Abstract
Purpose
To investigate molecular characteristics and antimicrobial susceptibility profiles of clinical isolates of Elizabethkingia in Shanghai, China.
Methods
Elizabethkingia isolates were collected in a university-affiliated hospital in 2012–2015 and 2017–2018. They were re-identified to species level by 16S rRNA gene and species-specific gene sequencing. Antimicrobial susceptibility testing, screening for metallo-beta-lactamase production, identification of antimicrobial resistance genes and pulsed-field gel electrophoresis (PFGE) were performed.
Results
Among 52 Elizabethkingia isolates, E. anophelis was the most prevalent species (67.3%), followed by E. meningoseptica (26.9%). High carriage rates of blaCME, blaBlaB and blaGOB genes were consistent with the poor in vitro activity of most β-lactams including carbapenems. Nevertheless, β-lactamase inhibitors increased susceptibility rates significantly for cefoperazone and piperacillin. Susceptibility rates for minocycline, tigecycline, rifampin and levofloxacin were 100%, 78.8%, 76.9% and 71.2%, respectively. Ser83Ile or Ser83Arg substitution in the DNA gyrase A unit was associated with resistance to fluoroquinolones. MIC50/MIC90 values of vancomycin and linezolid were 16/16 mg/L and 16/32 mg/L, respectively. Molecular typing showed twenty-one different types of PFGE and more than one indistinguishable isolates were observed in each of the eight subtypes.
Conclusion
Tetracyclines, tigecycline, β-lactam/β-lactamase inhibitor combinations, rifampin and fluoroquinolones demonstrated high rates of in vitro activity against clinical isolates of Elizabethkingia. Both genetic diversity and clonality were observed from this health-care facility. Our report provides potential alternative treatment options for Elizabethkingia infections.
Keywords: Elizabethkingia, antimicrobial susceptibility, molecular typing, multidrug resistance, resistant mechanism
Introduction
The genus Elizabethkingia is an infrequent Gram-negative non-fermenting bacillus and has recently emerged as a cause of life-threatening infections in humans, including meningitis, bacteremia, pneumonia and urinary tract infection.1–3 Importantly, neonatal meningitis was the most common presentation of Elizabethkingia infection in children while a variety of clinical manifestations were reported in immunocompromised patients.4 Furthermore, an increasing number of global cases of Elizabethkingia infection in recent years showed high morbidity and mortality, which reinforced the significance of early identification and treatment.4
Elizabethkingia has undergone several taxonomic changes since the first description in 1959. It was reclassified from genus Flavobacterium in 1994 and from genus Chryseobacterium in 2005.5 In addition to E. meningoseptica and E. miricola, several new species have been proposed in the last decade, including E. anophelis,6 E. bruuniana, E. ursingii, and E. occulta.7 E. anophelis and E. meningoseptica are most common among them.
The data from the SENTRY Antimicrobial Surveillance Program showed that C. meningoseptica represented only 0.1% (24/18,569) of the non-fermentative gram-negative bacilli in North America, Latin America, Europe and the Asia-Pacific region from 1997 to 2001.8 Despite the overall low isolation rate of clinical strains, healthcare–associated outbreaks attributable to Elizabethkingia species have been reported in Singapore,9 England10 and Taiwan11 since 2012. Moreover, two large-scale outbreaks were identified in the United States from 2014 to 2016, one causing significant mortality (6/10), and the second involving 65 individuals and resulting in 20 deaths (https://www.cdc.gov/Elizabethkingia/outbreaks/). Notably, the second outbreak occurred primarily in community settings, with the source of infection unclear.12
Elizabethkingia-related infections are complicated by the biofilm formation,13 intracellular invasion,14 and multidrug resistance of the strains, and thus one needs to be cautious in selecting appropriate antimicrobial drugs. Elizabethkingia isolates display intrinsic resistance to multiple β-lactams as a result of Ambler class A serine extended-spectrum β-lactamase (ESBL) gene blaCME15 and two chromosomal Ambler class B metallo-β-lactamase (MBL) genes, blaBlaB and blaGOB.16 It also confers resistance to quinolones due to mutations in DNA gyrase and/or topoisomerase IV genes.17
Given the limited epidemiological data on clinical isolates of Elizabethkingia in China, we investigated the molecular characteristics and antimicrobial susceptibility profiles of Elizabethkingia isolates in a university-affiliated hospital in Shanghai, China.
Materials and Methods
Identification of Elizabethkingia and Clinical Information of Patients
Non-duplicate isolates of Elizabethkingia were collected from a 1216-bed university-affiliated adult hospital in 2012–2018 with the exception of 2016 when isolates were missing. Elizabethkingia strains were preliminarily identified in the clinical laboratory from various clinical samples, such as specimens from respiratory tract, blood, urine, bile, exudate and indwelling needle. They were all included except those missing or dead. The hosts were inpatients and outpatients aged 18 years and older, and the departments included geriatrics, surgery, intensive care unit (ICU), neurology, infectious disease, general practice, hematology and thoracic surgery (Table 1).
Table 1.
Characteristics of 52 Patients with Elizabethkingia Colonization or Infection
| Age (years) | |
| Range | 18–96 |
| Mean±SD | 64±21 |
| Gender, n (%) | |
| Male | 36 (69.2) |
| Female | 16 (30.8) |
| Hospitalization duration (days), mean±SD | 39±40 |
| Comorbidity, n (%) | |
| Hypertension | 18 (34.6) |
| Diabetes mellitus | 7 (13.5) |
| Chronic obstructive pulmonary disease | 6 (11.5) |
| Cardiovascular disease | 5 (9.6) |
| End-stage renal disease | 4 (7.7) |
| Mechanical ventilation, n (%) | 29 (55.8) |
| Indwelling device, n (%) | 38 (73.1) |
| Central venous catheter | 28 (53.8) |
| Nasogastric tube | 22 (42.3) |
| Urinary catheter | 20 (38.5) |
| Surgical puncture or drain | 20 (38.5) |
| Surgery, n (%) | 20 (38.5) |
| Transplantation | 5 (9.6) |
| Chemoradiotherapy, n (%) | 3 (5.8) |
| Ward, n (%) | |
| Geriatrics | 14 (26.9) |
| Neurosurgery | 9 (17.3) |
| Surgery | 7 (13.5) |
| Intensive care unit | 7 (13.5) |
| Neurology | 5 (9.6) |
| Infectious disease | 4 (7.7) |
| General practice | 2 (3.8) |
| Hematology | 2 (3.8) |
| Thoracic surgery | 1 (1.9) |
| Outpatient | 1 (1.9) |
| Site of isolation, n (%) | |
| Respiratory tract | 45 (86.5) |
| Blood | 2 (3.8) |
| Urine | 2 (3.8) |
| Bile | 1 (1.9) |
| Exudate | 1 (1.9) |
| Indwelling needle | 1 (1.9) |
The isolates were re-identified to species level by PCR amplification and sequencing of the 16S rRNA gene followed by analysis using the EzTaxon server (http://www.ezbiocloud.net/, reference sequence: E. anophelis strain R26, GenBank accession number NZ_CP023401; E. meningoseptica type strain 13253, NZ_ASAN01000081; E. miricola DSM 14571, NZ_VNHK00000000.1; E. ursingii G4122, NZ_LNOK01000028),18,19 and by species-specific primers (E. anophelis-specific primers targeted lipid A-disaccharide synthase gene; E. meningoseptica-specific primers targeted a putative sodium-proton antiporter; E. miricola cluster-specific primers targeted urease gene ureG).20,21 The indistinguishable E. miricola cluster isolates were further confirmed by RNA polymerase subunit gene (rpoB) sequencing.21
Medical records of patients were retrospectively reviewed to acquire clinical information.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of antimicrobial agents were determined using the broth microdilution method and interpreted on the basis of the Clinical and Laboratory Standards Institute (CLSI) criteria for other non-Enterobacteriaceae for most of the antibiotics.22 The US FDA susceptibility breakpoints for Enterobacteriaceae were extrapolated for tigecycline. For rifampin, vancomycin and linezolid, the breakpoints for Staphylococcus spp. were applied.23,24
EDTA Combination Disk Test (EDT)
Imipenem discs and imipenem/0.5 M EDTA combination discs were used for the detection of the MBL phenotype as described previously.25 The test was considered to be positive if the diameter of the inhibition zone of the imipenem/EDTA disc was 7 mm larger than that of the imipenem disc alone.25
Identification of β-Lactamase Genes and Mutations in the Quinolone Resistance-Determining Regions (QRDRs)
All isolates were screened for ESBL gene blaCME and MBL genes, blaBlaB and blaGOB, as described previously.15,16,26 Mutations in the QRDRs of gyrA, gyrB, parC and parE genes were determined by PCR amplification and sequencing. Alignment was performed with the respective reference sequences in the GenBank database (NCBI reference sequence: E. anophelis strain NUHP1, NZ_CP007547.1; E. meningoseptica strain G4076, NZ_CP016376.1; E. miricola strain EM798-26, NZ_CP023746.1).17
Molecular Typing
Pulsed-field gel electrophoresis (PFGE) was performed with CHEF Mapper XA system (Bio-Rad, USA). The genomic DNA of Elizabethkingia was prepared in agarose blocks and digested with restriction enzyme XhoI. Salmonella enterica serotype Braenderup H9812, as a molecular size marker, was digested with XbaI. The DNA fragments were separated at 6.0 V/cm, 120° angle, temperature of 14°C, and switch times of 1 to 18 s for a total run time of 18 hrs. PFGE band profiles were analyzed using BioNumerics version 7.6 software. The similarity matrix was calculated by Dice’s coefficients with 1.5% optimization and 1.5% band matching tolerance, and a dendrogram was generated using the unweighted pair group method with arithmetic averages (UPGMA). Isolates with ≥95%, 85–95% and <85% similarity were considered as a PFGE subtype, a PFGE type and a different type, respectively.
Results
Identification and Prevalence of Elizabethkingia Isolates
A total of 57 (0.6%) non-duplicate Elizabethkingia isolates were collected among 8804 gram-negative non-fermenting bacillus isolates from a university-affiliated Hospital in 2012–2015 and 2017–2018.
Fifty-two isolates were confirmed to be Elizabethkingia isolates using 16S rRNA gene sequencing, including 35 (67.3%) isolates of E. anophelis, 14 (26.9%) isolates of E. meningoseptica and 2 (3.8%) isolates of E. miricola. One isolate had 99.6% similarity with E. ursingii strain G4122 and 99.3% similarity with E. miricola strain DSM 14,571 with 16S rRNA gene sequencing, but 99.9% similarity with E. miricola strain F13 (accession no: NZ_CP040450.1) with further rpoB gene sequencing. Therefore, this isolate was identified as E. miricola. All of the isolates were further confirmed with species-specific gene sequencing, which was consistent with the identification with 16S rRNA gene and rpoB gene sequencing.
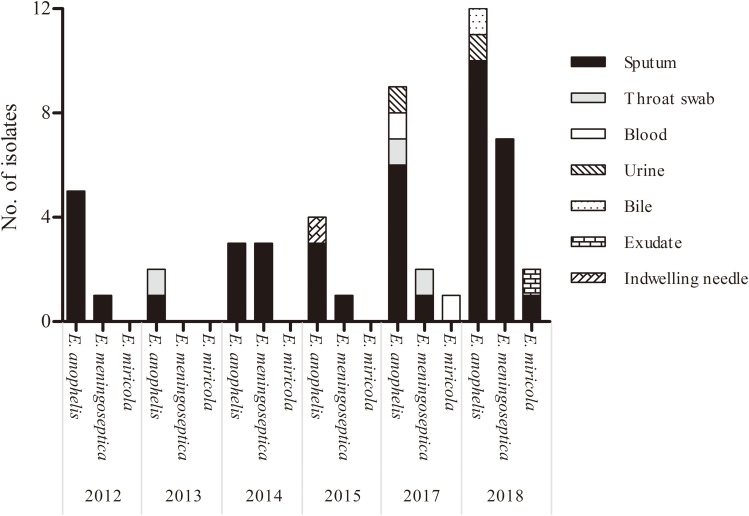
Most of the Elizabethkingia strains were isolated from the respiratory tract (45/52, 86.5%), followed by blood (2/52, 3.8%) and urine (2/52, 3.8%). An increase in numbers was observed from 6 (11.5%) isolates in 2012 to 12 (23.1%) isolates in 2017 and 21 (40.4%) in 2018 (Figure 1). Additionally, ten samples showed concomitant isolates of other bacterial species such as Stenotrophomonas maltophilia and Pseudomonas aeruginosa in sputum.
Figure 1.
The distribution of 52 Elizabethkingia isolates according to the year and site of isolation.
Clinical Characteristics of Patients
Of the 52 patients, 51 were hospitalized patients and one was outpatient (Table 1). Male accounted for 69.2% (36/52) while female constituted 30.8% (16/52). The mean age was 64 years old, with the range of 18–96 years old. Prolonged hospital stays (≥2 weeks) was observed in 46 patients. Geriatrics department was the most common ward (14/52, 26.9%), followed by neurosurgery department (9/52, 17.3%), surgery department (7/52, 13.5%) and ICU (7/52, 13.5%). Comorbidity was identified in all hospitalized patients, with hypertension as the most prevalent underlying disease (18/52, 34.6%) followed by diabetes mellitus (7/52, 13.5%), chronic obstructive pulmonary disease (6/52, 11.5%), cardiovascular disease (5/52, 9.6%) and end-stage renal disease (4/52, 7.7%). Besides, 20 (38.5%) patients had a history of surgery before Elizabethkingia was identified, and 5 (9.6%) underwent transplantation while 3 (5.8%) received chemoradiotherapy. Twenty-nine (55.8%) patients received mechanical ventilation and 38 (73.1%) patients had medical devices indwelled. The overall in-hospital mortality rate was 13.5% (7/52).
Antimicrobial Susceptibility Pattern
Multidrug resistance was observed in all 52 Elizabethkingia isolates (Table 2). All of them were resistant to aztreonam, ceftazidime, colistin, and all were non-susceptible to cefepime, carbapenems (imipenem and meropenem) and vancomycin. They also exhibited high resistance rates to linezolid, gentamicin, amikacin and trimethoprim-sulfamethoxazole (96.2%, 96.2%, 86.5% and 63.5%, respectively). Low susceptibility rates were observed for cefoperazone (5.8%) and piperacillin (46.2%) in contrast to the increased susceptibility when they were in combination with β-lactamase inhibitors, namely cefoperazone-sulbactam and piperacillin-tazobactam (88.5% and 86.5%, respectively).
Table 2.
Antimicrobial Susceptibilities of 52 Elizabethkingia Isolates Determined by the Broth Microdilution Method
| Antimicrobial Agents | Breakpoint (mg/L) | MIC (mg/L) | Number (%) of Isolates | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S | I | R | Range | 50% | 90% | S | I | R | |
| Piperacillin | ≤16 | 32–64 | ≥128 | 8 to >128 | 32 | 64 | 24 (46.2) | 23 (44.2) | 5 (9.6) |
| Piperacillin-tazobactam | ≤16/4 | 32/4–64/4 | ≥128/4 | 0.25/4 to >128/4 | 4/4 | 32/4 | 45 (86.5) | 7 (13.5) | 0 (0.0) |
| Cefoperazone | ≤16 | 32 | ≥64 | 16 to >128 | 32 | >128 | 3 (5.8) | 25 (48.1) | 24 (46.2) |
| Cefoperazone-sulbactam | ≤16/8 | 32/16 | ≥64/32 | 2/1–128/64 | 8/4 | 64/32 | 46 (88.5) | 1 (1.9) | 5 (9.6) |
| Ceftazidime | ≤8 | 16 | ≥32 | 64 to >128 | >128 | >128 | 0 (0.0) | 0 (0.0) | 52 (100.0) |
| Cefepime | ≤8 | 16 | ≥32 | 16 to >128 | 32 | >128 | 0 (0.0) | 8 (15.4) | 44 (84.6) |
| Aztreonam | ≤8 | 16 | ≥32 | >128 | >128 | >128 | 0 (0.0) | 0 (0.0) | 52(100.0) |
| Imipenem | ≤4 | 8 | ≥16 | 8 to >128 | 16 | 128 | 0 (0.0) | 6 (11.5) | 46 (88.5) |
| Meropenem | ≤4 | 8 | ≥16 | 8 to >128 | 32 | >128 | 0 (0.0) | 4 (7.7) | 48 (92.3) |
| Colistin | ≤2 | – | ≥4 | >128 | >128 | >128 | 0 (0.0) | 0 (0.0) | 52 (100.0) |
| Amikacin | ≤16 | 32 | ≥64 | 16 to >128 | 64 | >128 | 4 (7.7) | 3 (5.8) | 45 (86.5) |
| Gentamicin | ≤4 | 8 | ≥16 | 4 to >128 | 64 | >128 | 1 (1.9) | 1 (1.9) | 50 (96.2) |
| Ciprofloxacin | ≤1 | 2 | ≥4 | 0.25 to >128 | 1 | 128 | 26 (50.0) | 6 (11.5) | 20 (38.5) |
| Levofloxacin | ≤2 | 4 | ≥8 | 0.25–64 | 1 | 32 | 37 (71.2) | 1 (1.9) | 14 (26.9) |
| Rifampin | ≤1 | 2 | ≥4 | 0.5–64 | 1 | 8 | 40 (76.9) | 6 (11.5) | 6 (11.5) |
| Trimethoprim-sulfamethoxazole | ≤2/38 | - | ≥4/76 | 1/19–16/304 | 4/76 | 8/152 | 19 (36.5) | 0 (0.0) | 33 (63.5) |
| Tigecycline | ≤2 | 4 | ≥8 | 1–32 | 4 | 8 | 41 (78.8) | 8 (15.4) | 3 (5.8) |
| Doxycycline | ≤4 | 8 | ≥16 | 1–8 | 2 | 4 | 50 (96.2) | 2 (3.8) | 0 (0.00) |
| Minocycline | ≤4 | 8 | ≥16 | 0.25–4 | 0.5 | 1 | 52 (100.0) | 0 (0.0) | 0 (0.0) |
| Vancomycin | ≤4 | 8–16 | ≥32 | 8–32 | 16 | 16 | 0 (0.0) | 12 (23.1) | 40 (76.9) |
| Linezolid | ≤4 | - | ≥8 | 4–128 | 16 | 32 | 2 (3.8) | 0 (0.0) | 50 (96.2) |
Notes: In the combinations, the concentration of tazobactam was 4 mg/L constant. The ratio of cefoperazone to sulbactam was 2:1, and the ratio of trimethoprim to sulfamethoxazole was 1:19.
Abbreviations: MIC, minimal inhibitory concentration; S, susceptible; I, intermediate; R, resistant.
The susceptibility rates of Elizabethkingia isolates to ciprofloxacin, levofloxacin and rifampin were 50.0%, 71.2% and 76.9%, respectively. Minocycline was more active rather than doxycycline and tigecycline (susceptible rates, 100% versus 96.2% and 78.8%, respectively). MIC50/MIC90 values of vancomycin and linezolid against Elizabethkingia isolates were 16/16 mg/L and 16/32 mg/L, respectively.
MBL Phenotype and Genotype of Elizabethkingia Isolates
EDTA combination testing was positive for 52 Elizabethkingia isolates, suggesting the capability of MBL production. This was consistent with the results that these 52 isolates were non-susceptible to imipenem.
A total of 51 Elizabethkingia carried β-lactamase genes. Specifically, 49 Elizabethkingia isolates harbored the blaBlaB gene, 28 carried blaGOB gene and 36 had blaCME gene. Twenty-six Elizabethkingia isolates harbored the above three genes (Table 3). Two isolates were negative for MBL genotype detection.
Table 3.
Distribution of β-Lactamase Genes in 52 Elizabethkingia
| β-Lactamase Gene | No. of Isolates | EDTA Combination Disk Test |
|---|---|---|
| Negative | 1 | + |
| blaCME | 1 | + |
| blaBlaB | 14 | + |
| blaCME + blaBlaB | 8 | + |
| blaCME +blaGOB | 1 | + |
| blaBlaB + blaGOB | 1 | + |
| blaCME + blaBlaB+ blaGOB | 26 | + |
Single-nucleotide mutations in QRDR region of the gyrA gene were observed in 13 isolates which led to amino acid substitutions Ser83Ile in nine isolates and Ser83Arg in 4 isolates, conferring resistance to ciprofloxacin and levofloxacin. No non-synonymous alternations were detected in gyrB, parC or parE. One isolate of E. anophelis did not have amino acid substitutions in the QRDR region of the gyrA, gyrB, parC or parE, but showed high MICs of ciprofloxacin and levofloxacin (both 16 mg/L).
Genetic Relatedness and Retrospective Analysis
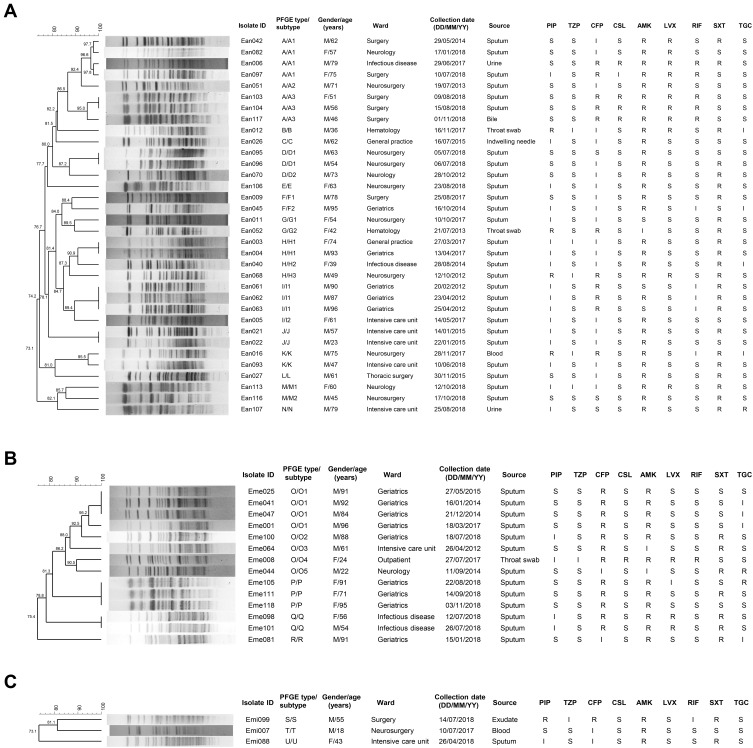
One isolate was resistant to XhoI digestion. The remaining 51 Elizabethkingia isolates were clustered into 21 different PFGE types designated A-T (Figure 2). Specifically, 34 E. anophelis isolates were grouped into 14 clusters, 14 E. meningoseptica isolates into 4 clusters while 3 E. miricola isolates into 3 clusters.
Figure 2.
Dendrogram of PFGE patterns of 52 Elizabethkingia isolates using the BioNumerics software. (A) Thirty-four E. anophelis isolates; (B) Fourteen E. meningoseptica isolates; (C) Three E. miricola isolates.
Abbreviations: M, male; F, female; PIP, piperacillin; TZP, piperacillin-tazobactam; CFP, cefoperazone; CSL, cefoperazone-sulbactam; AMK, amikacin; LVX, levofloxacin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; S, susceptible; I, intermediate; R, resistant.
Of all PFGE types, A and O types were the most frequent containing 8 isolates each. E. anophelis isolates in A type accounted for 23.5% (8/34), while E. meningoseptica isolates in O type made up 57.1% (8/14). Above all, there were indistinguishable isolates sharing 100% relatedness in each of 8 clusters (A3, D1, H1, I1, J, O1, P and Q subtypes), indicating clonal spread in this health-care facility. All of the identical clones in respective subtypes were collected from the sputum samples of patients in the same wards within 6 months, except that 2 indistinguishable isolates in the H1 subtype were recovered in departments of geriatrics and general practice, respectively. Meanwhile, the geriatrics department had the most isolates with identical clones (H1, I1, O1, P subtypes) spanning from 2012 to 2018. Similar antimicrobial susceptibility patterns of the above identical isolates were detected within subtypes. Mechanical ventilation and indwelling devices were also present among patients who had A3, D1, J, O1, P and Q subtypes. On the other hand, no epidemiological relationship was observed for closely related clones at 95% genetic similarity in the A1 and K subtypes. Possibly related strains in F, G and M types also exhibited variation of spacial or temporal distribution, which were clustered for ≥85% similarity values.
Discussion
Six species of the genus Elizabethkingia (E. anophelis, E. meningoseptica, E. miricola, E. bruuniana, E. ursingii and E. occulta) are emerging in children and immunocompromised patients with the growing emphasis on the pathogenicity for former four species.1,18 Nowadays, development of microbiological identification techniques makes it possible to recognize several emerging unusual bacteria that cause diseases mainly in immunocompromised patients. Actually, traditional identification systems with inferior discrimination power for infrequent species could lead to the misidentification, misdiagnosis, failure of therapy as well as underestimation of the incidence of the infection in the past.4 Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) Vitek mass spectrometry (VMS) and molecular identification techniques (16S rRNA, rpoB gene sequencing and whole-genome sequencing) emerge as advantageous tools for accurate identification of microbe with excellent discrimination, especially infrequent opportunistic bacteria, such as Elizabethkingia species,27 Cardiobacterium hominis28 and Kocuria marina.29
In this study, patients with comorbidity, indwelling devices, mechanical ventilation and prolonged hospital stays were vulnerable to Elizabethkingia colonization or infection. Geriatrics, neurosurgery, surgery and ICU departments were the top four departments. Prior studies also found that E. meningoseptica strains were commonly isolated from ICU in India30 and Taiwan.31 These data consistently suggest that Elizabethkingia has a predilection for infecting immunocompromised patients.
E. anophelis was the most prevalent species of Elizabethkingia followed by E. meningoseptica, while E. miricola isolates were rare in the collection. In fact, more evidence indicated the predominance of E. anophelis, rather than E. meningoseptica, among Elizabethkingia in clinical settings.18,20 In this study, we also found that the identification for Emi007 was ambiguous with 16S rRNA gene sequencing, which could be partly explained by the existence of multiple copies with different sequences and the hypervariable regions of 16S rRNA.32 It has been pointed out that the rpoB gene is a single copy gene and has a higher resolution of phylogenetic evolution than the 16S rRNA gene; thus, it could accurately distinguish Elizabethkingia strains at the species level.7,32 PFGE profiles showed finer resolution of clonal relationships, indicating that Emi007 was genetically more related to Emi099 than Emi088 with a similarity of 81.1%, thereby supporting its identity as E. miricola.
Genetic diversity and clonal dissemination were both suggested by PFGE typing. Although the genotypes of E. anophelis isolates were highly diverse, clonal spread was observed in several pairs of patients in the same or different departments. E. meningoseptica isolates from 2012 to 2018 were genetically homogenous (8/14 isolates in O type, 3/14 in P type, 2/14 in Q type) indicating clonal expansion and persistence in recent years. Previous reports have found that Elizabethkingia acquisition might be associated with water sources or water-related equipment, such as sinks and hand hygiene sink aerator within the hospital environment.33 Clonal dissemination could be mediated by the hands of hospital staff or patients.34 Therefore, it is necessary to reinforce hand hygiene and environmental cleaning when this genus is detected in the hospital.
The treatment for Elizabethkingia infection is challenging because Elizabethkingia isolates tend to display inherent resistance to antimicrobial agents, including aminoglycosides, most β-lactams and colistin. Multiple resistance genes and drug efflux systems in Elizabethkingia have been demonstrated by genomic and proteomic analysis.35,36 In this study, the high carriage rate of blaCME, blaBlaB and blaGOB genes was consistent with the broad-spectrum resistance to β-lactams including carbapenems. Nevertheless, piperacillin-tazobactam and cefoperazone-sulbactam were active in vitro against Elizabethkingia, in accordance with the results of a previous study where both combinations showed reasonable in vitro activity (70–85% and 65–80% of susceptibility rate, respectively) against 170 clinical Elizabethkingia isolates in China.37 Furthermore, treatment with combination therapy of piperacillin-tazobactam and trimethoprim-sulfamethoxazole or a fluoroquinolone was reported to be effective in paediatric patients with E. meningoseptica infections, with variable susceptibility of strains to piperacillin-tazobactam (100%), trimethoprim-sulfamethoxazole (78.6%) and fluoroquinolones (33.3–87.5%).38 The restoration of activity with β-lactam inhibitors may be explained by the production of ESBLs genes such as blaCME especially when MBLs were expressed at a low level, along with decreased outer membrane permeability involved in the carbapenem-resistant Elizabethkingia isolates.39 Despite the favorable activity of β-lactam/β-lactamase inhibitor combinations as a part of the combination regimen, there was little evidence over their use as monotherapy for patients. Therefore, their clinical efficacy needs further evaluation.
The reports varied on the susceptibility of Elizabethkingia isolates against fluoroquinolones.32 Favored in vitro activity of fluoroquinolones was observed in this study, and the susceptibility rate for levofloxacin was higher than that for ciprofloxacin. By contrast, fluoroquinolones exhibited poor activity against E. anophelis and E. meningoseptica while all E. miricola isolates were susceptible to levofloxacin and moxifloxacin in a hospital in South Korea.18 In another report, a significant difference was noted between the susceptibility rates of Elizabethkingia against ciprofloxacin and levofloxacin (9.8% and 52.2%, respectively).40 One possible explanation for such discrepancy was that levofloxacin containing the C-8 methoxy group exerted stronger antibacterial activity against fluoroquinolone-resistant bacteria that harbored gyrA mutation.41,42
Successful treatment has been described using fluoroquinolones for Elizabethkingia infection. In a retrospective clinical study, the quinolone group achieved a higher microbiological cure rate, higher clinical success rate and lower 14-day in-hospital mortality rate in patients with E. meningoseptica bacteraemia than the non-fluoroquinolone group did.43 In addition, higher 14-day mortality in patients with bacteraemia caused by levofloxacin-resistant E. meningoseptica compared with those with levofloxacin-susceptible strains was reported in Taiwan.44 Consequently, while it may be suitable to consider fluoroquinolones as a choice of empirical antimicrobial therapy for Elizabethkingia infection, early identification of fluoroquinolone resistance in Elizabethkingia isolates is of significant importance in tackling this multidrug-resistance pathogen.
We found the Ser83Ile or Ser83Arg substitution in GyrA in 13 fluoroquinolone-resistant clinical Elizabethkingia isolates, which was in accordant with previous reports.17 A study in Taiwan identified that non-synonymous alterations of additional sites in GyrA (positions 95 and 102) and GyrB (positions 425, 452, and 470) involved levofloxacin non-susceptibility as well.40 The AcrAB efflux pump also played a role in mediating fluoroquinolone resistance by increased expression in Elizabethkingia isolates.25
It remains controversial whether antibiotics active against Gram-positive cocci, such as vancomycin and linezolid, are useful in the fight against Elizabethkingia. In this study, MIC50/MIC90 values of vancomycin against Elizabethkingia isolates were 16/16 mg/L, as shown in a previous study.45 Interestingly, clinical cure was noted in patients with Elizabethkingia infection despite high MIC of vancomycin against Elizabethkingia (≥16 mg/L).23,46 Given the above conflicting data and scant information in literature, it may be necessary to establish the criteria for determining the in vitro antimicrobial susceptibility against Elizabethkingia.
Minocycline, doxycycline, tigecycline and rifampin were active against Elizabethkingia in vitro in this study. It is interesting that tigecycline, the derivative of minocycline, showed inferior antimicrobial activity, as reported in Taiwan.25 Overall, taking into account their limitation (poor distribution in tissue, for example), different pharmacokinetics characteristics and scanty supporting clinical evidence, their roles in treating patients with invasive Elizabethkingia infections need further evaluation.
Conclusion
Elizabethkingia isolates were still rare in this clinical setting since they tended to occur in immunocompromised patients and had a risk of misidentification through conventional methods. E. anophelis was the most prevalent species among them, most of which exhibited phylogenetic diversity. Putative circulating clones were discovered in E. anophelis and E. meningoseptica, indicating a potential risk of further dissemination. Elizabethkingia isolates displayed multidrug resistance characteristics. However, tetracyclines, tigecycline, β-lactam/β-lactamase inhibitor combinations, rifampin and fluoroquinolone demonstrated appealing in vitro activity against Elizabethkingia, and further clinical studies may be needed to determine their potential role in treating Elizabethkingia infection.
Acknowledgments
The authors thanked Dr. Yohei Doi for his critical review of the manuscript. The authors also thanked Mengyun Yin, Renru Han, Li Ding and Pei Li at Institute of Antibiotics for assistance and collaboration in laboratory work. This work was supported by grants from the National Natural Science Foundation of China (grant numbers 81872909 and 81673479). The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Ethics Approval
Verbal informed consent of patients was obtained after the approval of the Ethics Committee of Huashan Hospital, Fudan University, China (approval number: KY2019-544). The patient data were analyzed in anonymity.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lin JN, Lai CH, Yang CH, Huang YH, Lin HH. Elizabethkingia bruuniana infections in Humans, Taiwan, 2005–2017. Emerg Infect Dis. 2019;25(7):1412–1414. doi: 10.3201/eid2507.180768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank T, Gody JC, Nguyen LB, et al. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet. 2013;381(9880):1876. doi: 10.1016/S0140-6736(13)60318-9 [DOI] [PubMed] [Google Scholar]
- 3.Gajdacs M, Burian K, Terhes G. Resistance levels and epidemiology of non-fermenting gram-negative bacteria in urinary tract infections of inpatients and outpatients (RENFUTI): a 10-year epidemiological snapshot. Antibiotics (Basel). 2019;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dziuban EJ, Franks JL, So M, Peacock G, Blaney DD. Elizabethkingia in children: a comprehensive review of symptomatic cases reported from 1944 to 2017. Clin Infect Dis. 2018;67(1):144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim KK, Kim MK, Lim JH, Park HY, Lee ST. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int J Syst Evol Microbiol. 2005;55(Pt 3):1287–1293. [DOI] [PubMed] [Google Scholar]
- 6.Kampfer P, Matthews H, Glaeser SP, Martin K, Lodders N, Faye I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int J Syst Evol Microbiol. 2011;61(Pt 11):2670–2675. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson AC, Gulvik CA, Whitney AM, et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Van Leeuwenhoek. 2018;111(1):55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirby JT, Sader HS, Walsh TR, Jones RN. Antimicrobial susceptibility and epidemiology of a worldwide collection of Chryseobacterium spp: report from the SENTRY antimicrobial surveillance program (1997–2001). J Clin Microbiol. 2004;42(1):445–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teo J, Tan SY, Tay M, et al. First case of E anophelis outbreak in an intensive-care unit. Lancet. 2013;382(9895):855–856. [DOI] [PubMed] [Google Scholar]
- 10.Moore LS, Owens DS, Jepson A, et al. Waterborne Elizabethkingia meningoseptica in adult critical care. Emerg Infect Dis. 2016;22(1):9–17. doi: 10.3201/eid2201.150139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tai IC, Liu TP, Chen YJ, Lien RI, Lee CY, Huang YC. Outbreak of Elizabethkingia meningoseptica sepsis with meningitis in a well-baby nursery. J Hosp Infect. 2017;96(2):168–171. doi: 10.1016/j.jhin.2016.11.018 [DOI] [PubMed] [Google Scholar]
- 12.Perrin A, Larsonneur E, Nicholson AC, et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat Commun. 2017;8:15483. doi: 10.1038/ncomms15483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin PY, Chen HL, Huang CT, Su LH, Chiu CH. Biofilm production, use of intravascular indwelling catheters and inappropriate antimicrobial therapy as predictors of fatality in Chryseobacterium meningosepticum bacteraemia. Int J Antimicrob Agents. 2010;36(5):436–440. doi: 10.1016/j.ijantimicag.2010.06.033 [DOI] [PubMed] [Google Scholar]
- 14.Lin PY, Chiu CH, Chu C, Tang P, Su LH. Invasion of murine respiratory tract epithelial cells by Chryseobacterium meningosepticum and identification of genes present specifically in an invasive strain. New Microbiol. 2006;29(1):55–62. [PubMed] [Google Scholar]
- 15.Bellais S, Poirel L, Naas T, Girlich D, Nordmann P. Genetic-biochemical analysis and distribution of the Ambler class A beta-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob Agents Chemother. 2000;44(1):1–9. doi: 10.1128/AAC.44.1.1-9.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez LJ, Vila AJ. Carbapenem resistance in Elizabethkingia meningoseptica is mediated by metallo-beta-lactamase BlaB. Antimicrob Agents Chemother. 2012;56(4):1686–1692. doi: 10.1128/AAC.05835-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jian MJ, Cheng YH, Perng CL, Shang HS. Molecular typing and profiling of topoisomerase mutations causing resistance to ciprofloxacin and levofloxacin in Elizabethkingia species. Peerj. 2018;6:e5608. doi: 10.7717/peerj.5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han MS, Kim H, Lee Y, et al. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J Clin Microbiol. 2017;55(1):274–280. doi: 10.1128/JCM.01637-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon SH, Ha SM, Kwon S, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67(5):1613–1617. doi: 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chew KL, Cheng B, Lin R, Teo J. Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in Singapore. J Clin Microbiol. 2018;56:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenna D, Fuller A, Martin K, et al. rpoB gene sequencing highlights the prevalence of an E. miricola cluster over other Elizabethkingia species among UK cystic fibrosis patients. Diagn Microbiol Infect Dis. 2018;90(2):109–114. doi: 10.1016/j.diagmicrobio.2017.10.014 [DOI] [PubMed] [Google Scholar]
- 22.Lin JN, Lai CH, Yang CH, Huang YH, Lin HH. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia Anophelis. . J Antimicrob Chemother. 2018;73(9):2497–2502. doi: 10.1093/jac/dky197 [DOI] [PubMed] [Google Scholar]
- 23.Jean SS, Hsieh TC, Ning YZ, Hsueh PR. Role of vancomycin in the treatment of bacteraemia and meningitis caused by Elizabethkingia meningoseptica. Int J Antimicrob Agents. 2017;50(4):507–511. doi: 10.1016/j.ijantimicag.2017.06.021 [DOI] [PubMed] [Google Scholar]
- 24.Gajdacs M, Urban E. Epidemiology and resistance trends of Staphylococcus aureus isolated from vaginal samples: a 10-year retrospective study in Hungary. Acta Dermatovenerol Alp Pannonica Adriat. 2019;28(4):143–147. [PubMed] [Google Scholar]
- 25.Jian MJ, Cheng YH, Chung HY, et al. Fluoroquinolone resistance in carbapenem-resistant Elizabethkingia anophelis: phenotypic and genotypic characteristics of clinical isolates with topoisomerase mutations and comparative genomic analysis. J Antimicrob Chemother. 2019;74(6):1503–1510. doi: 10.1093/jac/dkz045 [DOI] [PubMed] [Google Scholar]
- 26.Yum JH, Lee EY, Hur SH, et al. Genetic diversity of chromosomal metallo-beta-lactamase genes in clinical isolates of Elizabethkingia meningoseptica from Korea. J Microbiol. 2010;48(3):358–364. doi: 10.1007/s12275-010-9308-5 [DOI] [PubMed] [Google Scholar]
- 27.Rahim GR, Gupta N. Elizabethkingia miricola: discrepancies in identification and antimicrobial susceptibilities. Diagn Microbiol Infect Dis. 2019;94(1):104. doi: 10.1016/j.diagmicrobio.2018.11.025 [DOI] [PubMed] [Google Scholar]
- 28.Wallet F, Loiez C, Decoene C, Courcol R. Rapid identification of Cardiobacterium hominis by MALDI-TOF mass spectrometry during infective endocarditis. Jpn J Infect Dis. 2011;64(4):327–329. [PubMed] [Google Scholar]
- 29.Pulcrano G, Balzaretti M, Grosini A, Piacentini V, Poddighe D. First report of Kocuria marina bloodstream infection unrelated to a central venous catheter: a mini-review on an emerging and under-recognized opportunistic pathogen. Infez Med. 2017;25(1):71–74. [PubMed] [Google Scholar]
- 30.Agarwal S, Kakati B, Khanduri S, Gupta S. Emergence of carbapenem resistant non-fermenting gram-negative bacilli isolated in an ICU of a tertiary care hospital. J Clin Diagn Res. 2017;11(1):C4–C7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko HK, Yu WK, Lien TC, et al. Intensive care unit-acquired bacteremia in mechanically ventilated patients: clinical features and outcomes. PLoS One. 2013;8(12):e83298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin JN, Lai CH, Yang CH, Huang YH. Elizabethkingia infections in humans: from genomics to clinics. Microorganisms. 2019;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi MH, Kim M, Jeong SJ, et al. Risk factors for Elizabethkingia acquisition and clinical characteristics of patients, South Korea. Emerg Infect Dis. 2019;25(1):42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yung CF, Maiwald M, Loo LH, et al. Elizabethkingia anophelis and association with tap water and handwashing, Singapore. Emerg Infect Dis. 2018;24(9):1730–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal A, Ravikumar R, Varun CN, et al. Global proteome profiling reveals drug-resistant traits in Elizabethkingia meningoseptica: an opportunistic nosocomial pathogen. Omics. 2019;23(6):318–326. [DOI] [PubMed] [Google Scholar]
- 36.Spengler G, Kincses A, Gajdacs M, Amaral L. New roads leading to old destinations: efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules. 2017;22:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen GX, Zhang R, Zhou HW. Heterogeneity of metallo-beta-lactamases in clinical isolates of Chryseobacterium meningosepticum from Hangzhou, China. J Antimicrob Chemother. 2006;57(4):750–752. [DOI] [PubMed] [Google Scholar]
- 38.Chan JC, Chong CY, Thoon KC, et al. Invasive paediatric Elizabethkingia meningoseptica infections are best treated with a combination of piperacillin/tazobactam and trimethoprim/sulfamethoxazole or fluoroquinolone. J Med Microbiol. 2019;68(8):1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodford N, Palepou MF, Babini GS, Holmes B, Livermore DM. Carbapenemases of Chryseobacterium (Flavobacterium) meningosepticum: distribution of blaB and characterization of a novel metallo-beta-lactamase gene, blaB3, in the type strain, NCTC 10016. Antimicrob Agents Chemother. 2000;44(6):1448–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin JN, Lai CH, Yang CH, Huang YH. Comparison of clinical manifestations, antimicrobial susceptibility patterns, and mutations of fluoroquinolone target genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis isolated in Taiwan. J Clin Med. 2018;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Y, Zhang W, Wang H, et al. Specific patterns of gyrA mutations determine the resistance difference to ciprofloxacin and levofloxacin in Klebsiella pneumoniae and Escherichia coli. BMC Infect Dis. 2013;13:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu T, Zhao X, Li X, et al. Enhancement of fluoroquinolone activity by C-8 halogen and methoxy moieties: action against a gyrase resistance mutant of Mycobacterium smegmatis and a gyrase-topoisomerase IV double mutant of Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45(10):2703–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang YC, Lin YT, Wang FD. Comparison of the therapeutic efficacy of fluoroquinolone and non-fluoroquinolone treatment in patients with Elizabethkingia meningoseptica bacteraemia. Int J Antimicrob Agents. 2018;51(1):47–51. [DOI] [PubMed] [Google Scholar]
- 44.Huang YC, Huang YW, Lin YT, Wang FD, Chan YJ, Yang TC. Risk factors and outcome of levofloxacin-resistant Elizabethkingia meningoseptica bacteraemia in adult patients in Taiwan. Eur J Clin Microbiol Infect Dis. 2017;36(8):1373–1380. [DOI] [PubMed] [Google Scholar]
- 45.Chang TY, Chen HY, Chou YC, Cheng YH, Sun JR. In vitro activities of imipenem, vancomycin, and rifampicin against clinical Elizabethkingia species producing BlaB and GOB metallo-beta-lactamases. Eur J Clin Microbiol Infect Dis. 2019;38(11):2045–2052. [DOI] [PubMed] [Google Scholar]
- 46.Lin PY, Chu C, Su LH, Huang CT, Chang WY, Chiu CH. Clinical and microbiological analysis of bloodstream infections caused by Chryseobacterium meningosepticum in nonneonatal patients. J Clin Microbiol. 2004;42(7):3353–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]