitochondria may undergo retrograde transport to be degraded by mitophagy in the soma.
TEMPORAL ORGANIZATION OF AUTOPHAGY IN NEURONS
Autophagy during neurodevelopment
Development of a functional nervous system requires neuronal differentiation, neurite outgrowth, neurite guidance, and formation of synaptic connections. Non-neuronal support cells such as astrocytes, microglia, and oligodendrocytes are also required for proper nervous system function and maintenance. All these cells and processes must be coordinated so that each neuron connects with the appropriate partners in the correct order. Autophagy is an intriguing candidate to modulate these neurodevelopmental steps.
Autophagy can act during neurodevelopment in neuronal precursors. Conditionally knocking out mTOR in GABAergic precursors increased autophagy in those cells and suppressed their proliferation, leading to a reduction in cortical interneurons (Ka et al. 2017). Thus, autophagy can modulate the first step in neuronal development, differentiation, or generation of the neurons themselves.
Autophagy also regulates axon outgrowth. In embryonic primary cortical neurons, depletion of Atg7 led to an increase in neurite length. Conversely, activation of autophagy with Rapamycin treatment resulted in a decrease in neurite length (Ban et al. 2013). In SH-SY5Y cells, an in vitro cell line model of neurons, induction of autophagy with Tri-ortho-cresyl phosphate (TOCP) also inhibited neurite outgrowth (Chen et al. 2013). In the converse experiment in chick embryonic DRG neurons, when axon outgrowth was blocked with cytochalasin-E, retrograde transport of axonal autophagosomes increased threefold (Hollenbeck & Bray 1987). These observations were confirmed in vivo in C. elegans; mutants for autophagy components displayed longer axons in the nociceptive sensory neuron PVD. Of note, however, many of the neurons examined did not display axon outgrowth phenotypes in the autophagy mutants. Thus, autophagy may regulate different stages of neurodevelopment in different neurons or may not affect the neurodevelopment of certain neurons at all (Stavoe et al. 2016). These subtle differences between neuron types may only become apparent in the context of intact nervous systems in vivo, in which neurons receive and respond to numerous cellular cues.
Finally, autophagy can also modulate synaptic formation. In a C. elegans interneuron, autophagy is required for synaptic vesicle clustering and active zone formation early during synaptogenesis. Autophagy component mutants displayed defects in active zone assembly and synaptic vesicle clustering in early larval stages, indicating that the observed defects were the result of improper synaptic formation instead of degeneration (Stavoe et al. 2016). Furthermore, mutations in selective autophagy genes did not phenocopy the bulk autophagy mutants (Stavoe et al. 2016), consistent with data indicating that nonselective autophagy is predominate in the axon. Similar results were observed in Drosophila. In atg1 (ULK1 in mammals) mutant animals, the NMJ displays reductions in both total NMJ area and number of synapses. As in worms, these defects in autophagy mutants were due to incorrect synaptic formation, not precocious degeneration or synaptic retraction (Wairkar et al. 2009). Similar NMJ undergrowth was observed in atg2 and atg18 (WIPIs in mammals) mutants. Additionally, overexpression of Atg1 increased NMJ synaptic bouton number in an autophagy-dependent manner (Shen & Ganetzky 2009). Taken together, these studies indicate that autophagy is an important regulator during neurodevelopment; autophagy negatively regulates axon outgrowth but positively regulates synaptic development. It is compelling to hypothesize that, at least for en passant synapses, these two processes may be related, with restrained axon outgrowth allowing for the proper deposition of synapses during neurodevelopment.
Autophagy in neuronal homeostasis
Autophagy is known to regulate cellular homeostasis in non-neuronal cells, especially in response to cellular stressors. Autophagy also acts to maintain neuronal homeostasis. Much of the evidence to support this derives from studies reducing one or more autophagy genes in neurons and observing protein accumulation or neurodegeneration. Conditional knock-out of Atg5 in mouse neural precursor cells led to partial loss of Purkinje cells and cerebral cortical pyramidal cells, and axonal swelling in numerous brain regions including cerebral cortex, hippocampus, and the nucleus gracilis. Neurons in a variety of brain regions, such as the cerebral cortex, hippocampus, striatum, and dorsal root ganglion, also exhibited accumulations of ubiquitin-positive inclusion bodies (Hara et al. 2006). Similarly, knockdown of Atg5 in M17 human neuroblastoma cells resulted in increased aggregations of α-synuclein oligomers (Yu et al. 2009).
Conditional knock-out of Atg7 in mouse neural precursors using the Nestin promoter resulted in neuronal death and accumulation of ubiquitin-positive structures and inclusion bodies in neurons in a variety of brain regions including cerebral cortex, Purkinje cells, and hippocampal pyramidal neurons (Komatsu et al. 2006). Likewise, conditionally knocking out Atg7 in Purkinje cells using the Pcp2 promoter in mice did not appear to affect dendrites, but did lead to neuronal and axonal dystrophy, degeneration of axon terminals, and eventual neuronal death (Komatsu et al. 2007b). Additionally, conditional knock-out of Atg7 in dopaminergic neurons using the DAT, EN, or TH promoters produced neuron loss, loss of striatal dopamine, accumulation of ubiquitin- and p62-containing inclusions, and accumulation of α-synuclein in vivo (Ahmed et al. 2012, Sato et al. 2018).
Neuron-specific knock-out of FIP200 in mice using the Nestin promoter triggered neuronal loss of Purkinje cells and granular cells, spongiosis in the cerebellar white matter, axonal and dendritic degeneration in Purkinje cells, axonal swelling, and accumulation of ubiquitin-positive aggregates (Liang et al. 2010). In complementary experiments, knockdown of FIP200 in Neuro-2a neuroblastoma cells led to neurite atrophy and apoptosis (Chano et al. 2007).
In transgenic mice overexpressing amyloid precursor protein (APP) and heterozygous for a Beclin1 deletion, synapses and dendrites degenerated in neocortex and hippocampus and layer II neurons of the entorhinal cortex were lost (Pickford et al. 2008). Analogously, knockdown of Beclin1 in M17 human neuroblastoma cells resulted in increased aggregations of α-synuclein oligomers (Yu et al. 2009). Conversely, overexpressing Beclin1 in B103 rat neuroblastoma cells overexpressing α-synuclein reduced accumulation of α-synuclein and restored neurite outgrowth. Furthermore, lentiviral expression of Beclin1 in the temporal cortex and hippocampus of α-synuclein transgenic mice reduced intraneuronal α-synuclein accumulation (Spencer et al. 2009). These experiments implicate Beclin1 in neuronal homeostasis, especially in the context of Alzheimer’s and Parkinson’s diseases.
FIP200, Beclin1, ATG5, and ATG7 all act early in autophagosome biogenesis; deletion of any of these genes results in failure to form autophagosomes. Thus, all of the studies discussed so far examined the effects of loss of autophagosome formation on neurons. However, inhibiting later stages of autophagy would allow initiation of autophagy, but disrupt complete sequestration and degradation of cargo. Knock-out of Epg5, which acts downstream from known Atg genes (Lu et al. 2011, Tian et al. 2010), yielded p62 aggregations and ubiquitin-positive inclusions in brain and spinal cord extracts, a reduction of pyramidal and motor neurons, axonal degeneration, and accumulation of TDP43 aggregates in neurons (Zhao et al. 2013).
Selective autophagy has also been implicated in neuronal homeostasis. Genetic studies indicate that FAM134B, an ERphagy receptor, is required to maintain neuronal homeostasis, as mutations cause a sensory and autonomic neuropathy known as HSAN-II (Kurth et al. 2009). Similarly, Fam134b−/− mice exhibit sensory neuropathy and an age-dependent loss of sensory axons, with impaired ER turnover apparent at the cellular level (Khaminets et al. 2015). Furthermore, the ER-associated proteins Atlastins function downstream of FAM134B (Liang et al. 2018). Of note, a loss-of-function mutation in the Atlastin ATL1 is causal for SPG3A, a hereditary spastic paraplegia (Namekawa et al. 2007).
Aggrephagy also plays a role in neuronal homeostasis. Conditional knock-out of aggrephagy receptor WDR81 using the Nestin promoter led to accumulation of p62 foci in cortical and striatal neurons (Liu et al. 2017). In humans, a missense mutation in WDR81 is associated with the rare neurodevelopmental condition known as CAMRQ for cerebellar ataxia, mental retardation, and disequilibrium syndrome (Gulsuner et al. 2011). Similarly, mice carrying a homozygous missense mutation in WDR81 exhibited Purkinje cell and photoreceptor cell death (Traka et al. 2013). These observations support a key role for WDR81 and other p62-interacting proteins (for example, see (Haidar et al. 2019)) in the maintenance of neuronal homeostasis, likely due to its role in aggrephagy within neurons. Together, these studies indicate the importance of the autophagy pathway in neuronal homeostasis, as loss of a variety of autophagosome biogenesis components resulted in aggregated proteins, neurite degeneration, and neuron loss.
Autophagy in neuronal activity, plasticity, and memory
Given the established role of autophagy in presynaptic compartments, an obvious experimental path was to determine the relationship between autophagy and synaptic activity. Upon NMDA stimulation of mouse cerebral microexplant cultures, the number of axonal GFP-LC3-positive puncta increased 2.5-fold. Although the number of autophagosomes increased in the axon, NMDA treatment did not appear to affect retrograde transport of autophagosomes (Katsumata et al. 2010). Similarly, in rat primary hippocampal neurons, stimulation with KCl or NMDA induced axonal autophagy (Shehata et al. 2012, Wang et al. 2015). Stimulation with KCl or chemical long-term potentiation also induced a higher LC3-II to LC3-I ratio, a common measure of autophagic flux, in primary embryonic rat hippocampal neurons (Glatigny et al. 2019).
Furthermore, inactivation of ATG7 selectively in dopamine neurons in mice enhanced evoked dopamine secretion and accelerated recovery. Similarly, rapamycin treatment, which induces autophagy, decreased synaptic vesicles in dopaminergic neurons in wild type mice, but not in ATG7-defective mice. Thus, these data suggest that autophagy acts in dopamine neurons to restrict synaptic transmission (Hernandez et al. 2012).
Autophagy also influences memory. Mice that had AAV-hippocampal injections of Fip200, Beclin1, or Atg12 shRNA displayed defects in hippocampal-associated memory performances, including novel object recognition and context-elicited freezing. Inducing autophagy in the hippocampus during training, but not twelve hours after training, enhanced performance in both memory paradigms. Conversely, acute inhibition of autophagy pharmacologically during training significantly decreased memory performances in either test. Consistent with these findings, inhibiting autophagy in primary rat hippocampal neurons or in vivo in the hippocampal dentate gyrus prevented an activity-dependent increase in dendritic spines (Glatigny et al. 2019).
Autophagy in aging
Whether protein levels of autophagy pathway components change with age is debated. In mouse hippocampus, VPS34, Beclin1, and ATG5 protein levels decreased between three and sixteen months of age. The LC3-II/LC3-I ratio also decreased with age (Glatigny et al. 2019). Similarly, in human brain, mRNA levels of Atg5, Atg7, Beclin1, and GABARAPL2 decrease with age; however, correlated protein level decreases were not assessed (Lipinski et al. 2010, Shibata et al. 2006). In Drosophila heads, dAtg8, dAtg2, and dAtg18 mRNA decreased with age (Simonsen et al. 2008). Conversely, transcriptome and proteome analysis in rat brain did not detect significant changes in autophagy pathway components between six months and 24 months (Ori et al. 2015).
Autophagosome biogenesis rates could change with age in neurons despite levels of autophagy proteins remaining constant with age. Post-translational modifications such as phosphorylation play important roles in the autophagy pathway. Altered levels of protein modification could alter the kinetics of autophagosome biogenesis independent of expression levels. Subcellular localization, as described above as especially important in neurons, could also affect rates of autophagosome biogenesis with age. In C. elegans, autophagy activity generally decreases with age. However, age appears to affect neuronal autophagy more variably than other tissues (Chang et al. 2017).
Modulating autophagy in neurons can impact longevity and lifespan. In Drosophila, Atg8a mutants exhibited decreased lifespans and increased ubiquitinated aggregates in neurons. Conversely, overexpression of dAtg8 in the CNS using the APPL-Gal4 driver prevented accumulation of protein aggregates and dramatically extended lifespan. However, driving dAtg8 overexpression pan-neuronally using earlier-expressing ELAV-Gal4 did not extend lifespan, suggesting that the age of autophagy ectopic induction is important (Simonsen et al. 2008). In C. elegans, inhibiting components of the initiation and nucleation complexes in neurons after reproduction ended extended lifespan, while inhibiting initiation or nucleation in pre-reproductive animals decreased lifespan. These data suggest that preventing the accumulation of partially formed autophagosomes is beneficial in post-reproductive animals (Wilhelm et al. 2017).
AUTOPHAGY IN DISEASE
The major neurodegenerative diseases associated with aging, including Alzheimer’s disease (AD), PD, Huntington’s disease (HD), ALS, and frontotemporal dementia (FTD) are all characterized by the accumulation of aggregated proteins and damaged mitochondria (Chu 2018). Observations from both experimental models and post-mortem analysis of human tissues suggest that dysfunction of autophagy may be a common contributor to the pathogenic process across these diseases (recently reviewed by (Boland et al. 2018, Chu 2018)). This hypothesis is further supported by the observation that mutations in many of the proteins involved in autophagy are sufficient to cause neurodevelopmental or neurodegenerative disease, including PINK1, Parkin, OPTN, TBK1, ATL1, SQSTM1, and WDR81 as discussed above, as well as ATG5, AP4, HTT, WIPI4, and DYNC1H1 (reviewed (Zhu et al. 2018)).
However, there is no consensus as of yet on what step or steps might be disrupted in the major neurodegenerative diseases such as AD, PD, or ALS. Studies to date have demonstrated aging or disease-associated deficits at multiple points in the pathway, including autophagosome biogenesis (De Pace et al. 2018, Rui et al. 2015, Stavoe & Holzbaur 2018), cargo-loading (Martinez-Vicente et al. 2010, Rudnick et al. 2017), intracellular transport (Nicolas et al. 2018, Wong & Holzbaur 2014b), and autophagosome-lysosome fusion or acidification (Lie & Nixon 2018, Nixon et al. 2005) across disease models. Deficits in intersecting pathways such as the ubiquitin-proteasome pathway and lysosomal degradation via chaperone-mediated autophagy are also likely to contribute to neurodegenerative disease (Scrivo et al. 2018). Further, it remains to be determined whether defects in neuronal autophagy are central, or if deficits in autophagy or related degradative pathways in support cells such as glia also contribute to age-related neurodegeneration.
It will be essential to fill in these gaps in order to develop effective therapeutic strategies. For example, non-selective axonal autophagy is differentially regulated than the selective removal of damaged mitochondria from the soma (Cornelissen et al. 2018, Devireddy et al. 2015, McWilliams et al. 2016, Sung et al. 2016). It is also unclear if autophagy in neurons responds to the same activators that induce autophagy in other tissues, such as starvation or inhibition of mTORC1 (Maday & Holzbaur 2016). However, the growing prevalence of neurodegeneration in our aging populations means that we must actively screen for therapeutic approaches now. (Boland et al. 2018) provide an extensive discussion of pharmacotherapies that have been tested or are currently being considered for the modulation of intracellular degradation pathways in the context of neurodegeneration.
FUTURE DIRECTIONS
Genetic, cellular and in vivo studies all highlight the key role for autophagy in maintaining cellular homeostasis in the neuron, and the importance of autophagy in neurodevelopment and neuronal function. While there has been considerable progress in the past few years, key questions remain. First, while the pathway for canonical autophagy is well-understood, we need to define the molecular components and cellular dynamics involved in more specific pathways, such as selective autophagy. For example, how is ER health maintained along the extended axon? Is ER membrane turned over continually, or only when damage is sensed? In regard to mitophagy, we now better understand the outlines of PINK1/Parkin mediated mitochondrial clearance, but there is good evidence for the participation of other parallel pathways to maintain mitochondrial quality control, and these need to be explored in more detail.
Next, we are only beginning to understand the effects of neuronal activity on autophagy, and how this may contribute to neuronal plasticity such as learning and memory. This leads directly to the next question – does neuronal autophagy change with aging? Does this affect our ability to learn or maintain memories, or does it have a more direct effect on neuronal homeostasis? If so, then how might a decline in autophagy contribute to neurodegeneration? Defects in autophagy have been implicated in all the major neurodegenerative diseases, including ALS, AD, PD, and HD, so further research on the underlying mechanisms is clearly necessary.
And finally, can autophagy and other degradative pathways in neurons be modulated therapeutically? There is evidence that neuronal autophagy is not regulated by the same pathways that regulate stress-induced autophagy in other tissues, so we need to identify the regulatory pathways involved and determine how they can be manipulated in vivo. The availability of small molecule effectors will allow us to determine if autophagy can be tuned to make neurons more resilient to the cellular stressors of aging, environmental toxins, or genetic risk factors, offering some hope for the future.
ACKNOWLEDGEMENTS
The authors thank members of the Holzbaur lab for thoughtful comments on the manuscript. This work was supported by the National Institutes of Health - AKHS was funded by K99 NS109286, and ELFH was funded by R37 NS060698.
REFERENCES
- Ahmed I, Liang Y, Schools S, Dawson VL, Dawson TM, Savitt JM. 2012. Development and Characterization of a New Parkinson’s Disease Model Resulting from Impaired Autophagy. J. Neurosci 32(46):16503–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL. 2014. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol. 206(5):655–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban B-K, Jun M-H, Ryu H-H, Jang D-J, Ahmad ST, Lee J-A. 2013. Autophagy Negatively Regulates Early Axon Growth in Cortical Neurons. Mol. Cell. Biol 33(19):3907–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Sheng M. 2011. Deconstruction for Reconstruction: The Role of Proteolysis in Neural Plasticity and Disease. Neuron. 69(1):22–32 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, et al. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171(4):603–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Yu WH, Corti O, Mollereau B, Henriques A, et al. 2018. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 17(9):660–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge MB. 1973. Fine structure of nerve fibers and growth cones of isolated sympathetic neurons in culture. J. Cell Biol. 56(3):713–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Zakaria HM, Simone A, Sheng ZH. 2012. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol 22(6):545–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E, Van Der Vaart A, Zhao M, Rieter E, Klionsky DJ, et al. 2012. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol 22(17):1545–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Kumsta C, Hellman AB, Adams LM, Hansen M. 2017. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife. 6:e18459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chano T, Okabe H, Hulette CM. 2007. RB1CC1 insufficiency causes neuronal atrophy through mTOR signaling alteration and involved in the pathology of Alzheimer’s diseases. Brain Res. 1168:97–105 [DOI] [PubMed] [Google Scholar]
- Chen JX, Sun YJ, Wang P, Long DX, Li W, et al. 2013. Induction of autophagy by TOCP in differentiated human neuroblastoma cells lead to degradation of cytoskeletal components and inhibition of neurite outgrowth. Toxicology. 310:92–97 [DOI] [PubMed] [Google Scholar]
- Chen ZW, Chang CS, Leil TA, Olsen RW. 2007. C-terminal modification is required for GABARAP-mediated GABA(A) receptor trafficking. J. Neurosci 27(25):6655–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng XT, Xie YX, Zhou B, Huang N, Farfel-Becker T, Sheng ZH. 2018. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol. 217(9):3127–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng XT, Zhou B, Lin MY, Cai Q, Sheng ZH. 2015. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J. Cell Biol. 209(3):377–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT. 2018. Mechanisms of selective autophagy and mitophagy: Implications for neurodegenerative diseases. Neurobiol. Dis 122(March 2018):23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, et al. 2015. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science (80-.). 347:1436–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Graybeal LL, Id SB, Thomas C, Bhattacharya S, Id DNC. 2018. Basal autophagy is required for promoting dendritic terminal branching in Drosophila sensory neurons. PLoS One. 13(11):1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen T, Vilain S, Vints K, Gounko N, Verstreken P, Vandenberghe W. 2018. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in drosophila. Elife. 7: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danieli A, Martens S. 2018. p62-mediated phase separation at the intersection of the ubiquitin-proteasome system and autophagy. J. Cell Sci. 131(19): [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pace R, Skirzewski M, Damme M, Mattera R, Mercurio J, et al. 2018. Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLoS Genet. 14(4): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy S, Liu A, Lampe T, Hollenbeck PJ. 2015. The Organization of Mitochondrial Quality Control and Life Cycle in the Nervous System In Vivo in the Absence of PINK1. J. Neurosci 35(25):9391–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragich JM, Kuwajima T, Hirose-Ikeda M, Yoon MS, Eenjes E, et al. 2016. Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife. 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, et al. 2011. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol 68(11):1440–46 [DOI] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ. 2014. The machinery of macroautophagy. Cell Res. 24(1):24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, et al. 2003. Blue Cheese Mutations Define a Novel, Conserved Gene Involved in Progressive Neural Degeneration. J. Neurosci 23(4):1254–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, et al. 2015. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci 18(5):631–36 [DOI] [PubMed] [Google Scholar]
- Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, et al. 2012. Disrupted Autophagy Leads to Dopaminergic Axon and Dendrite Degeneration and Promotes Presynaptic Accumulation of -Synuclein and LRRK2 in the Brain. J. Neurosci 32(22):7585–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M meng, Nirschl JJ, Holzbaur ELF. 2014. LC3 Binding to the scaffolding protein jip1 regulates processive dynein-driven transport of autophagosomes. Dev. Cell 29(5):577–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Presmanes EC, Pisoni GB, et al. 2016. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 18(11):1173–84 [DOI] [PubMed] [Google Scholar]
- George AA, Hayden S, Stanton GR, Brockerhoff SE. 2016. Arf6 and the 5’phosphatase of synaptojanin 1 regulate autophagy in cone photoreceptors. BioEssays. 38: [DOI] [PubMed] [Google Scholar]
- Glatigny M, Moriceau S, Rivagorda M, Ramos-Brossier M, Nascimbeni AC, et al. 2019. Autophagy Is Required for Memory Formation and Reverses Age-Related Memory Decline. Curr. Biol 29(3):435–48 [DOI] [PubMed] [Google Scholar]
- Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, et al. 2003. Parkin-deficient Mice Exhibit Nigrostriatal Deficits but not Loss of Dopaminergic Neurons. J. Biol. Chem 278(44):43628–35 [DOI] [PubMed] [Google Scholar]
- Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, et al. 2015. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc. Natl. Acad. Sci. U. S. A 112(28):E3699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Dikic I, Stolz A. 2018. ER-phagy at a glance. J. Cell Sci. 131(17): [DOI] [PubMed] [Google Scholar]
- Gulsuner S, Tekinay AB, Doerschner K, Boyaci H, Bilguvar K, et al. 2011. Homozygosity mapping and targeted genomic sequencing reveal the gene responsible for cerebellar hypoplasia and quadrupedal locomotion in a consanguineous kindred. Genome Res. 21(12):1995–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack TBB, Iuso A, Kremer LSS, Hartig M, Strom TMM, et al. 2016. Absence of the Autophagy Adaptor SQSTM1/p62 Causes Childhood-Onset Neurodegeneration with Ataxia, Dystonia, and Gaze Palsy. Am. J. Hum. Genet 99(3):735–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar M, Asselbergh B, Adriaenssens E, De Winter V, Timmermans J-P, et al. 2019. Neuropathy-causing mutations in HSPB1 impair autophagy by disturbing the formation of p62/SQSTM1 bodies. Autophagy. 00(00):1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, et al. 2010. Mitochondria Supply Membranes for Autophagosome Biogenesis during Starvation. Cell. 141(4):656–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495(7441):389–93 [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441(7095):885–89 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11(12):1433–37 [DOI] [PubMed] [Google Scholar]
- Hernandez D, Torres CA, Setlik W, Cebrián C, Mosharov EV., et al. 2012. Regulation of Presynaptic Neurotransmission by Macroautophagy. Neuron. 74(2):277–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck PJ. 1993. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell Biol. 121(2):305–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck PJ, Bray D. 1987. Rapidly transported organelles containing membrane and cytoskeletal components: Their relation to axonal growth. J. Cell Biol. 105:2827–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408(6811):488–92 [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. 2008. Beclin 1 Forms Two Distinct Phosphatidylinositol 3-Kinase Complexes with Mammalian Atg14 and UVRAG. Mol. Biol. Cell 19(12):5360–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 6(6):764–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, et al. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol 11(21):1680–85 [DOI] [PubMed] [Google Scholar]
- Ka M, Smith AL, Kim WY. 2017. MTOR controls genesis and autophagy of GABAergic interneurons during brain development. Autophagy. 13(8):1348–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150(6):1507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, et al. 2014. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J. Cell Biol. 205(2):143–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata K, Nishiyama J, Inoue T, Mizushima N, Takeda J, Yuzaki M. 2010. Dynein- and activity-dependent retrograde transport of autophagosomes in neuronal axons. Autophagy. 6(3):378–85 [DOI] [PubMed] [Google Scholar]
- Kauffman KJ, Yu S, Jin J, Mugo B, Nguyen N, et al. 2018. Delipidation of mammalian Atg8-family proteins by each of the four ATG4 proteases. Autophagy. 14(6):992–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, et al. 2015. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 522(7556):354–58 [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. 2001. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase y sorting in Saccharomyces cerevisiae. J. Cell Biol. 153(3):519–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Pisani A, Porter DR, Yamaguchi H, Tscherter A, et al. 2007. Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc. Natl. Acad. Sci 104(27):11441–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441(7095):880–84 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou Y shin, Ueno T, et al. 2007a. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell. 131(6):1149–63 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Iwata J, et al. 2007b. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. U. S. A 104(36):14489–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, et al. 2012. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2(5):120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. 2013. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 9(10):1491–99 [DOI] [PubMed] [Google Scholar]
- Kumar N, Leonzino M, Hancock-Cerutti W, Horenkamp FA, Li PQ, et al. 2018. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217(10):3625–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, et al. 2009. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat. Genet 41(11):1179–81 [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, et al. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 524(7565):309–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sato Y, Nixon RA. 2011. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J. Neurosci 31(21):7817–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leil TA. 2004. GABAA Receptor-Associated Protein Traffics GABAA Receptors to the Plasma Membrane in Neurons. J. Neurosci 24(50):11429–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, et al. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10(7):776–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Wang C, Peng X, Gan B, Guan JL. 2010. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem 285(5):3499–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Ahmed S, Corn JE. 2018. Atlastins remodel the endoplasmic reticulum for selective autophagy. J. Cell Biol. 217(10):3354–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie PPY, Nixon RA. 2018. Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol. Dis 122:94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MY, Cheng XT, Tammineni P, Xie Y, Zhou B, et al. 2017. Releasing Syntaphilin Removes Stressed Mitochondria from Axons Independent of Mitophagy under Pathophysiological Conditions. Neuron. 94(3):595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K, Stenmark H. 2006. Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119(4):605–14 [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, et al. 2010. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci 107(32):14164–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li Y, Wang X, Xing R, Liu K, et al. 2017. The BEACH-containing protein WDR81 coordinates p62 and LC3C to promote aggrephagy. J. Cell Biol. 216(5):1301–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Yang P, Huang X, Hu W, Guo B, et al. 2011. The WD40 Repeat PtdIns(3)P-Binding Protein EPG-6 Regulates Progression of Omegasomes to Autophagosomes. Dev. Cell 21(2):343–57 [DOI] [PubMed] [Google Scholar]
- Lystad AH, Ichimura Y, Takagi K, Yang Y, Pankiv S, et al. 2014. Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep. 15(5):557–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Holzbaur ELF. 2014. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev. Cell 30(1):71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Holzbaur ELF. 2016. Compartment-Specific Regulation of Autophagy in Primary Neurons. J. Neurosci 36(22):5933–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur ELF. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196(4):407–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, et al. 2010. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci 13(5):567–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, et al. 2010. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 465(7295):223–26 [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, et al. 2010. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190(4):511–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, et al. 2009. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11(4):385–96 [DOI] [PubMed] [Google Scholar]
- McWilliams TG, Prescott AR, Allen GFG, Tamjar J, Munson MJ, et al. 2016. Mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214(3):333–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, et al. 2018. Basal Mitophagy Occurs Independently of PINK1 in Mouse Tissues of High Metabolic Demand. Cell Metab. 27(2):439–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, et al. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116(9):1679–88 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, et al. 1998. A protein conjugation system essential for autophagy. Nature. 395(6700):395–98 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabey Y, et al. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152(4):657–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 15(3):1101–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AS, Holzbaur ELF. 2016. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc. Natl. Acad. Sci 113(24):E3349–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Bergman P, Dodgson S, Marcellin D, Claerr I, et al. 2018. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 19(9): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Hama Y, Izume T, Tamura N, Ueno T, et al. 2018. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217(11):3817–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Yen WL, Mari M, Cao Y, Xie Z, et al. 2012. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy. 8(5):780–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa M, Muriel MP, Janer A, Latouche M, Dauphin A, et al. 2007. Mutations in the SPG3A gene encoding the GTPase atlastin interfere with vesicle trafficking in the ER/Golgi interface and Golgi morphogenesis. Mol. Cell. Neurosci 35(1):1–13 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183(5):795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, et al. 2010a. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Kane LA, Hauser DN, Fearnley IM, Youle RJ. 2010b. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 6(8):1090–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch AL, Neufeld TP, Hays TS. 2017. A STRIPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J. Cell Biol. 216(2):441–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, et al. 2018. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron. 97(6):1268–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, et al. 2005. Extensive Involvement of Autophagy in Alzheimer Disease: An Immuno-Electron Microscopy Study. J. Neuropathol. Exp. Neurol 64(2):113–22 [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y. 2006. Assortment of Phosphatidylinositol 3-Kinase Complexes-Atg14p Directs Association of Complex I to the Pre-autophagosomal Structure in Saccharomyces cerevisiae. Mol. Biol. Cell 17(April):1527–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y 2014. Historical landmarks of autophagy research. Cell Res. 24(1):9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okerlund ND, Reimer RJ, Schneider K, Leal-Ortiz S, Montenegro-Venegas C, et al. 2017. Bassoon Controls Presynaptic Autophagy through Atg5. Neuron. 93(4):897–913 [DOI] [PubMed] [Google Scholar]
- Ori A, Toyama BH, Harris MS, Bock T, Iskar M, et al. 2015. Integrated Transcriptome and Proteome Analyses Reveal Organ-Specific Proteome Deterioration in Old Rats. Cell Syst. 1(3):224–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, et al. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy*[S]. J. Biol. Chem 282(33):24131–45 [DOI] [PubMed] [Google Scholar]
- Perez FA, Palmiter RD. 2005. Parkin-deficient mice are not a robust model of parkinsonism. Proc. Natl. Acad. Sci 102(6):2174–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, et al. 2008. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid β accumulation in mice. J. Clin. Invest 118(6):2190–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Vicinanza M, Ashkenazi A, Gratian MJ, Zhang Q, et al. 2018. The RAB11A-Positive Compartment Is a Primary Platform for Autophagosome Assembly Mediated by WIPI2 Recognition of PI3P-RAB11A. Dev. Cell 45(1):114–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AM. 2006. Presynaptic Terminals Independently Regulate Synaptic Clustering and Autophagy of GABAA Receptors in Caenorhabditis elegans. J. Neurosci 26(6):1711–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan L, Zhou C, Jin E, Kucharavy A, Zhang Y, et al. 2017. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature. 543(7645):443–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick ND, Griffey CJ, Guarnieri P, Gerbino V, Wang X, et al. 2017. Distinct roles for motor neuron autophagy early and late in the SOD1 G93A mouse model of ALS. Proc. Natl. Acad. Sci 114(39):E8294–8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui YN, Xu Z, Patel B, Chen Z, Chen D, et al. 2015. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 17(3):262–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, et al. 2013. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15(7):741–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Uchihara T, Fukuda T, Noda S, Kondo H, et al. 2018. Loss of autophagy in dopaminergic neurons causes Lewy pathology and motor dysfunction in aged mice. Sci. Rep 8(1):2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrivo A, Bourdenx M, Pampliega O, Cuervo AM. 2018. Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol. 17(9):802–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, Inokuchi K. 2012. Neuronal Stimulation Induces Autophagy in Hippocampal Neurons That Is Involved in AMPA Receptor Degradation after Chemical Long-Term Depression. J. Neurosci 32(30):10413–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Ganetzky B. 2009. Autophagy promotes synapse development in Drosophila. J. Cell Biol. 187(1):71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, et al. 2012. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep 2(1002): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, et al. 2006. Regulation of intracellular accumulation of mutant huntingtin by beclin 1. J. Biol. Chem 281(20):14474–85 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. 2008. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 4(2):176–84 [DOI] [PubMed] [Google Scholar]
- Sliter DA, Martinez J, Hao L, Chen X, Sun N, et al. 2018. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 561(7722):258–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, et al. 2018. CCPG1 Is a Non-canonical Autophagy CargoReceptor Essential for ER-Phagyand Pancreatic ER Proteostasis. Dev. Cell 44(2):217–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup SF, Kuenen S, Vanhauwaert R, Manetsberger J, Hernández-Díaz S, et al. 2016. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron. 92(4):829–44 [DOI] [PubMed] [Google Scholar]
- Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, et al. 2009. Beclin 1 Gene Transfer Activates Autophagy and Ameliorates the Neurodegenerative Pathology in alpha-Synuclein Models of Parkinson’s and Lewy Body Diseases. J. Neurosci 29(43):13578–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AKH, Hill SE, Hall DH, Colón-Ramos DA. 2016. KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Dev. Cell 38(2):171–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AKH, Holzbaur ELF. 2018. Expression of WIPI2B counteracts age-related decline in autophagosome biogenesis in neurons. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. 2008. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci 105(49):19211–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Tandarich LC, Nguyen K, Hollenbeck PJ. 2016. Compartmentalized Regulation of Parkin-Mediated Mitochondrial Quality Control in the Drosophila Nervous System In Vivo. J. Neurosci 36(28):7375–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20(21):5971–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, et al. 2014. Loss of mTOR-Dependent Macroautophagy Causes Autistic-like Synaptic Pruning Deficits. Neuron. 83(5):1131–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. 2004. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem 279(35):36268–76 [DOI] [PubMed] [Google Scholar]
- Tekirdag K, Cuervo AM. 2018. Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J. Biol. Chem 293(15):5414–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Li Z, Hu W, Ren H, Tian E, et al. 2010. C. elegans Screen Identifies Autophagy Genes Specific to Multicellular Organisms. Cell. 141(6):1042–55 [DOI] [PubMed] [Google Scholar]
- Traka M, Millen KJ, Collins D, Elbaz B, Kidd GJ, et al. 2013. WDR81 Is Necessary for Purkinje and Photoreceptor Cell Survival. J. Neurosci 33(16):6834–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. 2016. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science (80-.). 354(6315):1036–41 [DOI] [PubMed] [Google Scholar]
- van der Vaart A, Griffith J, Reggiori F. 2010. Exit from the Golgi Is Required for the Expansion of the Autophagosomal Phagophore in Yeast Saccharomyces cerevisiae. Mol. Biol. Cell 21(13):2270–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhauwaert R, Kuenen S, Masius R, Bademosi A, Manetsberger J, et al. 2017. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 36(10):1392–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wairkar YP, Toda H, Mochizuki H, Furukubo-Tokunaga K, Tomoda T, DiAntonio A. 2009. Unc-51 Controls Active Zone Density and Protein Composition by Downregulating ERK Signaling. J. Neurosci 29(2):517–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW. 1999. GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature. 397(6714):69–72 [DOI] [PubMed] [Google Scholar]
- Wang T, Martin S, Papadopulos A, Harper CB, Mavlyutov T a., et al. 2015. Control of Autophagosome Axonal Retrograde Flux by Presynaptic Activity Unveiled Using Botulinum Neurotoxin Type A. J. Neurosci 35(15):6179–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijdeven RH, Janssen H, Nahidiazar L, Janssen L, Jalink K, et al. 2016. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun 7: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm T, Byrne J, Medina R, Kolundžic E, Geisinger J, et al. 2017. Neuronal inhibition of the autophagy nucleation complex extends life span in post-reproductive C. Elegans. Genes Dev 31(15):1561–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler B, Faundez V, Maday S, Cai Q, Guimas Almeida C, Zhang H. 2018. The Endolysosomal System and Proteostasis: From Development to Degeneration. J. Neurosci 38(44):9364–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur EL. 2014a. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 111(42):E4439–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur ELF. 2014b. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci 34(4):1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Whiteus C, Xu CS, Hayworth KJ, Weinberg RJ, et al. 2017. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc. Natl. Acad. Sci 114(24):E4859–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. 2009. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem. J 417(3):747–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 5(8):1180–85 [DOI] [PubMed] [Google Scholar]
- Yu WH, Dorado B, Figueroa HY, Wang L, Planel E, et al. 2009. Metabolic activity determines efficacy of macroautophagic clearance of pathological oligomeric α-synuclein. Am. J. Pathol 175(2):736–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZQ, Ni T, Hong B, Wang HY, Jiang FJ, et al. 2012. Dual roles of Atg8 - PE deconjugation by Atg4 in autophagy. Autophagy. 8(6):883–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Puri R, Yang H, Lizzio MA, Wu C, et al. 2014. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife. 3: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G, Savova A, Danieli A, Romanov J, Tremel S, et al. 2018. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37(5): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhao YG, Wang X, Xu L, Miao L, et al. 2013. Mice deficient in epg5 exhibit selective neuronal vulnerability to degeneration. J. Cell Biol. 200(6):731–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, et al. 2009. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11(4):468–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Ma K, Gao R, Mu C, Chen L, et al. 2017. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 27(2):184–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Runwal G, Obrocki P, Rubinsztein DC. 2018. Autophagy in childhood neurological disorders. Dev. Med. Child Neurol. [DOI] [PubMed] [Google Scholar]