Abstract
Background
Foodstuffs with animal origins, particularly meat, are likely reservoirs of Helicobacter pylori.
Purpose
An existing survey was accompanied to assess phenotypic and genotypic profiles of antibiotic resistance and genotyping of vacA, cagA, cagE, iceA, oipA, and babA2 alleles amongst the H. pylori bacteria recovered from raw meat.
Methods
Six-hundred raw meat samples were collected and cultured. H. pylori isolates were tested using disk diffusion and PCR identification of antibiotic resistance genes and genotyping.
Results
Fifty-two out of 600 (8.66%) raw meat samples were contaminated with H. pylori. Raw ovine meat (13.07%) had the uppermost contamination. H. pylori bacteria displayed the uppermost incidence of resistance toward tetracycline (82.69%), erythromycin (80.76%), trimethoprim (65.38%), levofloxacin (63.46%), and amoxicillin (63.46%). All H. pylori bacteria had at least resistance toward one antibiotic, even though incidence of resistance toward more than eight antibiotics was 28.84%. Total distribution of rdxA, pbp1A, gyrA, and cla antibiotic resistance genes were 59.61%, 51.92%, 69.23%, and 65.38%, respectively. VacA s1a (84.61%), s2 (76.92%), m1a (50%), m2 (39.13%), iceA1 (38.46%), and cagA (55.76%) were the most generally perceived alleles. S1am1a (63.46%), s2m1a (53.84%), s1am2 (51.92%), and s2m2 (42.30%) were the most generally perceived genotyping patterns. Frequency of cagA-, oipA-, and babA2- genotypes were 44.23%, 73.07%, and 80.76%, respectively. A total of 196 combined genotyping patterns were also perceived.
Conclusion
The role of raw meat, particularly ovine meat, in transmission of virulent and resistant H. pylori bacteria was determined. VacA and cagA genotypes had the higher incidence. CagE-, babA2-, and oipA- H. pylori bacteria had the higher distribution. Supplementary surveys are compulsory to originate momentous relations between distribution of genotypes, antibiotic resistance, and antibiotic resistance genes.
Keywords: Helicobacter pylori, antibiotic resistance, genotyping, raw meat
Introduction
Meat of animals, particularly camel, caprine, ovine, bovine, and buffalo species, afford a bundle of nutrient components difficult to gain in diets with incomplete or no meat.1 Reversely, raw meat is not unavoidably safe, as evidenced by considerable rates of foodborne diseases accompanying with its consumption.2 Similarly, several outbreaks of foodborne diseases have been conveyed owing to the consumption of contaminated meat samples.2
Helicobacter pylori (H. pylori) is a microaerophilic and Gram-negative flagellated bacterium responsible for the occurrence of peptic ulcer disease, gastric adenocarcinoma, duodenal ulcer, type B gastritis, mucosa-associated lymphoid tissue (MALT) lymphoma, and gastric B-cell lymphoma.3 The main reservoir of H. pylori bacteria is the human stomach.3 In keeping with this, foods with animal origins, particularly meat, may play an imperative portion in transmission of H. pylori infections to humans.4,5 Foods with animal origins provide appropriate circumstances such as pH, moisture and activated water (AW) contents, and temperature for growth and survival of H. pylori.4,5 Additionally, the role of meat consumption as a risk factor for occurrence of H. pylori infections has been conveyed.6,7 Likewise, the bacterium has been recovered from diverse kinds of foods with animal origins.6,7
H. pylori infections are associated with the presence and activity of certain virulence markers such as Vacuolating Cytotoxin A (vacA). Cytotoxin Associated Gene A and E (cagA and cagE), Induced by Contact with the Epithelium Antigen (iceA), Outer Inflammatory Protein Antigen (oipA), and Blood Group Antigen-Binding Adhesin gene (babA).8,9 The vacA gene is polymorphic, containing mutable signals (type s1 or s2) and mid-regions (type m1 or m2). The s1 type is further alienated into s1a, s1b and s1c and the m1 into m1a and m1b alleles. The cagA gene is an indicator for the genomic pathogenicity island of c. 40 kb [cag pathogenicity island (cag-PAI)] and its activity is believed to be cooperated with interleukin 8 secretion, local inflammation, and severe and/or complicated occurrence of peptic ulcers and gastrointestinal disorders.8,9 CagE gene was found to serve as an improved biomarker of an intact cag-PAI in patients with severe gastrointestinal disorders.8,9 BabA2 gene mediates adherence of H. pylori to human Lewis b blood-group antigens on gastric epithelial cells.8,9 OipA is a significant virulence marker which is associated with clinically imperative presentation of peptic ulcers, such as enhanced interleukin-8 secretion and increased inflammation.8,9 The iceA gene was detected in the H. pylori recovered from patients with gastrointestinal disorders.8,9 There are at least two alleles of iceA:, iceA1, and iceA2.8,9 The relationship between H. pylori iceA1 and iceA2 and clinical outcomes has been addressed by some researchers.8,9 The presence of these alleles has been conveyed in different research conducted on diverse kinds of foods with animal origins.10 Genotyping using these virulence markers is considered as one of the best approaches to study the correlations between H. pylori isolates from different samples.
Antibiotic therapy is one of the best aspects of treatments of H. pylori infections. However, therapeutic choices have become slightly limited owing to the occurrence of resistance in some H. pylori strains.11,13 Recognized information revealed that H. pylori bacteria displayed the boost incidence of resistance toward diverse kinds of antibiotics such as tetracyclines, fluoroquinolone, aminoglycosides, penicillins, sulfonamides, and macrolides.11,13 The presence of certain antibiotic resistance genes, particularly rdxA, pbp1A, gyrA, and cla which encode resistance toward metronidazole, amoxicillin, fluoroquinolone, and clarithromycin antibiotic agents, respectively, is one of the most important reasons for occurrence of antibiotic resistance.14,15 Therefore, it is significant to know the exact phenotypic and genotypic patterns of antibiotic resistance of H. pylori bacteria recovered from foods with animal origins.
Data on the epidemiology and transmission of H. pylori is extremely significant in order to prevent its distribution and to identify high-risk populations. Considering the indistinct epidemiological aspects of H. pylori in meat, as a highly consumed foodstuff, an existing research was performed in order to assess the incidence, genotyping patterns and phenotypic and genotypic profiles of antibiotic resistance of the H. pylori bacteria recovered from raw meat samples of camel, caprine, ovine, bovine, and buffalo species.
Materials and Methods
Samples
From April to October 2018, a total 600 raw meat samples including bovine (n= 140), ovine (n=130), caprine (n= 130), buffalo (n= 100), and camel (n= 100) were arbitrarily collected from the butchers of diverse areas of Tehran province, Iran. All meat samples were collected from the femur muscle. Meat samples displayed natural physical (color, odor, pH, and density) constancy. Samples (40 g, in sterile glass bottles) were transported in ice-cooled flasks (at 4°C) to the laboratory within 2 hours after collection.
Isolation of Helicobacter pylori
Isolation of H. pylori bacteria was performed using the culture technique.17,18 Twenty-five grams of meat sample were applied for this resolve. Wilkins Chalgren anaerobe broth (Oxoid Ltd., Basingstoke, UK) was applied for this goal. Culture media were supplemented with 5% of horse serum (Sigma, St. Louis, MO), nalidixic acid (30 mg/L), vancomycin (10 mg/L), cycloheximide (100 mg/L), and trimethoprim (30 mg/L) (Sigma). Microaerophilic circumstances (5% oxygen, 85% nitrogen, and 10% CO2) was equipped using the MART system (MART system, Lichtenvoorde, The Netherlands). For comparison, a reference strain of H. pylori (ATCC 43504) was employed. Suspected colonies were then identified using colony morphology, Gram staining, and some biochemical tests such as urease, oxidase, catalase, and nitrate reduction.
DNA Extraction and 16S rRNA-Based Polymerase Chain Reaction (PCR) Confirmation
H. pylori isolates were additionally confirmed using the 16S rRNA-based PCR method. Colonies were sub-cultured on Wilkins Chalgren anaerobe broth supplemented with the same materials declared above.16,17 Genomic DNA was then extracted using a DNA extraction kit (Thermo Fisher Scientific, St. Leon-Rot, Germany). Technique was performed rendering to the factory guidelines. Purity (A260/A280) and concentration of extracted DNA were then plaid (NanoDrop, Thermo Scientific, Waltham, MA) and the DNA quality was scrutinized by electrophoresis. PCR was accompanied using a PCR thermal cycler (Eppendorf Co., Hamburg, Germany) rendering to the described procedure.18 H. pylori 26695 was used as positive, while sterile PCR grade water (Thermo Fisher Scientific) was used as negative controls.
Study the Antibiotic Resistance Pattern
Mueller–Hinton agar (Merck, Germany) was applied to assess the pattern of antibiotic resistance using the simple disk diffusion technique. Antibiotic resistance profile of H. pylori bacteria was researched toward dissimilar antibiotic agents (Oxoid, UK) using the guidelines of the Clinical and Laboratory Standards Institute (CLSI).19,20 Resistance patterns of bacteria were experienced toward levofloxacin (5 µg), ampicillin (10 µg), clarithromycin (2 µg), metronidazole (5 µg), streptomycin (10 µg), amoxicillin (10 µg), cefsulodin (30 µg), tetracycline (30 µg), erythromycin (5 µg), furazolidone (1 µg), trimethoprim (25 µg), rifampin (30 µg), and spiramycin (100 µg) (Oxoid). Positive controls (NCTC 13206 (CCUG 38770) and NCTC 13207 (CCUG 38772)) were accompanied in this experiment.
Study the Distribution of Antibiotic Resistance Genes and Genotyping Pattern
Distribution of antibiotic resistance genes and vacA, cagA, iceA, oipA, cagE, and babA2 genotypes of H. pylori bacteria were assessed rendering the preceding experiment.21,29 PCR circumstances were displayed in Table 1. Positive (SS1 (for cagA and cagE genotypes), 26,695 (for babA2, vacA, cagA, cagE, iceA genotypes and cla and rdxA antibiotic resistance genes), Tx30 (for vacA genotypes), J99 (for cagA and babA2), 88–23 (for cagA and vacA genotypes), 84–183 (for vacA and cagA genotypes), 43,504 (for vacA and iceA2 genotypes), 49,503 (for iceA1 genotypes), D0008 (for oipA genotype), 69A (for rdxA and pbp 1A antibiotic resistance gene), and RM92 (for gyrA antibiotic resistance gene), and negative (PCR grade water (Thermo Fisher Scientific)) controls were also accompanied in this experiment. Electrophoresis was addressed rendering previous experiments.21
Table 1.
Set of Primers and PCR Circumstances Applied for Detection of Antibiotic Resistance Genes and Genotyping of vacA, cagA, iceA, oipA, cagE, and babA2 Alleles
| Genes | Primer Sequence (5ʹ-3ʹ) | Size of Product (Bp) | Volume of PCR Reaction (50 µl) | PCR Programs | |
|---|---|---|---|---|---|
| VacA s1a | F: CTCTCGCTTTAGTAGGAGC R: CTGCTTGAATGCGCCAAAC |
213 | 5 µL PCR buffer 10X 1.5 mM MgCl2 200 µM dNTP (Thermo Fisher Scientific) 0.5 µM of each primers F & R 1.25 U Taq DNA polymerase (Thermo Fisher Scientific) 2.5 µL DNA template |
1 cycle: 95°C; 1 min. 32 cycle: 95°C; 45 s |
|
| VacA s1b | F: AGCGCCATACCGCAAGAG CTGCTTGAATGCGCCAAAC |
187 | |||
| VacA s1c | F: CTCTCGCTTTAGTGGGGYT R: CTGCTTGAATGCGCCAAAC |
213 | 64°C; 50 s 72°C; 70 s 1 cycle: 72°C; 5 min |
||
| VacA s2 | F: GCTAACACGCCAAATGATCC R: CTGCTTGAATGCGCCAAAC |
199 | |||
| VacA m1a | F: GGTCAAAATGCGGTCATGG R: CCATTGGTACCTGTAGAAAC |
290 | |||
| VacA m1b | F: GGCCCCAATGCAGTCATGGA R: GCTGTTAGTGCCTAAAGAAGCAT |
291 | |||
| VacA m2 | F: GGAGCCCCAGGAAACATTG R: CATAACTAGCGCCTTGCA |
352 | |||
| Cag A | F: GATAACAGCCAAGCTTTTGAGG R: CTGCAAAAGATTGTTTGGCAGA |
300 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 94°C; 1 min. 32 cycle: 95°C; 60 s 56°C; 60 s 72°C; 60 s 1 cycle: 72°C; 10 min |
|
| IceA | IceA1 | F: GTGTTTTTAACCAAAGTATC R: CTATAGCCASTYTCTTTGCA |
247 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 94°C; 1 min. 32 cycle: 94°C; 60 s 56°C; 60 s 72°C; 60 s 1 cycle: 72°C; 10 min |
| IceA2 | F: GTTGGGTATATCACAATTTAT R: TTRCCCTATTTTCTAGTAGGT |
229/334 | |||
| OipA | F: GTTTTTGATGCATGGGATTT R: GTGCATCTCTTATGGCTTT |
401 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 94°C; 1 min. 32 cycle: 94°C; 60 s 56°C; 60 s 72°C; 60 s 1 cycle: 72°C; 10 min |
|
| cagE | F: TTGAAAACTTCAAGGATAGGATAGAGC R: GCCTAGCGTAATATCACCATTACCC |
500 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 95°C; 4 min. 31 cycle: 95°C; 44 s 51°C; 45 s 72°C; 62 s 1 cycle: 72°C; 5 min |
|
| BabA2 | F: CCAAACGAAACAAAAAGCGT R: GCTTGTGTAAAAGCCGTCGT |
105–124 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 94°C; 1 min. 35 cycle: 94°C; 60 s 57°C; 45 s 72°C; 30 s 1 cycle: 72°C; 10 min |
|
| rdXA | Metronidazole | F: AATTTGAGCATGGGGCAGA R: GAAACGCTTGAAAACACCCCT |
581 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 95°C; 5 min. 40 cycle: 94°C; 30 s 55°C; 30 s 72°C; 60 s 1 cycle: 72°C; 10 min |
| pbp1A | Amoxicillin | F: GCGACAATAAGAGTGGCA R: TGCGAACACCCTTTTAAA T |
2,300 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 95°C; 3 min. 35 cycle: 95°C; 60 s 54°C; 60 s 72°C; 5 min 1 cycle: 72°C; 10 min |
| gyrA | Fluoroquinolone | F: TTTRGCTTATTCMATGAGCGT R: GCAGACGGCTTGGTARAATA |
2,300 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 94°C; 5 min. 35 cycle: 94°C; 30 s 47°C; 30 s 72°C; 30 s 1 cycle: 72°C; 5 min |
| cla | Clarithromycin | F: AGTCGGGACCTAAGGCGAG R: AGGTCCACCACGGGGTCTTG |
700 | 5 µL PCR buffer 10X 2 mM MgCl2 150 µM dNTP (Thermo Fisher Scientific) 0.75 µM of each primers F & R 1.5 U Taq DNA polymerase (Thermo Fisher Scientific) 3 µL DNA template |
1 cycle: 94°C; 5 min. 30 cycle: 94°C; 60 s 55°C; 60 s 72°C; 60 s 1 cycle: 72°C; 5 min |
Numerical Examination
Data were subjected to Microsoft office Excel (version 15; Microsoft Corp., Redmond, WA). Numerical examination was performed by means of the SPSS 21.0 numerical software (SPSS Inc., Chicago, IL). Chi-square test and Fisher’s exact two-tailed test were applied to measure any momentous relationship. P-value<0.05 was considered as a numerical momentous level.
Results
Table 2 embodies the incidence of H. pylori bacteria recovered from diverse kinds of raw meat samples. Fifty-two out of 600 (8.66%) raw meat samples were contaminated with H. pylori. Raw ovine (13.07%) samples had the uppermost contamination rate with H. pylori bacteria, while raw camel (3%) had the lowest. Numerical momentous variance was originated amid kinds of samples and incidence of H. pylori bacteria (P<0.05).
Table 2.
Incidence of H. pylori in Diverse Kinds of Raw Meat Samples
| Raw Milk Samples | No Samples Collected | N (%) of H. pylori Positive Samples* |
|---|---|---|
| Bovine | 140 | 8 (5.71) |
| Ovine | 130 | 17 (13.07) |
| Caprine | 130 | 15 (11.53) |
| Buffalo | 100 | 9 (9) |
| Camel | 100 | 3 (3) |
| Total | 600 | 52 (8.66) |
Note: *H. pylori isolates were also confirmed by the 16S rRNA-based PCR amplification.
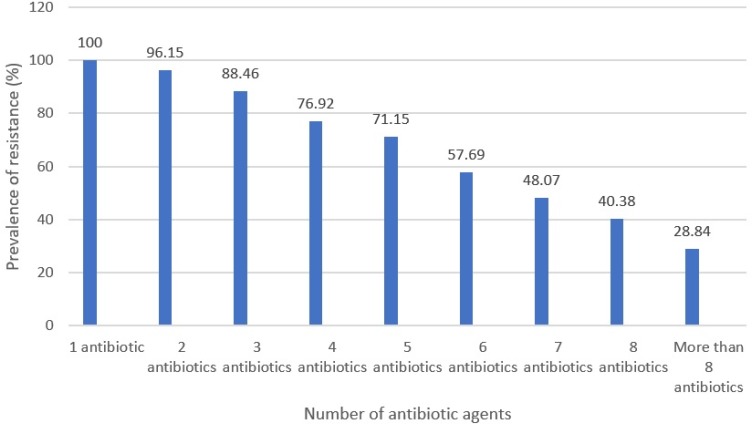
Table 3 embodies the antibiotic resistance pattern of H. pylori bacteria recovered from diverse kinds of raw meat samples. H. pylori bacteria displayed the uppermost incidence of resistance toward tetracycline (82.69%), erythromycin (80.76%), trimethoprim (65.38%), levofloxacin (63.46%), amoxicillin (63.46%), and clarithromycin (61.53%) antibiotic agents. H. pylori bacteria displayed the lowest incidence of resistance toward spiramycin (21.15%), furazolidone (25%), cefsulodin (38.46%), and rifampin (40.38%) antibiotic agents. H. pylori bacteria recovered from raw ovine meat samples displayed the most diverse incidence of resistance toward antibiotic agents. Numerical momentous variance was originated amid kinds of samples and incidence of antibiotic resistance of H. pylori bacteria (P<0.05). Figure 1 embodies the distribution of multi-drug resistant H. pylori bacteria recovered from diverse kinds of raw meat samples. All H. pylori bacteria recovered from raw meat samples had at least resistance toward one antibiotic agent, while incidence of resistance toward more than eight types of antibiotics was 28.84%.
Table 3.
Antibiotic Resistance Pattern of H. pylori Bacteria Isolated from Diverse Kinds of Raw Meat Samples
| Type of Raw Meat Samples (N H. Pylori Bacteria) | N (%) Isolates Resistant to Each Antibiotic | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Am10 | Clr2 | Er5 | Met5 | Lev5 | Tet30 | AMX 10 | Rif30 | Trp25 | Cef30 | S10 | Fzl1 | Spi100 | |
| Bovine (8) | 5 (62.50) | 4 (50) | 6 (75) | 3 (37.50) | 4 (50) | 7 (87.50) | 4 (50) | 3 (37.50) | 4 (50) | 3 (37.50) | 4 (50) | 2 (25) | 2 (25) |
| Ovine (17) | 15 (88.23) | 12 (70.58) | 15 (88.23) | 9 (52.94) | 12 (70.58) | 15 (88.23) | 12 (70.58) | 7 (41.17) | 13 (76.47) | 6 (35.29) | 11 (64.70) | 5 (29.41) | 4 (23.52) |
| Caprine (15) | 12 (80) | 10 (66.66) | 12 (80) | 10 (66.66) | 11 (73.33) | 12 (80) | 10 (66.66) | 6 (40) | 11 (73.33) | 6 (40) | 9 (60) | 3 (20) | 3 (20) |
| Buffalo (9) | 6 (66.66) | 5 (55.55) | 7 (77.77) | 4 (44.44) | 5 (55.55) | 7 (77.77) | 5 (55.55) | 4 (44.44) | 5 (55.55) | 4 (44.44) | 5 (55.55) | 2 (22.22) | 2 (22.22) |
| Camel (3) | 1 (33.33) | 1 (33.33) | 2 (66.66) | 1 (33.33) | 1 (33.33) | 2 (66.66) | 2 (66.66) | 1 (33.33) | 1 (33.33) | 1 (33.33) | 2 (66.66) | 1 (33.33) | – |
| Total (52) | 31 (59.61) | 32 (61.53) | 42 (80.76) | 27 (51.92) | 33 (63.46) | 43 (82.69) | 33 (63.46) | 21 (40.38) | 34 (65.38) | 20 (38.46) | 31 (59.61) | 13 (25) | 11 (21.15) |
Abbreviations: AM10, ampicillin (10 µg); CLR2, clarithromycin (2 µg); ER5, erythromycin (5 µg); Met5, metronidazole (5 µg); Lev5, levofloxacin (5 µg); Tet30, tetracycline (30 µg); AMX10, amoxicillin (10 µg); RIF30, rifampin (30 µg); TRP25, trimethoprim (25 µg); Cef30, cefsulodin (30 µg); S10, streptomycin (10 µg); FZL1, furazolidone (1 µg); Spi100, spiramycin (100 µg).
Figure 1.
Incidence of multi-drug resistant H. pylori bacteria isolated from raw meat samples.
Table 4 embodies the distribution of antibiotic resistance genes amongst the H. pylori bacteria recovered from diverse kinds of raw meat samples. Total distribution of rdxA, pbp1A, gyrA, and cla antibiotic resistance genes amongst the H. pylori bacteria recovered from diverse kinds of raw meat samples were 59.61%, 51.92%, 69.23%, and 65.38%, respectively. H. pylori bacteria recovered from raw ovine meat samples displayed the most diverse distribution of antibiotic resistance genes. Numerical momentous variance was originated amid kinds of samples and distribution of antibiotic resistance genes (P<0.05).
Table 4.
Distribution of Antibiotic Resistant Genes Amongst the H. pylori Bacteria Isolated from Diverse Kinds of Raw Meat Samples
| Type of Raw Meat Samples (N H. pylori Bacteria) | N (%) Isolates Harbored Each Antibiotic Resistant Gene | |||
|---|---|---|---|---|
| Metronidazole | Amoxicillin | Fluoroquinolone | Clarithromycin | |
| rdxA | pbp1A | gyrA | cla | |
| Bovine (8) | 4 (50) | 5 (62.50) | 4 (50) | 5 (62.50) |
| Ovine (17) | 11 (64.70) | 13 (76.47) | 13 (76.47) | 12 (70.58) |
| Caprine (15) | 10 (66.66) | 12 (80) | 12 (80) | 11 (73.33) |
| Buffalo (9) | 5 (55.55) | 6 (66.66) | 5 (55.55) | 5 (55.55) |
| Camel (3) | 1 (33.33) | 1 (33.33) | 2 (66.66) | 1 (33.33) |
| Total (52) | 31 (59.61) | 27 (51.92) | 36 (69.23) | 34 (65.38) |
Table 5 embodies the distribution of alleles amongst the H. pylori bacteria recovered from diverse kinds of raw meat samples. vacA s1a (84.61%), s2 (76.92%), m1a (50%), m2 (39.13%), iceA1 (38.46%), and cagA (55.76%) were the most generally perceived alleles amongst the H. pylori bacteria. Distribution of vacA s1c (7.69%) and m1b (21.15%) and iceA2 (7.69%) and babA2 (19.23%) alleles were lower than other detected genotypes. H. pylori bacteria recovered from raw ovine meat samples displayed the most diverse distribution of alleles. Numerical momentous variance was originated between type of samples and distribution of alleles of H. pylori bacteria (P<0.05). Furthermore, numerical momentous variance was originated amid distribution of cagA and cagE (P<0.01) and iceA1 and iceA2 (P<0.01) alleles.
Table 5.
Distribution of Alleles Amongst the H. pylori Bacteria Isolated from Diverse Kinds of Raw Meat Samples
| Type of Raw Meat Samples (N H. pylori Bacteria) | N (%) Isolates Harbor Each Genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VacA | CagA | IceA | OipA | CagE | BabA2 | ||||||||
| s1a | s1b | s1c | s2 | m1a | m1b | m2 | IceA1 | IceA2 | |||||
| Bovine (8) | 6 (75) | 1 (12.50) | – | 6 (75) | 7 (87.50) | 1 (12.50) | 6 (75) | 5 (62.50) | 4 (50) | 1 (12.50) | 2 (25) | 3 (37.50) | 2 (25) |
| Ovine (17) | 16 (94.11) | 6 (35.29) | 2 (11.76) | 15 (88.23) | 16 (94.11) | 5 (29.41) | 14 (82.35) | 12 (70.58) | 7 (41.17) | 2 (11.76) | 5 (29.41) | 4 (23.52) | 3 (17.64) |
| Caprine (15) | 13 (86.66) | 4 (26.66) | 1 (6.66) | 12 (80) | 13 (86.66) | 3 (20) | 10 (66.66) | 8 (53.33) | 5 (33.33) | 1 (6.66) | 4 (26.66) | 3 (20) | 3 (20) |
| Buffalo (9) | 6 (66.66) | 2 (22.22) | 1 (11.11) | 7 (77.77) | 8 (88.88) | 2 (22.22) | 5 (55.55) | 3 (33.33) | 3 (33.33) | – | 2 (22.22) | 2 (22.22) | 2 (22.22) |
| Camel (3) | 3 (100) | – | – | 2 (66.66) | 2 (66.66) | – | 1 (33.33) | 1 (33.33) | 1 (33.33) | – | 1 (33.33) | 1 (33.33) | – |
| Total (52) | 44 (84.61) | 13 (25) | 4 (7.69) | 40 (76.92) | 46 (50) | 11 (21.15) | 36 (39.13) | 29 (55.76) | 20 (38.46) | 4 (7.69) | 14 (26.92) | 13 (25) | 10 (19.23) |
Table 6 embodies the genotyping pattern of H. pylori bacteria recovered from diverse kinds of raw meat samples. S1am1a (63.46%), s2m1a (53.84%), s1am2 (51.92%), and s2m2 (42.30%) were the most generally perceived genotyping pattern of the vacA alleles of H. pylori bacteria recovered from diverse kinds of raw meat samples. Distribution of cagA-, oipA-, and babA2- genotypes were 44.23%, 73.07%, and 80.76%, respectively. We originated that 5.76% of H. pylori bacteria displayed iceA1/iceA2 genotyping pattern. S1cm1b (1.92%), s1cm2 (3.84%), s1cm1a (3.84%), and and s1bm1b (7.62%) had the lowest incidence amongst different genotyping patterns of H. pylori bacteria. H. pylori bacteria recovered from raw ovine meat samples displayed the most diverse distribution of genotypes.
Table 6.
Genotyping Pattern of H. pylori Bacteria Isolated from Diverse Kinds of Raw Meat Samples
| Type of Raw Meat Samples (N of H. pylori Bacteria) | Genotyping pattern (%) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s1am1a | s1am1b | s1am2 | s1bm1a | s1bm1b | s1bm2 | s1cm1a | s1cm1b | s1cm2 | s2m1a | s2m1b | s2m2 | CagA+ | CagA- | IceA1/IceA2 | OipA+ | OipA- | CagE+ | CagE- | BabA2+ | BabA2- | ||
| Bovine (8) | 4 (50) | 1 (12.50) | 4 (50) | 1 (12.50) | - | - | - | - | - | 3 (37.50) | 1 (12.50) | 3 (37.50) | 5 (62.50) | 3 (37.50) | 1 (12.50) | 2 (25) | 6 (75) | 3 (37.50) | 5 (62.50) | 2 (25) | 6 (75) | |
| Ovine (17) | 12 (70.58) | 3 (17.64) | 11 (64.70) | 3 (17.64) | 2 (11.76) | 3 (17.64) | 1 (5.88) | 1 (5.88) | 1 (5.88) | 11 (64.70) | 2 (11.76) | 10 (58.82) | 12 (70.58) | 5 (29.41) | 1 (5.88) | 5 (29.41) | 12 (70.58) | 4 (23.52) | 13 (76.47) | 3 (17.64) | 14 (82.35) | |
| Caprine (15) | 11 (73.33) | 2 (13.33) | 8 (53.33) | 2 (13.33) | 1 (6.66) | 2 (13.33) | 1 (6.66) | - | 1 (6.66) | 8 (53.33) | 1 (6.66) | 9 (60) | 8 (53.33) | 7 (46.66) | 1 (6.66) | 4 (26.66) | 11 (73.33) | 3 (20) | 12 (80) | 3 (20) | 12 (80) | |
| Buffalo (9) | 5 (55.55) | 1 (11.11) | 3 (21.42) | 1 (11.11) | 1 (11.11) | 1 (11.11) | - | - | - | 5 (55.55) | 1 (11.11) | 4 (44.44) | 3 (33.33) | 6 (66.66) | - | 2 (22.22) | 7 (77.77) | 2 (22.22) | 7 (77.77) | 2 (22.22) | 7 (77.77) | |
| Camel (3) | 1 (33.33) | - | 1 (33.33) | - | - | - | - | - | - | 1 (33.33) | - | 1 (33.33) | 1 (33.33) | 2 (66.66) | - | 1 (33.33) | 2 (66.66) | 1 (33.33) | 2 (66.66) | - | 3 (100) | |
| Total (52) | 33 (63.46) | 7 (13.46) | 27 (51.92) | 7 (13.46) | 4 (7.69) | 6 (11.53) | 2 (3.84) | 1 (1.92) | 2 (3.84) | 28 (53.84) | 5 (9.61) | 22 (42.30) | 29 (55.76) | 23 (44.23) | 3 (5.76) | 14 (26.92) | 38 (73.07) | 13 (25) | 39 (75) | 10 (19.23) | 42 (80.76) | |
Table 7 embodies the combined genotyping pattern of H. pylori bacteria recovered from diverse kinds of raw meat samples. We originated that s1a/cagA-/iceA1/oipA-/cagE-/babA- (32.69%), m1a/cagA-/iceA1/oipA-/cagE-/babA- (32.69%), s2/cagA-/iceA1/oipA-/cagE-/babA- (26.92%), s1a/cagA+/iceA1/oipA+/cagE-/babA- (23.07%), m1a/cagA+/iceA1/oipA+/cagE+/babA- (23.07%), m1a/cagA+/iceA1/oipA+/cagE-/babA- (23.07%), m2/cagA-/iceA1/oipA-/cagE-/babA- (23.07%), s1a/cagA+/iceA1/oipA+/cagE+/babA- (21.15%), s1a/cagA-/iceA1/oipA-/cagE+/babA- (21.15%), s1a/cagA-/iceA1/oipA+/cagE-/babA- (21.15%), s1b/cagA-/iceA1/oipA-/cagE-/babA- (21.15%), m1a/cagA-/iceA1/oipA-/cagE+/babA- (21.15%), m1a/cagA-/iceA1/oipA+/cagE-/babA- (21.15%), m1b/cagA-/iceA1/oipA-/cagE-/babA- (21.15%), s1a/cagA+/iceA1/oipA-/cagE+/babA- (19.23%), s2/cagA+/iceA1/oipA+/cagE-/babA- (19.23%), s2/cagA-/iceA1/oipA-/cagE+/babA- (19.23%), s2/cagA-/iceA1/oipA+/cagE-/babA- (19.23%), m1a/cagA+/iceA1/oipA-/cagE-/babA+ (19.23%), and m1a/cagA+/iceA1/oipA-/cagE+/babA- (19.23%) were the most generally perceived combined genotyping pattern of H. pylori bacteria recovered from diverse kinds of raw meat samples. Incidence of s1a/cagA+/iceA2/oipA+/cagE+/babA+, s1a/cagA+/iceA2/oipA+/cagE-/babA+, s1a/cagA-/iceA2/oipA+/cagE+/babA+, s1a/cagA+/iceA2/oipA-/cagE+/babA-, s1a/cagA-/iceA2/oipA+/cagE+/babA-, s1a/cagA-/iceA2/oipA+/cagE-/babA-, s1b/cagA+/iceA1/oipA+/cagE+/babA+, s1b/cagA-/iceA1/oipA-/cagE+/babA+, s1c/cagA+/iceA1/oipA+/cagE+/babA-, s1c/cagA+/iceA1/oipA+/cagE-/babA-, s1c/cagA-/iceA1/oipA-/cagE+/babA-, s2/cagA+/iceA2/oipA+/cagE+/babA-, s2/cagA+/iceA2/oipA+/cagE-/babA-, s2/cagA-/iceA2/oipA-/cagE-/babA-, m1a/cagA+/iceA2/oipA+/cagE+/babA+, m1a/cagA+/iceA2/oipA+/cagE-/babA+, m1a/cagA+/iceA2/oipA-/cagE+/babA-, m1a/cagA-/iceA2/oipA+/cagE+/babA-, m1b/cagA-/iceA1/oipA-/cagE+/babA+, m1b/cagA+/iceA2/oipA+/cagE-/babA-, m1b/cagA-/iceA2/oipA-/cagE-/babA-, m2/cagA+/iceA2/oipA+/cagE+/babA-, m2/cagA+/iceA2/oipA+/cagE-/babA-, and m2/cagA-/iceA2/oipA-/cagE-/babA- (1.92%) were lower than other detected combined genotyping patterns.
Table 7.
Combined Genotyping Pattern of H. pylori Bacteria Isolated from Diverse Kinds of Raw Meat Samples
| Combined Genotyping Patterns | Distribution* (%) |
|---|---|
| S1a/cagA+/iceA1/oipA+/cagE+/babA+ | 6 (11.53) |
| S1a/cagA+/iceA1/oipA+/cagE+/babA- | 11 (21.15) |
| S1a/cagA+/iceA1/oipA+/cagE-/babA+ | 8 (15.38) |
| S1a/cagA+/iceA1/oipA-/cagE+/babA+ | 7 (13.46) |
| S1a/cagA-/iceA1/oipA+/cagE+/babA+ | 8 (15.38) |
| S1a/cagA+/iceA1/oipA+/cagE-/babA- | 12 (23.07) |
| S1a/cagA+/iceA1/oipA-/cagE-/babA+ | 9 (17.30) |
| S1a/cagA+/iceA1/oipA-/cagE+/babA- | 10 (19.23) |
| S1a/cagA-/iceA1/oipA-/cagE+/babA- | 11 (21.15) |
| S1a/cagA-/iceA1/oipA-/cagE+/babA+ | 7 (13.46) |
| S1a/cagA-/iceA1/oipA+/cagE-/babA+ | 6 (11.53) |
| S1a/cagA-/iceA1/oipA+/cagE+/babA- | 9 (17.30) |
| S1a/cagA-/iceA1/oipA+/cagE-/babA- | 11 (21.15) |
| S1a/cagA-/iceA1/oipA-/cagE-/babA- | 17 (32.69) |
| S1a/cagA+/iceA2/oipA+/cagE+/babA+ | 1 (1.92) |
| S1a/cagA+/iceA2/oipA+/cagE+/babA- | 2 (3.84) |
| S1a/cagA+/iceA2/oipA+/cagE-/babA+ | 1 (1.92) |
| S1a/cagA+/iceA2/oipA-/cagE+/babA+ | – |
| S1a/cagA-/iceA2/oipA+/cagE+/babA+ | 1 (1.92) |
| S1a/cagA+/iceA2/oipA+/cagE-/babA- | 2 (3.84) |
| S1a/cagA+/iceA2/oipA-/cagE-/babA+ | – |
| S1a/cagA+/iceA2/oipA-/cagE+/babA- | 1 (1.92) |
| S1a/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| S1a/cagA-/iceA2/oipA-/cagE+/babA+ | – |
| S1a/cagA-/iceA2/oipA+/cagE-/babA+ | – |
| S1a/cagA-/iceA2/oipA+/cagE+/babA- | 1 (1.92) |
| S1a/cagA-/iceA2/oipA+/cagE-/babA- | 1 (1.92) |
| S1a/cagA-/iceA2/oipA-/cagE-/babA- | 3 (5.76) |
| S1b/cagA+/iceA1/oipA+/cagE+/babA+ | 1 (1.92) |
| S1b/cagA+/iceA1/oipA+/cagE+/babA- | 5 (9.61) |
| S1b/cagA+/iceA1/oipA+/cagE-/babA+ | 2 (3.84) |
| S1b/cagA+/iceA1/oipA-/cagE+/babA+ | 2 (3.84) |
| S1b/cagA-/iceA1/oipA+/cagE+/babA+ | 2 (3.84) |
| S1b/cagA+/iceA1/oipA+/cagE-/babA- | 6 (11.53) |
| S1b/cagA+/iceA1/oipA-/cagE-/babA+ | 3 (5.76) |
| S1b/cagA+/iceA1/oipA-/cagE+/babA- | 4 (7.69) |
| S1b/cagA-/iceA1/oipA-/cagE+/babA- | 5 (9.61) |
| S1b/cagA-/iceA1/oipA-/cagE+/babA+ | 1 (1.92) |
| S1b/cagA-/iceA1/oipA+/cagE-/babA+ | - |
| S1b/cagA-/iceA1/oipA+/cagE+/babA- | 3 (5.76) |
| S1b/cagA-/iceA1/oipA+/cagE-/babA- | 5 (9.61) |
| S1b/cagA-/iceA1/oipA-/cagE-/babA- | 11 (21.15) |
| S1b/cagA+/iceA2/oipA+/cagE+/babA+ | – |
| S1b/cagA+/iceA2/oipA+/cagE+/babA- | – |
| S1b/cagA+/iceA2/oipA+/cagE-/babA+ | – |
| S1b/cagA+/iceA2/oipA-/cagE+/babA+ | – |
| S1b/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| S1b/cagA+/iceA2/oipA+/cagE-/babA- | – |
| S1b/cagA+/iceA2/oipA-/cagE-/babA+ | – |
| S1b/cagA+/iceA2/oipA-/cagE+/babA- | – |
| S1b/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| S1b/cagA-/iceA2/oipA-/cagE+/babA+ | – |
| S1b/cagA-/iceA2/oipA+/cagE-/babA+ | – |
| S1b/cagA-/iceA2/oipA+/cagE+/babA- | – |
| S1b/cagA-/iceA2/oipA+/cagE-/babA- | – |
| S1b/cagA-/iceA2/oipA-/cagE-/babA- | – |
| S1c/cagA+/iceA1/oipA+/cagE+/babA+ | – |
| S1c/cagA+/iceA1/oipA+/cagE+/babA- | 1 (1.92) |
| S1c/cagA+/iceA1/oipA+/cagE-/babA+ | – |
| S1c/cagA+/iceA1/oipA-/cagE+/babA+ | – |
| S1c/cagA-/iceA1/oipA+/cagE+/babA+ | – |
| S1c/cagA+/iceA1/oipA+/cagE-/babA- | 1 (1.92) |
| S1c/cagA+/iceA1/oipA-/cagE-/babA+ | – |
| S1c/cagA+/iceA1/oipA-/cagE+/babA- | – |
| S1c/cagA-/iceA1/oipA-/cagE+/babA- | 1 (1.92) |
| S1c/cagA-/iceA1/oipA-/cagE+/babA+ | – |
| S1c/cagA-/iceA1/oipA+/cagE-/babA+ | – |
| S1c/cagA-/iceA1/oipA+/cagE+/babA- | – |
| S1c/cagA-/iceA1/oipA+/cagE-/babA- | – |
| S1c/cagA-/iceA1/oipA-/cagE-/babA- | 2 (3.84) |
| S1c/cagA+/iceA2/oipA+/cagE+/babA+ | – |
| S1c/cagA+/iceA2/oipA+/cagE+/babA- | – |
| S1c/cagA+/iceA2/oipA+/cagE-/babA+ | – |
| S1c/cagA+/iceA2/oipA-/cagE+/babA+ | – |
| S1c/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| S1c/cagA+/iceA2/oipA+/cagE-/babA- | – |
| S1c/cagA+/iceA2/oipA-/cagE-/babA+ | – |
| S1c/cagA+/iceA2/oipA-/cagE+/babA- | – |
| S1c/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| S1c/cagA-/iceA2/oipA-/cagE+/babA+ | – |
| S1c/cagA-/iceA2/oipA+/cagE-/babA+ | – |
| S1c/cagA-/iceA2/oipA+/cagE+/babA- | – |
| S1c/cagA-/iceA2/oipA+/cagE-/babA- | – |
| S2/cagA+/iceA1/oipA+/cagE+/babA+ | 5 (9.61) |
| S2/cagA+/iceA1/oipA+/cagE+/babA- | 9 (17.30) |
| S2/cagA+/iceA1/oipA+/cagE-/babA+ | 7 (13.46) |
| S2/cagA+/iceA1/oipA-/cagE+/babA+ | 6 (11.53) |
| S2/cagA-/iceA1/oipA+/cagE+/babA+ | 6 (11.53) |
| S2/cagA+/iceA1/oipA+/cagE-/babA- | 10 (19.23) |
| S2/cagA+/iceA1/oipA-/cagE-/babA+ | 8 (15.38) |
| S2/cagA+/iceA1/oipA-/cagE+/babA- | 9 (17.30) |
| S2/cagA-/iceA1/oipA-/cagE+/babA- | 10 (19.23) |
| S2/cagA-/iceA1/oipA-/cagE+/babA+ | 5 (9.61) |
| S2/cagA-/iceA1/oipA+/cagE-/babA+ | 4 (7.69) |
| S2/cagA-/iceA1/oipA+/cagE+/babA- | 7 (13.46) |
| S2/cagA-/iceA1/oipA+/cagE-/babA- | 10 (19.23) |
| S2/cagA-/iceA1/oipA-/cagE-/babA- | 14 (26.92) |
| S2/cagA+/iceA2/oipA+/cagE+/babA+ | – |
| S2/cagA+/iceA2/oipA+/cagE+/babA- | 1 (1.92) |
| S2/cagA+/iceA2/oipA+/cagE-/babA+ | – |
| S2/cagA+/iceA2/oipA-/cagE+/babA+ | – |
| S2/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| S2/cagA+/iceA2/oipA+/cagE-/babA- | 1 (1.92) |
| S2/cagA+/iceA2/oipA-/cagE-/babA+ | – |
| S2/cagA+/iceA2/oipA-/cagE+/babA- | – |
| S2/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| S2/cagA-/iceA2/oipA-/cagE+/babA+ | – |
| S2/cagA-/iceA2/oipA+/cagE-/babA+ | – |
| S2/cagA-/iceA2/oipA+/cagE+/babA- | – |
| S2/cagA-/iceA2/oipA+/cagE-/babA- | – |
| S2/cagA-/iceA2/oipA-/cagE-/babA- | 1 (1.92) |
| M1a/cagA+/iceA1/oipA+/cagE+/babA+ | 6 (11.53) |
| M1a/cagA+/iceA1/oipA+/cagE+/babA- | 12 (23.07) |
| M1a/cagA+/iceA1/oipA+/cagE-/babA+ | 8 (15.38) |
| M1a/cagA+/iceA1/oipA-/cagE+/babA+ | 7 (13.46) |
| M1a/cagA-/iceA1/oipA+/cagE+/babA+ | 8 (15.38) |
| M1a/cagA+/iceA1/oipA+/cagE-/babA- | 12 (23.07) |
| M1a/cagA+/iceA1/oipA-/cagE-/babA+ | 10 (19.23) |
| M1a/cagA+/iceA1/oipA-/cagE+/babA- | 10 (19.23) |
| M1a/cagA-/iceA1/oipA-/cagE+/babA- | 11 (21.15) |
| M1a/cagA-/iceA1/oipA-/cagE+/babA+ | 7 (13.46) |
| M1a/cagA-/iceA1/oipA+/cagE-/babA+ | 6 (11.53) |
| M1a/cagA-/iceA1/oipA+/cagE+/babA- | 9 (17.30) |
| M1a/cagA-/iceA1/oipA+/cagE-/babA- | 11 (21.15) |
| M1a/cagA-/iceA1/oipA-/cagE-/babA- | 17 (32.69) |
| M1a/cagA+/iceA2/oipA+/cagE+/babA+ | 1 (1.92) |
| M1a/cagA+/iceA2/oipA+/cagE+/babA- | 2 (3.84) |
| M1a/cagA+/iceA2/oipA+/cagE-/babA+ | 1 (1.92) |
| M1a/cagA+/iceA2/oipA-/cagE+/babA+ | – |
| M1a/cagA-/iceA2/oipA+/cagE+/babA+ | 2 (3.84) |
| M1a/cagA+/iceA2/oipA+/cagE-/babA- | 2 (3.84) |
| M1a/cagA+/iceA2/oipA-/cagE-/babA+ | – |
| M1a/cagA+/iceA2/oipA-/cagE+/babA- | 1 (1.92) |
| M1a/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| M1a/cagA-/iceA2/oipA-/cagE+/babA+ | – |
| M1a/cagA-/iceA2/oipA+/cagE-/babA+ | - |
| M1a/cagA-/iceA2/oipA+/cagE+/babA- | 1 (1.92) |
| M1a/cagA-/iceA2/oipA+/cagE-/babA- | 2 (3.84) |
| M1a/cagA-/iceA2/oipA-/cagE-/babA- | 3 (5.76) |
| M1b/cagA+/iceA1/oipA+/cagE+/babA+ | 2 (3.84) |
| M1b/cagA+/iceA1/oipA+/cagE+/babA- | 5 (9.61) |
| M1b/cagA+/iceA1/oipA+/cagE-/babA+ | 2 (3.84) |
| M1b/cagA+/iceA1/oipA-/cagE+/babA+ | 2 (3.84) |
| M1b/cagA-/iceA1/oipA+/cagE+/babA+ | 2 (3.84) |
| M1b/cagA+/iceA1/oipA+/cagE-/babA- | 6 (11.53) |
| M1b/cagA+/iceA1/oipA-/cagE-/babA+ | 3 (5.76) |
| M1b/cagA+/iceA1/oipA-/cagE+/babA- | 4 (7.69) |
| M1b/cagA-/iceA1/oipA-/cagE+/babA- | 5 (9.61) |
| M1b/cagA-/iceA1/oipA-/cagE+/babA+ | 1 (1.92) |
| M1b/cagA-/iceA1/oipA+/cagE-/babA+ | – |
| M1b/cagA-/iceA1/oipA+/cagE+/babA- | 4 (7.69) |
| M1b/cagA-/iceA1/oipA+/cagE-/babA- | 5 (9.61) |
| M1b/cagA-/iceA1/oipA-/cagE-/babA- | 11 (21.15) |
| M1b/cagA+/iceA2/oipA+/cagE+/babA+ | – |
| M1b/cagA+/iceA2/oipA+/cagE+/babA- | – |
| M1b/cagA+/iceA2/oipA+/cagE-/babA+ | – |
| M1b/cagA+/iceA2/oipA-/cagE+/babA+ | – |
| M1b/cagA-/iceA2/oipA+/cagE+/babA+ | - |
| M1b/cagA+/iceA2/oipA+/cagE-/babA- | 1 (1.92) |
| M1b/cagA+/iceA2/oipA-/cagE-/babA+ | – |
| M1b/cagA+/iceA2/oipA-/cagE+/babA- | – |
| M1b/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| M1b/cagA-/iceA2/oipA-/cagE+/babA+ | – |
| M1b/cagA-/iceA2/oipA+/cagE-/babA+ | – |
| M1b/cagA-/iceA2/oipA+/cagE+/babA- | – |
| M1b/cagA-/iceA2/oipA+/cagE-/babA- | – |
| M1b/cagA-/iceA2/oipA-/cagE-/babA- | 1 (1.92) |
| M2/cagA+/iceA1/oipA+/cagE+/babA+ | 4 (7.69) |
| M2/cagA+/iceA1/oipA+/cagE+/babA- | 9 (17.30) |
| M2/cagA+/iceA1/oipA+/cagE-/babA+ | 7 (13.46) |
| M2/cagA+/iceA1/oipA-/cagE+/babA+ | 6 (11.53) |
| M2/cagA-/iceA1/oipA+/cagE+/babA+ | 5 (9.61) |
| M2/cagA+/iceA1/oipA+/cagE-/babA- | 9 (17.30) |
| M2/cagA+/iceA1/oipA-/cagE-/babA+ | 8 (15.38) |
| M2/cagA+/iceA1/oipA-/cagE+/babA- | 9 (17.30) |
| M2/cagA-/iceA1/oipA-/cagE+/babA- | 9 (17.30) |
| M2/cagA-/iceA1/oipA-/cagE+/babA+ | 4 (7.69) |
| M2/cagA-/iceA1/oipA+/cagE-/babA+ | 4 (7.69) |
| M2/cagA-/iceA1/oipA+/cagE+/babA- | 7 (13.46) |
| M2/cagA-/iceA1/oipA+/cagE-/babA- | 9 (17.30) |
| M2/cagA-/iceA1/oipA-/cagE-/babA- | 12 (23.07) |
| M2/cagA+/iceA2/oipA+/cagE+/babA+ | – |
| M2/cagA+/iceA2/oipA+/cagE+/babA- | 1 (1.92) |
| M2/cagA+/iceA2/oipA+/cagE-/babA+ | – |
| M2/cagA+/iceA2/oipA-/cagE+/babA+ | – |
| M2/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| M2/cagA+/iceA2/oipA+/cagE-/babA- | 1 (1.92) |
| M2/cagA+/iceA2/oipA-/cagE-/babA+ | – |
| M2/cagA+/iceA2/oipA-/cagE+/babA- | – |
| M2/cagA-/iceA2/oipA+/cagE+/babA+ | – |
| M2/cagA-/iceA2/oipA-/cagE+/babA+ | – |
| M2/cagA-/iceA2/oipA+/cagE-/babA+ | – |
| M2/cagA-/iceA2/oipA+/cagE+/babA- | – |
| M2/cagA-/iceA2/oipA+/cagE-/babA- | – |
| M2/cagA-/iceA2/oipA-/cagE-/babA- | 1 (1.92) |
Note: *Distribution was achieved based on the total numbers of 52 H. pylori isolates.
Discussion
H. pylori is a common bacterium with considerable clinical rank. About 50% of the world’s population have been infected with H. pylori bacteria. Despite the boost occurrence of infection, the main reservoir of the bacterium and the routes of infections are still unspecified.30 Furthermore, bacterial transmission between persons ensues through the oral–oral and oral–fecal routes.30 However, oral–fecal transmission has additional implications, since H. pylori may occur in food and water supplies subsequent to fecal contamination.30 Additionally, isolation of H. pylori from raw vegetables,16,31 meat,32,33 salads,16,34 ready to eat foods,35,36 and milk37,38 proposes that foodstuffs may act as vehicles for transmission of H. pylori to human community.
The current survey was carried out in order to assess the incidence, phenotypic and genotypic pattern of antibiotic resistance and genotyping profile of vacA, cagA, cagE, iceA, oipA, and babA alleles of the H. pylori bacteria recovered from raw camel, caprine, ovine, bovine, and buffalo meat samples. The contamination rate of H. pylori in bovine, ovine, caprine, buffalo, and camel meat samples was 5.71%, 13.07%, 11.53%, 9%, and 3%, respectively. Despite the higher importance of meat as a food which is served as so many kinds of undercooked products and therefore its higher risk of contamination with H. pylori, scarce data are available in this field. Saeidi and Sheikhshahrokh39 stated that the incidence of H. pylori bacteria amongst the raw cow, sheep, goat, buffalo, and camel meat samples were 25%, 37%, 22%, 28%, and 14%, respectively. Gilani et al40 stated that the incidence of H. pylori bacteria amongst the hamburger and minced meat samples were 1.42% and 12.50%, respectively. Additionally, H. pylori DNA was detected in 44% and 36% of ready-to-eat raw tuna meat and raw chicken samples, respectively.36 Moreover, Hemmatinezhad et al10 conveyed that the incidence of H. pylori bacteria amongst the 550 ready-to-eat food samples was 13.45% in which olive salad (36%), restaurant salad (30%), fruit salad (28%), and soup (22%) were the most generally contaminated samples. Similarly, Ghorbani et al31 stated that the incidence of H. pylori bacteria amongst the 300 foodstuffs were 20%, in which the incidence of contamination of ready to eat fish, ham, chicken sandwich, vegetable sandwich, meat sandwich and minced meat samples were 15%, 8.33%, 5%, 45%, 20%, and 32%, respectively. Finally, Talimkhani and Mashak41 represented that the incidence of H. pylori bacteria in raw bovine, ovine, and caprine meat samples were 4%, 10%, and 8%, respectively. We originated that ovine meat was the most routinely contaminated samples. Similarly, Saeidi and Sheikhshahrokh (2016),40 Talimkhani and Mashak,41 Momtaz et al,42 and Elhariri et al43 stated the higher incidence of H. pylori in ovine sources. Likewise, Rahimi and Kheirabadi 44 stated that the incidence of H. pylori bacteria in raw bovine, ovine, caprine, buffalo, and camel milk samples were 1.41%, 12.20%, 8.70%, 23.40%, and 3.60%, respectively. Higher incidence of H. pylori in raw ovine meat samples may be owing to the more appropriate circumstances existing in ovine meat, such as higher fat and protein contents and water activity and also optimum pH. Additionally, ovine meat may have a higher qualification for growth and survival of H. pylori bacteria. Furthermore, variances in the feed of ovine with other animal species may affect the incidence rate of H. pylori existing in their meat. Using thorns and thistles in deserts and living away from humans and the polluted environments are the most likely reasons for the lower incidence of H. pylori in camel meat. Lower incidence of H. pylori in raw camel meat was also conveyed.17,37,44
Resistance toward human and animal-based antibiotic agents was studied in the current research. H. pylori bacteria displayed the high incidence of resistance toward tetracycline, erythromycin, trimethoprim, levofloxacin, amoxicillin, and clarithromycin antibiotic agents. Resistance toward metronidazole, amoxicillin, levofloxacin, and clarithromycin were accompanied by the presence of rdxA, pbp1A, gyrA, and cla antibiotic resistance genes. Considerable incidence of resistance toward human-based antibiotics including erythromycin, metronidazole, levofloxacin, clarithromycin, amoxicillin, cefsulodin, furazolidone, rifampin, and spiramycin in H. pylori bacteria characterized their anthropogenic origin. Thus, this finding can indirectly prove that the H. pylori bacteria were transmitted from infected humans to meat samples through cross-contamination and meat manipulation in slaughterhouses. Extreme, unlawful, and forbidden prescription of antibiotic agents in medicine and also veterinary caused a significant occurrence of antibiotic resistance. Diverse research on India, Iran, Taiwan, China, Nigeria, Thailand, Senegal, Saudi Arabia, Egypt, Brazil, Colombia, and Argentina showed that H. pylori bacteria displayed a high incidence of resistance toward tetracyclines, aminoglycosides, penicillins, metronidazole, fluoroquinolones, and macrolides,45 which is similar to our findings. Recent surveys revealed that the incidence of resistance of H. pylori bacteria recovered from foodstuffs toward metronidazole, erythromycin, clarithromycin, amoxicillin, tetracycline, levofloxacin trimethoprim, furazolidone, and spiramycin antibiotic agents had ranges between 27.27–89.18%, 53.73–80.64%, 72.72–94.59%, 63.63–90.32%, 36.48–58.06%, 34.32–63.63%, 9.09–29.03%, and 9.09–16.12%, respectively.10,17,40,46,47 Despite the boost in importance of detection of antibiotic resistance genes, there were no previously published data on the detection of rdxA, pbp1A, gyrA, and cla antibiotic resistance genes in H. pylori bacteria recovered from foodstuffs. However, their detection has been done in H. pylori bacteria recovered from human clinical specimens.14,48,51
The final part of our survey focused on the genotyping of vacA, cagA, cagE, iceA, oipA, and babA alleles of the H. pylori bacteria. We also originated that vacA s1a, s2, m1a, and m2, and iceA1 and cagA, s1am1a, s2m1a, s1am2, s2m2, and s1a/cagA-/iceA1/oipA-/cagE-/babA-, m1a/cagA-/iceA1/oipA-/cagE-/babA-, s2/cagA-/iceA1/oipA-/cagE-/babA-, s1a/cagA+/iceA1/oipA+/cagE-/babA-, m1a/cagA+/iceA1/oipA+/cagE+/babA-, m1a/cagA+/iceA1/oipA+/cagE-/babA-, m2/cagA-/iceA1/oipA-/cagE-/babA-, s1a/cagA+/iceA1/oipA+/cagE+/babA-, s1a/cagA-/iceA1/oipA-/cagE+/babA-, s1a/cagA-/iceA1/oipA+/cagE-/babA-, s1b/cagA-/iceA1/oipA-/cagE-/babA-, m1a/cagA-/iceA1/oipA-/cagE+/babA-, m1a/cagA-/iceA1/oipA+/cagE-/babA-, m1b/cagA-/iceA1/oipA-/cagE-/babA-, s1a/cagA+/iceA1/oipA-/cagE+/babA-, s2/cagA+/iceA1/oipA+/cagE-/babA-, s2/cagA-/iceA1/oipA-/cagE+/babA-, s2/cagA-/iceA1/oipA+/cagE-/babA-, m1a/cagA+/iceA1/oipA-/cagE-/babA+, and m1a/cagA+/iceA1/oipA-/cagE+/babA- were the most generally perceived genotypes amongst the H. pylori bacteria. The boost incidence of vacA, cagA, iceA1, oipA, cagE and babA2 genotypes was also conveyed in the H. pylori bacteria recovered from clinical samples of human and animal species.42,52,54 Khaji et al55 conveyed that vacA s1a (91.66%), m1a (61.61%), s2 (36.66%), and m2 (31.66%) were the most generally perceived genotypes amongst the H. pylori bacteria recovered from raw milk of animal species. They also showed that s1am1a (41.66%), s2m1a (25%), s1am2 (16.66%), and s2m2 (13.33%) were the most generally perceived genotyping patterns amongst the H. pylori isolates. Ranjbar et al (2018)47 conveyed that vacA s1a (83.58%), m1a (80.59%), s2 (77.61%) and m2 (68.65%), cagA (73.13%) and babA2 (44.77%) were the most generally perceived genotypes amongst the H. pylori bacteria recovered from diverse kinds of raw milk samples. They showed that the distribution of s1am1a, s2m1a, s1am2 and s2m2 genotyping patterns and cagA-, oipA-, and babA2- genotypes were 56.71%, 56.71%, 43.28%, and 43.28% and 26.86%, 62.68%, and 55.22%, respectively. Additionally, amongst all of the detected combined genotypes, s1a/cagA+/iceA1/oipA−/babA2- (28.35%), m1a/cagA+/iceA1/oipA−/babA2- (28.35%), s2/cagA+/iceA1/oipA−/babA2- (26.86%), s1a/cagA+/iceA1/oipA−/babA2+ (25.37%), m1a/cagA+/iceA1/oipA−/babA2+ (25.37%), s2/cagA+/iceA1/oipA−/babA2+ (23.88%), s1a/cagA+/iceA1/oipA+/babA2- (22.38%), and m2/cagA +/iceA1/oipA−/babA2+ (22.38%) had the uppermost distribution. Hemmatinezhad et al56 stated that vacA s1a (78.37%), vacA m2(75.67%), vacA m1a (51.35%), and cagA (41.89%) alleles, s1am2 (70.27%), s1am1a (39.18%), and m1am2 (31.08%) genotypes, and s1a/cagA+/iceA1/oipA− (12.16%), s1a/cagA+/iceA1/oipA+ (10.81%), s1a/cagA−/iceA1/oipA+ (10.81%), s1b/cagA+/iceA1/oipA− (9.45%), m2/cagA+/iceA1/oipA+ (9.45%), m2/cagA+/iceA1/oipA− (9.45%), m2/cagA−/iceA1/oipA+ (9.45%), and m2/cagA−/iceA1/oipA− (9.45%) combined genotypic patterns were the most generally perceived in the H. pylori bacteria recovered from ready to eat food. According to Talimkhani and Mashak,41 vacA s1a (87.50%), vacA m1a (87.50%), vacA s2 (82.50%), cagA (80%), and vacA m2 (62.50%) alleles and s1am1a (62.50%), s2m1a (55%), s1am2 (50%), s2m2 (45%), and m1am2(42.50%) genotypes were the most generally perceived in H. pylori bacteria recovered from meat, milk, and vegetable samples. In studies conducted by Gilani et al40,56 s1am1a, s1am1b, and s2m1a were the most generally perceived genotypes amongst the H. pylori bacteria recovered from raw meat and meat products. There were no previousdata on detection of cagE genotypes amongst the H. pylori bacteria recovered from food samples. The presence of vacA, iceA, oipA, cagA, cagE and babA2 genotypes in the H. pylori isolates may cause certain facilities for bacterial adhesion to gastric epithelial cells, interleukin-8 and −10 and cytotoxin secretion and occurrence of inflammation, vacuolization, apoptosis of gastric epithelial cells, and even peptic ulceration in individuals who consume studied contaminated meat samples.
Absolutely, impact of food-borne microbes, particularly bacteria, in occurrence of food-borne diseases has been measured in Iran and diverse surveys have been conducted in this field.57–74
Conclusions
In conclusion, we documented extensive delivery of virulent and resistant H. pylori bacteria in raw camel, caprine, ovine, bovine, and buffalo meat samples. Boost incidence of H. pylori bacteria in raw meat magnifies that raw meat, particularly raw ovine meat, may be the natural reservoirs of H. pylori. We also originated that vacA, cagA, iceA, and babA2 alleles were predominant amongst the H. pylori isolates. In keeping with this, cagE-, babA2-, and oipA- H. pylori bacteria had the higher distribution. Similarities in the genotyping pattern of H. pylori bacteria between numerous meat sources signify their same route of contamination. H. pylori isolates displayed a high incidence of resistance toward tetracycline, erythromycin, trimethoprim, levofloxacin, amoxicillin, and clarithromycin (61.53%) antibiotic agents. The phenotypic pattern of antibiotic resistance was also confirmed by the genotypic pattern, with considerable distribution of rdxA, pbp1A, gyrA, and cla antibiotic resistance genes. Furthermore, the high incidence of multi-drug resistant H. pylori bacteria displays that raw meat of animal species may be a reservoir of antibiotic resistant H. pylori. Further research should be performed to determine the probable relationships between the presence of genotypes, antibiotic resistance, and antibiotic resistance genes. Additionally, conduction of comprehensive research is essential to determine molecular genetic homology of H. pylori bacteria recovered from raw meat of animal species and those of human clinical specimens.
Acknowledgments
The authors would like to thank Dr. Manouchehr Momeni Shahraki, Dr. Mojtaba Masoudimanesh, and Dr. Amirthossein Sheikhshahrokh for their assistance in laboratory-based examinations.
Ethics Criteria
The study was approved by the Ethical Council of Research of the Faculty of Veterinary Medicine, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran.
Disclosure
The authors report no conflicts of interest.
References
- 1.Wyness L. The role of red meat in the diet: nutrition and health benefits. Proc Nutr Soc. 2016;75(3):227–232. doi: 10.1017/S0029665115004267 [DOI] [PubMed] [Google Scholar]
- 2.Omer MK, Alvarez-Ordonez A, Prieto M, Skjerve E, Asehun T, Alvseike OA. A systematic review of bacterial foodborne outbreaks related to red meat and meat products. Foodborne Pathog Dis. 2018;15(10):598–611. doi: 10.1089/fpd.2017.2393 [DOI] [PubMed] [Google Scholar]
- 3.Crowe SE. Helicobacter pylori infection. New England J Med. 2019;380(12):1158–1165. doi: 10.1056/NEJMcp1710945 [DOI] [PubMed] [Google Scholar]
- 4.Quaglia N, Dambrosio A. Helicobacter pylori: a foodborne pathogen? World J Gastroenterol. 2018;24(31):3472–3487. doi: 10.3748/wjg.v24.i31.3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zamani M, Vahedi A, Maghdouri Z, Shokri-Shirvani J. Role of food in environmental transmission of Helicobacter pylori. Caspian J Int Med. 2017;8(3):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Hecke T, Van Camp J, De Smet S. Oxidation during digestion of meat: interactions with the diet and helicobacter pylori gastritis, and implications on human health. Comp Rev Food Sci Food Safe. 2017;16(2):214–233. doi: 10.1111/1541-4337.12248 [DOI] [PubMed] [Google Scholar]
- 7.Xia Y, Meng G, Zhang Q, et al. Dietary patterns are associated with Helicobacter pylori infection in Chinese adults: a cross-sectional study. Sci Report. 2016;6:32334. doi: 10.1038/srep32334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sedaghat H, Moniri R, Jamali R, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2, and oipA genotypes in patients with upper gastrointestinal diseases. Iran J Microbiol. 2014;6(1):14. [PMC free article] [PubMed] [Google Scholar]
- 9.Akeel M, Shehata A, Elhafey A, et al. Helicobacter pylori vacA, cagA and iceA genotypes in dyspeptic patients from southwestern region, Saudi Arabia: distribution and association with clinical outcomes and histopathological changes. BMC Gastroenterol. 2019;19(1):16. doi: 10.1186/s12876-019-0934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemmatinezhad B, Momtaz H, Rahimi E. VacA, cagA, iceA and oipA genotypes status and antimicrobial resistance properties of Helicobacter pylori isolated from various types of ready to eat foods. Ann Clin Microbiol Antimicrob. 2016;15(1):2. doi: 10.1186/s12941-015-0115-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki S, Esaki M, Kusano C, Ikehara H, Gotoda T. Development of Helicobacter pylori treatment: how do we manage antimicrobial resistance? World J Gastroenterol. 2019;25(16):1907. doi: 10.3748/wjg.v25.i16.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterol. 2018;155(5):1372–1382. doi: 10.1053/j.gastro.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yousefi-Avarvand A, Vaez H, Tafaghodi M, Sahebkar AH, Arzanlou M, Khademi F. Antibiotic resistance of Helicobacter pylori in Iranian children: a systematic review and meta-analysis. Microb Drug Res. 2018;24(7):980–986. doi: 10.1089/mdr.2017.0292 [DOI] [PubMed] [Google Scholar]
- 14.Zerbetto De Palma G, Mendiondo N, Wonaga A, et al. Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from Buenos Aires City. Microb Drug Res. 2017;23(3):351–358. doi: 10.1089/mdr.2015.0361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashemi S, Sheikh A, Goodarzi H, et al. Genetic basis for metronidazole and clarithromycin resistance in Helicobacter pylori strains isolated from patients with gastroduodenal disorders. Infect Drug Res. 2019;12:535–543. doi: 10.2147/IDR.S192942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atapoor S, Dehkordi FS, Rahimi E. Detection of Helicobacter pylori in various types of vegetables and salads. Jundishapur J Microbiol. 2014;7(5):e10013. doi: 10.5812/jjm [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mousavi S, Dehkordi FS, Rahimi E. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw milk and unpasteurized dairy products in Iran. J Anim Toxin Trop Dis. 2014;20(1):51. doi: 10.1186/1678-9199-20-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho S-A, Hoyle J, Lewis F, et al. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29(11):2543–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews J. BSAC Disc Diffusion Method Antimicrob. 2.1.4 ed. Birmingham, UK: British Society for Antimicrobial Chemotherapy; 2003. [Google Scholar]
- 20.CLSI. Clinical and Laboratory Standard Institute In: Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. Wayne, PA: National Committee for Clinical Laboratory Standards; 2015:M45. [Google Scholar]
- 21.Yamazaki S, Yamakawa A, Okuda T, et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J Clin Microbiol. 2005;43(8):3906–3916. doi: 10.1128/JCM.43.8.3906-3916.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Chi DS, Laffan JJ, et al. Comparison of cytotoxin genotypes of Helicobacter pylori in stomach and saliva. Digest Dis Sci. 2002;47(8):1850–1856. doi: 10.1023/A:1016417200611 [DOI] [PubMed] [Google Scholar]
- 23.Peek JR, Thompson SA, Donahue JP, et al. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physic. 1998;110(6):531–544. [PubMed] [Google Scholar]
- 24.Sheu B, Sheu S, Yang H, Huang A, Wu -J-J. Host gastric Lewis expression determines the bacterial density of Helicobacter pylori in babA2 genopositive infection. Gut. 2003;52(7):927–932. doi: 10.1136/gut.52.7.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomasini ML, Zanussi S, Sozzi M, Tedeschi R, Basaglia G, De Paoli P. Heterogeneity of cag genotypes in Helicobacter pylori isolates from human biopsy specimens. J Clin Microbiol. 2003;41(3):976–980. doi: 10.1128/JCM.41.3.976-980.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debets-Ossenkopp Y, Sparrius M, Kusters J, Kolkman J, Vandenbroucke-Grauls C. Mechanism of clarithromycin resistance in clinical isolates of Helicobacter pylori. FEMS Microb Lett. 1996;142(1):37–42. doi: 10.1111/j.1574-6968.1996.tb08404.x [DOI] [PubMed] [Google Scholar]
- 27.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Molecular Biol. 1975;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2 [DOI] [PubMed] [Google Scholar]
- 28.Tankovic J, Lascols C, Sculo Q, Petit J-C, Soussy C-J. Single and double mutations in gyrA but not in gyrB are associated with low-and high-level fluoroquinolone resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2003;47(12):3942–3944. doi: 10.1128/AAC.47.12.3942-3944.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Versalovic J, Shortridge D, Kibler K, et al. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40(2):477–480. doi: 10.1128/AAC.40.2.477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dore MP, Sepulveda AR, El-Zimaity H, et al. Isolation of Helicobacter pylori from sheep—implications for transmission to humans. Am J Gastroenterol. 2001;96(5):1396. [DOI] [PubMed] [Google Scholar]
- 31.Ghorbani F, Gheisari E, Dehkordi FS. Genotyping of vacA alleles of Helicobacter pylori strains recovered from some Iranian food items. Trop J Pharm Res. 2016;15(8):1631–1636. doi: 10.4314/tjpr.v15i8.5 [DOI] [Google Scholar]
- 32.Mard SA, Khadem Haghighian H, Sebghatulahi V, Ahmadi B. Dietary factors in relation to Helicobacter pylori infection. Gastroenterol Res Pract. 2014;2014:1–5. doi: 10.1155/2014/826910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevenson TH, Bauer N, Lucia LM, Acuff GR. Attempts to isolate Helicobacter from cattle and survival of Helicobacter pylori in beef products. J Food Prot. 2000;63(2):174–178. doi: 10.4315/0362-028X-63.2.174 [DOI] [PubMed] [Google Scholar]
- 34.Yahaghi E, Khamesipour F, Mashayekhi F, et al. Helicobacter pylori in vegetables and salads: genotyping and antimicrobial resistance properties. Biomed Res Int. 2014;2014:1–11. doi: 10.1155/2014/757941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poms RE, Tatini SR. Survival of Helicobacter pylori in ready-to-eat foods at 4 C. Int J Food Microbiol. 2001;63(3):281–286. doi: 10.1016/S0168-1605(00)00441-4 [DOI] [PubMed] [Google Scholar]
- 36.Meng X, Zhang H, Law J, Tsang R, Tsang T. Detection of Helicobacter pylori from food sources by a novel multiplex PCR assay. J Food Safe. 2008;28(4):609–619. doi: 10.1111/j.1745-4565.2008.00135.x [DOI] [Google Scholar]
- 37.Esmaeiligoudarzi D, Tameshkel FS, Ajdarkosh H, Arsalani M, Sohani MH, Behnod V. Prevalence of Helicobacter pyloriinIranian milk and dairy products using culture and ureC based-PCR techniques. Biomed Pharmacol J. 2015;8(1):179–183. doi: 10.13005/bpj/597 [DOI] [Google Scholar]
- 38.Talaei R, Souod N, Momtaz H, Dabiri H. Milk of livestock as a possible transmission route of Helicobacter pylori infection. Gastroenterol Hepatol Bed Bench. 2015;8(Suppl1):S30. [PMC free article] [PubMed] [Google Scholar]
- 39.Saeidi E, Sheikhshahrokh A. VacA genotype status of Helicobacter pylori isolated from foods with animal origin. Biomed Res Int. 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilani A, Razavilar V, Rokni N, Rahimi E. VacA and cagA genotypes status and antimicrobial resistance properties of Helicobacter pylori strains isolated from meat products in Isfahan province, Iran. Iran J Vet Res. 2017;18(2):97. [PMC free article] [PubMed] [Google Scholar]
- 41.Talimkhani A, Mashak Z. Prevalence and genotyping of Helicobacter pylori isolated from meat, milk and vegetable in Iran. Jundishapur J Microbiol. 2017;10:11. doi: 10.5812/jjm [DOI] [Google Scholar]
- 42.Momtaz H, Dabiri H, Souod N, Gholami M. Study of Helicobacter pylori genotype status in cows, sheep, goats and human beings. BMC Gastroenterol. 2014;14(1):61. doi: 10.1186/1471-230X-14-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elhariri M, Hamza D, Elhelw R, Hamza E. Occurrence of cagA+ vacA s1a m1 i1 Helicobacter pylori in farm animals in Egypt and ability to survive in experimentally contaminated UHT milk. Sci Reports. 2018;8(1):14260. doi: 10.1038/s41598-018-32671-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahimi E, Kheirabadi EK. Detection of Helicobacter pylori in bovine, buffalo, camel, ovine, and caprine milk in Iran. Foodborne Pathog Dis. 2012;9(5):453–456. doi: 10.1089/fpd.2011.1060 [DOI] [PubMed] [Google Scholar]
- 45.Hunt R, Xiao S, Megraud F, et al. Helicobacter pylori in developing countries. J Gastrointestin Liver Dis. 2011;20(3):299–304. [PubMed] [Google Scholar]
- 46.Ranjbar R, Yadollahi Farsani F, Safarpoor Dehkordi F. Antimicrobial resistance and genotyping of vacA, cagA, and iceA alleles of the Helicobacter pylori strains isolated from traditional dairy products. J Food Saf. 2019;39(2):12594. doi: 10.1111/jfs.12594 [DOI] [Google Scholar]
- 47.Ranjbar R, Farsani FY, Dehkordi FS. Phenotypic analysis of antibiotic resistance and genotypic study of the vacA, cagA, iceA, oipA and babA genotypes of the Helicobacter pylori strains isolated from raw milk. Antimicrob Res Infect Control. 2018;7(1):115. doi: 10.1186/s13756-018-0409-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lauener F, Imkamp F, Lehours P, et al. Genetic determinants and prediction of antibiotic resistance phenotypes in Helicobacter pylori. J Clin Med. 2019;8(1):53. doi: 10.3390/jcm8010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amin M, Shayesteh AA, Serajian A, Goodarzi H. Assessment of metronidazole and clarithromycin resistance among helicobacter pylori isolates of Ahvaz (Southwest of Iran) during 2015-2016 by phenotypic and molecular methods. Jundishapur J Microbiol. 2019;12(4):e80156. [Google Scholar]
- 50.Zhang Y, Zhao F, Kong M, et al. Validation of a high-throughput multiplex genetic detection system for Helicobacter pylori identification, quantification, virulence, and resistance analysis. Front Microbiol. 2016;7:1401. doi: 10.3389/fmicb.2016.01401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manal D, Ahmed E-S, Maged E-G, et al. Detection of antimicrobial resistance genes ofHelicobacter pyloristrainsto clarithromycin, metronidazole, amoxicillin and tetracycline among Egyptian patients. Egyp J Med Hum Genet. 2018;19:417–423. doi: 10.1016/j.ejmhg.2018.01.004 [DOI] [Google Scholar]
- 52.Dabiri H, Jafari F, Baghaei K, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, oipA, iceA, babA2 and babB genotypes in Iranian dyspeptic patients. Microb Pathog. 2017;105:226–230. doi: 10.1016/j.micpath.2017.02.018 [DOI] [PubMed] [Google Scholar]
- 53.Torkan S, Shahreza MHS. VacA, CagA, IceA and OipA genotype status of Helicobacter pylori isolated from biopsy samples from Iranian dogs. Trop J Pharm Res. 2016;15(2):377–384. doi: 10.4314/tjpr.v15i2.22 [DOI] [Google Scholar]
- 54.Podzorski RP, Podzorski DS, Wuerth A, Tolia V. Analysis of the vacA, cagA, cagE, iceA, and babA2 genes in Helicobacter pylori from sixty-one pediatric patients from the Midwestern United States. Diagn Microbiol Infect Dis. 2003;46(2):83–88. doi: 10.1016/S0732-8893(03)00034-8 [DOI] [PubMed] [Google Scholar]
- 55.Khaji L, Banisharif G, Alavi I. Genotyping of the Helicobacter pylori isolates of raw milk and traditional dairy products. Microbiol Res. 2017;8(2):43–46. doi: 10.4081/mr.2017.7288 [DOI] [Google Scholar]
- 56.Gilani A, Razavilar V, Rokni N, Rahimi E. VacA and cagA genotypes of Helicobacter pylori isolated from raw meat in Isfahan province, Iran. Vet Res Forum. 2017;8(1):75–80. [PMC free article] [PubMed] [Google Scholar]
- 57.Dehkordi FS, Barati S, Momtaz H, Ahari SNH, Dehkordi SN. Comparison of shedding, and antibiotic resistance properties of Listeria monocytogenes isolated from milk, feces, urine, and vaginal secretion of bovine, ovine, caprine, buffalo, and camel species in Iran. Jundishapur J Microbiol. 2013;6(3):284. [Google Scholar]
- 58.Dehkordi FS, Parsaei P, Saberian S, et al. Prevalence study of theileria annulata by comparison of four diagnostic techniques in southwest Iran. Bulgar J Vet Med. 2012;15(2):123–130. [Google Scholar]
- 59.Hemmatinezhad B, Khamesipour F, Mohammadi M, Safarpoor Dehkordi F, Mashak Z. Microbiological Investigation of O‐Serogroups, Virulence Factors and Antimicrobial Resistance Properties of Shiga Toxin‐Producing E scherichia Coli Isolated from Ostrich, Turkey and Quail Meats. J Food Safe. 2015;35(4):491––500.. [Google Scholar]
- 60.Madahi H, Rostami F, Rahimi E, Dehkordi FS. Prevalence of enterotoxigenic Staphylococcus aureus isolated from chicken nugget in Iran. Jundishapur J Microbiol. 2014;7(8 ):e10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Momtaz H, Davood Rahimian M, Safarpoor Dehkordi F. Identification and characterization of Yersinia enterocolitica isolated from raw chicken meat based on molecular and biological techniques. J App Poultry Res. 2013;22(1):137–145 [Google Scholar]
- 62.Rahimi E, Sepehri S, Dehkordi FS, Shaygan S, Momtaz H. Prevalence of Yersinia species in traditional and commercial dairy products in Isfahan Province, Iran. Jundishapur J Microbiol. 2014;7(4):e9249. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dehkordi FS, Khamesipour F, Momeni M. Brucella abortus and Brucella melitensis in Iranian bovine and buffalo semen samples: The first clinical trial on seasonal, Senile and geographical distribution using culture, conventional and real-time polymerase chain reaction assays. Kafkas Univ Vet Fak Derg. 2014;20(6):821––828.. [Google Scholar]
- 64.Dehkordi FS, Haghighi N, Momtaz H, Rafsanjani MS, Momeni M. Conventional vs real-time PCR for detection of bovine herpes virus type 1 in aborted bovine, buffalo and camel foetuses. Bulgarian J Vet Med.2013;16(2):102–111. [Google Scholar]
- 65.Rahimi E, Yazdanpour S, Dehkordi F. Detection of Toxoplasma gondii antibodies in various poultry meat samples using enzyme linked immuno sorbent assay and its confirmation by polymerase chain reaction. J Pure Appl Microbiol.2014;8(1):421––427.. [Google Scholar]
- 66.Dehkordi FS, Valizadeh Y, Birgani T, Dehkordi K. Prevalence study of Brucella melitensis and Brucella abortus in cow's milk using dot enzyme linked immuno sorbent assay and duplex polymerase chain reaction. J Pure Appl Microbiol. 2014;8:1065––1069.. [Google Scholar]
- 67.Ranjbar R, Seif A, Safarpoor Dehkordi F. Prevalence of Antibiotic Resistance and Distribution of Virulence Factors in the Shiga Toxigenic Escherichia coli Recovered from Hospital Food. Jundishapur J Microbiol. 2019;12(5 ):e82659. [Google Scholar]
- 68.Nejat S, Momtaz H, Yadegari M, Nejat S, Safarpour Dehkordi F, Seasonal Khamesipour F., Geographical, Age and Breed Distributions of Equine Viral Arteritis in Iran. Kafkas Univ Vet Fak Derg. 2015; 21(1):111––116.. [Google Scholar]
- 69.Ranjbar R, Safarpoor Dehkordi F, Sakhaei Shahreza MH, Rahimi E. Prevalence, identification of virulence factors, O-serogroups and antibiotic resistance properties of Shiga-toxin producing Escherichia coli strains isolated from raw milk and traditional dairy products. Antimicrob Res Infect Control. 2018;7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Momtaz H, Safarpoor Dehkordi F, Taktaz T, Rezvani A, Yarali S. Shiga Toxin-Producing Escherichia coli Isolated from Bovine Mastitic Milk: Serogroups, Virulence Factors, and Antibiotic Resistance Properties . Sci World J. 2012;2012:618709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dehkordi AH, Khaji L, Shahreza MS, et al. One-year prevalence of antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus recovered from raw meat. Trop Biomed 2017;34(2):396––404.. [PubMed] [Google Scholar]
- 72.Safarpoor Dehkordi F, Gandomi H, Akhondzadeh Basti A, Misaghi A, Rahimi E. Phenotypic and genotypic characterization of antibiotic resistance of methicillin-resistant Staphylococcus aureus isolated from hospital food. Antimicrob Res Infect Control. 2017;6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Momtaz H, Dehkordi FS, Rahim E., Asgarifar A, Momeni M. Virulence genes and antimicrobial resistance profiles of Staphylococcus aureus isolated from chicken meat in Isfahan province, Iran. J Appl Poult Res. 2013;22(4):913––921.. [Google Scholar]
- 74.Ranjbar R, Masoudimanesh M, Dehkordi FS, Jonaidi-Jafari N, Shiga Rahimi E. (Vero)-toxin producing Escherichia coli isolated from the hospital foods; virulence factors, o-serogroups and antimicrobial resistance properties. Antimicrob Res Infect Control. 2017;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]