Abstract
Huntington’s disease (HD) is a neurodegenerative disease caused by a CAG repeat expansion in the Huntingtin gene (HTT), translated into a Huntingtin protein with a polyglutamine expansion. There is preferential loss of medium spiny neurons within the striatum and cortical pyramidal neurons. Pridopidine is a small molecule showing therapeutic potential in HD preclinical and clinical studies. Pridopidine has nanomolar affinity to the sigma-1 receptor (sigma-1R), which is located predominantly at the endoplasmic reticulum (ER) and mitochondrial associated ER membrane, and activates neuroprotective pathways. Here we evaluate the neuroprotective effects of pridopidine against mutant Huntingtin toxicity in mouse and human derived in vitro cell models. We also investigate the involvement of the sigma-1 receptor in the mechanism of pridopidine. Pridopidine protects mutant Huntingtin transfected mouse primary striatal and cortical neurons, with an EC50 in the mid nanomolar range, as well as HD patient-derived induced pluripotent stem cells (iPSCs). This protection by pridopidine is blocked by NE-100, a purported sigma-1 receptor antagonist, and not blocked by ANA-12, a reported TrkB receptor antagonist. 3PPP, a documented sigma-1 receptor agonist, shows similar neuroprotective effects. Genetic knock out of the sigma-1 receptor dramatically decreases protection from pridopidine and 3PPP, but not protection via brain derived neurotrophic factor (BDNF). The neuroprotection afforded by pridopidine in our HD cell models is robust and sigma-1 receptor dependent. These studies support the further development of pridopidine, and other sigma-1 receptor agonists as neuroprotective agents for HD and perhaps for other disorders.
Keywords: Neuroprotection, Huntingtin toxicity, Huntington’s disease, Mutant-huntingtin, Pridopidine, 3PPP, Sigma-1 receptor, Primary neurons, Patient-derived induced pluripotent stem cells
1. Introduction
Huntington’s disease (HD) is a progressive, autosomal dominant neurodegenerative disorder caused by an expanded trinucleotide CAG repeat within exon 1 of the Huntingtin gene (HTT); the expanded repeat produces a mutant Huntingtin protein with an extended polyglutamine repeat. The disease affects movements, emotions and cognition (Ross et al., 2014; McColgan and Tabrizi, 2018). Medium spiny neurons (MSNs) within the striatum and cortical pyramidal neurons are preferentially affected in HD (McColgan and Tabrizi, 2018; Saudou and Humbert, 2016). At a molecular level, BDNF levels are reduced in HD brains. The BDNF pathway is critical for neuronal survival and is believed to play a role in HD pathogenesis (Zuccato and Cattaneo, 2007). MSNs are particularly vulnerable to a decrease of BDNF levels both in vivo (Baydyuk et al., 2011) and in vitro when differentiated from patient-derived iPSCs (HD iPSC Consortium, 2012).
Recent efforts seek to slow disease progression using Huntingtin specific antisense oligonucleotide and antibody approaches (Rodrigues and Wild, 2017). Since these methods may have difficulty accessing the central nervous system, small molecule approaches remain viable.
Pridopidine is a small molecule, which shows potential as a disease-modifying HD treatment in both clinical trials and preclinical studies. Clinical trials to date have shown pridopidine to be safe with some efficacy in improving patient movements (de Yebenes et al., 2011; Huntington Study Group HART Investigators, 2013). Some benefits observed in pridopidine preclinical studies include: decreased mutant Huntingtin aggregate size in striatal tissues (Squitieri et al., 2015), preserved MSN synaptic spine density and normalized calcium homeostasis (Ryskamp et al., 2016), memory improvement (Sahlholm et al., 2018), and increased expression of general cell survival pathways like brain derived neurotrophic factor (BDNF) and AKT (Geva et al., 2016).
Originally developed as a dopamine stabilizer, pridopidine has micromolar affinity for the dopamine D2 receptor (Dyhring et al., 2010). Recent studies show pridopidine to be more potent at the sigma-1 receptor, with nanomolar affinity, preferentially binding at physiologically relevant doses (Sahlholm et al., 2013; Sahlholm et al., 2015). The sigma-1 receptor is a highly conserved 25 kDa transmembrane protein encoded by the SIGMAR1 gene. It is located at the endoplasmic reticulum (ER) and mitochondrial associated ER membrane, as well as the nuclear envelope and plasma membrane (Hayashi and Su, 2007; Su et al., 2016).
The sigma-1 receptor is shown to be neuroprotective against many aspects of toxicity of neurodegenerative diseases including excitotoxicity, ER stress and mitochondrial dysfunction (Nguyen et al., 2015). Sigma-1 receptor agonists have been tested in models of stroke (Allahtavakoli and Jarrott, 2011), Alzheimer’s disease (Lahmy et al., 2013), Parkinson’s disease (Francardo et al., 2017), Amyotrophic Lateral Sclerosis (Peviani et al., 2013) and HD (Geva et al., 2016).
Additionally, several drugs commonly used in neuropsychiatry (Donepezil, Ifenprodil, SSRIs – for a review see Albayrak and Hashimoto, 2017) have been shown to have sigma-1 receptor binding properties in addition to their designed targets reinforcing the idea of a potential wide neuromodulating effect of the receptor activation.
Here we use mouse and human derived in vitro cell models to determine if pridopidine can protect against mutant Huntingtin toxicity; we utilize pharmacological and genetic tools to ascertain if this protection is mediated via the sigma-1 receptor.
2. Material and methods
2.1. Mice
Mice were handled in accordance with Johns Hopkins Institutional Animal Care and Use Committee guidelines. CD1 pregnant female mice were obtained from Charles River Laboratories. Sigma-1 receptor knockout and wild type pregnant female mice were obtained from TEVA Pharmaceutical Industries. The sigma-1 receptor knockout mice have a TF0499 gene trap mutation between exon 2 and 3 of the SIGMAR1 gene on chromosome 4.
2.2. Drugs/compounds
Pridopidine was received directly from TEVA Pharmaceutical Industries. Pridopidine was used at concentrations ranging from 0.001 to 1 μM for the dose response experiment and at 1 μM for all the other experiments. BDNF, 3PPP, ANA-12, and NE-100 were purchased from Millipore Sigma. 3PPP was used at concentrations ranging from 0.01 to 100 μM for the dose response experiment and at 1 μM for all the other experiments. BDNF was used at 20 ng/mL (HD iPSC Consortium, 2012). Antagonists ANA-12 (IC50 = 50 nM; Cazorla et al., 2011) and NE-100 were used at 1 μM (IC50 = 5 nM; Okuyama et al., 1993).
2.3. Primary neuron cultures
Primary striatal and cortical neurons were prepared from E15 to E17 embryos per our protocol (Arbez et al., 2017). Cells were plated on 24-well plates at a density of 106 cell/mm2 in Neurobasal medium with added 2% B27, 2 mM Glutamax and 1% PenStrep. The cells were incubated at 37 °C and 5% CO2. All cell culture supplies were purchased from Corning and culture medias from ThermoFisher.
2.4. Transfection of primary neurons
At day in vitro (DIV) five, using Lipofectamine 2000 (ThermoFisher), neurons were co-transfected with Htt-N586 plasmids, with either 22Q or 82Q, and GFP (Htt N586: GFP ratio of 10:1). Transfections were performed per our protocol (Arbez et al., 2017), with all treatments beginning immediately after transfection. The transfection rate was determined to be around 5% with a percentage of co-transfection of GFP and Htt N586 higher than 95%.
2.5. Induced pluripotent stem cell cultures
Generation and characterization of control (CTR 21n2) and HD (HD 109n1 and HD 50n5) iPSC lines were per a published protocol (HD iPSC Consortium, 2017; Mattis et al., 2015). To differentiate the iPSCs into neural progenitors, colonies were grown on Matrigel in TeSR media (feeder-free) and later converted into non-adherent colonies, called “EZ-spheres”, using a published protocol (HD iPSC Consortium, 2012). Briefly these colonies were lifted using 2 mg/mL Collagenase IV solution in DMEM, and propagated in Stemline NSC medium (Millipore Sigma) supplied with bFGF, EGF (100 ng/mL each, Peprotech) and 5 μg/mL heparin (Millipore Sigma) in non-adherent tissue culture flasks. The “EZ-spheres” were mechanically chopped weekly for propagation. To differentiate the neural progenitors into medium spiny neuron phenotypes, the “EZ-spheres” were first moved into Neural Induction Medium (NIM: 1% N2 in DMEM:F12, ThermoFisher-Invitrogen) for 5 days and then BDNF (20 ng/mL; Peprotech) was added for 2 days. For the following 21 days, cells were differentiated in NIM with 20 ng/mL BDNF (20 ng/mL; Peprotech), rhSHH (200 ng/mL; Peprotech), and DKK1 (100 ng/mL, Peprotech) to promote a rostral forebrain fate. Cells were matured into medium spiny neuron phenotypes in NIM supplied with 20 ng/mL BDNF, dibutyryl cyclic AMP (dbcAMP, 0.5 mM; Millipore Sigma) and valproic acid (VPA, 0.5 mM; Millipore Sigma) for 14 days. Medium was half-changed three times per week or as needed. The cells were plated into 48-well Matrigel coated plates on day 26 of differentiation and fixed with 4% paraformaldehyde (Electron Microscopy Sciences) solution in PBS for 30 min on day 42 of differentiation for nuclear condensation toxicity assays and immunostainings.
2.6. Representative primary neuron images
Primary striatal neurons from CD1 mice (plated and transfected with GFP as previously stated) were stained with a solution of 0.2 μg/mL Hoechst 3342 (Millipore Sigma) in PBS for 10 min at room temperature. The plate was then washed once with PBS before being imaged using a Keyence BZ-X700 microscope at 20× objective.
2.7. Nuclear condensation cell death assay
Nuclear condensation experiments were performed in primary striatal and cortical neurons following a published protocol (Arbez et al., 2017; Watkin et al., 2014). 48 h after transfection, cells were fixed with a solution of 4% paraformaldehyde in PBS for 30 min. Then nuclei were stained for 15–20 min using a solution of 0.2 μg/mL Hoechst 3342 (Millipore Sigma) in PBS. Automated picture acquisition was performed on a Zeiss Axiovert 200 inverted microscope at 10X objective, and automated quantification of nuclear intensity in transfected cells was performed using Volocity software (Perkin Elmer). Expression of the different Htt N586 constructs used was previously showed to be equal by immunocytochemistry in primary neurons and by western blot in HEK cells (Watkin et al., 2014). Cells were considered dead when their relative nuclear intensity was higher than the average intensity plus two standard deviations. 4 wells per condition were used per experiment and all the experiments were reproduced 3 to 6 times each. A total of 5000 to 10,000 cells were quantified per condition. Statistical analysis was performed using ANOVA with a Bonferroni post-hoc test.
To induce toxicity in the iPSCs we utilized a published BDNF-withdrawal protocol (HD iPSC Consortium, 2012; HD iPSC Consortium, 2017). iPSCs were plated into 48-well plates and differentiated into medium spiny neuron phenotypes as indicated above. After 42 days of total differentiation time, the medium was replaced with BDNF-free NIM. For each experimental treatment, the NIM was supplemented with each compound before being applied to the cells for 48 h. As a positive control 20 ng/mL BDNF was supplemented back into the NIM. After 48 h of treatment, the plate was fixed using 4% paraformaldehyde for 10 min before being washed with PBS and stained for 15–20 min using a solution of 0.2 μg/mL Hoechst 3342 (Millipore Sigma). To avoid clumped iPSC islets, 10 images per well were manually taken at random using a Zeiss Axiovert 200 inverted microscope at 10x objective. Quantification of nuclear intensity was performed as described above for mouse primary neurons. Statistical analysis was performed using ANOVA with a SNK post-hoc test.
2.8. Live cell imaging cell death assay
Primary cortical neurons from CD1, sigma-1 receptor wild type (WT) and sigma-1 receptor knockout (Sigma1R-KO) mice were prepared and transfected as previously stated. 24 h after transfection, neurons expressing GFP were randomly chosen and automatically imaged every 20 min over the course of 10 h on a Zeiss Axiovert 200 inverted microscope. Neuronal morphology was blindly quantified using ImageJ software. Manual classification of cell death was performed using morphological criteria. Cells were considered dead when they showed complete neuronal projection disappearance, soma fragmentation, or soma shape change from oval to completely circular. An arbitrary morphological score was assigned as either 100 for alive cells or 0 for dead cells in each frame. The mean morphological score of all the cells was calculated for each time point. Survival analysis was performed using a Gehan-Breslow test to determine a statistical difference between groups, followed by an All Pairwise Multiple Comparison (Holm-Sidak method) to identify the differences between groups.
2.9. Mitochondrial potential assay
Mitochondrial potential was measured as previously described (Arbez et al., 2017). Primary cortical neurons were transfected and treated as described above. 24 h after transfection, neurons were loaded with 20 nM Tetramethyl rhodamine methyl ester (TMRM) for 45 min at 37 °C in the incubator. After rinsing 3 times with PBS to remove the excess dye, the medium was replaced with complete Neurobasal medium without phenol red for imaging. Neurons were imaged live on a Zeiss Axiovert 200 inverted microscope. Measure of the average TMRM intensity in the soma of transfected cells was quantified using Image J. Intensity measurements were normalized to the intensity of untreated controls.
2.10. Immunocytochemistry
After fixation, cells were permeabilized with PBS containing 0.2% Triton X-100 for 10 min, then treated with PBS containing 0.2% Triton X-100 and 10% BSA as a blocking buffer for 2 h. Anti-MAP2 antibody (rabbit, 1:500, Cell signaling technology, ref. #4542) was incubated overnight at 4 °C in PBS containing 10% BSA. Cells were rinsed in PBS and incubated for 2 h with anti-rabbit CY3-conjugated antibody (1:200, Jackson Laboratories, ref. #711-165-152). Nuclei were stained with Hoechst 33342 and after washing twice in PBS, cells were mounted onto slides and imaged using a Keyence BZ-X700 microscope at 40X objective.
2.11. Immunophenotyping
For immunophenotyping, iPSC differentiated to MSN-like phenotypes on 12 mm coverslips were fixed with 4% PFA in PBS for 15 min at room temperature, permeabilized with PBS containing 0.5% Triton X-100 for 10 min, washed with PBS and incubated with 10% Normal Goat Serum (NGS, Sigma) in PBS for blocking for 1 h. Then, cells were co-incubated with Anti-MAP2 antibody (clone AP20, mouse, 1:500, EMD Millipore, ref. #MAB3418) and Anti-DARPP32 (rabbit, 1:100, Santa Cruz biotech, ref. #SC-11365) or Anti-Bcl11b (Ctip2, rabbit, 1:200, Santa Cruz biotech, ref. #SC-365988) overnight at 4 °C in PBS containing 10% NGS. Next morning, cells were rinsed three times with PBS and co-incubated for 1 h with goat anti-mouse Alexa488-conjugated (1:1000, Cell Signaling technology, ref. #4408) and goat anti-rabbit Alexa555-conjugated antibodies (1:1000, Cell Signaling technology, ref. #4413) in 10% NGS in PBS. Nuclei were stained with Hoechst 33342 and after washing twice in PBS, cells were mounted onto slides and imaged using a Keyence BZ-X700 microscope at 40× objective.
2.12. CellTiter-Glo luminescent cell viability assay
HD and control iPSCs were differentiated toward striatal neural cells for 48 days, dissociated with TrypLE solution (ThermoFisher), and replated into Matrigel (BD) coated 96-well plates, 104 cells/well. On day 6 after re-plating, the cells were transferred to BDNF-free NIM. For each experimental treatment, NIM was supplemented with each compound before being applied to the cells for 48 h. As a positive control 20 ng/mL BDNF was supplemented back into the NIM. Relative intracellular [ATP] values in cell extracts were measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, G7571) according to manufacturer’s instructions. Luminescence was measured on SpectraMax 340PC microplate reader (Molecular Devices) and the values were normalized to the amount of dsDNA per well determined by QuantiFluor dsDNA System (Promega) according to the manufacturer’s instructions. Statistical analysis was performed using One-way ANOVA for at least three independent experiments.
3. Results
3.1. Pridopidine protects mouse primary striatal and cortical neurons against mutant Huntingtin induced toxicity
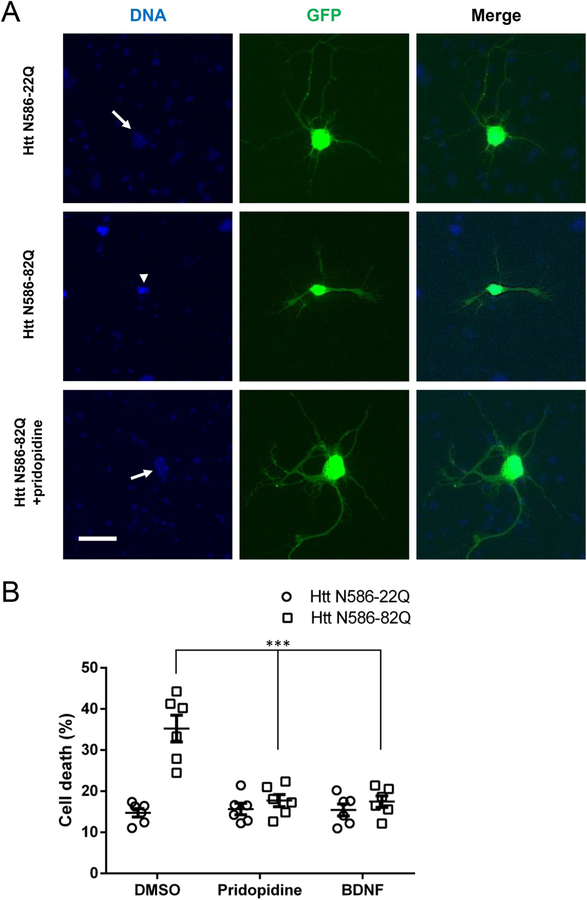
To evaluate the neuroprotective effects of pridopidine, we first tested a CD1 mouse primary neuron model of HD toxicity. We co-transfected primary striatal neurons with GFP and a plasmid expressing the first 586 amino acids of the Huntingtin protein (Htt) containing 22 glutamines (control, Htt N586-22Q) or 82 glutamines (mutant, Htt N586-82Q). There was robust and reproducible cell death caused by the mutant plasmid compared to control. This cell death was characterized by condensation of the nucleus, detected with Hoechst DNA staining, and fragmentation or disappearance of dendrites and axons, detected with the transfected GFP (Fig. 1A). Htt N586-82Q transfected striatal neurons treated with pridopidine showed preservation of healthy neuronal phenotype compared to their untreated counterparts. Quantitatively, 1 μM pridopidine protected Htt N586-82Q transfected striatal neurons as well as 20 ng/mL BDNF in nuclear condensation assays (Fig. 1B).
Fig. 1. Pridopidine protects striatal neurons from mutant Huntingtin induced toxicity.
(A) Representative images of GFP transfected striatal neurons. Cells were transfected as stated, treated for 48 h, fixed using 4% paraformaldehyde in PBS, and stained with Hoechst. Healthy non-condensed nuclei (arrow) and apoptotic condensed nuclei (arrowhead) are indicated. Scale bar: 25 μm.
(B) Quantification of nuclear condensation assay in striatal neurons. CD1 primary striatal neurons transfected at DIV5 were treated with 1 μM pridopidine or 20 ng/mL BDNF in the culture media for 48 h before nuclei were stained with Hoechst. Quantification of nuclear staining intensity in transfected cells was performed using Volocity. Results presented as individual values plus means ± S.E. of the percentage of dead cells. *** p < .001 vs DMSO, ANOVA with Bonferroni post-hoc test. (n = 6 independent neuronal preparations).
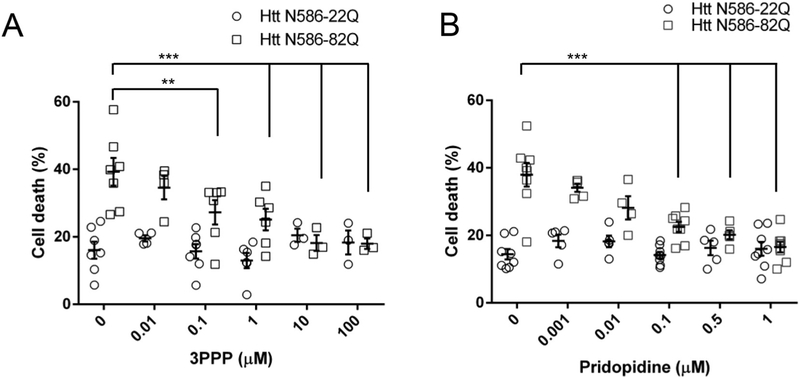
We then shifted our model from striatal to cortical neurons, to assess the neuroprotection of pridopidine across multiple concentrations. Using the nuclear condensation assay to detect cell death, we find striatal and cortical neurons to be comparable. The shift in cell type was a suitable technical convenience to obtain more cells and provided statistically relevant results across varied treatment conditions. Given the high affinity of pridopidine to the sigma-1 receptor we also tested 3PPP, a sigma-1 receptor agonist (Fig. 2A). 3PPP protected Htt N586-82Q transfected cortical neurons in a dose-dependent manner with maximum protection occurring between 10 and 100 μM and an EC50 of about 1 μM. We observed a significant dose-dependent protection from pridopidine with concentrations as low as 100 nM; with complete protection occurring at 1 μM and EC50 in the mid nanomolar range (Fig. 2B). Pridopidine and 3PPP did not elicit significant cellular toxicity at high concentrations, as illustrated by their effect on Htt N586-22Q transfected neurons.
Fig. 2. In nuclear condensation assays, pridopidine protects cortical neurons from mutant Huntingtin toxicity.
(A) Quantification of 3PPP treated cortical neuron cell death. Transfected CD1 primary cortical neurons were treated with a given 3PPP concentration in the culture media for 48 h before nuclei were stained with Hoechst. Quantification of nuclear staining intensity in transfected cells was performed using Volocity. Results presented as individual values plus means ± S.E. of the percentage of dead cells. ** p < .01 vs Htt-82Q untreated cells, *** p < .001 vs Htt-82Q untreated cells, ANOVA with Bonferroni post-hoc test. (n = 8 independent neuronal preparations).
(B) Quantification of pridopidine treated cortical neuron cell death. Transfected CD1 primary cortical neurons were treated with a given pridopidine concentration in the culture media for 48 h before nuclei were stained with Hoechst. Quantification of nuclear staining intensity in transfected cells was performed using Volocity. Results presented as individual values plus means ± S.E. of the percentage of dead cells. *** p < .001 vs Htt-82Q untreated cells, ANOVA with Bonferroni post-hoc test. (n = 8 independent neuronal preparations).
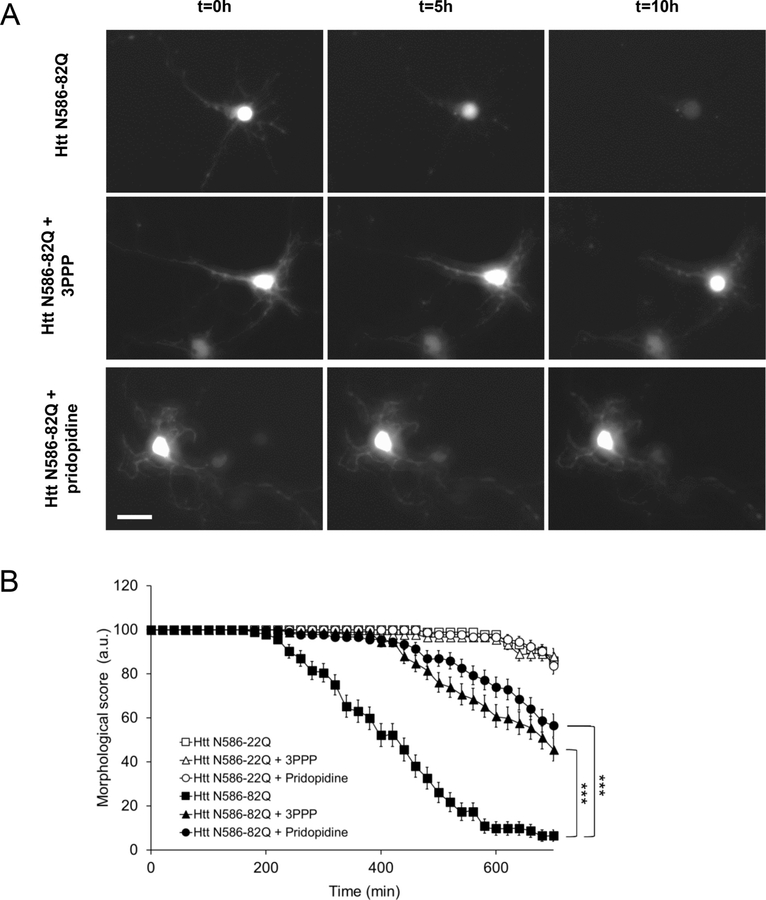
To confirm the neuroprotection by pridopidine, we also employed a live cell imaging assay of cell death. 24 h after treatment we tracked randomly selected GFP transfected neurons across 10 h, characterizing cell death by the fragmentation and disappearance of neuronal projections (Fig. 3A). The number of healthy untreated Htt N586-82Q transfected neurons progressively declined over 10 h (Fig. 3B). When the same cells were treated with either 1 μM 3PPP or 1 μM pridopidine, healthy neuronal morphology was notably conserved over time. Neurons transfected with the control Htt N586-22Q plasmid were unaffected by 3PPP and pridopidine treatments, and showed no significant increase in toxicity when left untreated.
Fig. 3. In live cell imaging assays, pridopidine protects cortical neuron morphology from mutant Huntingtin toxicity.
(A) Representative images of GFP transfected neurons after treatment with 1 μM pridopidine or 1 μM 3PPP at 0, 5 and 10 h of live cell imaging. Scale bar: 20 μm.
(B) Quantification of cortical neuron death in live cell imaging assay. Transfected CD1 primary cortical neurons were tracked over 10 h using automated picture acquisition performed on a Zeiss Axiovert 200 inverted microscope. Manual classification of cell death was performed, based on neuronal projection disappearance and soma shape change from oval to circular. *** p < .001 vs Htt-82Q untreated cells. Survival analysis was performed using a Gehan-Breslow test to determine a statistical difference between groups, followed by an All Pairwise Multiple Comparison (Holm-Sidak method) to identify the differences between groups. (n = 92 independent cells from 3 independent neuronal preparations).
3.2. The neuroprotection of pridopidine is blocked by a sigma-1 receptor antagonist
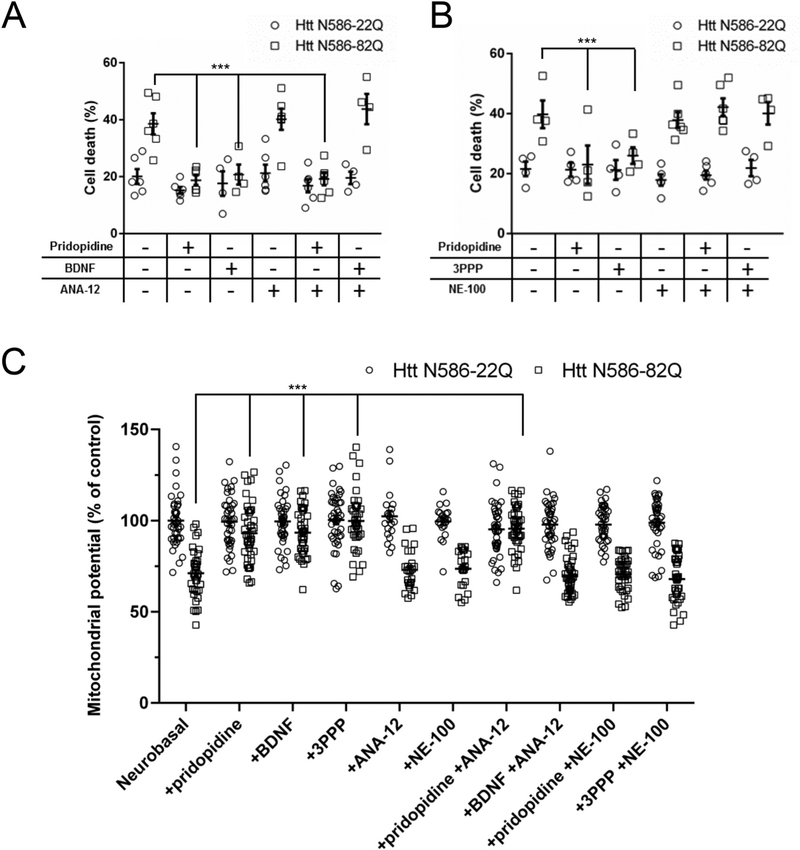
We next focused on elucidating the molecular mechanism of pridopidine in our CD1 primary cortical neuron model. Given the reported ability of pridopidine to increase BDNF expression, we investigated the mechanistic necessity of the sigma-1 receptor and the TrkB receptor activated BDNF pathway in the actions of pridopidine. To pharmacologically block the BDNF pathway, we used the TrkB receptor antagonist ANA-12. Alone 1 μM ANA-12 conferred no additional protection or toxicity, as demonstrated by the Htt N586-22Q transfected cells (Fig. 4A). The Htt N586-82Q transfected neurons were rescued by BDNF and this protection was blocked by ANA-12. By contrast, when ANA-12 was added in combination with pridopidine, it did not block the protection conferred by pridopidine. We also used NE-100, a documented sigma-1 receptor antagonist. NE-100 also showed no protective or toxic effects when alone in culture at 1 μM (Fig. 4B). When added in combination with 3PPP, NE-100 blocked all protection. In a strikingly similar way, NE-100 in combination with pridopidine markedly blocked protection of the Htt N586-82Q transfected neurons.
Fig. 4. A sigma-1 receptor antagonist blocks pridopidine-dependent protection of cortical neurons.
Quantification of cell death in neurons treated with pridopidine in combination with a TrkB (ANA-12 1 μM (A)) or sigma-1 receptor (NE-100 1 μM (B)) antagonist. Transfected CD1 primary cortical neurons were treated with a given compound combination in the culture media for 48 h before nuclei were stained with Hoechst. Quantification of nuclear staining intensity in transfected cells was performed using Volocity. Results presented as individual values plus means ± S.E. of the percentage of dead cells. *** p < .001 vs Htt-82Q untreated cells. ANOVA with Bonferroni post-hoc test. (n = 6 independent neuronal preparations).
(C) Quantification of mitochondrial potential using TMRM staining, Average intensity of staining in the soma of transfected cells was measured after 24 h of the indicated treatments. * p < .05 vs Htt N86-82Q by ANOVA with Bonferroni post-hoc test. (n = 4 independent neuronal preparations for a total of 40 independent cells).
To confirm further the results obtained with the nuclear condensation assay and the live cell imaging, we reproduced the experiments using a mitochondrial potential sensitive dye (Arbez et al., 2017). When transfected with mHtt, primary neurons showed a decrease of the mitochondrial potential (Fig. 4C). This failure of the mitochondria can be measured 24 h after transfection, preceding the nuclear condensation. Treatment with 1 μM pridopidine, 1 μM 3PPP or 20 ng/mL BDNF protected the mitochondria against depolarization. This protection could be blocked by NE-100 for pridopidine and 3PP and by ANA-12 for BDNF (Fig. 4C).
3.3. The effects of pridopidine on patient-derived induced pluripotent stem cells
We also investigated if the protection by pridopidine was reproducible in human neural cells. We repeated pharmacological testing of pridopidine in established lines of unaffected control (21n2) and HD (109n1, 50n5) patient-derived induced pluripotent stem cells (iPSCs) differentiated toward a medium spiny neuron phenotype (HD iPSC Consortium, 2012; HD iPSC Consortium, 2017). These cells endogenously contain a Huntingtin gene with either a control CAG length of 21 repeats (21n2) or an HD expanded CAG length of 109 (109n1) or 50 repeats (50n5), eliminating the need to transfect exogenous plasmids to model HD.
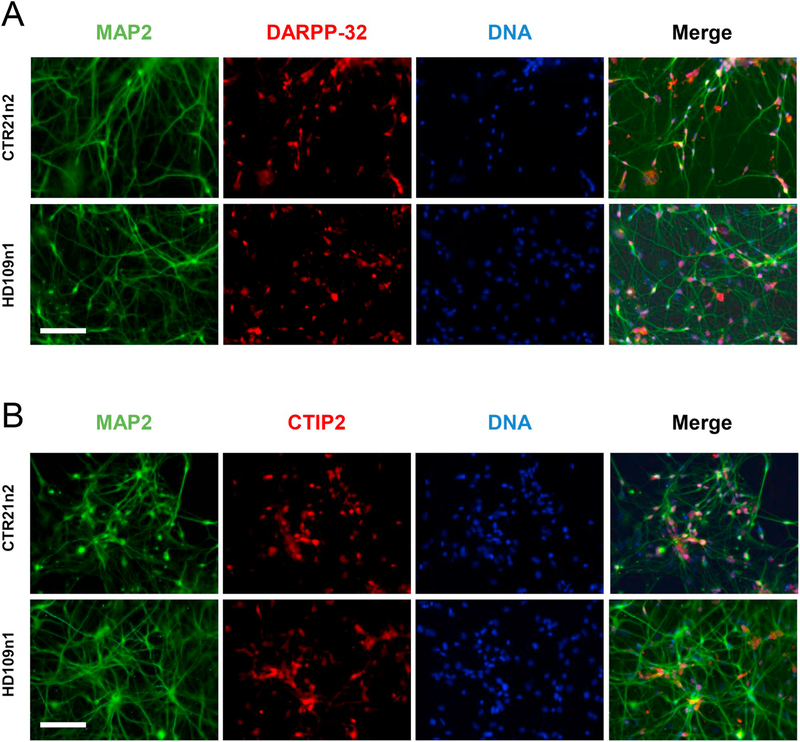
Differentiation of the iPSCs was done according to our established protocols. The HD and control iPSCs were differentiated into medium spiny neuron-like phenotypes over 42 days and then tested for expression of specific markers. In addition to the expression of pan neuronal marker like MAP2, differentiated iPSCs were also for a large part positive for specific markers of medium spiny neurons DARPP-32 (Fig. 5A) and CTIP2 (Fig. 5B).
Fig. 5. Characterization of differentiated patient-derived induced pluripotent stem cells.
(A) Representative images of differentiated control (CTR21n2) and HD (HD109n1) iPSCs. Cells were stained with Hoechst to visualize nuclei and MAP2 to visualize projections and the medium spiny neuron marker DARPP-32. Scale bar: 150 μm.
(B) Representative images of differentiated control (CTR21n2) and HD (HD109n1) iPSCs. Cells were stained with Hoechst to visualize nuclei and MAP2 to visualize projections and the medium spiny neuron marker CTIP2. Scale bar: 150 μm.
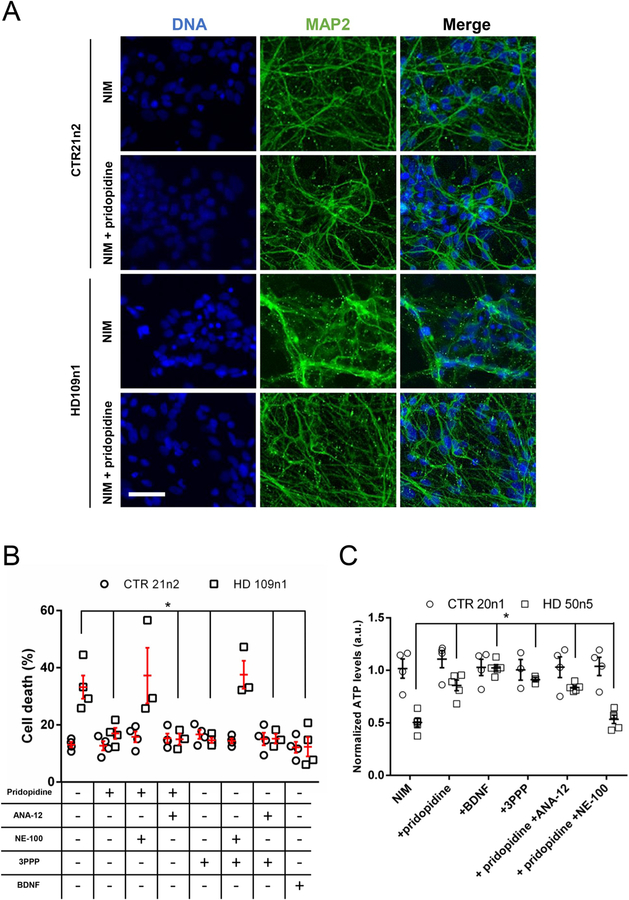
Toxicity was induced using a published BDNF-withdrawal protocol (HD iPSC Consortium, 2012; HD iPSC Consortium, 2017). transferred into BDNF-free Neural Induction Medium (NIM) for 48 h. When stained with MAP2, most cells showed reactivity. BDNF removal did not affect the cell morphology of the control line but induced some fragmentation and aggregation in the HD cells (Fig. 6A, middle column), as illustrated by an increased number of MAP2 puncta and more diffuse staining. This effect of BDNF withdrawal was prevented by a treatment with 1 μM pridopidine. The toxic effect of the BDNF removal on the HD iPSCs was also be visible with nuclear staining. After treatment, HD cells presented a higher number of condensed and bright nuclei (Fig. 6A, left column).
Fig. 6. Pridopidine protects patient-derived induced pluripotent stem cells via the sigma-1 receptor.
(A) Representative images of differentiated control (CTR21n2) and HD (HD109n1) iPSCs in NIM and treated with pridopidine. Cells were stained with Hoechst to visualize nuclei and MAP2 to visualize projections. Scale bar: 100 μm.
(B) Cell death quantification in iPSCs treated with 1 μM pridopidine or 20 ng/mL BDNF and either 1 μM ANA-12 or 1 μM NE-100. Cells were treated with a given compound combination in NIM for 48 h before being stained and assayed. Quantification of nuclear intensity in transfected cells was performed using Volocity. Results presented as individual values plus means ± S.E. of the percentage of dead cells. * p < .05 vs HD 109n1 untreated cells, ANOVA with SNK post-hoc test. (n = 4 independent 48-well plate preparations).
(C) Quantification of ATP levels using CellTiter-glo assay. Results presented as individual values plus means ± S.E. of normalized ATP levels. * p < .05 vs HD 50n5 untreated cells, ANOVA with SNK post-hoc test. (n = 3 to 5 independent 48-well plate preparations).
Toxicity was analyzed using nuclear condensation assay. Supplementing BDNF back into the NIM at 20 ng/mL provided a positive control and healthy standard nuclear phenotype against which the protection of pridopidine was compared. While control 21n2 neurons remained unaffected by BDNF-withdrawal, pridopidine treatment significantly rescued toxicity observed in the HD 109n1 neurons (Fig. 6B). Adding 1 μM ANA-12 in combination with pridopidine did not block protection of the 109n1 neurons. When 1 μM NE-100 was added in combination with pridopidine, however, all protection was abolished. 3PPP produced noticeably similar rescue of the HD 109n1 neurons which was also blocked by NE-100.
To verify our toxicity results, we then performed a CellTiter Glo Luminescent Cell Viability Assay (Promega) for HD and control iPSCs differentiated to MSNs. The assay measures ATP levels using automated plate reader and allows to normalize results to double-stranded DNA (dsDNA) content of the same wells of 96-well plates measured with the QuantiFluor assay (Promega) containing a fluorescent dye that allows quantification of small amount of dsDNA. Using this system, we showed that, just like for toxicity measured by nuclear condensation, the control cells are not sensitive to a 48 h BDNF withdrawal whereas the cells containing the expanded Htt demonstrate significant decrease of their ATP levels (Fig. 6C). When NIM was supplemented with 1 μM pridopidine, 1 μM 3PPP or 20 ng/mL BDNF, HD cells were protected from BDNF-withdrawal induced toxicity measured by the CellTiter Glo assay. As in our toxicity experiments performed with nuclear condensation assay, the protective effect of pridopidine was blocked by a sigma1-receptor antagonist, NE-100, but not by a TrkB receptor antagonist ANA-12 (Fig. 6C). These experiments were performed using HD 50n5 cells with 50 CAG repeats in mutant Htt demonstrating also that pridopidine protection from cell toxicity is efficient in models with high as well as less expanded and more common in HD patients repeat number.
3.4. The sigma-1 receptor is required for pridopidine-dependent protection of cortical neurons against mutant Huntingtin toxicity
To validate our pharmacological results, we utilized a mouse model with a genetic, constitutive knockout of the sigma-1 receptor. The published strain contains a TF0499 vector gene trap mutation between exons 2 and 3 of the SIGMAR1 gene. Constitutive sigma-1 receptor knockout mice are known to be viable and show no overt phenotypic abnormalities (Langa et al., 2003). Knockout primary cortical neurons (Sigma1R-KO) were simultaneously tested with neurons from mice with an identical genetic background lacking the gene trap mutation (WT).
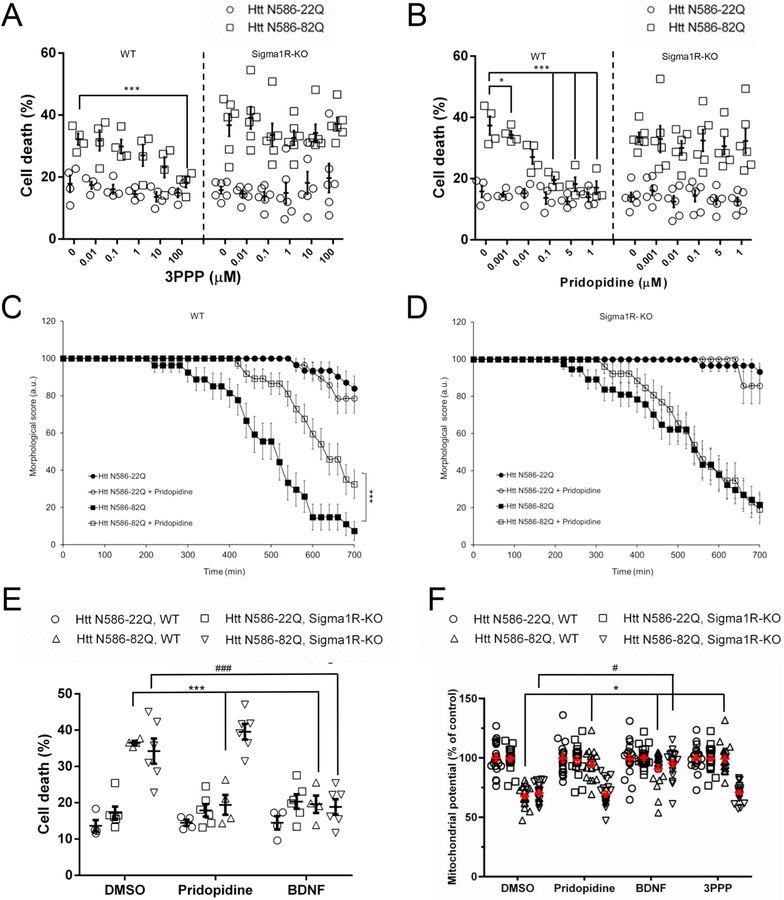
Treating WT Htt N586-82Q transfected neurons with 1 μM 3PPP resulted in complete dose-dependent protection up to 100 μM (Fig. 7A). This protection was notably absent in the Sigma1R-KO neurons. Treatment of WT Htt N586-82Q neurons with pridopidine reproduced a dose-dependent protection comparable to our initial CD1 mouse experiments (Fig. 7B). The protection provided by pridopidine was then eliminated in the Sigma1R-KO neurons. With these mice, we also repeated live cell imaging assays. Across 10 h, WT Htt N586-82Q transfected neurons treated with pridopidine maintained a healthy morphology (Fig. 7C). Sigma1R-KO neurons, on the contrary, showed no protection by pridopidine (Fig. 7D). In nuclear condensation assays, BDNF protected both WT and Sigma1R-KO cortical neurons from mutant Huntingtin toxicity the genetic knockout of the sigma-1 receptor abrogated all protection by pridopidine (Fig. 7E).
Fig. 7. Genetic knock-out of the sigma-1 receptor abolishes the protection of cortical neurons by pridopidine treatment.
(A-B) Cell death quantification of 3PPP (A) and pridopidine (B) treated sigma-1 receptor wild type and knockout cortical neurons. Sigma-1 receptor wild type (WT) and knockout (Sigma1R-KO) mouse primary cortical neurons were assayed and quantified as described. Results presented as individual values plus means ± S.E. of the percentage of dead cells. ** p < .01 vs Htt-82Q untreated cells, *** p < .001 vs Htt-82Q untreated cells, ANOVA with Bonferroni post-hoc test. (n = 4 independent WT neuronal preparations and n = 6 independent Sigma1R-KO neuronal preparations).
(C-D) Live cell imaging assay of a treatment with 1 μM pridopidine in WT (C) and Sigma1R-KO (D) cortical neurons. Transfected mouse primary cortical neurons were tracked and cell death was quantified as described. *** p < .001 vs Htt-82Q untreated cells. Survival analysis was performed using a Gehan-Breslow test to determine a statistical difference between groups, followed by an All Pairwise Multiple Comparison (Holm-Sidak method) to identify the differences between groups. (n = 70 independent cells from 3 independent neuronal preparations).
(E) Quantification of cell death in WT and Sigma1R-KO primary cortical neurons treated with 1 μM pridopidine or 20 ng/mL BDNF. Automated picture acquisition and quantification of nuclear intensity performed as described. Results presented as individual values plus means ± S.E. of the percentage of dead cells. *** p < .001 vs Htt-82Q, WT and #p < .05 vs HttN86–82Q, Sigma1R-KO. ANOVA with Bonferroni post-hoc test. (n = 4 independent WT neuronal preparations and n = 6 independent Sigma1R-KO neuronal preparations).
(F) Quantification of mitochondrial potential using TMRM staining, Average intensity of staining in the soma of transfected WT and Sigma1R-KO neurons was measured after 24 h of the indicated treatments. * p < .05 vs Htt N86-82Q, WT and # p < .05 vs Htt N86-82Q, Sigma1R-KO by ANOVA with Bonferroni post-hoc test. (n = 3 independent neuronal preparations for a total of 20 independent cells).
Finally, to confirm the results in the Sigma1R-KO neurons, we also measured the mitochondrial potential in these cells. When transfected with mHtt, primary neurons from the WT also showed a decrease of the mitochondrial potential that could be prevented by a treatment with 1 μM pridopidine, 1 μM 3PPP or 20 ng/mL BDNF (Fig. 7F). In contrast, Sigma1R-KO neurons showed the same decrease of mitochondrial potential induced by mHtt but were not protected by a treatment with either pridopidine or 3PPP.
4. Discussion
In this study, we assessed the protection of pridopidine in mouse striatal and cortical neurons, transfected with an N-terminal fragment of the Huntingtin protein. Our study also marks the first test of pridopidine in HD patient-derived induced pluripotent stem cells differentiated into medium spiny neuron-like phenotypes. Through pharmacological and genetic tools, we have examined the involvement of the sigma-1 receptor in pridopidine neuroprotection. We find that pridopidine protects both mouse and human cells from mutant Huntingtin toxicity via a sigma-1 receptor dependent mechanism.
Pridopidine is protective at high potency, with an EC50 about 50 to 100 times more than the sigma-1 receptor agonist 3PPP. The primary neuron model we utilized is an overexpression system, transiently transfecting high levels of exogenous mutant Huntingtin protein fragment, causing robust and relatively rapid cell death. It is suggested that full-length mutant Huntingtin protein must be proteolytically cleaved in cells to yield significant toxicity, and that only N-terminal fragments with expanded polyglutamine repeats demonstrate toxicity in HD models (Waldron-Roby et al., 2012).
To avoid the limitations that our mouse model may have concerning translation of results to humans, we also used our HD and control patient induced pluripotent stem cell model. This system expresses endogenous levels of mutant Huntingtin protein without the need for exogenous plasmid transfection. Reduction of BDNF is believed to play a role in HD pathogenesis, and BDNF is consistently highly protective against mutant Huntingtin (Gharami et al., 2008; Perez-Navarro et al., 2000; Saudou and Humbert, 2016; Simmons et al., 2017; Zuccato and Cattaneo, 2007); therefore we used BDNF-withdrawal to induce toxicity in these patient-derived neurons. We found the protection by pridopidine in human neurons to be comparable to the protection found in our tests of mouse neurons.
In vitro, relatively acute pridopidine treatments seem to substantially benefit HD neurons. A 2015 study revealed that only 6 h of pridopidine treatment improved STHdh111/111 HD knock-in mouse striatal neurons, decreasing apoptosis and increasing phosphorylation of the pro-survival kinase ERK (Squitieri et al., 2015). Overall our results from 48 h of pridopidine treatment are consistent with this study. Direct application of pridopidine to neurons in vitro has highly potent neuroprotective effects.
Previous studies have suggested that the sigma-1 receptor antagonist NE-100 blocks the protection of pridopidine (Geva et al., 2016; Sahlholm et al., 2018; Squitieri et al., 2015). The sigma-1 receptor is also often linked to the BDNF pathway (Ruscher and Wieloch, 2015). Chronic pridopidine treatment in vivo enhances BDNF protein and mRNA levels (Geva et al., 2016; Kusko et al., 2018; Squitieri et al., 2015). The TrkB receptor antagonist ANA-12 did not affect protection by pridopidine in both of our cell models. By contrast, protection by pridopidine was blocked with NE-100.
In our model, the neuroprotective effect of pridopidine may be mediated by the sigma 1 receptor via pathways not related to enhancing BDNF secretion or by activation of pathways downstream of the TrkB receptor, simply because primary neurons do not produce much BDNF in culture. It is interesting to see that in the iPSC model, while ANA-12 did not block the neuroprotective effect of pridopidine either, there was some variability suggesting it had some minor effect. It is possible that iPSC culture is more heterogeneous and may contain cells that are capable to produce BDNF in response to pridopidine stimulation and that the absence of BDNF during the experiment make these smalls change of BDNF production more visible. However, to be able to reach a definitive conclusion, it will be necessary to repeat these experiments with more HD iPS lines with a range of CAG-repeat length.
All these experiments, however, relied on pharmacological tools and tested few antagonistic compounds. Therefore, to substantiate our results, we used a sigma-1 receptor knockout mouse model. In the knockout neurons, we observed similar neuroprotection due to pridopidine (as well as continued protection by BDNF, independent of pridopidine). Our results support the idea that the mechanism of pridopidine is sigma-1 receptor dependent.
Future directions also warrant further investigation of the sigma-1 receptor when testing the efficacy of pridopidine in HD. Although we used pharmacological tools to examine the sigma-1 receptor in our human neural cells, there is a need for a genetic knockout model to further validate those results. Such a model would rule-out possible off-target effects of our reagents. More detailed mechanistic results could also be determined by observing the subcellular localization of the sigma-1 receptor with and without pridopidine treatments. The protein is known to translocate within the cell to enact modulatory functions (Tsai et al., 2014). A Parkinson’s disease model has visualized this protein translocation in response to treatment with an agonist using simple histology (Francardo et al., 2017).
Sigma-1 receptor protein levels are upregulated in late but not early HD brains (Ryskamp et al., 2016). The only other examination of the sigma-1 receptor in HD human cells used HeLa cells transfected with N-terminal mutant Huntingtin (Miki et al., 2015). This study suggested that the sigma-1 receptor helped degrade abnormal proteins because of its localization at nuclear inclusions. Taken together, these findings suggest that the sigma-1 receptor is upregulated in HD as a compensatory mechanism. There are multiple splice variants of the sigma-1 receptor, which vary in expression across brain regions (Shioda et al., 2012). Future studies might investigate what variants comprise the upregulated sigma-1 receptor levels in HD.
The neuroprotection afforded by pridopidine in our HD cell models is robust and sigma-1 receptor dependent. These studies support the further development of pridopidine, and other sigma-1 receptor agonists as neuroprotective agents for HD and other disorders.
Acknowledgements
This work was supported by the National Institutes of Health: Hopkins Post-baccalaureate Research Education Program (PREP) grant NIH R25GM109441 (CRE).
This work was funded by Teva Pharmaceuticals Industries.
Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS086452.
References
- Albayrak Y, Hashimoto K, 2017. Sigma-1 receptor agonists and their clinical implications in neuropsychiatric disorders. Adv. Exp. Med. Biol 964, 153–161. [DOI] [PubMed] [Google Scholar]
- Allahtavakoli M, Jarrott B, 2011. Sigma-1 receptor ligand PRE-084 reduced infarct volume, neurological deficits, pro-inflammatory cytokines and enhanced anti-inflammatory cytokines after embolic stroke in rats. Brain Res. Bull 85, 219–224. [DOI] [PubMed] [Google Scholar]
- Arbez N, et al. , 2017. Post-translational modifications clustering within proteolytic domains decrease mutant huntingtin toxicity. J. Biol. Chem 292, 19238–19249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baydyuk M, et al. , 2011. Chronic deprivation of TrkB signaling leads to selective late-onset nigrostriatal dopaminergic degeneration. Exp. Neurol 228, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazorla M, et al. , 2011. Identification of a low-molecular weight TrkB antagonist with anxiolytic and antidepressant activity in mice. J. Clin. Invest 121, 1846–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Yebenes JG, et al. , 2011. Pridopidine for the treatment of motor function in patients with Huntington’s disease (MermaiHD): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 10, 1049–1057. [DOI] [PubMed] [Google Scholar]
- Dyhring T, et al. , 2010. The dopaminergic stabilizers pridopidine (ACR16) and (−)-OSU6162 display dopamine D(2) receptor antagonism and fast receptor dissociation properties. Eur. J. Pharmacol 628, 19–26. [DOI] [PubMed] [Google Scholar]
- Francardo V, et al. , 2017. Neuroprotection and neurorestoration as experimental therapeutics for Parkinson’s disease. Exp. Neurol 298, 137–147. [DOI] [PubMed] [Google Scholar]
- Geva M, et al. , 2016. Pridopidine activates neuroprotective pathways impaired in Huntington disease. Hum. Mol. Genet 25, 3975–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharami K, et al. , 2008. Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington’s disease phenotypes in mice. J. Neurochem 105, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Su TP, 2007. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131, 596–610. [DOI] [PubMed] [Google Scholar]
- HD iPSC Consortium, 2012. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell 11, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HD iPSC Consortium, 2017. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat. Neurosci 20, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington Study Group HART Investigators, 2013. A randomized, double-blind, placebo-controlled trial of pridopidine in Huntington’s disease. Mov. Disord 28, 1407–1415. [DOI] [PubMed] [Google Scholar]
- Kusko R, et al. , 2018. Large-scale transcriptomic analysis reveals that pridopidine reverses aberrant gene expression and activates neuroprotective pathways in the YAC128 HD mouse. Mol. Neurodegener 13, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy V, et al. , 2013. Blockade of tau hyperphosphorylation and Abeta(1)(−)(4)(2) generation by the aminotetrahydrofuran derivative ANAVEX2–73, a mixed muscarinic and sigma(1) receptor agonist, in a nontransgenic mouse model of Alzheimer’s disease. Neuropsychopharmacology 38, 1706–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langa F, et al. , 2003. Generation and phenotypic analysis of sigma receptor type I (sigma 1) knockout mice. Eur. J. Neurosci 18, 2188–2196. [DOI] [PubMed] [Google Scholar]
- Mattis VB, et al. , 2015. HD iPSC-derived neural progenitors accumulate in culture and are susceptible to BDNF withdrawal due to glutamate toxicity. Hum. Mol. Genet 24, 3257–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColgan P, Tabrizi SJ, 2018. Huntington’s disease: a clinical review. Eur. J. Neurol 25, 24–34. [DOI] [PubMed] [Google Scholar]
- Miki Y, et al. , 2015. Sigma-1 receptor is involved in degradation of intranuclear inclusions in a cellular model of Huntington’s disease. Neurobiol. Dis 74, 25–31. [DOI] [PubMed] [Google Scholar]
- Nguyen L, et al. , 2015. Role of sigma-1 receptors in neurodegenerative diseases. J. Pharmacol. Sci 127 (1), 17–29. [DOI] [PubMed] [Google Scholar]
- Okuyama S, et al. , 1993. NE-100, a novel sigma receptor ligand: in vivo tests. Life Sci. 53 (PL285–90). [DOI] [PubMed] [Google Scholar]
- Perez-Navarro E, et al. , 2000. Brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4/5 prevent the death of striatal projection neurons in a rodent model of Huntington’s disease. J. Neurochem 75, 2190–2199. [DOI] [PubMed] [Google Scholar]
- Peviani M, et al. , 2013. Neuroprotective effects of the Sigma-1 receptor (S1R) agonist PRE-084, in a mouse model of motor neuron disease not linked to SOD1 mutation. Neurobiol. Dis 62, 218–232. [DOI] [PubMed] [Google Scholar]
- Rodrigues FB, Wild EJ, 2017. Clinical trials corner: September 2017. J. Huntingtons Dis 6, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, et al. , 2014. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol 10, 204–216. [DOI] [PubMed] [Google Scholar]
- Ruscher K, Wieloch T, 2015. The involvement of the sigma-1 receptor in neurode-generation and neurorestoration. J. Pharmacol. Sci 127, 30–35. [DOI] [PubMed] [Google Scholar]
- Ryskamp D, et al. , 2016. The sigma-1 receptor mediates the beneficial effects of pridopidine in a mouse model of Huntington disease. Neurobiol. Dis 97, 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlholm K, et al. , 2013. The dopamine stabilizers ACR16 and (−)-OSU6162 display nanomolar affinities at the sigma-1 receptor. Mol. Psychiatry 18, 12–14. [DOI] [PubMed] [Google Scholar]
- Sahlholm K, et al. , 2015. Pridopidine selectively occupies sigma-1 rather than dopamine D2 receptors at behaviorally active doses. Psychopharmacology 232, 3443–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlholm K, et al. , 2018. Pridopidine reverses phencyclidine-induced memory impairment. Front. Pharmacol 9, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, Humbert S, 2016. The biology of huntingtin. Neuron 89, 910–926. [DOI] [PubMed] [Google Scholar]
- Shioda N, et al. , 2012. Expression of a truncated form of the endoplasmic reticulum chaperone protein, sigma1 receptor, promotes mitochondrial energy depletion and apoptosis. J. Biol. Chem 287, 23318–23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DA, et al. , 2017. Neurotrophin receptor signaling as a therapeutic target for Huntington’s disease. CNS Neurol. Disord. Drug Targets 16, 291–302. [DOI] [PubMed] [Google Scholar]
- Squitieri F, et al. , 2015. Pridopidine, a dopamine stabilizer, improves motor performance and shows neuroprotective effects in Huntington disease R6/2 mouse model. J. Cell. Mol. Med 19, 2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TP, et al. , 2016. The Sigma-1 receptor as a pluripotent modulator in living systems. Trends Pharmacol. Sci 37, 262–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, et al. , 2014. Sigma-1 receptor chaperones in neurodegenerative and psychiatric disorders. Expert Opin. Ther. Targets 18, 1461–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron-Roby E, et al. , 2012. Transgenic mouse model expressing the caspase 6 fragment of mutant huntingtin. J. Neurosci 32 (1), 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkin EE, et al. , 2014. Phosphorylation of mutant huntingtin at serine 116 modulates neuronal toxicity. PLoS One 9, e88284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E, 2007. Role of brain-derived neurotrophic factor in Huntington’s disease. Prog. Neurobiol 81, 294–330. [DOI] [PubMed] [Google Scholar]