Summary
Paratrajectura longcementglandatus Amin, Heckmann et Ali, 2018 (Transvenidae) was recently described from two species of percid fishes collected from the marine territorial waters of Iraq and Iran in the Persian Gulf. The genus Paratrajectura Amin, Heckmann et Ali, 2018 is a close relative to transvenid genera Trajectura Pichelin et Crib, 2001 and Transvena Pichelin et Crib, 2001. Morphologically, Paratrajectura is characterised by having apical proboscis cone, long, tubular cement glands, short lemnisci, prominent roots on all proboscis hooks, subterminal female gonopore, and males with long pre-equatorial testes. Molecular studies of P. longcementglandatus using 18S rDNA and cox1 genes compared with available data of members of other families of Echinorhynchida showed that P. longcementglandatus is grouped with species of the genus Transvena forming a clade within the family Transvenidae.
Keywords: Spiny headed transvenid worms, 18S rDNA, cox1, phylogeny, Perciformes, Middle East
Introduction
Pichelin & Cribb (2001) described the family Transvenidae with two genera: monotypic Transvena with T. annulospinosa Pichelin et Cribb, 2001, and Trajectura with two species, T. ikedai (Machida, 1992) and T. perinsolens Pichelin et Cribb, 2001. Specimens of the two genera were recovered from wrasses (Labridae, Perciformes) in the Pacific off southern Australia and southern Japan. Lisitsyna et al. (2019) described two other species of the family Transvenidae, namely Transvena pichelinae Lisitsyna, Kudlai, Cribb et Smit, 2019, and Pararhadinorhynchus sodwanensis Lisitsyna, Kudlai, Cribb et Smit, 2019 from the marine fishes from the Sodwana Bay, South Africa. The other genus of this family, Paratrajectura, was established by Amin et al. (2018). It comprises one species Paratrajectura longcementglandatus Amin, Heckmann et Ali, 2018, which was described on the basis of worms from the Japanese threadfin bream Nemipterus japonicus Bloch (Nemipteridae) and the tigertooth croacker, Otolithes ruber Bloch et Schneider (Sciaenidae, Perciformes) caught in the marine territorial waters of Iraq and Iran, the Persian Gulf (Amin et al., 2018). The genus Paratrajectura is characterised by having apical proboscis cone, long, tubular cement glands, short lemnisci, prominent roots on all proboscis hooks, subterminal female gonopore, and males with long pre-equatorial testes.
While, several studies have been published about sequence data for acanthocephalans including two Transvena spp. (Westram et al., 2011; Garcia-Varela, et al., 2013; Pinacho-Pinacho et al., 2014; Lisitsyna et al., 2019), no sequence data has been published for P. longcementglandatus whose phylogenetic relationship with other acanthocephalans and related families was unknown. In this paper, we report the molecular profile of P. longcementglandatus, validate its generic affiliations, and explore its evolutionary relationships with related and other species and taxa based on partial 18S rDNA and cox1 genes.
Materials and Methods
DNA extraction and PCR amplification
For extraction of genomic DNA, five adult worms of P. longcementglandatus were washed with sterile distilled water several times to remove the ethanol residuals. Total DNA was extracted using Qiagen DNeasy Blood and Tissue kit (Qiagen Inc., Valencia, California, USA) according to manufacturer’s instructions and kept at −20 °C until use.
PCR reactions were performed in 30 μL volumes containing 2 × red PCR premix (Ampliqon, Odense, Denmark), 20 pmol of each primer and 3 μL of extracted DNA. The partial 18S rRNA gene was amplified using the forward primer (5′-AGATTAAGCCATGCATG-CGTAAG-3′) and reverse primer (5′- ACCCACCGAATCAAGAAA-GAG-3′). Also, primers used for the amplification of the partial mitochondrial cytochrome oxidase subunit1 (cox1) gene were COI-F (5′-AGTTCTAATCATAARGATATYGG-3′) and COI-R (5′-TAAACT-TCAGGGTGACCAAAAAATCA-3′) (Folmer et al., 1994). PCR conditions for 18S rRNA gene amplification included of an initial denaturing step of 95 °C for 5 min and 35 cycles followed by denaturing step at 95 °C for 30 s, annealing step of 61 °C for 30 s, and 60 s of extension at 72 °C, and 72 °C for 7 min as a final extension. The thermal PCR profiles for cox1 gene consisted of initial denaturation at 95 °C for 6 minutes followed by 35 cycles of 95 °C for 30 s (denaturation), 55 °C for 30 s (annealing), and at 72 °C for 60 s (extension) with a final extension of 72 °C for 6 minutes. PCR products were analysed on 1.5 % agarose gel and visualized with UV transluminator. Next, the PCR products were sequenced in both directions using the same PCR primers with ABI 3130 sequencer.
The obtained sequence results were manually edited and trimmed using Chromas software v.2.01 (Technelysium Pty Ltd., Brisbane, Queensland, Australia). Next, generated sequences were compared with GenBank submitted sequences using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih gov/). Also, Clustal W method of Bioedit software v.7.0.9 was used for multiple sequence alignment (Larkin et al., 2007). The sequences of 18S rRNA and cox1 genes were submitted to GenBank database (Accession Numbers: MK770616 for 18S rRNA and MK770615 for cox1)
Phylogenetic analysis
The phylogenetic tree was constructed using Maximum-Likelihood model and Tamura-3-parameter model by Molecular and Evolution Genetic Analysis software v.6 (MEGA 6). The reliability of topology of the tree was supported with Bootstrap value based on 1000 replications. The whole scientific names of acanthocephalan species, names of host species, localities, and GenBank accession numbers used in the phylogenetic analysis are listed in Table 1.
Table 1.
Acanthocephalan species represented in the phylogenetic analysis with their family, host species, GenBank accession numbers, locations, and references.
| Species | Host | GenBank Acc. no. 185rDNA |
GenBank Acc. no. cox1 |
Location | Reference |
|---|---|---|---|---|---|
| Gymnorhadinorhynchidae | |||||
| Gymnorhadinorhynchus sp. | Regalecus russelii | MK014866 | MK012665 | Japan | Steinauer et al. (2019) |
| Gymnorhadinorhynchus decapteri (Braicovich, Lanfranchi, Farber, Marvaldi, Luque et Timi, 2014) | Decapterus punctatus | KJ590123 | KJ590125 | Brazil | Braicovich et al. (2014) |
| Cavisomidae | |||||
| Neorhadinorhynchus nudus (Harada, 1938) | Auxis thazard | - | MG757444 | China | Li et al. (2018) |
| Filisoma bucerium (Van Cleave, 1940) | Kyphosus elegans | AF064814 | DQ089722 | Na* | García-Varela et al. (2000), García-Varela and Nadler (2006) |
| Filisoma rizalinum (Tubangui et Masilungan, 1946) | Scatophagus argus | JX014229 | - | Indonesia | Verweyen et al. (2011) |
| Rhadinorhynchidae | |||||
| Rhadinorhynchus laterospinosus (Amin, Heckmann et Van Ha, 2011) | Auxis rochei | MK457183 | MK572744 | Vietnam | Amin et al. (2019a) |
| Rhadinorhynchus sp. | Sciaenidae | AY062433 | DQ089712 | Na | García-Varela et al. (2002), García-Varela and Nadler (2006) |
| Serrasentis sagittifer (Linton, 1889) | Lutjanus sebae | - | MF134296 | Australia | Barton et al. (2018) |
| Serrasentis sagittifer | Johnius coitor | JX014227 | - | Indonesia | Verweyen et al. (2011) |
| Serrasentis nadakali (George et Nadakal, 1978) | Na | KC291715 | KC291713 | Na | Paul et al. (unpublished) |
| Gorgorhynchoides bullock (Cable et Mafarachisi, 1970) | Eugerres plumieri | AY830154 | DQ089715 | Na | Garcia-Varela and Nadler (2005, 2006) |
| Rhadinorhynchus lintoni (Cable et Linderoth, 1963) | Selar Crumrn- opht halmus | JX014224 | - | USA | Verweyen et al. (2011) |
| Rhadinorhynchus pristis (Rudolphi, 1802) | Selar Crumrn- opht halmus | JX014226 | - | USA | Verweyen et al. (2011) |
| Transvenidae | |||||
| Paratrajectura longcementglandatus (Amin, Heckmann et Ali, 2018) | Percid fishes | MK770616 | MK770615 | Marine waters of Iraq and Iran | Present study |
| Transvena annulospinosa (Pichelin et Cribb, 2001) | Anampses neoguinaicus | AY830153 | DQ089711 | Na | Garcia-Varela and Nadler (2005, 2006) |
| Transvena pichelinae sp. n. (Lisitsyna, 2019) | Thalassoma purpureum | MN105736, MN105737 | MN 104895, MN 104896 | South Africa | Lisitsyna et al. (2019) |
| Pararhadinorhynchus sodwanensis sp. n. (Lisitsyna, 2019) | Pomadasys furcatus | MN 105738 | - | South Africa | Lisitsyna et al. (2019) |
| Pararhadinorhynchus sp. | Siganus fuscescens | HM545903 | - | China | Wang et al. (unpublished) |
| Echinorhynchidae | |||||
| Pseudoacanthocephalus toshimai (Nakao, 2016) | Rana pirica | LC129278 | LC100044 | Japan | Nakao (2016) |
| Pseudoacanthocephalus lucidus (Van Cleave, 1925) | Rana ornativentris | LC129279 | LC100057 | Japan | Nakao (2016) |
| Acanthocephalus lucii (Müller, 1776) | Perca fluviatilis | AY830152 | Na | Garcia-Varela and Nadler (2005), Benesh et al. (2006) | |
| Acanthocephalus lucii | Perca fluviatilis | AM039837 | England | Garcia-Varela and Nadler (2005), Benesh et al. (2006) | |
| Acanthocephalus anguillae (Müller, 1780) | Perca fluviatilis | AM039865 | Austria | Benesh et al. (2006) | |
| Acanthocephalus dirus (Van Cleave, 1931) | Asellus aquaticus | AY830151 | DQ089718 | Na | Garcia-Varela and Nadler (2005, 2006) |
| Acanthocephalus clavula (Dujardin, 1845) | Perca fluviatilis | AM039866 | Ireland | Benesh et al. (2006) | |
| Acanthocephalus nanus (Van Cleave, 1925) | Cynops pyrrhogaster | LC129889 | Japan | Nakao (2016) | |
| Echinorhynchus salmonis (Müller, 1784) | Coregonus lavaretus | KP261017 | Finland | Wayland et al. (2015) | |
| Echinorhynchus gadi (Müller, 1776) | Na | AY218123 | AY218095 | Na | Giribet et al. (2004) |
| Echinorhynchus bothniensis (Zdzitowiecki et Valtonen, 1987) | Osmerus eperlanus | KP261018 | Finland | Wayland et al. (2015) | |
| Echinorhynchus truttae (Schrank, 1788) | Thymallus thymallus | AY830156 | DQ089710 | Na | Garcia-Varela and Nadler (2005, 2006) |
| Echinorhynchus brayi (Wayland, Sommerville et Gibson, 1999) | Pachycara crassiceps | KP261015 | Atlantic Ocean: Porcupine Seabight | Wayland et al. (2015) | |
| Echinorhynchus cinctulus (Porta, 1905) | Lota lota | KP261014 | Finland | Wayland et al. (2015) | |
| Pomphorhynchidae | |||||
| Longicollum pagrosomi (Yamaguti, 1935) | Pagrus major | LC195887 | - | Japan | Mekata et al. (unpublished) |
| Longicollum pagrosomi | Oplegnathus fasciatus | - | KY490048 | China | Li et al. (2017) |
| Pomphorhynchus zhoushanensis (Li, Chen, Amin et Yang, 2017) | Oplegnathus fasciatus | - | KY490045 | China | Li et al. (2017) |
| Pomphorhynchus bulbocolli (Linkins, 1919) | Lepomis macrochirus | - | DQ089709 | Na | Garcia-Varela and Nadler (2006) |
| Pomphorhynchus purhepechus (García-Varela, Mendoza-Garfias, Choudhury et Pérez-Ponce de León, 2017) | Moxostoma austrinum | - | KY911281 | Na | Garcia-Varela et al. (2017) |
| Tenuiproboscis sp. | Epinephelus malabaricus | - | JF694273 | Na | Vijayan et al. (unpublished) |
| Pomphorhynchus laevis (Zoega in Müller, 1776) | Gammarus pulex | AY423346 | AY423348 | France | Perrot-Minnot (2004) |
| Pomphorhynchus tereticollis (Rudolphi, 1809) | Gammarus pulex | AY423347 | AY423351 | France | Perrot-Minnot (2004) |
| Arhythmacanthidae | |||||
| Acanthocephaloides propinquus (Dujardin, 1845) | Gobius bucchichii | AY830149 | DQ089713 | Na | Garcia-Varela and Nadler (2005, 2006) |
| Diplosentidae | |||||
| Sharpilosentis peruviensis (Lisitsyna, Scholz et Kuchta, 2015) | Duopalatinus cf. peruanus | - | KP967562 | Peru | Lisitsyna et al. (2015) |
| Illiosentidae | |||||
| Dollfusentis chandleri (Golvan, 1969) | Na | - | DQ320484 | Na | Baker and Sotka(unpublished) |
| Dentitruncus truttae (Sinzar, 1955) | Salmo trutta | JX460865 | JX460877 | Croatia | Vardić Smrzlić et al. (2013) |
| Illiosentis sp. | Na | AY830158 | DQ089705 | Na | Garcia-Varela and Nadler (2005, 2006) |
| Leptorhynchoides thecatus (Linton, 1891) | Lepomis cyanellus | AF001840 | DQ089706 | Na | Near et al. (1998), Garcia-Varela and Nadler (2006) |
| Pseudoleptorhynchoides lamothei (Salgado-Maldonado, 1976) | Ariopsis guatemalenis | EU090950 | EU090949 | Na | Near et al. (1998), Garcia-Varela and Nadler (2006) |
| Koronacantha pectinaria (Van Cleave, 1940) | Microlepidotus brevipinnis | AF092433 | DQ089707 | Na | García-Varela and Nadler (2005, 2006) |
| Koronacantha Mexicana (Monks et Pérez-Ponce de León, 1996) | Haemulopsis leuciscus | AY830157 | DQ089708 | Na | García-Varela and Nadler (2005, 2006) |
| Neoechinorhynchidae (Outgroup) | |||||
| Neoechinorhynchus brentnickoli (Monks, Pulido-Flores and Violante- González, 2011) | Dormitator latifrons | - | JN830849 | Na | Pinacho-Pinacho et al. (2012) |
| Neoechinorhynchus saginata (Van Cleave & Bangham, 1949) | Na | - | DQ089704 | Na | García-Varela and Nadler (2006) |
| Floridosentis mugilis (Machado Filho, 1951) | Na | AF064811 | DQ089723 | Na | García-Varela and Nadler (2006), García-Varela et al. (2000) |
| Neoechinorhynchus crassus (Van Cleave, 1919) | Na | KU363969 | - | Iran | Dadar and Adel (unpublished) |
| Neoechinorhynchus pseudemydis (Cable and Hopp, 1954) | Capoeta aculeata | KU363973 | - | Iran | Dadar and Adel (unpublished) |
*Na = not available
Ethical Approval and/or Informed Consent
The authors declare compliance with all relevant ethical standards.
Results
The specimens of P. longcementglandatus successfully presented amplifications of about 1234 bp for the 18S rDNA gene and 664 bp for the cox1 gene. Comparisons of the18S rDNA and cox1 sequences from this parasite with other available acanthocephalan sequences in GenBank, using multiple sequence alignment, showed that it had the highest similarity with T. annulospinosa based on18S rDNA (98 %) and cox1 (77 %) genes. The 18S rDNA dataset (1129 nt) included 26 sequences for species of seven families within the Echinorhynchida and the novel sequence of P. longcementglandatus. The cox1 dataset (538 nt) included 39 sequences for species of nine families of Echinorhynchida and the sequence of P. longcementglandatus.
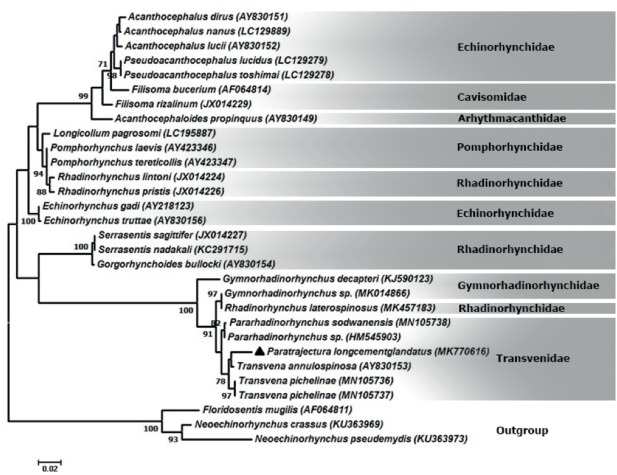
The phylogenetic reconstruction based on the partial sequence spanning the 18S rDNA showed that our sequence of P. longcementglandatus is clustered with Transvena annulospinosa (AY830153), T. pichelinae (MN105736 and MN105737), P. sodwanensis (MN105738) and an unidentified species of Pararhadinorhynchus (HM545903) with strong support forming a clade of the family Transvenidae. Also, the species of Gymnorhadinorhynchus sp. (MK014866) (Gymnorhadinorhynchidae) and Rhadinorhynchus laterospinosus (MK457183) (Rhadinorhynchidae) are very closely related with the family Transvenidae in the tree with 100 % of bootstrap support. The sequence of Gymnorhadinorhynchus decapteri (KJ590123) (Gymnorhadinorhynchidae) is located at the basal position to the members of the clade. Other families of the order Echinorhynchida including Rhadinorhynchidae, Pomphorhynchidae, Cavisomidae, Arhythmacanthidae and Echinorhynchidae located in a major sister clade (Fig. 1). Inter-generic differences are noted between P. longcementglandatus and T. annulospinosa, T. pichelinae, P. sodwanensis and Pararhadinorhynchus sp. from Transvenidae based on partial 18S rDNA sequence were 2.4 % (18 nt), 2.8 % (21 nt), 2.9 % (22 nt) and 2.9 % (22 nt), respectively. According to phylogenetic analyses based on the cox1 gene, our sequence of P. longcementglandatus (MK770615) is grouped with T. annulospinosa (DQ089711) and T. pichelinae (MN104895 and MN104896) with strong support in a clade of the family Transvenidae. The species of G. decapteri (KJ590125) and Gymnorhadinorhynchus sp. (MK012665) (Gymnorhadinorhynchidae), Neorhadinorhynchus nudus (MG757444) (Cavisomidae), R. laterospinosus (MK572744) and Rhadinorhynchus sp. (DQ089712) (Rhadinorhynchidae), appear as a sister group of the family Transvenidae (Fig. 2). The interspecific divergence between P. longcementglandatus and T. annulospinosa, T. pichelinae based on partial cox1 gene was 23.4 % (141 nt), 27.3 % (144 nt), respectively.
Fig. 1.
Phylogenetic tree based on the Maximum likelihood analysis using 18S rDNA sequence of Paratrajectura longcementglandatus of current study and sequences of the closest-related members of the order Echinorhynchida deposited in the GenBank. Outgroup: Floridosentis mugilis, Neoechinorhynchus pseudemydis and N. crassus. Bootstrap values lower than 70 are omitted.
Fig. 2.
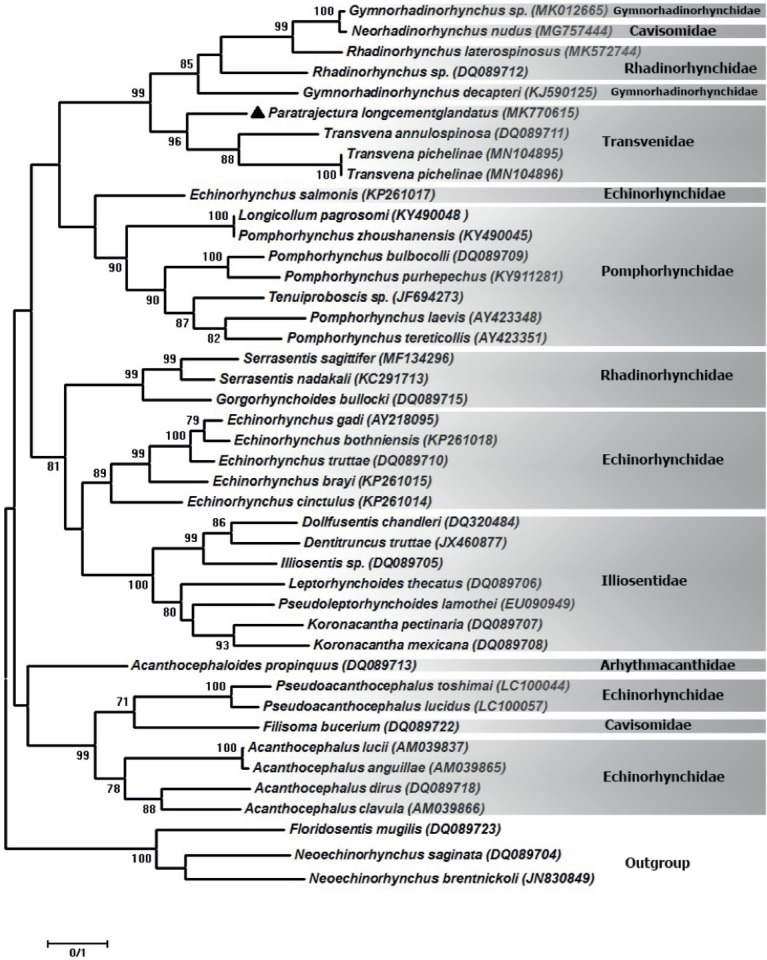
Phylogenetic reconstruction based on the Maximum likelihood analysis using partial region of the cox1 sequence of Paratrajectura longcementglandatus of current study and sequences of the closest-related members of order Echinorhynchida deposited in the GenBank. Outgroup: Floridosentis mugilis, Neoechinorhynchus saginata and N. brentnickoli. Bootstrap values lower than 70 are omitted.
Discussion
Recently, molecular methods are applied for species identification, classification and phylogenetic analysis of acanthocephalan species (García-Varela et al., 2002). To date, molecular profile has been provided for few species of the family Transvenidae including T. annulospinosa, T. pichelinae, P. sodwanensis and Pararhadinorhynchus sp. (Pichelin & Cribb, 2001; Lisitsyna et al., 2019). In the current study, phylogenetic relationships of P. longcementglandatus as another genus of this family is described based on partial 18S rDNA and cox1 genes determining relationships with other acanthocephalan families.
This study showed that the interspecific variation between P. longcementglandatus and species of Transvena based on partial 18S rDNA was 2.4 % – 2.8 % (18 – 21 nt) and between it and species of Pararhadinorhynchus was 2.9 % (22 nt). Also based on cox1 gene, inter-generic variations between P. longcementglandatus and T. annulospinosa was 23.4 % (141 nt). These results illustrate that sequence differences between the genera of the family based on cox1 gene is higher than18S rDNA and it is appropriate to consider for taxonomic studies at the generic level.
The phylogenetic analysis of the 18S rDNA sequence (Fig. 1) showed that P. longcementglandatus is grouped in a highly supported clade with T. annulospinosa (AY830153), T. pichelinae (MN105736 and MN105737), P. sodwanensis (MN105738) and Pararhadinorhynchus sp. (HM545903) forming a clade of the family Transvenidae. In the clade, the family Transvenidae grouped close to R. laterospinosus (MK457183) and G. decapteri (KJ590123) (Gymnorhadinorhynchidae). Our phylogenetic tree for 18S rDNA is similar to those of García-Varela et al. (2002) and Lisitsyna et al. (2019) where the family Transvenidae grouped close to different species of Rhadinorhynchus (Rhadinorhynchidae) and Gymnorhadinorhynchus (Gymnorhadinorhynchidae).
Our phylogenetic analysis of the cox1 gene (Fig. 2) confirmed that P. longcementglandatus is grouped with T. annulospinosa (DQ089711) and T. pichelinae (MN104895 and MN104896) which made the clade of the family Transvenidae with good statistical support. Also, the families Rhadinorhynchidae, Gymnorhadinorhynchidae and Cavisomidae appear as a sister group with the clade of family Transvenidae. Other families of Echinorhynchida such as Pomphorhynchidae, Echinorhynchidae, Cavisomidae, Illiosentidae, Rhadinorhynchidae, Gymnorhadinorhynchidae, Diplosentidae and Arhythmacanthidae are well separated in the later clade. In the present study, the higher level of variation in cox1 gene compared to the 18S rDNA gene provides better resolution of the relationships within closely related taxa. While Amin et al. (2019a) presented relationships in their analysis of Rhadinorhynchus based on cox1 sequences, it was not clearer than 18S rDNA due to the lack of sufficient sequences of this gene in GenBank.
One of the most commonly used molecular markers for classification of acanthocephalans is the small subunit from RNA ribosomal gene or 18S rRNA. This gene displays a slow evolution rate and is highly conserved. It was used to infer phylogenetic relationships among the major classes of Acanthocephala (García-Varela & Pérez-Ponce de León, 2015). Most of phylogenetic studies of acanthocephalans similar to this research showed that 18S rDNA sequences appear to be suitable marker for phylogenies among acanthocephalans (García-Varela et al., 2000; Near, 2002; Herlyn et al., 2003; Verweyen et al., 2011; Amin et al., 2019b). Also, cox1 gene is commonly used for phylogenetic studies and to recognize and establish species limits in acanthocephalans (Guillen-Hernández et al., 2008; Alcántar-Escalera et al., 2013; García-Varela, et al., 2013). The present study confirmed that this gene has high genetic diversity among genera of the family and other families of Echinorhynchida which would be more particularly useful for phylogenetic analysis.
Finally, the genetic data collected in the current study provide a better understanding of the taxonomic status of P. longcementglandatus. Sequence variations within the family Transvenidae and among other families of Echinorhynchida based on cox1 gene is higher than18S rDNA that can be useful for achieving a proper assessment of biodiversity. More sequence data from other geographical isolates using more gene targets will be useful for exploring the phylogenetic relationships among species. On the other hand, using of molecular tools for identification of acanthocephalan species is still scarce due to the lack of sequences of different genera of acanthocephalans in GenBank (Amin et al., 2013; Salgado-Maldonado, 2013; Weaver & Smales, 2013; Amin et al., 2014; Smales, 2014; Gomes et al., 2015; Steinauer & Nickol, 2015). More molecular studies are recommended in order to elucidate acanthocephalans classification.
Acknowledgements
This project was supported by the Department of Biology, Brigham Young University (BYU), Provo, Utah and by an Institutional Grant from the Parasitology Center, Inc. (PCI), Scottsdale, Arizona.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Alcántar-Escalera F., GarcÍ-Varela M., E. Vázquez-Domínguez E., G. Pérez-Ponce De León G.. Using DNA barcoding to link cystacanths and adults of the acanthocephalan Polymorphus brevis in central Mexico. Mol. Ecol. Res. 2013;13(6):1116–1124. doi: 10.1111/1755-0998.12090. [DOI] [PubMed] [Google Scholar]
- Amin O.M., Gholami Z., Akhlaghi M., Heckmann R.A.. The description and host-parasite relationships of a new quadrigyrid species (Acanthocephala) from the Persian tooth-carp, Aphanius farsicus (Actinoptreygii: Cyprinodontidae) in Iran. J. Parasitol. 2013;99(2):257–264. doi: 10.1645/GE-3247.1. [DOI] [PubMed] [Google Scholar]
- Amin O.M., Heckmann R.A., Ali A.H.. The finding of pacific transvenid acanthocephalan in the Arabian Gulf, with the description of Paratrajectura longcementglandatus n. gen., n. sp. from perciform fishes and emendation of Transvenidae. J. Parasitol. 2018;104(1):39–51. doi: 10.1645/17-84. [DOI] [PubMed] [Google Scholar]
- Amin O.M., R. A. Heckmann R.A., Dallarés S., Constenla M., Ha N.V.. Morphological and molecular description of Rhadinorhynchus laterospinosus Amin, Heckmann & Ha, 2011 (Acanthocephala, Rhadinorhynchidae) from marine fish off the Pacific coast of Vietnam. Parasite. 2019a;26:14. doi: 10.1051/parasite/2019015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin O.M., Heckmann R.A., Ha N.V.. Acanthocephalans from fishes and amphibians in Vietnam, with descriptions of five new species. Parasite. 2014;21:53. doi: 10.1051/parasite/2014052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin O.M., Heckmann R.A., Sharifdini M., Albayati N.Y.. Moniliformis cryptosaudi n. sp. (Acanthocephala: Moniliformidae) from the long-eared hedgehog Hemiechinus auritus (Gmelin) (Erinaceidae) in Iraq; A case of incipient cryptic speciation related to M. saudi in Saudi Arabia. Acta Parasitol. 2019b;64(1):195–204. doi: 10.2478/s11686-018-00021-9. [DOI] [PubMed] [Google Scholar]
- Barton D.P., Smales L., Morgan T.J.A.. A redescription of Serrasentis sagittifer (Rhadinorhynchidae: Serrasentinae) from Rachycentron canadum (Rachycentridae) with Comments on its biology and its relationship to other species of Serrasentis. J. Parasitol. 2018;104(2):117–132. doi: 10.1645/17-94. [DOI] [PubMed] [Google Scholar]
- Benesh D.P., Hasu T., Suomalainen L.R., Valtonen E.T., Tiirola M.. Reliability of mitochondrial DNA in an acanthocephalan: the problem of pseudogenes. Int. J. Parasitol. 2006;36(2):247–254. doi: 10.1016/j.ijpara.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Braicovich P.E., Lanfranchi A.L., Farber M.D., Marvaldi A.E., Luque J.L., Timi J.T.. Genetic and morphological evidence reveals the existence of a new family, genus and species of Echinorhynchida (Acanthocephala) Folia Parasitol. 2014;61(4):377–384. [PubMed] [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R.. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3(5):294–299. [PubMed] [Google Scholar]
- Garfías-Varela M., Cumings M.P., Pérez-Ponce De León G., Gardner S.L., Laclette J.P.. Phylogenetic analysis based on 18S ribosomal RNA gene sequences supports the existence of class Polyacanthocephala (Acanthocephala) Mol. Phylogenet. Evol. 2002;23(2):288–292. doi: 10.1016/S1055-7903(02)00020-9. [DOI] [PubMed] [Google Scholar]
- Garfías-Varela M., Pérez-Ponce De León G. Morand S., Krasnov B., Littlewood D. Parasite Diversity and Diversification: Evolutionary Ecology Meets Phylogenetics. Cambridge: Cambridge University Press; 2015. Advances in the classification of acanthocephalans: Evolutionary history and evolution of the parasitism; pp. 182–201. [DOI] [Google Scholar]
- Garfías-Varela M., Pérez-Ponce De León G., Aznar F.J., Nadler S.A.. Phylogenetic relationship among genera of Polymorphidae (Acanthocephala), inferred from nuclear and mitochondrial gene sequences. Mol. Phylogenet. Evol. 2013;68(2):176–184. doi: 10.1016/j.ympev.2013.03.029. [DOI] [PubMed] [Google Scholar]
- Garfías-Varela M., Pérez-Ponce De León G., De La Torre P., Cummings M.P., Sarma S., Laclette J.P.. Phylogenetic relationships of Acanthocephala based on analysis of 18S ribosomal RNA gene sequences. J. Mol. Evol. 2000;50(6):532–540. doi: 10.1007/s002390010056. [DOI] [PubMed] [Google Scholar]
- Garfías-Varela M., Pinacho-Pinacho C. D.. Uribe A. L. S. Mendoza-Garfías B.. First record of the intermediate host of Pseudocorynosoma constrictum Van Cleave, 1918 (Acanthocephala: Polymorphidae) in Central Mexico. Comp. Parasitol. 2013;80(2):171–179. doi: 10.1654/4612.1. [DOI] [Google Scholar]
- Garfías-Varela M., Nadler S.A.. Phylogenetic relationships of Palaeacantocephala (Acanthocephala) inferred from SSU and LSU rDNA gene sequences. J. Parasitol. 2005;91(6):1401–1409. doi: 10.1645/GE-523R.1. [DOI] [PubMed] [Google Scholar]
- Garfías-Varela M., Nadler S.A.. Phylogenetic relationships among Syndermata inferred from nuclear and mitochondrial gene sequences. Mol. Phylogenetics Evol. 2006;40(1):61–72. doi: 10.1016/j.ympev.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Garfías-Varela M., Mendoza-Garfias B., Choudhury A., Pérez-Ponce De León G.. Morphological and molecular data for a new species of Pomphorhynchus Monticelli, 1905 (Acanthoce-phala: Pomphorhynchidae) in the Mexican redhorse Moxostoma austrinum Bean (Cypriniformes: Catostomidae) in central Mexico. Syst. Parasitol. 2017;94(9):989–1006. doi: 10.1007/s11230-017-9756-y. [DOI] [PubMed] [Google Scholar]
- Giribet G., Sørensen M.V., Funch P., Kristensen R.M., Sterrer W.. Investigations into the phylogenetic position of Micrognathozoa using four molecular loci. Cladistics. 2004;20(1):1–13. doi: 10.1111/j.1096-0031.2004.00004.x. [DOI] [PubMed] [Google Scholar]
- Gomes A.P.N., Olifiers N., Souza J.G., Barbosa H.S., D’andrea P.S., Maldonado A.. A new acanthocephalan species (Archiacanthocephala: Oligacanthorhynchidae) from the crab-eating fox Cerdocyon thous in the Brazilian pantanal wetlands. J. Parasitol. 2015;101(1):74–80. doi: 10.1645/13-321.1. [DOI] [PubMed] [Google Scholar]
- Guillén-Hernández S., Garfías-Varela M., Pérez-Ponce De León G.. First record of Hexaglandula corynosoma (Travassos, 1915) Petrochenko, 1958 (Acanthocephala: Polymorphidae) in intermediate and definitive hosts in Mexico. Zootaxa. 2008;1873(1873):61–68. doi: 10.5281/zenodo.184057. [DOI] [Google Scholar]
- Herlyn H., Piskurek O., Schmitz J., Ehlers U., Zischler H.. The syndermatan phylogeny and the evolution of acanthocephalan endoparasitism as inferred from 18S rDNA sequences. Mol. Phylogenet. Evol. 2003;26(1):155–164. doi: 10.1016/S1055-7903(02)00309-3. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R, Mcgettigan, P.A., Mcwilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. Vol. 23. Oxford, England: 2007. pp. 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li L., Chen H.-X., Amin O.M., Yang Y.. Morphologicalvariability and molecular characterization of Pomphorhynchus zhoushanensis sp. nov. (Acanthocephala: Pomphorhynchidae), with comments on the systematic status of Pomphorhynchus Monticelli, 1905. Parasitol. Int. 2017;66(5):693–698. doi: 10.1016/j.parint.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Li L., Chen H.-X., Yang Y.. Morphological and molecular study of Neorhadinorhynchus nudus (Harada, 1938) (Acanthocephala: Cavisomidae) from Auxis thazard Lacepede (Perciformes: Scombridae) in the South China Sea. Acta Parasitol. 2018;63(3):479–485. doi: 10.1515/ap-2018-0057. [DOI] [PubMed] [Google Scholar]
- Lisitsyna O.I., Kudlai O., Cribb T.H., Smit N.J.. Three new species of acanthocephalans (Palaeacanthocephala) from marine fishes collected off the East Coast of South Africa. Folia Parasitol. 2019;66:012. doi: 10.14411/fp.2019.012. [DOI] [PubMed] [Google Scholar]
- Nakao M.. Pseudoacanthocephalus toshimai sp. nov. (Palaeacanthocephala: Echinorhynchidae), a common acanthocephalan of anuran and urodelan amphibians in Hokkaido, Japan, with a finding of its intermediate host. Parasitol. Int. 2016;65(4):323–332. doi: 10.1016/j.parint.2016.03.011. [DOI] [PubMed] [Google Scholar]
- Near T.J.. Acanthocephalan phylogeny and the evolution of parasitism. Integr. Comp. Biol. 2002;42(3):668–677. doi: 10.1093/icb/42.3.668. [DOI] [PubMed] [Google Scholar]
- Near J.T., Garey J.R., Nadler S.A.. Phylogenetic relationships of the acanthocephala inferred from 18S ribosomal DNA sequences. Mol. Phylogenet. Evol. 1998;10(3):287–298. doi: 10.1006/mpev.1998.0569. [DOI] [PubMed] [Google Scholar]
- Perrot-Minnot M.-J.. Larval morphology, genetic divergence, and contrasting levels of host manipulation between forms of Pomphorhynchus laevis (Acanthocephala) Int. J. Parasitol. 2004;34(1):45–54. doi: 10.1016/j.ijpara.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Pichelin S., Cribb T.H.. The status of the Diplosentidae (Acanthocephala: Palaeacanthocephala) and a new family of acanthocephalans from Australian wrasses (Pisces: Labridae) Folia Parasitol. 2001;48:289–303. doi: 10.14411/fp.2001.047. [DOI] [PubMed] [Google Scholar]
- Pinacho-Pinacho C. D., Pérez-Ponce De León G., Garfías-Varela M.. Description of a new species of Neoechinorhynchus (Acanthocephala: Neoechinorhynchidae) a parasite of Dormitator latifrons from Southwestern Mexico based on morphological and molecular characters. Parasitol. Int. 2012;61(4):634–644. doi: 10.1016/j.parint.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Pinacho-Pinacho C. D., Sereno-Uribe A. L., Garfías-Varela M.. Morphological and molecular data reveal a new species of Neoechinorhynchus (Acanthocephala: Neoechinorhynchidae) from Dormitator maculatus in the Gulf of Mexico. Parasitol. Int. 2014;63(6):763–771. doi: 10.1016/j.parint.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Salgado-Maldonado G.. Redescription of Neoechinorhynchus Neoechinorhynchus golvani Salgado-Maldonado, 1978 (Acanthocephala: Neoechinorhynchidae) and description of a new species from freshwater cichlids (Teleostei: Cichlidae) in Mexico. Parasitol. Res. 2013;112(5):1891–1901. doi: 10.1007/s00436-013-3374-7. [DOI] [PubMed] [Google Scholar]
- Smales L.. The genus Rhadinorhynchus (Acanthocephala: Rhadinorhynchidae) from marine fish in Australia with the description of four new species. Acta Parasitol. 2014;59(4):721–736. doi: 10.2478/s11686-014-0305-4. [DOI] [PubMed] [Google Scholar]
- Steinauer M.L., Garcia-Vedrenne A.E., Weinstein S.B., Kuris A.M.. Acanthocephalan parasites of the oarfish, Regalecus russelii (Regalecidae), with a description of a new species of Gymnorhadinorhynchus (Acanthocephala: Gymnorhadinorhynchidae) J. Parasitol. 2019;105(1):124–132. [PubMed] [Google Scholar]
- Steinauer M.L., Nickol B.B.. Revision of Leptorhynchoides thecatus (Acanthocephala: Illiosentidae), with morphometric analysis and description of six new species. J. Parasitol. 2015;101(2):193–212. doi: 10.1645/14-644.1. [DOI] [PubMed] [Google Scholar]
- Vardić Smrzlić I., Damir V., Damir K., Zrinka D., Emil G., Helena C., Emin T.. Molecular characterisation and infection dynamics of Dentitruncus truttae from trout Salmo trutta and Oncorhynchus mykiss in Krka River, Croatia. Vet. Parasitol. 2013;197(3 – 4):604–613. doi: 10.1016/j.vetpar.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Verweyen L., Klimpel S., Palm H.W.. Molecular phylogeny of the Acanthocephala (class Palaeacanthocephala) with a paraphyletic assemblage of the orders Polymorphida and Echinorhynchida. PLOS One. 2011;6:e28285. doi: 10.1371/journal.pone.0028285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayland M.T., Vainio J.K., Gibson D.I., Herniou E.A., Littlewood D.T.J., Väinölä R.. The systematics of Echinorhynchus Zoega in Müller, 1776 (Acanthocephala, Echinorhynchidae) elucidated by nuclear and mitochondrial sequence data from eight European taxa. ZooKeys. 2015;484:25–52. doi: 10.3897/zookeys.484.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weave r H. J., Smales L.R.. Filisoma filiformis n. sp.(Echinorhynchida: Cavisomidae), a new species of Acanthocephala from Kyphosus spp. (Perciformes: Kyphosidae) from the South Pacific, and a key to the genus Filisoma. Comp. Parasitol. 2013;80(1):33–39. doi: 10.1654/4571.1. [DOI] [Google Scholar]
- Westram A.M., Baumgartner C., Keller I., Jokela J.. Are cryptic host species also cryptic to parasites? Host specificity and geographical distribution of acanthocephalan parasites infecting freshwater. Gammarus. Infect. Genet. Evol. 2011;11(5):1083–1090. doi: 10.1016/j.meegid.2011.03.024. [DOI] [PubMed] [Google Scholar]