Amyotrophic lateral sclerosis (ALS) is a fatal motor neuron degenerative disease. TDP-43 (TAR DNA-binding protein 43) and FUS (fused in sarcoma) are aggregation-prone RNA-binding proteins that in ALS can mislocalize to the cytoplasm of affected motor neuron cells, often forming cytoplasmic aggregates in the process. Such mislocalization and aggregation are implicated in ALS pathology, though the mechanism(s) of TDP-43 and FUS cytoplasmic toxicity remains unclear.
KEYWORDS: Cdc48, VCP, endocytosis, TDP-43, FUS, ALS, FTLD, autophagy
ABSTRACT
Amyotrophic lateral sclerosis (ALS) is a fatal motor neuron degenerative disease. TDP-43 (TAR DNA-binding protein 43) and FUS (fused in sarcoma) are aggregation-prone RNA-binding proteins that in ALS can mislocalize to the cytoplasm of affected motor neuron cells, often forming cytoplasmic aggregates in the process. Such mislocalization and aggregation are implicated in ALS pathology, though the mechanism(s) of TDP-43 and FUS cytoplasmic toxicity remains unclear. Recently, we determined that the endocytic function aids the turnover (i.e., protein degradation) of TDP-43 and reduces TDP-43 toxicity. Here, we identified that Cdc48 and Ubx3, a Cdc48 cofactor implicated in endocytic function, regulates the turnover and toxicity of TDP-43 and FUS expressed in Saccharomyces cerevisiae. Cdc48 physically interacts and colocalizes with TDP-43, as does VCP, in ALS patient tissue. In yeast, FUS toxicity also depends strongly on endocytic function but not on autophagy under normal conditions. FUS expression also impairs endocytic function, as previously observed with TDP-43. Taken together, our data identify a role for Cdc48/VCP and endocytic function in regulating TDP-43 and FUS toxicity and turnover. Furthermore, endocytic dysfunction may be a common defect affecting the cytoplasmic clearance of ALS aggregation-prone proteins and may represent a novel therapeutic target of promise.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disease characterized by progressive motor neuron degeneration, muscle weakness, and fatal paralysis due to respiratory failure, which typically occurs 3 to 5 years after diagnosis. ALS is mostly a sporadic disease, with ≤10% of cases exhibiting a hereditary component (1–3). Many ALS-linked mutations affect proteins involved in RNA metabolism or protein clearance pathways (2), though a mechanistic understanding of the disease pathology remains unclear, and no effective treatment for ALS currently exists.
In ≥97% of ALS patients, a classic motor neuron hallmark of disease is relocalization from the nucleus to the cytoplasm of the RNA binding protein TDP-43 (TAR DNA-binding protein 43), which also often forms ubiquitinated cytoplasmic protein aggregates (inclusion bodies) in motor neuron cells. In approximately 1% of familial ALS patients lacking a TDP-43 pathology, a similar behavior is exhibited by FUS (fused in sarcoma), another RNA binding protein (2). The cytoplasmic mislocalization and aggregation of FUS are more commonly observed in another related neurodegenerative disease, frontotemporal lobar dementia (FTLD), where about 10% of patients exhibit FUS aggregates. The remaining 90% of FTLD patients exhibit aggregates of either TDP-43 (45%) or the microtubule-associated protein Tau (45%) (2). TDP-43 and FUS possess similar domains, including RNA recognition motifs (RRMs), glycine-rich domains, and/or prion-like domains, and have been proposed to affect multiple steps of mRNA metabolism (4, 5).
Under conditions of cellular stress, both TDP-43 and FUS relocalize into cytoplasmic stress granules (SGs) (6, 7), which are dynamic mRNA-protein assemblies implicated in regulating mRNA function (8, 9). Given this and in light of the fact that ALS-associated mutations generally reduce SG dynamics, SGs have been theorized to promote the formation of the TDP-43 and FUS aggregates that are observed in ALS patients (10, 11), though recent studies suggest that SG-independent aggregation mechanisms also likely exist (12–14). Regardless, various studies indicate that TDP-43 and FUS aggregates or simply the excessive cytoplasmic localization of TDP-43 or FUS may result in a toxic gain of function that leads to motor neuron degeneration (15–21). While the mechanism of such toxicity remains unclear, preventing aggregation and/or enhancing the clearance of cytoplasmic TDP-43 and FUS is of considerable therapeutic interest. This approach has shown promise in various ALS models, in which the prevention of SG assembly or the upregulation of cytoplasmic protein turnover (i.e., degradation) has been tested (22–26).
Recently, we and others demonstrated that under normal growth conditions, TDP-43 toxicity, aggregation, and protein turnover in Saccharomyces cerevisiae and human cell line models depend strongly on endocytic function (26, 27); such effects were largely independent of autophagic or proteasomal contributions. Defects in endocytic function increased TDP-43 protein levels, aggregation, and toxicity, whereas genetic enhancement of endocytosis rates suppressed these phenotypes. Defects in endocytic function also exacerbated motor neuron dysfunction in a TDP-43 fly model of ALS, whereas endocytic enhancement suppressed such dysfunction (26). Finally, expression of cytoplasmic TDP-43, particularly aggregation-prone forms in human cells, correlated with impairment of the endocytosis function itself (26). Key questions remain and include the following: how is endocytosis-dependent TDP-43 clearance (which needs to be better defined) achieved, how does cytoplasmic TDP-43 inhibit endocytosis, and is endocytic inhibition observed with other genetic ALS models?
Cdc48 and its human homolog, valosin-containing protein (VCP; p97), are type II AAA+ ATPase chaperones that act as segregases of ubiquitinated proteins. Cdc48/VCP removes ubiquitinated proteins from complexes or membranes and often (though not always) aids the targeting of said proteins for degradation by autophagic or proteasomal means (28, 29). Cdc48/VCP functions in many diverse cellular processes, including the DNA damage response, cell cycle control, autophagy, proteolytic turnover, endocytosis, and SG clearance (29–33). Specificity for Cdc48/VCP function is typically derived from interactions with various cofactors that help recruit Cdc48/VCP to distinct ubiquitinated substrates (28, 29).
VCP is mutated in a small fraction of ALS patients (1% of patients with sporadic and familial ALS) and in most patients with inclusion body myopathy with early-onset Paget’s disease and frontotemporal dementia (IBMPFD) (34, 35). As in ALS patients, patients with IBMPFD also commonly exhibit cytoplasmic TDP-43 aggregates in affected cells (e.g., muscle cells, frontal cortex neurons) (34, 35). Additionally, the neurodegenerative effects of VCP mutants in flies depend upon TDP-43 cytoplasmic aggregation (36). Finally, disease alleles of VCP lead to the accumulation of cytoplasmic TDP-43 aggregates in human cells (32) and mouse models (37). Given these observations, we were curious to define the link between Cdc48/VCP function and the accumulation of pathological protein aggregates in ALS, FTLD, and IBMPFD, focusing on whether Cdc48/VCP aids TDP-43 and FUS turnover and, if so, by what mechanism.
Several studies have shown strong links between cytoplasmic TDP-43 and FUS protein localization and aggregation and neuron loss (15, 16, 38). However, the mechanism by which cells clear these apparently toxic cytoplasmic aggregates is still unclear. In this report, we demonstrate that, like cytoplasmic TDP-43 expression, cytoplasmic FUS expression impairs endocytosis rates in yeast. Interestingly, inhibition of Cdc48 and Ubx3 (a Cdc48 endocytosis-promoting cofactor) leads to defects in FUS and TDP-43 turnover and increased toxicity, while inhibition of VCP causes the accumulation of the TDP-43 and FUS proteins and increases TDP-43 cytoplasmic aggregation. Additionally, Cdc48 physically interacts with and colocalizes with TDP-43. Finally, TDP-43 cytoplasmic aggregates colocalize with VCP in ALS patient tissue. Taken together, these data suggest a role for Cdc48/VCP-facilitated endocytosis in regulating TDP-43 and FUS turnover and thus toxicity. This suggests that endocytic dysfunction may represent a novel therapeutic target for numerous devastating neurodegenerative diseases characterized by TDP-43 and FUS cytoplasmic pathology.
RESULTS
Cdc48 colocalizes with and mediates TDP-43 degradation and toxicity.
VCP disease alleles are associated with cytoplasmic relocalization and the aggregation of TDP-43 (32, 36, 37), and mutant VCP toxicity in a fly model of ALS depends upon the cytoplasmic accumulation of TDP-43 (36). However, the mechanistic details of why VCP impairment affects cytoplasmic TDP-43 accumulation remain unclear. We began to address this using an established yeast TDP-43 model (18), focusing on interactions with the yeast VCP homolog, Cdc48.
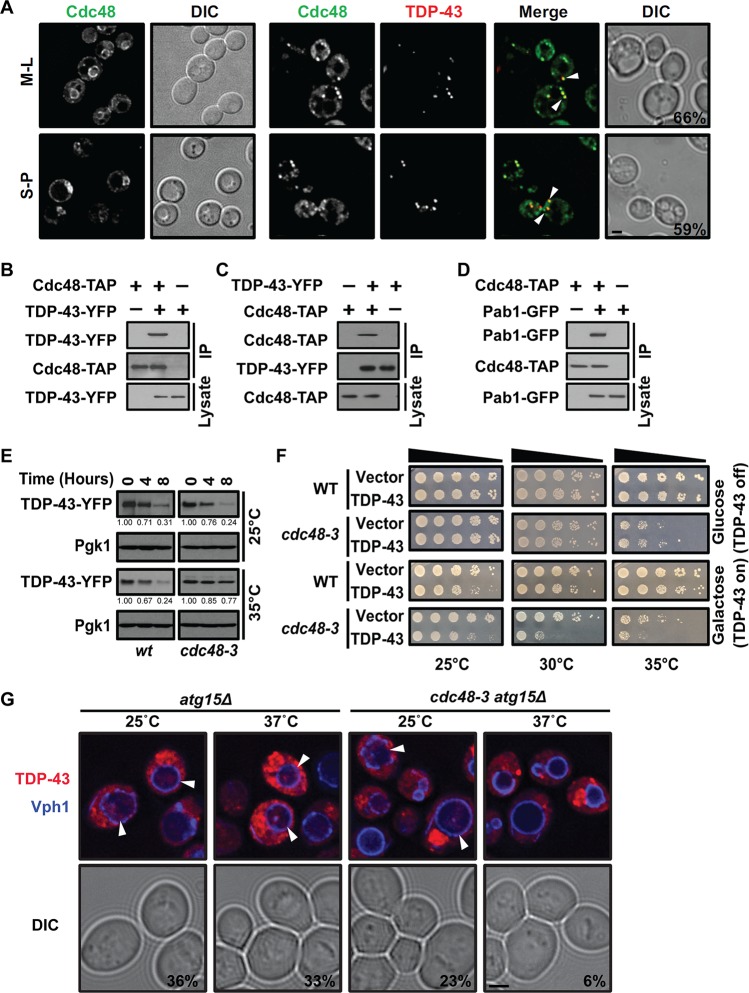
First, the localization of TDP-43 and Cdc48 was examined. Interestingly, expression of TDP-43 induced the relocalization of Cdc48 from a predominantly nuclear region (mostly the perinuclear region in mid-log-phase cells) into cytoplasmic foci that exhibited approximately 60% colocalization with TDP-43 foci under mid-log-phase and stationary-phase conditions (Fig. 1A). Consistent with colocalization in foci, reciprocal immunoprecipitation of tandem affinity purification (TAP)-tagged Cdc48 and yellow fluorescent protein (YFP)-tagged TDP-43 revealed a robust interaction between Cdc48 and TDP-43 (Fig. 1B and C); this is consistent with prior VCP-TDP-43 coimmunoprecipitation data from human cell culture and brain tissue (39). In yeast, TDP-43 cytoplasmic foci colocalize with SGs, as has been previously shown (40), and we also confirmed this with various SG marker proteins (data not shown). Consistent with this, Cdc48-TAP immunoprecipitation also revealed a robust interaction with Pab1 (a core SG component)-tagged green fluorescent protein (GFP) (Fig. 1D). Taken together, our data suggest that Cdc48 colocalizes with and physically interacts with TDP-43 and Pab1, likely at least partially within an SG context.
FIG 1.
Cdc48 interacts with TDP-43 and regulates TDP-43 turnover and toxicity. (A) A strain with endogenously expressed Cdc48-GFP alone (left) and a strain with Cdc48-GFP transformed with a plasmid expressing TDP-43–mRuby2 (right) were imaged in both mid-log phase (M-L) and stationary phase (S-P). Arrowheads and percentages indicate Cdc48 and TDP-43 colocalization. Bar, 2 μm. (B) A strain with endogenously expressed Cdc48-TAP was transformed with an empty vector or a plasmid expressing TDP-43–YFP, and then TAP immunoprecipitation was performed. (C) As described in the legend to panel B but with GFP immunoprecipitation, i.e., reciprocal pulldown. (D) A strain with Cdc48-TAP and untagged TDP-43 was transformed with an empty vector or Pab1-GFP, and the interaction was determined. (E) WT and cdc48-3 mutant strains were treated with 0.2 mg/ml cycloheximide (CHX) for the indicated amounts of time at either 25°C or 35°C. TDP-43 protein levels were detected and normalized to the level of the Pgk1 loading control, as indicated by the values beneath the gels. (F) Serial dilution growth assay of WT and cdc48-3 mutant strains transformed with the empty vector or a plasmid expressing GAL1-regulated TDP-43–YFP at different temperatures. (G) atg15Δ and atg15Δ cdc48-3 strains were transformed with TDP-43–mCherry and Vph1-BFP (to highlight the vacuolar membrane). The percentages quantify the vacuolar accumulation of the TDP-43–mCherry signal in vacuoles, with arrowheads indicating examples. Bar, 2 μm. DIC, differential interference contrast; IP, immunoprecipitation.
Next, we tested whether Cdc48 affects TDP-43 stability, given the many roles that Cdc48 plays in protein turnover. Since Cdc48 is an essential gene, we turned to a commonly used temperature-sensitive (ts) mutant, the cdc48-3 mutant, in which the protein encoded by cdc48-3 possesses two missense point mutations, P257L and R387K, that inhibit Cdc48’s ATPase activity (41). The protein degradation rate was measured by blocking new protein synthesis. At the permissive temperature of 25°C, no change in TDP-43 turnover rate was observed in wild-type (WT) or cdc48-3 mutant strains. However, at 35°C, which strongly impairs Cdc48-3 function, TDP-43 turnover rates were significantly decreased in the mutant strain relative to those in the WT (Fig. 1E). TDP-43 toxicity, evidenced by reduced cell growth by TDP-43 transformants relative to that by control strains transformed with the empty vector, was also exacerbated by the inactivation of Cdc48 (Fig. 1F), consistent with the findings of a previous study (42). Specifically, this was evident at temperatures that modestly (30°C) and strongly (35°C) inhibit Cdc48-3 function. Finally, we examined the effect of inactivating Cdc48 function on TDP-43 vacuolar targeting by utilizing an atg15Δ mutant background. Atg15 is a vacuolar lipase whose absence facilitates the detection of substrates targeted to vacuoles by vesicular trafficking means (43, 44). We also switched to the use of an mCherry-tagged form of TDP-43 to avoid the loss of YFP fluorescence in the acidic vacuolar compartment and utilized blue fluorescent protein (BFP)-tagged Vph1 (a vacuolar ATPase subunit) as a marker of vacuolar membranes. Importantly, while robust targeting of TDP-43–mCherry to the vacuole was observed under high-optical-density (OD) growth conditions in the atg15Δ strain both at 25°C and at 37°C, less targeting was visible in the atg15Δ cdc48-3 strain, even at the permissive temperature of 25°C and particularly at the nonpermissive temperature of 37°C (Fig. 1G). Together, these data demonstrate that Cdc48 regulates TDP-43 protein stability, toxicity, and vacuolar targeting in yeast.
Cdc48 may regulate TDP-43 toxicity in part by an endocytosis-related mechanism.
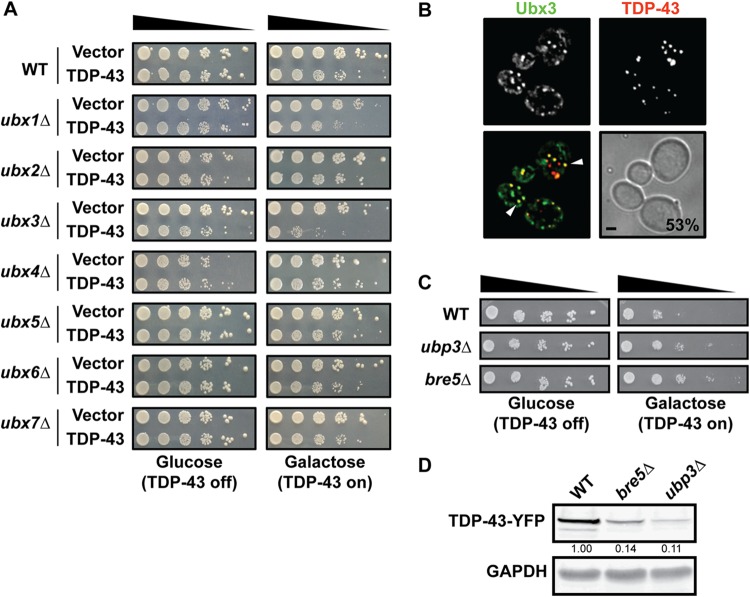
The Cdc48 specificity of function is largely determined by the use of cofactor proteins that help recruit it to its targets. In yeast, members of the Ubx family are well-described examples of such proteins (45). Thus, we first examined TDP-43 toxicity in a range of Ubx-null strains (Fig. 2A). Here and in all other spotting assays, we performed induction with 0.25% galactose rather than the more standard 2% concentration, as this results in a more modest toxicity, related to lower protein expression levels (data not shown); this aids in the identification of mutants enhancing TDP-43 (or FUS) toxicity. Importantly, only deletion of UBX3, which functions along with Cdc48 in yeast endocytosis (46), clearly exhibited enhanced TDP-43 toxicity (Fig. 2A). No clear effect on TDP-43 toxicity was observed with any other Ubx mutants, including those with a deletion of UBX1 (SHP1) or UBX2, each of which facilitates Cdc48 function in autophagy and proteasomal turnover (47, 48) (Fig. 2A). These findings are consistent with those of our prior work that endocytosis plays a greater role than autophagy in regulating TDP-43 toxicity and turnover in yeast during normal growth (26). Finally, like Cdc48, Ubx3 foci localized with TDP-43 foci at a frequency of 53% (Fig. 2B).
FIG 2.
Cdc48 cofactors Ubx3, Bre5, and Ubp3 affect TDP-43 toxicity. (A) Growth assay of WT and indicated isogenic null mutant strains transformed with an empty vector or a plasmid expressing GAL1-regulated TDP-43–YFP. (B) A strain with Ubx3-GFP was transformed with a plasmid expressing TDP-43–mRuby2, and images were taken at mid-log phase. Percentages and arrowheads indicate Ubx3 and TDP-43 colocalization. Bar, 2 μm. (C) As described for panel A with the indicated null strains. (D) Western blot of steady-state TDP-43–YFP levels in the indicated strains; the quantities beneath the gel, determined by normalization to the level for the GAPDH loading control, are based on 3 biological replicates.
We also examined additional gene deletion strains whose genes either are known Cdc48 cofactors or have been linked to Cdc48 function (49). Notably, deletion of UBP3 and BRE5 suppressed TDP-43 toxicity (Fig. 2C). Ubp3 is a deubiquitinase whose activity depends on the cofactor Bre5, which also harbors an RNA binding domain. Deubiquitinases play many roles in ubiquitin metabolism and are generally thought to stabilize proteins by antagonizing ubiquitin-mediated turnover (50). We thus examined TDP-43 steady-state protein levels in the ubp3Δ and bre5Δ strains and saw a striking decrease in TDP-43 abundance in both strains (Fig. 2D). In principle, TDP-43 being more highly ubiquitinated may facilitate its turnover and reduce its toxicity via multiple mechanisms, including via the proteasome and autophagy, though our previous study revealed almost no impact of blocking these pathways on TDP-43 steady-state levels (26).
Together, these findings and the Ubx3 data suggest that Cdc48 regulation of TDP-43 involves an endocytosis-promoting function.
FUS toxicity and turnover depend on endocytic function and Cdc48.
FUS shares many properties in common with TDP-43, including a propensity to form aggregates in ALS and other neurodegenerative diseases, including FTLD (2). A physical interaction between FUS and VCP has also been described (51). A genetic link between FUS and VCP, in which the knockdown of Cabeza (an FUS ortholog) in Drosophila alters Ter94 expression (VCP ortholog), has also been reported (52). However, evidence that the toxicity of FUS (when expressed at physiological or high levels) or its turnover is regulated by VCP or, indeed, by endocytosis in general has not been described. To determine if our findings with TDP-43 (26) were more generalizable to other aggregation-prone disease-associated proteins, particularly those linked to ALS, we next determined if FUS toxicity and turnover could be regulated by endocytosis and Cdc48 as well.
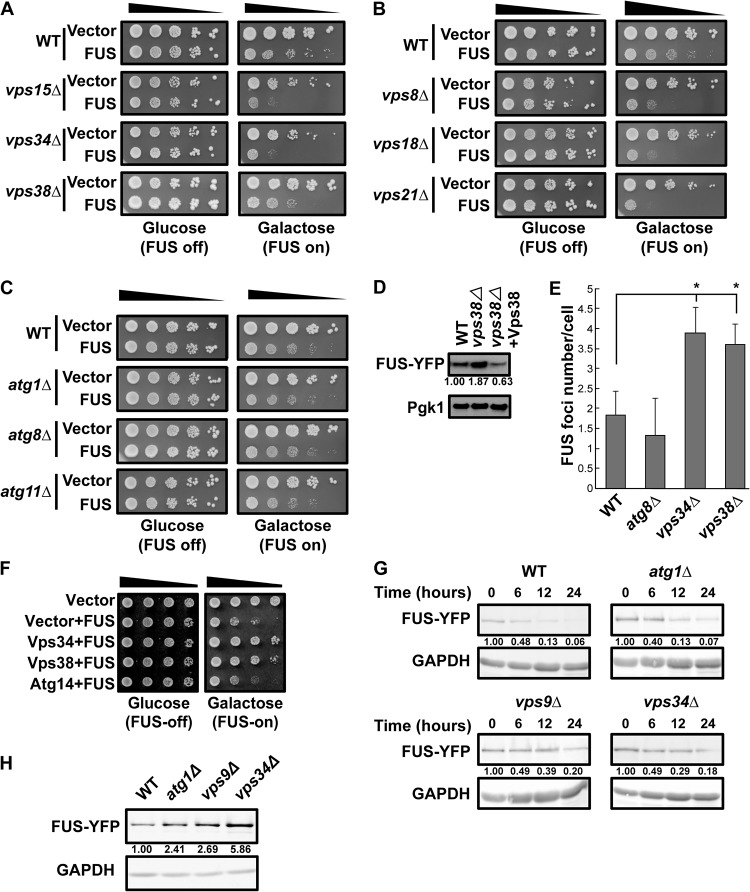
Several findings suggest that FUS protein levels and, thus, toxicity are regulated by endocytosis-dependent turnover in yeast like TDP-43 protein levels are (26). First, deletion of phosphatidylinositol 3-kinase (PI3K) complex subunit VPS15, VPS34, or VPS38, each of which impairs endocytosis, enhanced FUS toxicity (Fig. 3A), a finding similar to that of our prior work with TDP-43 (26). Additionally, deletion of several other genes that facilitate the maturation of endocytic compartments, including VPS8 (encoding the class C core vacuole/endosome tethering [CORVET] complex), VPS18 (encoding the CORVET/homotypic fusion and vacuole protein sorting [HOPS] complex), and VPS21 (encoding the Rab5 protein) (53), also exacerbated FUS toxicity in yeast (Fig. 3B). In contrast, deletion of core (ATG1 and ATG8) and selective (ATG11) autophagy genes had no effect on FUS toxicity (Fig. 3C). Second, deletion of VPS38, the PI3K complex endocytosis-specific subunit, induced FUS accumulation, which could be suppressed by Vps38 reexpression via a plasmid (Fig. 3D). Third, FUS focus formation also increased in vps34Δ and vps38Δ null strains, while showing no increase over the WT in an atg8Δ strain (Fig. 3E). Fourth, overexpression of VPS34 and VPS38, but not that of ATG14 (a PI3K complex autophagy-specific subunit gene), rescued FUS-induced cell toxicity (Fig. 3F). This partly mirrors our prior observation that Vps38 overexpression rescues TDP-43-induced cell toxicity, which correlates with increased endocytic activity (26). Finally, the FUS protein showed no stabilization in an atg1Δ strain background but showed modest stabilization in vps9Δ (Rab5 guanine exchange factor [GEF]) and vps34Δ strain backgrounds (Fig. 3G). Interestingly, FUS steady-state protein levels were strongly elevated in vps34Δ cells compared with those in WT cells (mirroring the findings presented in Fig. 3D), with smaller increases also being evident in both the vps9Δ and atg1Δ strains (Fig. 3H). Taken together, these findings suggest that in yeast, the endocytic function is a key regulator of FUS toxicity and turnover, while autophagy may also play a small role in turnover.
FIG 3.
FUS toxicity, aggregation, and turnover are regulated by endocytosis in yeast. (A to C) Serial dilution growth assay of the indicated strains expressing an empty vector or transformed with a plasmid expressing GAL1-regulated FUS-YFP. The strains for which the results are shown in panels A and B are impaired in endocytosis, while the strains for which the results are shown in panel C are impaired in autophagy. (D) Level of FUS-YFP in the indicated strains. The plasmid expressing Vps38-mRuby2 was used for rescue. (E) FUS-YFP focus formation in the indicated strains, based on 3 biological replicates. *, P < 0.05 by analysis of variance with Dunnett’s post hoc test. (F) WT cells with plasmids expressing the empty vector, the vector plus FUS-YFP, Vps34-mRuby2 and FUS-YFP, and Vps38-mRuby2 and FUS-YFP were tested by a growth assay. (G) FUS-YFP protein stability assays following GAL1 transcriptional shutoff (glucose addition). FUS-YFP levels were quantified following normalization to the level for the GAPDH loading control, with the normalized amounts being indicated beneath the gels. (H) Representative Western blot of steady-state levels of FUS-YFP in the indicated strains.
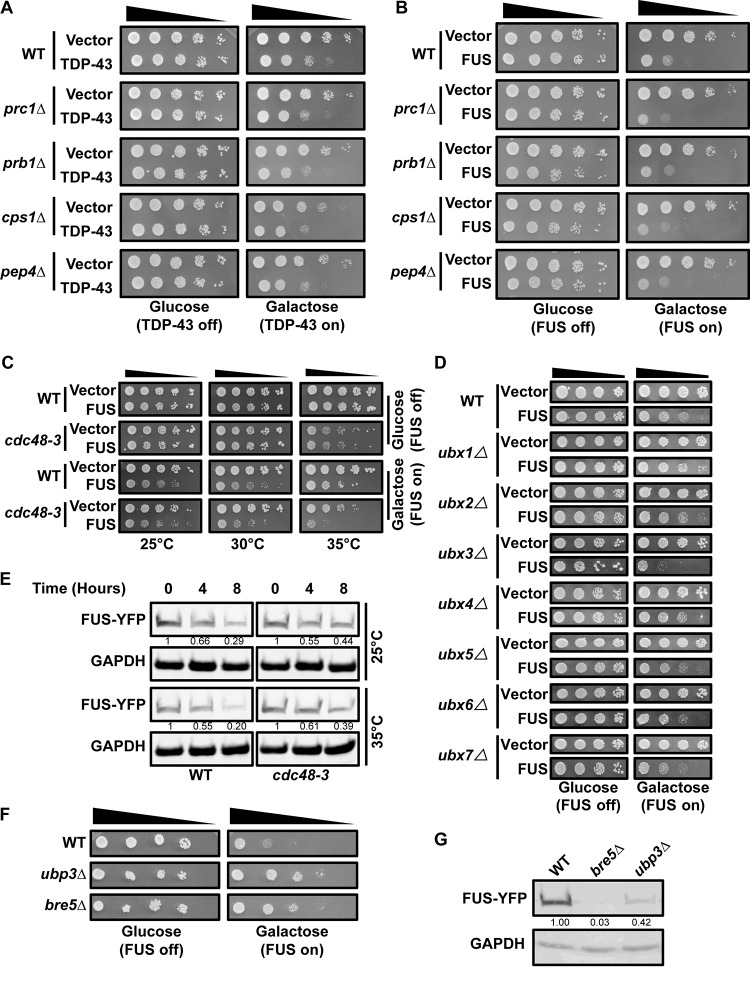
In our prior work (26), any endocytosis-related mutant that increased steady-state TDP-43 protein levels and/or increased TDP-43 stability also increased TDP-43 toxicity. Higher levels of transcriptionally driven expression of TDP-43 also increased toxicity in yeast (26). However, despite increased TDP-43 protein stability in vacuolar protease mutants (26), TPD-43 toxicity in vacuolar protease mutants was not altered from that in WT cells (Fig. 4A). We observed the same phenomenon for FUS (Fig. 4B). Thus, FUS and TDP-43 toxicity likely depends on cytoplasmic localization, and neither protein is presumably toxic if it accumulates within vacuoles.
FIG 4.
Cdc48 regulates FUS similarly to TDP-43. (A, B) Serial dilution growth assay of vacuolar protease mutant null strains expressing an empty vector or a plasmid expressing GAL1-regulated TDP-43–YFP (A) or FUS-YFP (B). (C) Serial dilution growth assay of the cdc48-3 strain at permissive (25°C), modest (30°C), and strongly inhibiting (35°C) temperatures expressing an empty vector or a plasmid expressing GAL1-regulated FUS-YFP. (D) Serial dilution growth assays with the indicated strains expressing an empty vector or a plasmid expressing GAL1-regulated FUS-YFP. (E) FUS-YFP protein stability in the WT and cdc48-3 strains following GAL1 transcriptional shutoff (glucose addition) at either 25°C or 35°C. The quantities indicated beneath the gels are based on 2 biological replicates following normalization to the level for the GAPDH loading control. (F) As described for panel D. (G) Western blot of steady-state FUS-YFP levels in the indicated strains; quantities are based on 3 biological replicates following normalization to the level for the GAPDH loading control.
We next determined whether Cdc48 plays a role in the regulation of FUS and found results similar to those observed with TDP-43. First, partial and strong inhibition of Cdc48 at 30°C and 35°C, respectively, enhanced FUS toxicity in the cdc48-3 strain, particularly at 35°C (Fig. 4C). Second, of all Ubx-null strains examined, only deletion of UBX3, which functions in endocytosis (46), clearly increased FUS toxicity (Fig. 4D). Third, the inactivation of Cdc48 at 35°C also led to a modest increase in FUS protein stability (Fig. 4E). Interestingly, as with TDP-43, the bre5Δ and ubp3Δ strains also suppressed FUS toxicity and FUS protein levels at steady state (Fig. 4F and G).
Finally, repeating our growth assays in WT and cdc48-3 strains with GAL1-driven expression of GFP alone revealed no toxic effects of GFP expression (data not shown), consistent with the prior findings of other labs (17, 42); this argues that the conclusions drawn from the findings of our growth assay with FUS-YFP and TDP-43–YFP are not artifacts.
In summary, these data suggest that FUS toxicity and turnover, like those with TDP-43, are likely regulated in part by a Cdc48-facilitated endocytic process.
The endocytosis rate is regulated by Cdc48, TDP-43, and FUS.
TDP-43 expression in yeast and human cells impairs endocytosis rates, and TDP-43 toxicity and protein accumulation are suppressed by driving endocytosis rates by VPS38 (yeast) or RAB5 (human cell) overexpression (26). Having made similar toxicity and protein level observations for FUS in yeast (Fig. 3D and F), we were curious if FUS, like TDP-43, also inhibits endocytosis. We were also curious if defects in endocytosis caused by Cdc48 inactivation using the cdc48-3 ts allele would be epistatic to, additive with, or synergistic with the effects of TDP-43 or FUS expression.
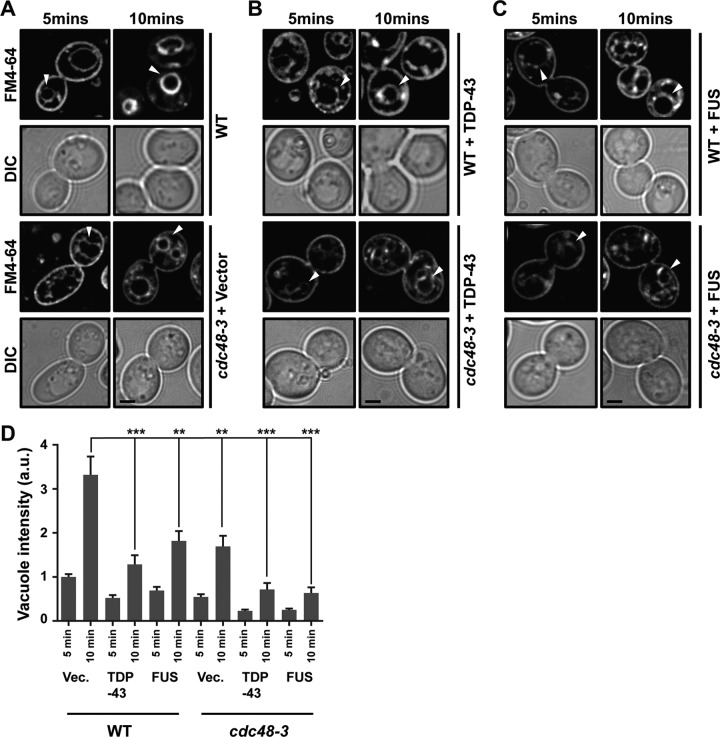
Yeast endocytosis rates were measured by following the uptake of an extracellular dye, FM4-64, which is endocytosed and ultimately stains vacuolar membranes. The faster and more intensely that vacuolar membrane staining occurs, the better that endocytosis is functioning (54). In WT cells, vacuole staining was clearly visible by 5 min and intense by 10 min (Fig. 5A and D). Like TDP-43 (26), expression of FUS also impaired endocytosis rates, evident by reduced vacuolar staining over the same time period (Fig. 5B to D). Consistent with previous findings for mammalian cells (31, 33) and yeast cells (46), inactivation of Cdc48 also impaired endocytosis rates to an extent similar to that seen with FUS expression, though the impairment was not quite as striking as that seen with TDP-43 expression (Fig. 5). Combining TDP-43 or FUS expression with Cdc48 inactivation led to an additive inhibitory effect on endocytosis that was significant only in FUS-expressing cells (Fig. 5B to D). Taken together, these results suggest that both FUS and TDP-43 inhibit endocytosis in a manner at least partly independent of the blocking of Cdc48 function.
FIG 5.
The endocytosis rate is regulated by Cdc48, TDP-43, and FUS. (A) WT and cdc48-3 cells were cultured at 35°C for 2 h and then stained with FM4-64 dye (8 μM) for the time periods indicated and examined. Arrowheads indicate vacuolar staining. The persistence of plasma membrane staining in cdc48-3 cells and reduced vacuolar membrane staining relative to that in the WT indicates an endocytic defect. (B, C) As described for panel A, except with the additional expression of TDP-43–YFP (B) or FUS-YFP (C). (A to C) Bars, 2 μm. (D) Analysis of vacuolar staining intensity of the images in panels A to C. The results are averages from two biological replicates. a.u. absorbance units; Vec., vector. P values were determined by analysis of variance with Tukey’s post hoc test (only comparisons to the WT at 10 min are shown). **, P < 0.01; ***, P < 0.001. Data are shown as the mean ± SEM.
VCP regulates endocytosis and TDP-43/FUS turnover in human cells.
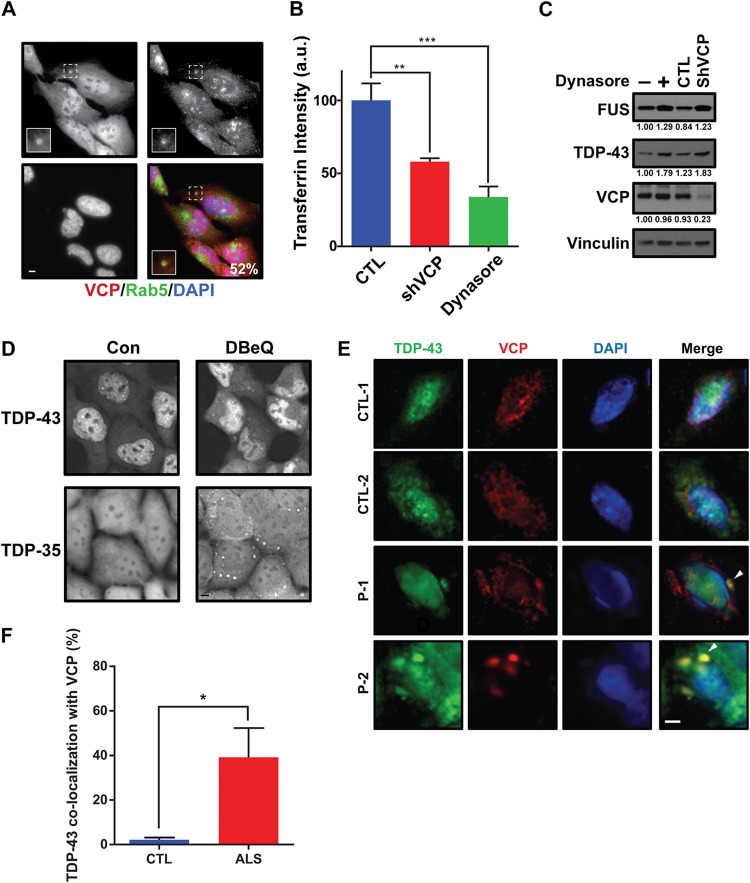
We next turned our focus to VCP in human cells. VCP is a known regulator of endocytosis, localizing to and affecting early endosome size (33), and ultimately affects endocytosis trafficking rates and endocytic substrate turnover in lysosomes (31, 33). First, we examined if endogenous VCP localized with endogenous Rab5, an early endosomal marker. U-2 OS cells most commonly showed diffuse VCP signals in the cytoplasm and nucleus, though a small percentage of cells exhibited cytoplasmic foci that colocalized with Rab5 cytoplasmic foci about 50% of the time (Fig. 6A). More extensive VCP colocalization with Rab5 was also previously observed in COS7 cell lines expressing a constitutively active Rab5 Q79L mutant that drives early endosome enlargement (33). Next, the endocytic uptake of fluorescently labeled transferrin (55) was performed to measure clathrin-dependent endocytosis rates in WT and VCP knockdown cells. Dynasore, a small-molecule inhibitor of dynamin, was used as a positive control of endocytic impairment (56). As expected, VCP knockdown cells exhibited impaired endocytosis, albeit to a degree less than that seen in Dynasore-treated cells (Fig. 6B); this finding is consistent with similar data obtained for COS7 cells using Eer1, a VCP inhibitor (33). These data confirm that VCP positively regulates endocytic function in our cell line model.
FIG 6.
VCP regulates TDP-43 and FUS in mammalian cells. (A) U-2 OS cells were immunostained for endogenous VCP and Rab5; DAPI staining indicates the nucleus; the percentage value indicates the number of VCP cytoplasmic foci colocalized with Rab5 cytoplasmic foci. (Insets) Enlarged images of the squares with dashed borders. Bar, 5 μm. (B) A transferrin uptake assay was performed in control cells (CTL) and HEK293A cells in which VCP was knocked down with VCP-specific shRNA (shVCP). Dynasore (40 μM, 24 h) was utilized as a positive control for endocytosis inhibition. P values were determined by Student’s paired two-tailed t test. **, P < 0.01; ***, P < 0.001. Data are shown as the mean ± SEM. (C) HEK293A cells either were treated with 40 μM dynasore for 24 h or expressed a stably integrated VCP-specific shRNA, followed by determination of endogenous TDP-43 and FUS protein levels; quantities are normalized to vinculin levels, with the normalized values being indicated beneath the gels. (D) HEK293A cells expressing stably integrated TDP-43–GFP or TDP-35–GFP were examined under normal growth conditions or following DBeQ treatment (10 μM, 1 h). Bar, 5 μm. Con, control. (E, F) Fluorescence immunohistochemistry of control (CTL) and ALS patient (P) frontal cortex tissue with TDP-43, VCP, and DAPI and quantification of the percentage of cytoplasmic TDP-43 foci that colocalize with VCP foci. Tissues from two control patients and two ALS patients were examined. The arrowhead indicates VCP colocalization with TDP-43. Bar, 5 μm. *, P < 0.05 by Student’s unpaired two-tailed t test. Data are shown as the mean ± SEM.
We next determined if endogenous steady-state TDP-43 and FUS levels in HEK293A cells were affected when endocytosis was inhibited. Both Dynasore treatment and knockdown of VCP increased steady-state TDP-43 and FUS levels, mirroring findings in yeast (Fig. 6C). Using HEK293A cell lines with lentivirus-integrated TDP-43–GFP (26), we also examined whether VCP inactivation using the specific inhibitor N2,N4-dibenzylquinazoline-2,4-diamine (DBeQ) altered the localization or aggregation of WT TDP-43 or TDP-35 (in which TDP-35 consists of TDP-43 with an N-terminally truncated aggregation-prone allele that mislocalizes to the cytoplasm). Strikingly, in both cases, TDP-43 cytoplasmic aggregation was markedly increased (Fig. 6D); whether this reflects the loss of VCP functions in endocytosis or other proteostatic mechanisms remains unclear. Finally, using immunofluorescence, we examined VCP colocalization with TDP-43 aggregates in frontal cortex tissue from ALS disease and control patients. This revealed that VCP is significantly localized with TDP-43 aggregates in ALS tissue samples relative to its localization in control patients (Fig. 6E and F), again mirroring Cdc48 colocalization with TDP-43 in yeast.
In summary, our data suggest that the previously reported genetic interactions between VCP and TDP-43 (36) and the relocalization of TDP-43 to cytoplasmic foci in VCP mutant contexts (32, 36, 37) may reflect a role for VCP in facilitating cytoplasmic TDP-43 turnover, at least in part via an endocytic mechanism. Our yeast data suggest that Cdc48/VCP may also act similarly on cytoplasmic FUS. These findings are relevant to therapeutic development not only for ALS but also for other TDP-43/FUS proteinopathies, including FTLD and particularly IBMPFD, where VCP mutations account for most disease cases.
DISCUSSION
Several pieces of evidence suggest that the effect of Cdc48 and VCP on TDP-43 and FUS toxicity and turnover may depend on an endocytosis-promoting function. First, inactivation of Cdc48 (46) or VCP (33) or expression of either FUS or TDP-43 impairs endocytosis (Fig. 5 and 6). Second, combining the expression of either of these aggregation-prone proteins with Cdc48-3 inactivation further exacerbated endocytosis inhibition (Fig. 5) and TDP-43/FUS toxicity (Fig. 1F and 4C). Third, TDP-43 and FUS toxicity was increased in ubx3Δ strains (Fig. 2A and 4D), which promotes endocytosis (46). In contrast, deletion of all other Ubx Cdc48 cofactors, including Ubx1 and Ubx2, which function in autophagic and proteasomal turnover pathways (47, 48), had no effect on TDP-43 or FUS toxicity. Interestingly, Ubx3 also forms part of a membrane-associated E3 ubiquitin ligase complex that is found throughout the endocytic pathway but that is particularly found at vacuolar membranes, where it is implicated in the ESCRT-dependent turnover of vacuolar membrane transporter proteins in the vacuole itself (57). Fourth, TDP-43 and FUS showed no increase in cellular toxicity in yeast when autophagy was blocked but did show an increase in cellular toxicity when endocytosis was inhibited (26) (Fig. 3A to C). Finally, mutations in VCP associated with disease can specifically affect interactions with the endocytosis-promoting cofactor UBXD1 (31), though, notably, defects in autophagy have also been reported for such mutants (58). However, this may also reflect defects in endocytosis, given the interconnected nature of the endocytic and autophagic pathways in human cells (59). Taken together, these data argue that at least part of the mechanism by which Cdc48 and VCP mediate TDP-43 and FUS protein levels and their toxicity is by facilitating turnover by a mechanism dependent on endocytosis activity.
VCP and Cdc48 have been linked to several distinct aspects of endocytic function (31, 33). For example, VCP interacts with and decreases the formation of EEA1 (early endosome-associated antigen 1) oligomers. EEA1 promotes the fusion of endocytic vesicles into early endosomes; thus, the increased EEA1 oligomerization may explain the increase in early endosome size that accompanies VCP knockdown or chemical inhibition (33). VCP inhibition and disease mutations also impair endolysosomal sorting and/or the eventual turnover of membrane-associated proteins, such as epidermal growth factor receptor (EGFR) and caveolar 1 (CAV-1), which itself functions in caveola-mediated endocytosis (31, 60). A role for a VCP cofactor, UBXD1, in CAV-1 endolysosomal sorting and turnover was also identified (31). VCP also robustly interacts with clathrin in human cells (61), while in yeast, the Cdc48 cofactor Ubx3 interacts with clathrin and functions in clathrin-mediated endocytosis, as does Cdc48 (46).
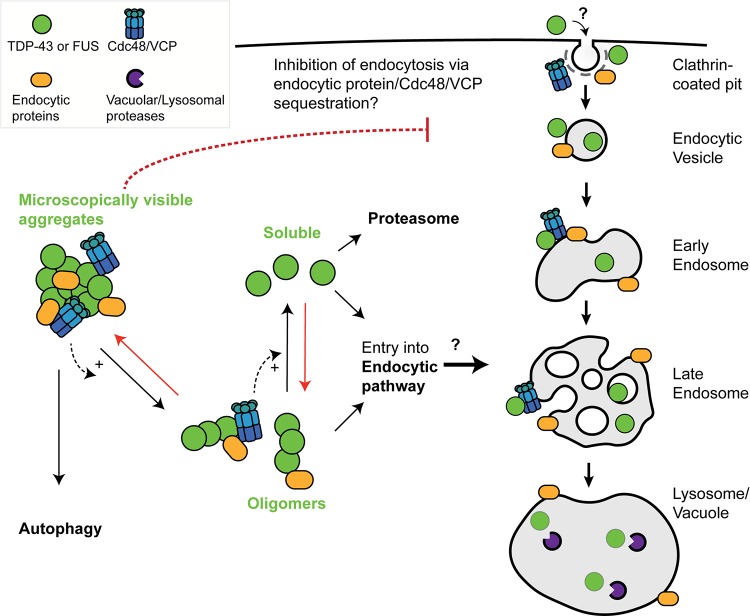
We propose that TDP-43 and FUS can be degraded in lysosomes/vacuoles by an endocytosis-dependent trafficking mechanism stimulated by Cdc48/VCP function. A key question is, how might Cdc48 and VCP promote TDP-43 and FUS turnover via endocytosis? We propose two general models (Fig. 7), which are not mutually exclusive. First, as described above and reviewed elsewhere (28, 30), Cdc48/VCP may drive various steps in the endocytic pathway. If TDP-43 and FUS can enter endocytic compartments independently of a physical interaction with Cdc48/VCP en route to clearance in vacuoles/lysosomes, then Cdc48/VCP impairment and the resulting endocytic downregulation may indirectly result in TDP-43 and FUS accumulation.
FIG 7.
Speculative model of TDP-43 and FUS engagement with endocytosis and Cdc48/VCP function. In yeast, the toxicity, turnover, and aggregation of both TDP-43 and FUS depend on endocytosis function (26; this study). We suggest that large microscopically visible aggregates of TDP-43 and FUS are most likely cleared from the cytoplasm by autophagy, with oligomeric or soluble forms of TDP-43 and FUS being subject to clearance by endocytic or proteasomal means. If true, given our yeast toxicity data, in which endocytic but not autophagic mutants enhance TDP-43 and FUS toxicity, this would suggest that oligomeric rather than microscopically visible forms of TDP-43 and FUS are the toxic cellular species. Cdc48/VCP, which localizes in microscopically visible TDP-43 aggregates, may facilitate TDP-43 and FUS conversion into oligomeric and soluble forms and/or facilitate entry of TDP-43 and FUS into the endocytic pathway. Red arrows broadly represent antagonistic aggregation-promoting processes, which may include cellular stress, impaired proteostasis, RNA metabolism defects, and perturbation of nuclear-cytoplasmic trafficking, all of which are implicated in ALS pathology. Finally, sequestration of Cdc48/VCP (and various endocytic proteins) within TDP-43 or FUS aggregates may contribute to the observed inhibition of endocytosis rates caused by TDP-43 or FUS expression, owing to various roles described for Cdc48/VCP in the endocytic pathway. Alternatively, TDP-43 and FUS may directly impair endocytic trafficking by affecting endocytic vesicle invagination, fusion, or other trafficking processes. How TDP-43 and FUS inhibit endocytosis and the means by which TDP-43 and FUS enter the endocytic pathway remain mechanistically undefined.
Alternatively, a second possibility is that Cdc48/VCP directly interacts with TDP-43 and FUS aggregates and promotes a remodeling event (e.g., aggregate disruption, targeting endocytic vesicles) that facilitates TDP-43 and FUS trafficking via the endocytic pathway, resulting in their turnover in lysosomes/vacuoles. Supporting this, Cdc48 physically interacts with TDP-43 (Fig. 1B) and colocalizes, as does Ubx3, with TDP-43 aggregates (Fig. 1A and 2B). However, whether these aggregates are active sites of TDP-43 clearance activity or whether they may form part of the mechanism by which TDP-43 impairs endocytosis activity, possibly by sequestration of endocytic factors, including Cdc48, remains unclear. Interestingly, TDP-43 and FUS cytoplasmic aggregates are typically ubiquitinated (62, 63), thus making them likely substrates for Cdc48/VCP activity. In addition, Cdc48/VCP cofactors may even facilitate TDP-43 or FUS ubiquitination events themselves, given the role of Ubx3 in an E3 ubiquitin ligase complex described previously and the fact that multiple VCP cofactors interact with E3 ubiquitin ligases (64).
Cdc48/VCP could also promote the disassembly and turnover of TDP-43/FUS aggregates by the proteasome or autophagy. However, our prior TDP-43 data from studies with yeast (26) and similar work by the Braun lab (27) argue against a significant role for the proteasome or autophagy in clearing TDP-43 in yeast under normal growth conditions. Similarly, for FUS, no significant impact of impaired autophagy on FUS toxicity was observed (Fig. 3C), though a minor role in turnover was seen (Fig. 3H). A key area of future research will be to determine in greater mechanistic detail how VCP promotes TDP-43 and FUS turnover in an endocytic function-dependent manner.
An intriguing result was the observation that ubp3Δ and bre5Δ yeast strains suppressed TDP-43 and FUS toxicity. The Ubp3/Bre5 complex has been identified as a regulator of numerous vesicular trafficking pathways, including as a negative regulator of mitophagy (65) and a positive effector of ribophagy (66), the cytoplasm-to-vacuole trafficking pathway (67), and endoplasmic reticulum-Golgi apparatus transport (68). In yeast, Ubp3 and Bre5 also localize within SGs (69) and facilitate the assembly of stationary-phase induced SGs (70); indeed, Ubp3 and Bre5 are homologs of USP10 and G3BP1, respectively, which are also key regulators of mammalian SG assembly (71, 72). Thus, a formal possibility (the subject of our other unpublished work) is that impaired SG assembly may limit TDP-43 aggregation, accumulation, and toxicity. Alternatively, the loss of Bre5 and Ubp3 function may also reflect the altered regulation of various autophagic trafficking pathways or the increased turnover of proteins by the proteasome.
Defects in endocytosis have been associated with various neurodegenerative diseases, including Alzheimer’s disease and Parkinson’s disease (73–76). Furthermore, aggregation-prone neurodegenerative disease protein clearance via endocytic function-dependent means has also been previously suggested. Specifically, α-synuclein degradation in human cell models occurs at least partly by the endolysosomal pathway, independently of autophagy and proteasomal turnover (77), while late defects in endocytic trafficking lead to the accumulation of poly(Q) Huntingtin aggregates in yeast Huntingtin models (78). Several genes associated with ALS onset, including ALS2, FIG4, and CHMP2B, also function in endocytosis (discussed in reference 26). Additionally, C9ORF72, the most commonly mutated gene identified in ALS, also shares DENN domain (differentially expressed in normal and neoplastic cells) similarly with Rab GEF proteins and exhibits colocalization with Rab5. In addition, depletion of C9ORF72 inhibits endocytosis rates in neuroblastoma cells (79), accelerates the degeneration of induced motor neurons (iMNs) derived from ALS patients, and correlates with endocytic trafficking defects (80). Strikingly, genetic and pharmacological interventions that enhanced endocytic trafficking in the iMN model and C9ORF72 mouse models also suppressed ALS-associated degeneration phenotypes (80). Thus, we speculate that endocytic dysfunction may promote the accumulation of cytoplasmic TDP-43 and FUS, leading to aggregation. This may be a key part of the pathology of ALS and related degenerative diseases characterized by TDP-43 and/or FUS inclusion bodies and, thus, an attractive target for therapeutic development.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Untransformed yeast strains were cultured at 30°C with yeast extract-peptone-dextrose (YPD) medium. Strains transformed with plasmids were grown in Sabouraud dextrose (SD) medium with appropriate nutrients for plasmid selection. In liquid culture, strains expressing GAL1-driven TDP-43 and FUS from plasmids (or vector controls) were initially cultured overnight in either 2% sucrose (if starter cultures were to be used for spotting assays) or 0.25% galactose and 1.75% sucrose (if starter cultures were to be used for microscopy assays) and then backdiluted to an optical density at 600 nm (OD600) of 0.1 in the identical medium, followed by growth at 30°C to mid-log phase (OD600, 0.3 to 0.6); for the analyses for which the results are shown in Fig. 1G, cells were instead inoculated at an OD600 of 0.2 and cultured at 25°C for 20 h to an OD600 of approximately 2.5 to 3 in 0.25% galactose, rather than 2% galactose, which ensured the more modest expression and toxicity of TDP-43 (26) and FUS (data not shown). The more modest expression and toxicity of TDP-43 and FUS facilitate the identification of enhancer mutants. Transformation was performed by a standard lithium acetate method. All strains and plasmids used in this study are listed in Table S1 in the supplemental material.
Yeast serial dilution assays.
Yeast strains transformed with plasmids expressing TDP-43, FUS, or the empty vector were grown overnight as described above, backdiluted to an OD600 of 0.2, serially diluted (1/6 dilutions), and spotted onto identical agar media. All images are representative of those from a minimum of three biological replicates imaged on the same day.
Culture of HEK293A cells, HEK293A cells expressing TDP-43–GFP, and TDP-35–GFP, and HEK293A cells in which VCP is knocked down.
HEK293A cell culture and the generation of cell lines into which TDP-43–GFP-, TDP-35–GFP-, and VCP-specific short hairpin RNA (shRNA) were stably integrated have been previously described (26). Briefly, HEK293A cells with lentivirus-integrated TDP-43–GFP or TDP-43 made to express a 35-kDa TDP-43 fragment (TDP-35–GFP; amino acids 90 to 414, which is a cytoplasmic aggregation-prone allele, with the shorter sequence mimicking that after cleavage by caspases [21]) were cultured at 37°C in Dulbecco modified Eagle medium (catalog number SH30022.01; HyClone) with 10% fetal bovine serum (catalog number 26140079; Gibco), and 50 U/ml penicillin and 50 μg/ml streptomycin (catalog number 15140-122; Gibco). DBeQ (a VCP inhibitor; catalog number A862910; Apexbio Technology) was added at a concentration of 10 μM for 1 h prior to live cell DeltaVision microscopy (see below). Dynasore (40 μM) was added to the culture for 24 h prior to lysis and Western blot analyses for endogenous TDP-43, FUS, VCP, and vinculin levels.
U-2 OS cell culture and immunohistochemistry.
U-2 OS cells were grown, fixed, and processed for immunohistochemistry as previously described (81). Anti-Rab5 (catalog number C8B1; Cell Signaling) and anti-VCP (catalog number 11433; Abcam) were utilized at 1:100 and 1:200 dilutions, respectively and labeled with Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin and Alexa Fluor 647-conjugated goat anti-mouse immunoglobulin secondary antibodies (Thermo Fisher Scientific), respectively, at a dilution of 1:1,000. Slides were mounted with ProLong Gold antifade DAPI (4′,6-diamidino-2-phenylindole) mounting medium (catalog number P36931; Molecular Probes) to stain the nuclei.
Patient tissue immunofluorescence.
Based on an Abcam paraffin-embedded immunohistochemistry staining protocol, samples were dewaxed in xylene (2 times for 3 min each time) and in xylene mixed 1:1 with 100% ethanol (once for 3 min) and then fixed with successive incubations in 100% (2 times for 3 min each time), 90% (3 min), 80% (3 min), 70% (3 min), and 50% (3 min) ethanol and distilled water for 10 min. Antigen was retrieved by keeping tissue slices in boiled sodium citrate buffer for 20 min. Endogenous peroxidases were quenched with 3% H2O2 for 30 min. Each slice was then blocked with 5% goat serum for 1 h and then incubated with anti-TDP-43 (catalog number 10782-2-AP; Proteintech) and anti-VCP (catalog number 11433; Abcam) at 1:200 dilutions overnight at 4°C. Following 4 washes with Tris-buffered saline–Tween 20 (TBS-T), each slice was incubated with an appropriate Alexa Fluor-conjugated secondary antibody (Thermo Fisher Scientific). The slices were then washed 4 times in TBS-T and mounted with ProLong Gold antifade DAPI mounting medium (catalog number P36931; Molecular Probes) to stain the nuclei.
Fluorescence microscopy.
Most fluorescence microscopy methods have previously been described in detail (32). Briefly, early yeast stationary-phase cells (OD600, >3.0) and mid-log-phase cells (OD600, 0.3 to 0.6) and fixed human cells/tissues (see above) were examined using a DeltaVision Elite microscope. The data were analyzed by use of the Fiji platform (82). At least 50 cells (yeast or human) were analyzed per biological replicate, and all yeast images shown are representative of those from a minimum of 3 biological replicates. All quantitative analyses (e.g., colocalization frequency, vacuole protein accumulation) were quantified in a blind manner to avoid bias, and the results are summarized in Table S1.
Western blotting and protein stability assays.
Western blotting was conducted as described previously (83). Primary antibodies were as follows: anti-GFP (catalog number 902602; BioLegend), anti-TDP-43 (catalog number 60019-2-Ig; Proteintech), antivinculin (catalog number V4505; Sigma-Aldrich), anti-Pgk1 (catalog number ab113687; Abcam), anti-VCP (catalog number ab11433; Abcam), anti-TAP (catalog number CAB1001; Thermo Fisher Scientific), and anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase; catalog number MA5-15738; Invitrogen). For TDP-43 and FUS protein stability assays in yeast, expression was halted by addition of either cycloheximide at 0.2 mg/ml (translational shutoff) or additional 2% glucose (GAL1 transcriptional shutoff) to the growth media, as indicated in the figure legends. The results of quantification of key Western blotting data sets are listed in Table S1.
Endocytosis rate assays.
Mid-log-phase yeast cells (OD600, 0.3 to 0.6) were harvested and incubated on ice for 10 min and then suspended in YPD containing 8 μM FM4-64 (catalog number T3166; Thermo Fisher Scientific) at 25°C and imaged at the times indicated in the figures. Specifically, at least 50 vacuolar membranes were quantified per biological replicate by first collapsing two 0.4 μM z-slices in the most central focal plane of the yeast cells and then manually drawing a region of interest around the vacuole, thresholding until all vacuolar membrane pixels were selected, followed by calculation of the average pixel intensity. This approach was required due to the variability in the vacuole membrane size and shape and the occasional discontinuous FM4-64 labeling, which led to errors in automated quantitation. All images were quantified in a blind manner to avoid bias. For mammalian cells, transferrin assays were conducted per Thermo Fisher Scientific product guidelines. Briefly, HEK293A cells were kept on ice for 10 min, washed with live cell imaging solution (catalog number A14291DJ; Thermo Fisher Scientific), and then incubated with 25 μg/ml fluorescent transferrin (catalog number T23362; Thermo Fisher Scientific) at 37°C for 15 min. The cells were then washed again with live cell imaging solution, imaged, and quantified for intracellular fluorescence using the Fiji platform.
Immunoprecipitation analyses.
Ten-milliliter cultures of appropriately transformed yeast with suitable selective SD medium were grown to mid-log growth phase (OD600, 0.3 to 0.6), and the cell pellets were obtained by centrifugation at 13,000 × g and 4°C, harvested, and resuspended in 1 ml of radioimmunoprecipitation assay cell lysis buffer. A 50% volume of glass beads (catalog number 11079105; BioSpec Products) was added, and the cells were vortexed for 20 s before being incubated on ice; this was repeated two more times. The cells were centrifuged at 13,000 × g for 15 min to separate unlysed cells and insoluble cell debris. The supernatant was harvested, and the protein concentration was assessed for normalization via the Bradford assay. Supernatants were incubated with magnetic Dynabeads (Thermo Fisher Scientific) labeled with either anti-TAP antibody or anti-GFP antibody for 2 h at 4°C, followed by three bead washes in lysis buffer. Bound proteins were eluted with gel loading buffer, boiled in a standard SDS gel loading buffer at 95°C for 5 min, and then subjected to standard Western blot analyses.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Southern Arizona VA health care agency and the VA Repository for providing us with ALS and control patient tissue.
This work was supported by start-up funds to J.R.B. from the University of Arizona, support from the NIH (grant NIH RO1-GM1145664), a starter grant (grant 16-IIP-266) from the ALS Association, and support from the Frick Foundation.
J.R.B. conceived the study; J.R.B., G.L., and A.B. designed the experiments; G.L., A.B., A.N.W., F.P., E.B., and A.B. performed the experiments; J.R.B., G.L., A.B., A.N.W., F.P., E.B., and A.B. analyzed the data; J.R.B. wrote the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Boille S, Vande Velde C, Cleveland DW, Boillée S, Vande Velde C, Cleveland DW. 2006. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron 52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Ling S-C, Polymenidou M, Cleveland DW. 2013. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renton AE, Chiò A, Traynor BJ. 2014. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagier-Tourenne C, Polymenidou M, Cleveland DW. 2010. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ederle H, Dormann D. 2017. TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett 591:1489–1507. doi: 10.1002/1873-3468.12646. [DOI] [PubMed] [Google Scholar]
- 6.Baron DM, Kaushansky LJ, Ward CL, Sama RRK, Chian R-J, Boggio KJ, Quaresma AJC, Nickerson JA, Bosco DA. 2013. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener 8:30. doi: 10.1186/1750-1326-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu-Yesucevitz L, Bilgutay A, Zhang Y-J, Vanderweyde T, Vanderwyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B. 2010. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchan JR. 2014. MRNP granules. Assembly, function, and connections with disease. RNA Biol 11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Ramaswami M, Taylor JP, Parker R. 2013. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li YR, King OD, Shorter J, Gitler AD. 2013. Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogler TO, Wheeler JR, Nguyen ED, Hughes MP, Britson KA, Lester E, Rao B, Betta ND, Whitney ON, Ewachiw TE, Gomes E, Shorter J, Lloyd TE, Eisenberg DS, Taylor JP, Johnson AM, Olwin BB, Parker R. 2018. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563:508–513. doi: 10.1038/s41586-018-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes N, Eshleman N, Buchan JR. 2018. Stress granules and ALS: a case of causation or correlation? Adv Neurobiol 20:173–212. doi: 10.1007/978-3-319-89689-2_7. [DOI] [PubMed] [Google Scholar]
- 14.Gasset-Rosa F, Lu S, Yu H, Chen C, Melamed Z, Guo L, Shorter J, Da Cruz S, Cleveland DW. 2019. Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102:339–357.e7. doi: 10.1016/j.neuron.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scekic-Zahirovic J, Sendscheid O, El Oussini H, Jambeau M, Sun Y, Mersmann S, Wagner M, Dieterlé S, Sinniger J, Dirrig-Grosch S, Drenner K, Birling M-C, Qiu J, Zhou Y, Li H, Fu X-D, Rouaux C, Shelkovnikova T, Witting A, Ludolph AC, Kiefer F, Storkebaum E, Lagier-Tourenne C, Dupuis L. 2016. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J 35:1077–1097. doi: 10.15252/embj.201592559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. 2010. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci 30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. 2011. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. 2008. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A 105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, Hipp MS. 2016. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 20.Cascella R, Capitini C, Fani G, Dobson CM, Cecchi C, Chiti F. 2016. Quantification of the relative contributions of loss-of-function and gain-of-function mechanisms in TAR DNA-binding protein 43 (TDP-43) proteinopathies. J Biol Chem 291:19437–19448. doi: 10.1074/jbc.M116.737726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y-J, Xu Y-F, Cook C, Gendron TF, Roettges P, Link CD, Lin W-L, Tong J, Castanedes-Casey M, Ash P, Gass J, Rangachari V, Buratti E, Baralle F, Golde TE, Dickson DW, Petrucelli L. 2009. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci U S A 106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, Pulst SM, Taylor JP, Rigo F, Gitler AD. 2017. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H-J, Raphael AR, Ladow ES, McGurk L, Weber RA, Trojanowski JQ, Lee V-Y, Finkbeiner S, Gitler AD, Bonini NM. 2014. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet 46:152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G, Court FA, van Zundert B, Hetz C. 2013. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 9:1308–1320. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 25.Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, Shaw C, Chandran S, Finkbeiner S. 2014. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol 10:677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G, Coyne AN, Pei F, Vaughan S, Chaung M, Zarnescu DC, Buchan JR. 2017. Endocytosis regulates TDP-43 toxicity and turnover. Nat Commun 8:2092. doi: 10.1038/s41467-017-02017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leibiger C, Deisel J, Aufschnaiter A, Ambros S, Tereshchenko M, Verheijen BM, Büttner S, Braun RJ. 2018. TDP-43 controls lysosomal pathways thereby determining its own clearance and cytotoxicity. Hum Mol Genet 27:1593–1607. doi: 10.1093/hmg/ddy066. [DOI] [PubMed] [Google Scholar]
- 28.Xia D, Tang WK, Ye Y. 2016. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene 583:64–77. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer H, Weihl CC. 2014. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci 127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bug M, Meyer H. 2012. Expanding into new markets—p97 in endocytosis and autophagy. J Struct Biol 179:78–82. doi: 10.1016/j.jsb.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Ritz D, Vuk M, Kirchner P, Bug M, Schütz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, Glatter T, Gstaiger M, Aebersold R, Weihl CC, Meyer H. 2011. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol 13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchan JR, Kolaitis R-M, Taylor JP, Parker R. 2013. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramanathan HN, Ye Y. 2012. The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res 22:346–359. doi: 10.1038/cr.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weihl CC, Pestronk A, Kimonis VE. 2009. Valosin-containing protein disease: inclusion body myopathy with Paget’s disease of the bone and fronto-temporal dementia. Neuromuscul Disord 19:308–315. doi: 10.1016/j.nmd.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Ortiz CJ, Hoshino H, Cheng D, Liu-Yescevitz L, Blurton-Jones M, Wolozin B, LaFerla FM, Kitazawa M. 2013. Neuronal-specific overexpression of a mutant valosin-containing protein associated with IBMPFD promotes aberrant ubiquitin and TDP-43 accumulation and cognitive dysfunction in transgenic mice. Am J Pathol 183:504–515. doi: 10.1016/j.ajpath.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, Lee V-Y, Forman MS, Taylor JP. 2010. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci 30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nalbandian A, Llewellyn KJ, Badadani M, Yin HZ, Nguyen C, Katheria V, Watts G, Mukherjee J, Vesa J, Caiozzo V, Mozaffar T, Weiss JH, Kimonis VE. 2013. A progressive translational mouse model of human valosin-containing protein disease: The VCPR155H/+ mouse. Muscle Nerve 47:260–270. doi: 10.1002/mus.23522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brettschneider J, Arai K, Del Tredici K, Toledo JB, Robinson JL, Lee EB, Kuwabara S, Shibuya K, Irwin DJ, Fang L, Van Deerlin VM, Elman L, McCluskey L, Ludolph AC, Lee VMY, Braak H, Trojanowski JQ. 2014. TDP-43 pathology and neuronal loss in amyotrophic lateral sclerosis spinal cord. Acta Neuropathol 128:423–437. doi: 10.1007/s00401-014-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gitcho MA, Strider J, Carter D, Taylor-Reinwald L, Forman MS, Goate AM, Cairns NJ. 2009. VCP mutations causing frontotemporal lobar degeneration disrupt localization of TDP-43 and induce cell death. J Biol Chem 284:12384–12398. doi: 10.1074/jbc.M900992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elden AC, Kim H-J, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rüb U, Auburger G, Trojanowski JQ, Lee V-Y, Van Deerlin VM, Bonini NM, Gitler AD. 2010. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. 2011. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell 41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, Fang X, Shorter J, Krogan NJ, Finkbeiner S, Farese RV, Gitler AD. 2012. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet 44:1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epple UD, Suriapranata I, Eskelinen EL, Thumm M. 2001. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol 183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ. 2001. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem 276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuberth C, Buchberger A. 2008. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci 65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farrell KB, Grossman C, Di Pietro SM. 2015. New regulators of clathrin-mediated endocytosis identified in Saccharomyces cerevisiae by systematic quantitative fluorescence microscopy. Genetics 201:1061–1070. doi: 10.1534/genetics.115.180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen E-L, Thumm M. 2010. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol 190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuberth C, Richly H, Rumpf S, Buchberger A. 2004. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep 5:818–824. doi: 10.1038/sj.embor.7400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baek GH, Cheng H, Choe V, Bao X, Shao J, Luo S, Rao H. 2013. Cdc48: a Swiss Army knife of cell biology. J Amino Acids 2013:183421. doi: 10.1155/2013/183421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reyes-Turcu FE, Ventii KH, Wilkinson KD. 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang T, Jiang X, Chen G, Xu J. 2015. Interaction of amyotrophic lateral sclerosis/frontotemporal lobar degeneration-associated fused-in-sarcoma with proteins involved in metabolic and protein degradation pathways. Neurobiol Aging 36:527–535. doi: 10.1016/j.neurobiolaging.2014.07.044. [DOI] [PubMed] [Google Scholar]
- 52.Azuma Y, Tokuda T, Shimamura M, Kyotani A, Sasayama H, Yoshida T, Mizuta I, Mizuno T, Nakagawa M, Fujikake N, Ueyama M, Nagai Y, Yamaguchi M. 2014. Identification of ter94, Drosophila VCP, as a strong modulator of motor neuron degeneration induced by knockdown of Caz, Drosophila FUS. Hum Mol Genet 23:3467–3480. doi: 10.1093/hmg/ddu055. [DOI] [PubMed] [Google Scholar]
- 53.Balderhaar HJ, Kleine Ungermann C. 2013. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci 126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 54.Vida TA, Emr SD. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barysch SV, Jahn R, Rizzoli SO. 2010. A fluorescence-based in vitro assay for investigating early endosome dynamics. Nat Protoc 5:1127–1137. doi: 10.1038/nprot.2010.84. [DOI] [PubMed] [Google Scholar]
- 56.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Li M, Koshi T, Emr SD. 2015. Membrane-anchored ubiquitin ligase complex is required for the turnover of lysosomal membrane proteins. J Cell Biol 211:639–652. doi: 10.1083/jcb.201505062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao T-P, Dantuma NP, Taylor JP. 2010. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tooze SA, Abada A, Elazar Z. 2014. Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb Perspect Biol 6:a018358. doi: 10.1101/cshperspect.a018358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirchner P, Bug M, Meyer H. 2013. Ubiquitination of the N-terminal region of caveolin-1 regulates endosomal sorting by the VCP/p97 AAA-ATPase. J Biol Chem 288:7363–7372. doi: 10.1074/jbc.M112.429076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pleasure IT, Black MM, Keen JH. 1993. Valosin-containing protein, VCP, is a ubiquitous clathrin-binding protein. Nature 365:459–462. doi: 10.1038/365459a0. [DOI] [PubMed] [Google Scholar]
- 62.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee V-Y. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 63.Deng H-X, Zhai H, Bigio EH, Yan J, Fecto F, Ajroud K, Mishra M, Ajroud-Driss S, Heller S, Sufit R, Siddique N, Mugnaini E, Siddique T. 2010. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann Neurol 67:739–748. doi: 10.1002/ana.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. 2008. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1α turnover. Cell 134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Müller M, Kötter P, Behrendt C, Walter E, Scheckhuber CQ, Entian K-D, Reichert AS. 2015. Synthetic quantitative array technology identifies the Ubp3-Bre5 deubiquitinase complex as a negative regulator of mitophagy. Cell Rep 10:1215–1225. doi: 10.1016/j.celrep.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 66.Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, Dargemont C. 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep 11:548–554. doi: 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baxter BK, Abeliovich H, Zhang X, Stirling AG, Burlingame AL, Goldfarb DS. 2005. Atg19p ubiquitination and the cytoplasm to vacuole trafficking pathway in yeast. J Biol Chem 280:39067–39076. doi: 10.1074/jbc.M508064200. [DOI] [PubMed] [Google Scholar]
- 68.Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C. 2003. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol 5:661–667. doi: 10.1038/ncb1003. [DOI] [PubMed] [Google Scholar]
- 69.Rinnerthaler M, Lejskova R, Grousl T, Stradalova V, Heeren G, Richter K, Breitenbach-Koller L, Malinsky J, Hasek J, Breitenbach M. 2013. Mmi1, the yeast homologue of mammalian TCTP, associates with stress granules in heat-shocked cells and modulates proteasome activity. PLoS One 8:e77791. doi: 10.1371/journal.pone.0077791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nostramo R, Varia SN, Zhang B, Emerson MM, Herman PK. 2016. The catalytic activity of the Ubp3 deubiquitinating protease is required for efficient stress granule assembly in Saccharomyces cerevisiae. Mol Cell Biol 36:173–183. doi: 10.1128/MCB.00609-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takahashi M, Higuchi M, Matsuki H, Yoshita M, Ohsawa T, Oie M, Fujii M. 2013. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol Cell Biol 33:815–829. doi: 10.1128/MCB.00763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schreij AMA, Fon EA, McPherson PS. 2016. Endocytic membrane trafficking and neurodegenerative disease. Cell Mol Life Sci 73:1529–1545. doi: 10.1007/s00018-015-2105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu A, Shibata Y, Shah B, Calamini B, Lo DC, Morimoto RI. 2014. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc Natl Acad Sci U S A 111:E1481–E1490. doi: 10.1073/pnas.1321811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nixon RA. 2005. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging 26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 76.Wang X, Huang T, Bu G, Xu H. 2014. Dysregulation of protein trafficking in neurodegeneration. Mol Neurodegener 9:31. doi: 10.1186/1750-1326-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tofaris GK, Kim HT, Hourez R, Jung J-W, Kim KP, Goldberg AL. 2011. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc Natl Acad Sci U S A 108:17004–17009. doi: 10.1073/pnas.1109356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meriin AB, Zhang X, Alexandrov IM, Salnikova AB, Ter-Avanesian MD, Chernoff YO, Sherman MY. 2007. Endocytosis machinery is involved in aggregation of proteins with expanded polyglutamine domains. FASEB J 21:1915–1925. doi: 10.1096/fj.06-6878com. [DOI] [PubMed] [Google Scholar]
- 79.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RAK, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, King AE, Atkin JD. 2014. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi Y, Lin S, Staats KA, Li Y, Chang W-H, Hung S-T, Hendricks E, Linares GR, Wang Y, Son EY, Wen X, Kisler K, Wilkinson B, Menendez L, Sugawara T, Woolwine P, Huang M, Cowan MJ, Ge B, Koutsodendris N, Sandor KP, Komberg J, Vangoor VR, Senthilkumar K, Hennes V, Seah C, Nelson AR, Cheng T-Y, Lee S-J, August PR, Chen JA, Wisniewski N, Hanson-Smith V, Belgard TG, Zhang A, Coba M, Grunseich C, Ward ME, van den Berg LH, Pasterkamp RJ, Trotti D, Zlokovic BV, Ichida JK. 2018. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med 24:313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol 431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 82.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu G, Yu F-X, Kim Y, Meng Z, Naipauer J, Looney D, Liu X, Gutkind J, Mesri E, Guan K-L. 2015. Kaposi sarcoma-associated herpesvirus promotes tumorigenesis by modulating the Hippo pathway. Oncogene 34:3536–3546. doi: 10.1038/onc.2014.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.