HBO1 (MYST2/KAT7) is essential for histone 3 lysine 14 acetylation (H3K14ac) but is dispensable for H4 acetylation and DNA replication in mouse tissues. In contrast, previous studies using small interfering RNA (siRNA) knockdown in human cell lines have suggested that HBO1 is essential for DNA replication. To determine if HBO1 has distinctly different roles in immortalized human cell lines and normal mouse cells, we performed siRNA knockdown of HBO1.
KEYWORDS: histone acetylation, HBO1, MYST2, KAT7, chromatin
ABSTRACT
HBO1 (MYST2/KAT7) is essential for histone 3 lysine 14 acetylation (H3K14ac) but is dispensable for H4 acetylation and DNA replication in mouse tissues. In contrast, previous studies using small interfering RNA (siRNA) knockdown in human cell lines have suggested that HBO1 is essential for DNA replication. To determine if HBO1 has distinctly different roles in immortalized human cell lines and normal mouse cells, we performed siRNA knockdown of HBO1. In addition, we used CRISPR/Cas9 to generate 293T, MCF7, and HeLa cell lines lacking HBO1. Using both techniques, we show that HBO1 is essential for all H3K14ac in human cells and is unlikely to have a direct effect on H4 acetylation and only has minor effects on cell proliferation. Surprisingly, the loss of HBO1 and H3K14ac in HeLa cells led to the secondary loss of almost all H4 acetylation after 4 weeks. Thus, HBO1 is dispensable for DNA replication and cell proliferation in immortalized human cells. However, while cell proliferation proceeded without HBO1 and H3K14ac, HBO1 gene deletion led to profound changes in cell adhesion, particularly in 293T cells. Consistent with this phenotype, the loss of HBO1 in both 293T and HeLa principally affected genes mediating cell adhesion, with comparatively minor effects on other cellular processes.
INTRODUCTION
HBO1 (MYST2/KAT7) is a member of the MYST family of lysine acetyltransferases, a large family of chromatin regulators characterized by a conserved lysine acetyltransferase domain, which also includes TIP60 (KAT5), MOZ (MYST3/KAT6A), QKF (MORF/KAT6B), and MOF (MYST1/KAT8) (1). HBO1 (histone acetyltransferase binding to ORC) was originally identified in a yeast two-hybrid screen using the human-origin recognition complex subunit 1 (ORC1) protein as bait, suggesting a direct interaction between HBO1 and ORC1 (2). The assembly of ORC together with MCM2 to -7 (MCM2-7), CDC6/CDC18, and CDT1 forms the DNA prereplication complex, which confers a license for the initiation of DNA replication and acts as a checkpoint ensuring that DNA replication occurs only once per cell cycle (3). As well as interacting with ORC1, HBO1 has also been proposed to interact with other prereplication complex members, including MCM2 (4) and CDT1 (5). Furthermore, HBO1 has also been shown to acetylate ORC2, MCM2, CDC6, and geminin in cell-free acetylation assays (6). The direct physical interaction between HBO1 and members of the DNA prereplication complex has led to the widely accepted notion that HBO1 is essential for DNA replication and cell proliferation (7).
Studies investigating the role of HBO1 in DNA replication and cell proliferation have utilized RNA interference (RNAi) knockdown to generate HBO1-deficient immortalized human cell lines. For instance, short hairpin RNA (shRNA) knockdown of HBO1 in 293T cells resulted in a large accumulation of cells in the G2/M phase of the cell cycle, suggesting a role for HBO1 in normal cell cycle progression (8). Indeed, when these HBO1-deficient 293T cells were used in a proliferation assay spanning 3 days, the cells stopped growing altogether (8). shRNA knockdown of HBO1 in MCF7 cells (8) and HeLa cells (9) resulted in a dramatic decrease in the percentage of bromodeoxyuridine (BrdU)-positive cells in the S phase of the cell cycle. Both shRNA and small interfering RNA (siRNA) knockdown of HBO1 in HeLa cells caused a significant shift in MCM2 and MCM6 localization from the chromatin fraction to the cytoplasmic fraction, leading the authors of those studies to suggest that HBO1 is essential for MCM2-7 loading and normal DNA replication licensing (6, 9).
Apart from ascribing a role for HBO1 in cell proliferation and DNA replication, these in vitro RNAi knockdown studies also sought to determine specific HBO1 histone acetylation targets. It was reported previously that HBO1 siRNA and shRNA knockdown caused significant reductions in histone 4 lysine 5 acetylation (H4K5ac), H4K8ac, and H4K12ac but not H4K16ac or H3ac in HeLa cells (10) and 293T cells (8). Furthermore, HBO1 siRNA knockdown caused a drastic reduction in global H4 acetylation in HeLa and MCF7 cells (11). By chromatin immunoprecipitation (ChIP) analysis, HBO1 was found to associate with mammalian origins of replication in HEK293, HeLa, and ATCC CCL-156 lymphoblastoid cell lines (5). HBO1 has also been reported to enhance CDT1-dependent rereplication and is proposed to act as a coactivator of CDT1 at replication origins in HeLa, HEK293, HepG2, and MCF10a cell lines (5). Additionally, siRNA knockdown of HBO1 resulted in a loss of H4 but not H3 acetylation at origins of replication in HeLa cells under normal (10) and stress (12) conditions.
In addition to RNAi knockdown studies, the coexpression of HBO1 and JADE1 led to a significant increase in H4 acetylation in 293T cells (13) and HeLa cells (10) while stimulating MCM complex loading (10). HBO1 was also found to predominantly acetylate histone H4 in cell-free assays (11). Taken together, there is a substantial body of literature to suggest that HBO1 functions as an essential H4 acetyltransferase and is indispensable for DNA replication and cell proliferation. However, these results were predominantly derived from in vitro experiments utilizing immortalized cell lines primarily of cancerous origin.
In contrast, we have previously shown that HBO1 is not essential for DNA replication or cell proliferation and that HBO1 is critical for specifically mediating global H3K14ac rather than H4 acetylation during mouse development (14) and T cell development (15). In addition, we demonstrated that Hbo1-null primary mouse embryonic fibroblasts (MEFs) displayed a 10-fold reduction in H3K14ac, while acetylation levels at H3K9, H4K5, H4K8, H4K12, and H4K16 remained unchanged or were increased (14). Hbo1 knockout (KO) MEFs also displayed normal proliferation rates and cell cycle profiles similar to those of wild-type (WT) controls. Hbo1 knockout embryos die at midgestation, not as a result of DNA replication defects but rather due to a failure to express developmental patterning genes adequately. Thus, it appears that HBO1-mediated H3K14ac is essential for the robust de novo activation of key embryonic patterning genes during early embryonic development but not for DNA replication or cell proliferation.
The substantial literature suggesting that HBO1 is important for cell cycle progression in human cancer cell lines, in contrast to our finding of normal proliferation in normal mouse cells, suggested the exciting possibility that the function of HBO1 differs between cancer and normal cells and that cancer cells are “addicted” to HBO1 for fundamental cellular processes such as sustained proliferation and survival. This is important for investment in the development of new anticancer therapeutics that target KAT6A/B and HBO1 by inhibiting their enzymatic activity (16, 17). To investigate this enticing possibility, we used CRISPR/Cas9-mediated insertion and deletion mutations (indels) to remove all HBO1 protein in human-derived 293T, MCF7, and HeLa cell lines. These cell lines were chosen as they were extensively used in previous RNAi knockdown approaches investigating HBO1 function.
We found that mutation of the HBO1 locus in 293T, MCF7, and HeLa cells did not result in cell cycle arrest. Instead, the cell lines continued to proliferate without HBO1 throughout our proliferation assays and incorporated BrdU. The primary histone acetylation mark lost after mutating the HBO1 locus was H3K14ac, and this was common to all three cell lines. Interestingly, we observed cell adhesion defects in the cell lines tested. Examining the more acute effects of a reduction in HBO1 using siRNA knockdown of HBO1 with the same siRNA reported previously by Miotto and Struhl (5, 18) or an alternative siRNA molecule, we were unable to reproduce a cell cycle arrest phenotype. Transfection with either siRNA reproducibly reduced HBO1 protein levels but had no effect on H4 acetylation and rather led to a reduction in H3K14 acetylation. We conclude that HBO1 is essential for H3K14ac but not for DNA replication or cell proliferation in 293T, MCF7, and HeLa cells. Thus, we demonstrate that the effects on DNA synthesis and the histone acetylation function of HBO1 in immortalized human cell lines do not differ substantially from its role in mouse embryonic development and normal mouse cells.
RESULTS
CRISPR/Cas9 mediates highly efficient loss of HBO1 in human cell lines.
To determine if HBO1 is essential for cell proliferation, DNA replication, and H4 acetylation in human cell lines, we utilized an inducible CRISPR/Cas9 system (19) to mutate the HBO1 locus in 293T, MCF7, and HeLa cell lines.
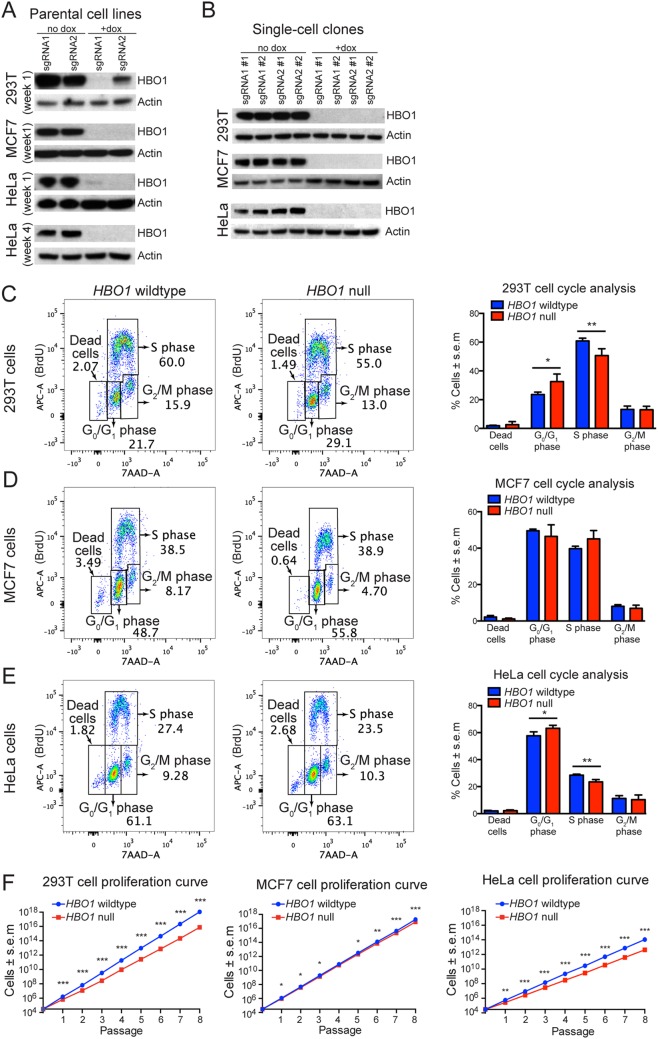
293T, MCF, and HeLa cell lines were transduced with the Cas9 and single guide RNA (sgRNA) vectors coding for either of two HBO1 sgRNAs (sgRNA1 or sgRNA2) (see Fig. S1 in the supplemental material). The guides were selected to mutate the MYST lysine acetyltransferase domain of HBO1. The constitutively expressed green fluorescent protein (GFP) and mCherry selection markers allow for efficient fluorescence-activated cell sorting (FACS) and enrichment of double-positive cells. Subsequently, cells were cultured in medium supplemented with 1 μg/ml doxycycline (Dox) for 3 days to induce HBO1 sgRNA expression to guide CRISPR/Cas9-mediated indel formation within the MYST domain of the HBO1 gene. These “parental cell lines” were then (i) used for assessing the extent of HBO1 protein loss after HBO1 sgRNA induction, (ii) used for determining the levels of histone acetylation after the loss of HBO1, and (iii) subjected to single-cell FACS to derive single-cell-derived clonal cell lines (clonal cell lines) that lack HBO1 protein. Clonal cell lines were expanded to characterize their individual HBO1 mutation and used for subsequent experiments. Non-Dox-treated parental cell lines with intact HBO1 protein were also sorted to establish clonal cell lines with wild-type HBO1 loci, which were used as controls.
Western immunoblotting experiments show that Dox induction of HBO1 sgRNA1 expression in 293T parental cell lines resulted in HBO1 protein levels that were almost undetectable, while HBO1 sgRNA2 expression resulted in a significant but less efficient reduction in HBO1 protein (Fig. 1A). HBO1 sgRNA expression in MCF7 cells led to undetectable levels of HBO1 protein (Fig. 1A). Analysis of HeLa cells showed a similar response to HBO1 sgRNA induction, with HBO1 protein being reduced to almost undetectable levels, as assessed by Western immunoblot analysis in parental cell lines at weeks 1 and 4 after Dox treatment (Fig. 1A).
FIG 1.
HBO1 is not essential for DNA replication or sustained cell proliferation. (A) 293T, MCF7, and HeLa parental cells were treated with Dox for 3 days to induce sgRNA expression and assessed for HBO1 protein levels 1 week later. HeLa parental cells were also assessed for HBO1 protein levels 4 weeks after the initial Dox treatment. Each lane represents one parental cell line transduced with HBO1 sgRNA1 or sgRNA2. (B) 293T, MCF7, and HeLa clonal cell lines were assessed for HBO1 protein levels approximately 4 to 6 weeks after the initial Dox treatment. Each lane represents one clonal cell line transduced with HBO1 sgRNA1 or sgRNA2. (C to E) Flow cytometry quantification of BrdU and 7-AAD in cells, which distinguishes phases of the cell cycle in 293T, MCF7, and HeLa clonal cell lines. (F) Cumulative proliferation curves of 293T, MCF7, and HeLa clonal cell lines. Thirty thousand cells were seeded during the start of each passage interval. Clonal cell lines are the same as the ones presented in panel B and Fig. 3F to K. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data are presented as means ± standard errors of the means for 4 clonal cell lines, i.e., 2 each transduced with HBO1 sgRNA1 or sgRNA2. Numerical cell cycle phase assessment and cell proliferation rates per passage interval for individual clonal cell lines are shown in Fig. S2 in the supplemental material.
All parental cell lines consist of a mixed population of cells harboring a diverse variety of CRISPR/Cas9-induced indels and various extents of HBO1 depletion. To obtain a homogenous cell population with defined CRISPR/Cas9-induced indels resulting in a complete loss of HBO1 protein (HBO1-null cells), we sorted single cells derived from parental cell lines and expanded these into clonal cell lines. This procedure requires single cells to proliferate without the benefit of a community effect and takes approximately 4 weeks. The successful derivation of clonal cell lines lacking HBO1 indicates that HBO1 is not essential for even clonal proliferation of cells. Western immunoblotting confirmed a complete lack of HBO1 protein in selected Dox-treated clonal 293T, MCF7, and HeLa cell lines (Fig. 1B). Two clonal lines for each sgRNA, sgRNA1 and sgRNA2, for each of the 3 cell lines, 12 clonal cell lines altogether, were selected for further experiments.
In order to determine the nature of the indels in all 12 clonal cell lines, we used a next-generation sequencing approach (19) and found that almost all indels led to frameshift mutations and premature stop codons in the HBO1 locus (Fig. S1). Based on these next-generation sequencing data (Fig. S1) and the lack of detectable HBO1 protein (Fig. 1), we concluded that our selected Dox-treated 293T, MCF7, and HeLa clonal cell lines were HBO1-null cells. These same HBO1-null cells were used in all subsequent experiments.
To ensure that any phenotypic observations in our experiments were due to HBO1 mutations rather than off-target effects, we examined predicted off-target sites for the two HBO1 sgRNAs. The top 5 potential off-target sites based on sgRNA sequence match/mismatch, and, thus, the likelihood of being targeted, were situated in different genomic locations for the two HBO1 sgRNAs (sgRNA1 and -2) and were all outside known genes, with the exception of one pseudogene, which is not expressed (Tables S1 and S2). The frequencies of off-target indels in these top 5 potential off-target sites for each sgRNA in all clonal cell lines are displayed in Table S3. While some off-target indels were observed, these did not occur in all clonal cell lines, were not the same for the two HBO1 sgRNAs, and did not target known expressed genes. Therefore, the only mutations in common among all 12 clonal cell lines, which were derived from 3 independent cell types, are indels in the HBO1 locus (Fig. S1). We conclude that any common phenotype exhibited by all 12 HBO1-mutated clonal cell lines could be attributed to the loss of HBO1.
Loss-of-function mutations in HBO1 alter cell cycle dynamics but do not affect sustained DNA replication, proliferation, or cell survival.
To determine if the loss of HBO1 resulted in a failure of DNA replication or altered cell cycle kinetics, as reported previously (5, 6, 8–10), we subjected HBO1-null and HBO1 wild-type 293T, MCF7, and HeLa clonal cell lines to BrdU/7-aminoactinomycin D (7-AAD) FACS cell cycle analysis (Fig. 1C to E). Cells were cultured in medium supplemented with BrdU before staining with anti-BrdU allophycocyanin (APC)-conjugated antibodies to directly determine DNA synthesis activity, while 7-AAD was used to measure DNA content. We found that clonal HBO1-null 293T, MCF7, and HeLa cell lines efficiently incorporated BrdU (Fig. 1C to E), indicating that HBO1 is not essential for DNA synthesis. Quantification of cells in different phases of the cell cycle demonstrated an increase in cells in G0/G1 and a corresponding decrease of cells in the S phase of the cell cycle for 293T (16.8% decrease; P < 0.01) and HeLa (17.4% decrease; P < 0.01) HBO1-null clonal cell lines (Fig. 1C and E). There were no significant cell cycle changes in HBO1-null MCF7 cells (Fig. 1D). There were no significant increases in dead cells in any HBO1-null cell line, indicating that HBO1 was not essential for cell survival.
To determine if HBO1 was essential for the long-term sustained proliferation and survival of 293T, MCF7, and HeLa cells, we monitored clonal cell lines for 8 serial passages, with passage intervals of approximately 1 week. Thirty thousand cells were seeded at the start of each passage interval. Cumulative cell counts over 8 weeks showed that HBO1-null cells proliferated more slowly than controls in all three cell types (Fig. 1F). During individual passage intervals, HBO1-null MCF7 cells did not display major proliferation defects, whereas HBO1-null 293T and HeLa cells showed reduced proliferation compared to control cells (Fig. S2). HBO1-null 293T clonal cell lines displayed the largest reduction in cell proliferation, followed by HeLa and then MCF7 single-cell clonal cell lines.
RNA interference knockdown of HBO1 does not affect cell proliferation or cell cycle dynamics.
The CRISPR/Cas9 system is efficient in producing specific and reproducible null mutations in HBO1 (Fig. S1); however, it is a relatively slow process, and our initial analysis was performed after 1 week. Hence, there is a possibility that a rapid reduction in HBO1 protein levels has an acute effect on DNA replication and cell cycle progression.
To investigate the possibility that acute downregulation of HBO1 has an effect different from that of genetic deletion, we performed siRNA knockdown using the same siRNA previously reported by Miotto and Struhl (5, 18). These authors describe experiments using siRNA 1 and siRNA 2; however, only one siRNA sequence is reported, as the two sequences shown are the forward and reverse oligonucleotides that form one double-stranded siRNA (18). The extent of siRNA knockdown was reported by Miotto and Struhl in 2006 (18) and in 2008 (5). We used the sequences GGCUAAGCCAGAGUUCUCATT and UGAGAACUCUGGCUUAGCCTG (18) to generate the double-stranded siRNA; we term this HBO1 siRNAMiotto. In addition, we purchased a second siRNA, HBO1 siRNAs253 (Thermo Fisher siRNA s253), which is a validated siRNA against exon 3 of human HBO1.
In our CRISPR experiments, HeLa cells showed a more prominent effect on proliferation than MCF7 cells. As HeLa cells were used by Miotto and Struhl in 2008 (5), 2010 (10), and 2011 (12), we examined HeLa cell cycle parameters after transfection with nonsilencing RNA, HBO1 siRNAMiotto, and HBO1 siRNAs253 as well as with untreated and Lipofectamine-only controls. Western blot analysis showed that both HBO1 siRNAMiotto and HBO1 siRNAs253 were similarly effective in reducing HBO1 protein levels (Fig. 2A) and reduced HBO1 protein at 48 and 72 h relative to nonsilencing control siRNA-treated, Lipofectamine-only-treated, and untreated control cells.
FIG 2.
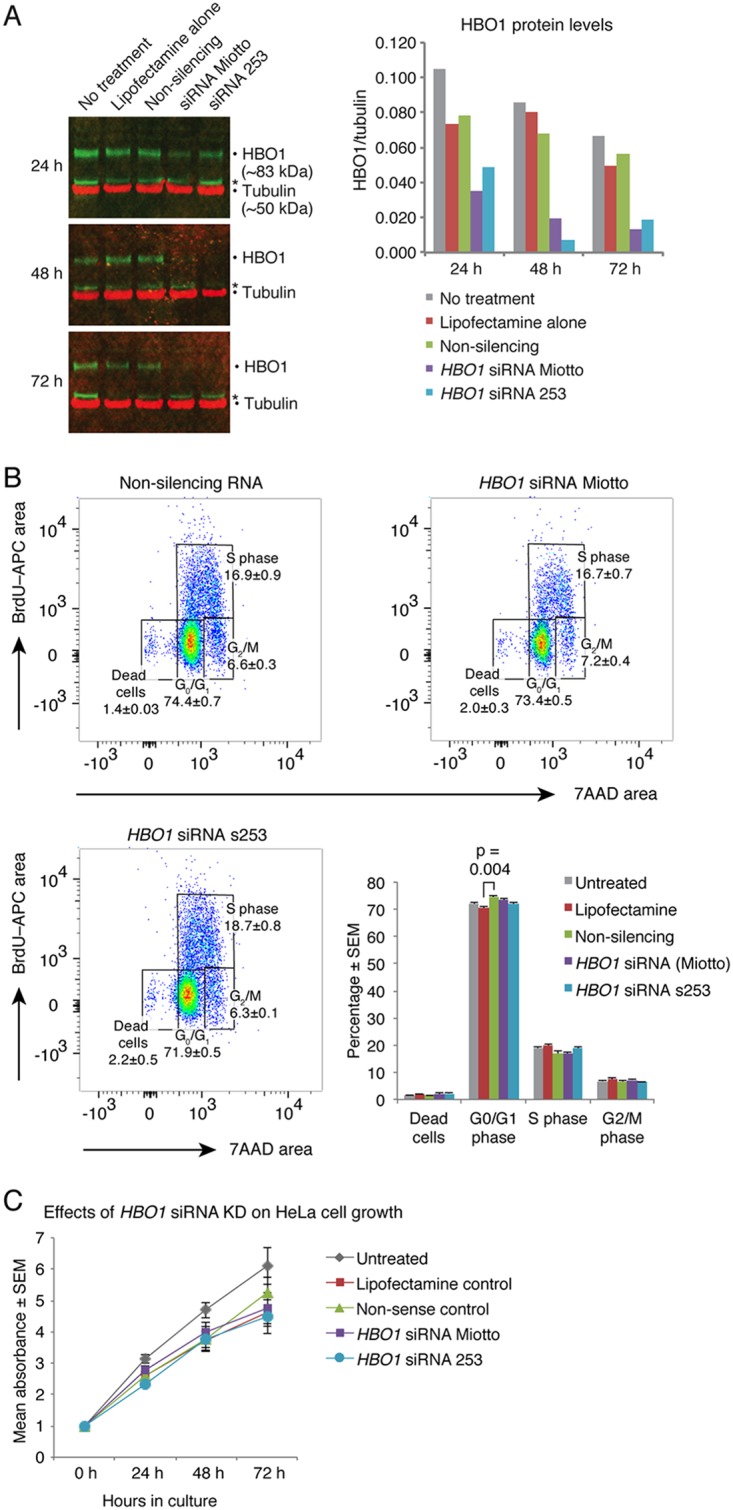
Acute reduction in HBO1 does not lead to perturbation of HeLa cell cycle parameters. (A) HBO1 siRNAMiotto and HBO1 siRNAs253 are effective in reducing HBO1 protein levels. (B) Flow cytometry quantification of BrdU and 7-AAD in HeLa cells 72 h after HBO1 knockdown using siRNA. Note that there is no difference in the rates of the nucleotide analogue BrdU during S phase comparing untreated cells, cells treated with Lipofectamine alone, or cells with Lipofectamine-mediated transfection of nonsilencing RNA, HBO1 siRNAMiotto, and HBO1 siRNAs253. (C) Treatment of HeLa cells with siRNAs targeting HBO1 mRNA does not impair proliferation. KD, knockdown.
No differences in cell cycle parameters were seen among these 5 treatment groups except for a slight reduction in G0/G1 when the Lipofectamine control was compared to the nonsilencing siRNA control (Fig. 2B). In particular, BrdU uptake, indicating the efficiency of DNA synthesis, was unaffected between treatment groups. Furthermore, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays showed no statistically significant differences in cell proliferation, although there was a slight reduction in cell proliferation between all cultures that were treated with Lipofectamine (with or without any siRNA) and untreated cells (Fig. 2C).
Loss-of-function mutations in HBO1 alter chromatin dynamics during replication.
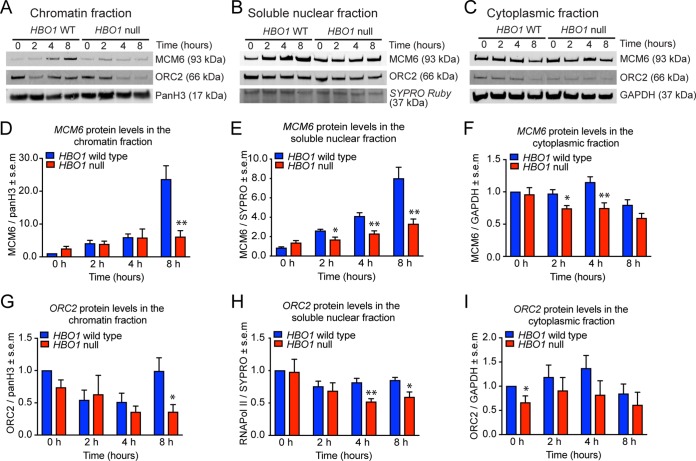
To assess the efficiency of ORC2 and MCM6 loading onto chromatin during the cell cycle, HeLa cells with an HBO1 deletion and control cells were synchronized using nocodazole-induced arrest and release, and the chromatin-enriched, soluble nuclear, and soluble cytoplasmic fractions were then assessed by Western blotting and densitometry. As reported previously by Iizuka and colleagues (6), we observed normal levels of MCM6 in the chromatin-enriched fraction 0 h, 3 h, and 4 h after release from nocodazole but a pronounced decrease at 8 h (Fig. 3A). Unlike observations by Iizuka and colleagues, this reduction in chromatin-associated MCM6 was not accompanied by a corresponding increase in the cytoplasmic fraction (Fig. 3A to F). Instead, MCM6 protein levels were reduced in the soluble cytoplasmic and nuclear fractions at time points preceding the reduction in chromatin-associated MCM6, namely, from 2 h (cytoplasmic and soluble nuclear fractions) versus 8 h (chromatin associated). Also, in contrast to the results reported by Iizuka and colleagues, who observed no effect on ORC2, we observed that the protein levels of OCR2 were similarly affected by the lack of HBO1, with reductions in the cytoplasm and soluble nuclear fractions preceding the reduction in the chromatin-enriched fraction (Fig. 3A, C, and G to I). Like ORC2 and MCM6, the levels of a number of other nuclear and cytoplasmic proteins commonly used as loading controls were also reduced in the absence of HBO1 (Fig. S3). Overall, these data suggest that the reduction in replication-licensing protein levels in the absence of HBO1 is not restricted to the chromatin or nuclear fraction but is also detectable in the cytoplasm. Furthermore, the reduction in protein levels appears to affect a range of proteins, including cytoplasmic and nuclear proteins. It should be noted that these cells were originally expanded from single cells for 4 to 6 weeks and then proliferated at a steady rate throughout 8 passages, and they were clearly viable at the end of the experiment. This indicates that HBO1 is not essential for sustained cell proliferation, survival, or DNA replication.
FIG 3.
Cytoplasmic, soluble nuclear, and chromatin-bound protein levels in HBO1-deleted HeLa cells compared to control cells. HeLa cells were synchronized for 16 h with 50 ng/ml nocodazole and released from nocodazole-induced cell cycle arrest for 2, 4, or 8 h or harvested without release at 0 h, as indicated, and fractionated into chromatin-enriched, soluble nuclear, and soluble cytoplasmic fractions. Lysed proteins were separated by SDS-PAGE, blotted, and subjected to immunodetection of specific proteins, as indicated. (A to C) Representative images of Western blots. (D to F) Densitometry results of MCM6 protein levels normalized to values for wild-type cells at 0 h and relative to loading controls. (G to I) Densitometry results of ORC2 protein levels normalized to values for wild-type cells at 0 h and relative to loading controls. Data are displayed as means ± standards errors of the means for eight experiments (incorporating 2 control cell lines for each of 2 sgRNAs [sgRNA1 and sgRNA2] and 4 HBO1-deleted cell lines for each of the 2 sgRNAs) and were analyzed by Student’s t test. Related data are displayed in Fig. S3 in the supplemental material, including protein levels of other proteins affected by the loss of HBO1.
Loss of HBO1 alters cell morphology.
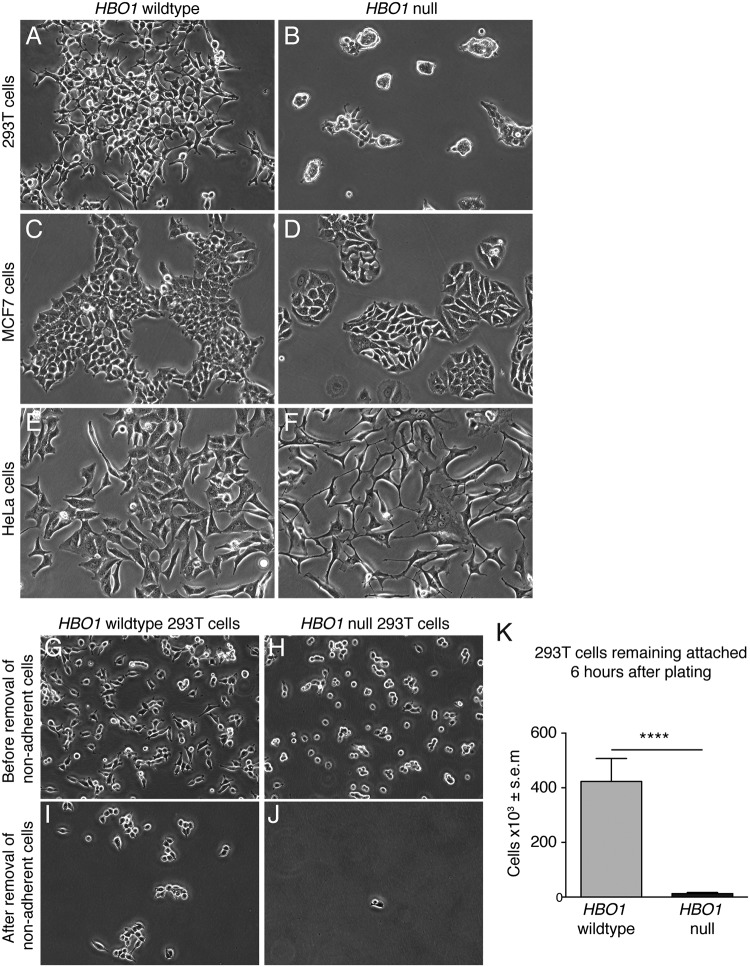
We observed significant changes in cell morphology in HBO1-null 293T and HeLa clonal cell lines (Fig. 4). Compared to HBO1 wild-type 293T cells, all HBO1-null 293T clonal cell lines grew in cell clusters upon seeding and failed to attach efficiently and spread out across culture dishes efficiently (Fig. 4A versus Fig. 4B). HBO1-null MCF7 cells showed only subtle morphological differences (Fig. 4C versus Fig. 4D), while HBO1-null HeLa clonal cell lines featured distinct cellular processes and tended to grow with a noticeably slender morphology compared to HBO1 wild-type control cell lines (Fig. 4E versus Fig. 4F). HBO1-null morphologies were consistent between HBO1 sgRNA1- and sgRNA2-transduced cells in all three cell types. We hypothesized that the failure of HBO1-null 293T cells to attach efficiently may be due to cell adhesion defects. To test the cell adhesion characteristics of the 293T cells, we seeded 2 million cells for 6 h before gentle removal of nonadherent cells with phosphate-buffered saline (PBS) and subsequently counted the remaining adherent cells. We observed a significant reduction in attached HBO1-null 293T cells compared to HBO1 wild-type control cell lines (Fig. 4G to J). In the absence of HBO1, there was a 33-fold reduction in the number of cells that adhered after 6 h of culture (Fig. 4K). The major defect in cell adhesion to the cell culture substrate suggests the possibility that the reduced cell number after passage observed by others (8) was not due to defects in DNA replication but rather was due to a failure to attach after passaging.
FIG 4.
HBO1 depletion alters cellular morphology. (A to F) Cell morphology of 293T, MCF7, and HeLa single-cell clonal lines. (G to K) 293T cells were cultured for 6 h before removal of nonadherent cells and then counted. ****, P < 0.0001.
HBO1 is essential for H3K14ac.
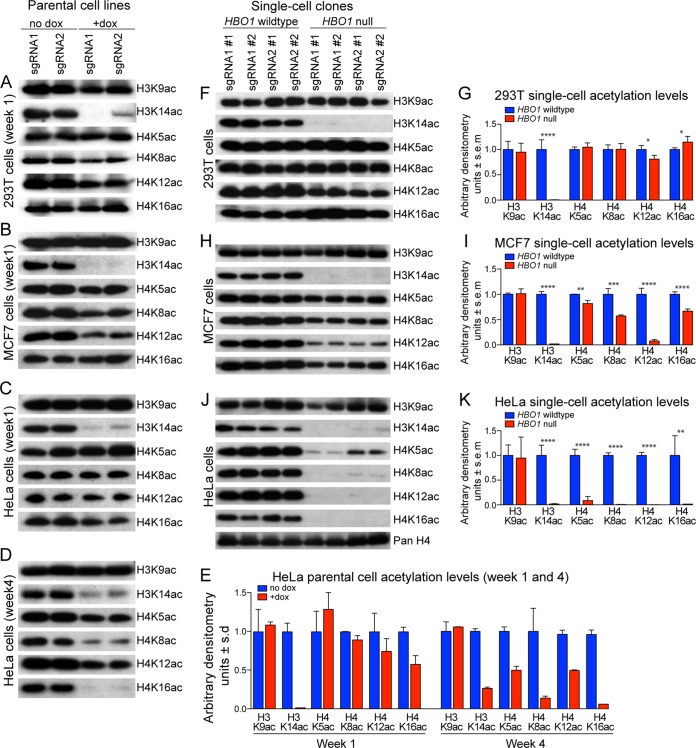
The loss of HBO1 correlated with a significant reduction in H3K14ac in all six parental cell lines (two sgRNAs and three cell types) (Fig. 5A to E). Loading controls are displayed in Fig. S4. In addition, the use of another antibody raised against acetylated H3K14 (catalogue number 070353; Merck Millipore) confirmed the loss of H3K14ac in HBO1-null 293T, MCF7, and HeLa cells (data not shown). Thus, the primary loss of H3K14ac as a result of a lack of HBO1 protein has been validated with different antibodies detecting the same modified histone residues in different cell types.
FIG 5.
HBO1 is essential for H3K14ac. (A to D) 293T, MCF7, and HeLa parental cells were treated with Dox for 3 days and assessed for histone acetylation levels 1 week (A to C) or 4 weeks (D) after Dox treatment. (E) HeLa parental cell acetylation levels were quantified by densitometry. Ponceau S staining was conducted as a loading control for the respective Western blots (see Fig. S4 in the supplemental material). Each lane represents one parental cell line transduced with HBO1 sgRNA1 or sgRNA2. HBO1 protein levels in the respective parental cell lines are shown in Fig. 1A. (F to K) 293T, MCF7, and HeLa clonal cell lines were assessed for histone acetylation levels approximately 4 to 6 weeks after the initial Dox treatment. Histone acetylation levels were quantified by densitometry in panels G, I, and K. Ponceau S staining was conducted as a loading control for the respective Western blots (Fig. S4). Each lane represents one clonal cell line transduced with HBO1 sgRNA1 or sgRNA2. HBO1 protein levels in the respective clonal cell lines are shown in Fig. 1B. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Data are presented as means ± standard deviations from 2 parental cell lines, each transduced with HBO1 sgRNA1 or -2, in panel E. Data are presented as means ± standard errors of the means from 4 clonal cell lines, i.e., 2 each transduced with HBO1 sgRNA1 or sgRNA2, in panels G, I, and K.
In 293T and MCF7 cells, we detected modest reductions in H4K8ac and H4K12ac, whereas H3K9, H4K5, and H4K16 acetylation levels were relatively unchanged (Fig. 5A and B). Notably, HBO1 sgRNA2 was less efficient in reducing HBO1 protein in 293T parental cells (Fig. 1A), and this was associated with more residual H3K14ac in these cells (Fig. 5A), suggesting a direct correlation between HBO1 protein levels and H3K14ac levels. HeLa parental cell lines displayed a reduction in only H3K14ac, while H3K9, H4K5, H4K8, H4K12, and H4K16 acetylation levels remained relatively unchanged 1 week after sgRNA induction (Fig. 5C and E). Interestingly, after 4 weeks of culture, these HeLa parental cell lines displayed significant reductions in H3K14ac, H4K8ac, and H4K16ac, while H4K5ac and H4K12ac were more modestly reduced (Fig. 5D and E). On the other hand, H3K9ac remained relatively unchanged (Fig. 5D and E).
Consistent with the findings observed in parental cell lines, HBO1-null 293T clonal cell lines displayed a dramatic reduction in H3K14ac and a modest reduction in H4K12ac. However, H3K9ac, H4K5ac, and H4K8ac levels were unaffected (Fig. 5F and G and Fig. S5). Interestingly, the H4K16ac level appears slightly increased in these HBO1-null cell lines, similarly to Hbo1-null MEFs (14). HBO1-null MCF7 clonal cell lines showed dramatic reductions in H3K14ac and H4K12ac as well as smaller reductions in H4K8ac and H4K16ac (Fig. 5H and I). In contrast, H3K9ac and H4K5ac remained relatively unchanged. Similarly, HBO1-null HeLa clonal cell lines showed dramatic reductions in H3K14ac but also in H4K5ac, H4K8ac, H4K12ac, and H4K16ac, while H3K9ac remained relatively unchanged (Fig. 5J and K). In addition, anti-pan-H4 Western immunoblotting confirmed that total histone H4 levels remained unchanged in HBO1-null HeLa clonal cell lines (further loading controls are shown in Fig. S5). Importantly, HBO1 sgRNA1 and sgRNA2 were consistent in producing the same histone acetylation changes in the respective cell lines at both the parental and single-cell clonal levels.
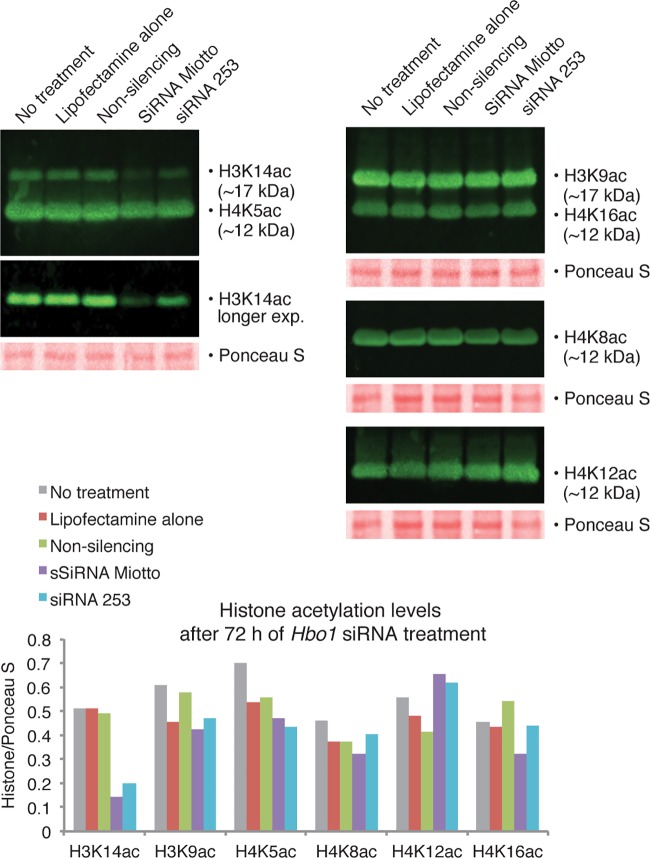
Acute loss of HBO1 leads to acute loss of H3K14ac but not H4ac.
To determine the acute effects of the loss of HBO1, we examined acetylation levels in HeLa cells after HBO1 siRNA knockdown. We transfected HeLa cells with nonsilencing RNA, HBO1 siRNAMiotto, and HBO1 siRNAs253 using Lipofectamine and compared histone acetylation levels with those in Lipofectamine-alone-treated and untreated controls. Histone acetylation and HBO1 protein levels were assayed 24 h, 48 h, and 72 h after transfection. As noted above (Fig. 2), HBO1 protein levels were reduced by both siRNA molecules by 24 h, and the maximum reduction in HBO1 was achieved at 48 h. Western immunoblotting revealed a decrease in the level of H3K14ac, with the maximum relative reduction occurring 72 h after transfection with siRNAMiotto and siRNAs253 (Fig. 6), which was already evident at 24 and 48 h (Fig. S5). Only minor and inconsistent differences in H4K5, H4K8, H4K12, and H4K16 acetylation levels were seen between treatment groups after transfection with silencing siRNAs compared to nonsilencing RNA or Lipofectamine-alone-treated and untreated controls (Fig. 6 and Fig. S5).
FIG 6.
HBO1 siRNA-mediated knockdown reduces H3K14ac levels. Untreated HeLa cells, HeLa cells treated with Lipofectamine alone, or HeLa cells with Lipofectamine-mediated transfection of nonsilencing RNA, HBO1 siRNAMiotto, and HBO1 siRNAs253 were assayed for histone lysine acetylation. Transfection of HBO1 siRNAMiotto and HBO1 siRNAs253 resulted in reduced levels of H3K14ac but no changes in H3K9ac or H4K5ac, H4K8ac, H4K12ac, or H4K16ac. Western blots were performed 72 h after treatment. Western blots for the 24-h and 48-h time points are displayed in Fig. S5 in the supplemental material.
Loss of HBO1 leads to deregulation of gene expression, particularly genes controlling cell adhesion.
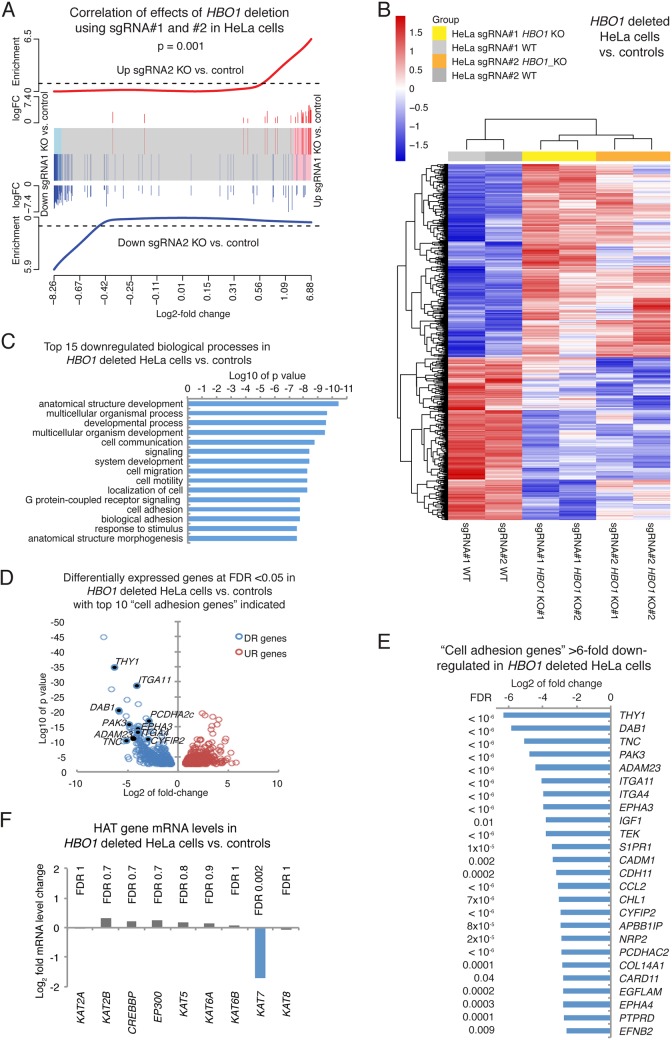
Two clonal HBO1-deleted cell lines per sgRNA (sgRNA1 and -2) and per cell type (293T and HeLa) as well as one clonal control cell line per sgRNA (sgRNA1 and -2) and per cell type (293T and HeLa) were used for RNA isolation, library construction, and RNA sequencing (RNA-Seq). Despite the use of clonal cell lines, which has the tendency to reveal differences between individual subclones (20), the genotypes segregated by multidimensional scaling in dimension 1 for the HeLa cells and in dimension 2 for the 293T cells, with differences between sgRNA1 and -2 accounting for dimension 2 in the HeLa cells and dimension 1 in the 293T cells (Fig. S6). Results between the effects of HBO1 deletion caused by sgRNA1 and -2 were positively correlated for HeLa cells (P = 0.001) (Fig. 7A) and 293T cells (P = 0.03) (Fig. S6). Subsequently, the combined data obtained with sgRNA1 and sgRNA2 were analyzed to focus on gene expression changes caused by HBO1 deletion and supported by both sgRNAs.
FIG 7.
Cell adhesion genes are downregulated in the absence of HBO1. Shown are RNA-Seq results for HeLa cells with an HBO1 deletion using sgRNA1 and sgRNA2 versus control cells (A to F) and combined analysis of sgRNA1 and sgRNA2 effects (B to F). (A) Barcode plot showing a high correlation between genes up- and downregulated as a consequence of the HBO1 deletion using sgRNA1 compared to sgRNA2 in HeLa cells. log FC, log fold change. (B) Heat map of genes differentially expressed between HBO1 knockout (KO) (guide 1, clones 1 and 2; guide 2, clones 1 and 2) and control HeLa cell (WT) clones induced for the expression of guide 1 or guide 2 in the absence of Cas9. (C) Biological process gene ontology terms enriched in genes downregulated in HBO1-null and control HeLa cells. (D) Volcano plot displaying log10 P values over log2 fold expression differences between HBO1-null and control HeLa cells, with downregulated (DR) “cell adhesion genes” indicated. UR, upregulated. (E) Cell adhesion genes >6-fold downregulated in HBO1-null and control HeLa cells. Two clonal control cell lines (one each for sgRNA1 and -2) and 4 clonal HBO1-null cell lines (two each for sgRNA1 and -2) were used. Data were analyzed by RNA sequencing as described in Materials and Methods. Genes differentially expressed with an FDR of <0.05 were considered significant. Related data on 293T and HeLa cells are displayed in Fig. S6 and S7 in the supplemental material. All data are provided in Tables S4 and S5. (F) Histone acetyltransferase (HAT) gene expression, apart from HBO1, is not changed.
The lack of HBO1 resulted in the up- and downregulation of more than 800 genes, with false discovery rates (FDRs) of <0.05 for both cell types (856 genes in 293T and 818 in HeLa cells) (Fig. 7B, Fig. S6 and S7, and Table S4). Genes associated with a number of gene ontology (GO) annotation terms were affected in both 293T cells and HeLa cells (Table S5), including cell adhesion genes in HeLa cells (Fig. 7C to E and Table S5) and 293T cells (Fig. S7 and Table S5), congruent with our finding that cell adhesion is severely affected in both cell types (Fig. 4). Cell adhesion genes that were downregulated more than 6-fold in HeLa cells included alpha integrin (IGTA11 and IGTA4), tenascin (TNC), protocadherin (PCDHA2C), and ephrin receptor (EPHA3 and EPHA4) genes (Fig. 7E and Table S4). Other biological processes enriched for genes downregulated in HBO1 KO HeLa cells included cell communication, cell migration, and cell motility (Fig. 7C and Table S5). Gene upregulation was less pronounced in amplitude (Table S5), and upregulated biological processes included signal transduction and organ development (Fig. S7).
The cell cycle regulator gene CDKN1A was upregulated approximately 2-fold, in agreement with the moderate reduction in cell proliferation (Fig. 1), suggesting an increase in cells undergoing cell cycle arrest after deletion of HBO1. However, this did not result in complete cell cycle arrest (Fig. 2), and all three human cell lines continued to grow exponentially (Fig. 1). The loss of HBO1 function did not affect the expression of other histone acetyltransferase genes (Fig. 7F).
DISCUSSION
Utilizing the same cell lines as the ones used to study HBO1 function in previous reports, we have shown that HBO1 is not, in fact, required for DNA replication or cell proliferation. In contrast, we show that HBO1 is required for global H3K14ac in 293T, MCF7, and HeLa cells. These results were obtained both in an acute-reduction model (siRNA knockdown) and after the generation of loss-of-function mutations (CRISPR/Cas9). Our results do not exclude a minor, indirect role for HBO1 in cell cycle progression, as might be expected given its major role in transcription (14); indeed, we saw an attenuated response to cell cycle arrest and release, with lower levels of both MCM6 and ORC2 in the nuclear fraction. However, these results demonstrate that HBO1 is not essential for DNA replication. The successful derivation of clonal cell lines lacking HBO1 indicates that HBO1 is not essential even for the stringent test of clonal proliferation from a single sorted cell. Furthermore, we were unable to demonstrate any effect of acute HBO1 knockdown on DNA synthesis, cell cycle progression, or the acetylation of H4 using either the siRNA reported by Miotto and Struhl in 2006 (18) and 2008 (5) or an unrelated but equally effective siRNA.
Our work is in conflict with previous reports suggesting that HBO1 is essential for DNA replication and H4 acetylation. All 6 of our Dox-treated parental cell lines and their 12 HBO1-null clonal daughter cell lines showed a consistent, and complete, loss of H3K14ac. Interestingly, HBO1-null HeLa cells also showed a delayed but almost complete loss of H4K5ac, H4K8ac, H4K12ac, and H4K16ac along with H3K14ac. It should be noted that reductions in H3K14ac occurred first (week 1), before decreases in H4 acetylation (week 4). Similarly, a large reduction in H3K14ac was observed in MCF7 parental cell lines before reductions in H4 acetylation in clonal cell lines. This suggests that the primary consequence resulting from a loss of HBO1 is a reduction in H3K14ac, while reductions in H4 acetylation appear to be a secondary consequence. This may indicate a dependence of other histone acetyltransferases on HBO1 H3K14ac activity, specifically in HeLa and MCF7 cells. It is indeed intriguing and novel that HBO1-null HeLa cells are viable and proliferate normally with an almost complete lack of H3K14 and H4 acetylation, particularly as H3K14ac is the second most abundant acetylation modification in the nucleus of human cells, with >15% of sites acetylated (21).
We note that the secondary decreases in H4 acetylation observed in our study are limited to MCF7 and HeLa cells, which are of cancerous origin. This could suggest that cancerous cells may be indirectly dependent on HBO1 for normal levels of H4 acetylation. Interestingly, the secondary decreases in H4 acetylation in our HBO1-null HeLa cells included H4K16ac. H4K16 is acetylated by MOF during development (22) and in human cell lines, including HeLa cells (23), suggesting an interaction between HBO1 and MOF function. It should also be noted that previous studies investigating HBO1 acetylation activity by shRNA and siRNA knockdown in 293T cells (8) and HeLa cells (10) used a panel of anti-acetylated-histone antibodies, including an antibody raised against diacetylated H3, which detects both acetylated H3K9 and H3K14. Interestingly, H3K9ac, catalyzed by MOZ (24, 25) and GCN5/PCAF (26), increases in the absence of H3K14ac in embryonic fibroblasts (14). Thus, any decreases in H3K14ac may have been masked by normal or elevated levels of H3K9ac. However, this does not account for the relatively unchanged H4 acetylation levels observed in our HBO1-null 293T cells. With regard to cell-free assays showing a correlation between HBO1 and H4 acetylation (11), it should be noted that histone acetyltransferases display altered and greater histone residue specificity when assayed as part of their native protein complex than in cell-free acetylation assays (27–29).
Proliferation studies using HBO1 depletion by shRNA in 293T cells assessed cell growth only during 3 days of culture, during which HBO1-deficient cells were reported to stop growing altogether, leading to the suggestion that HBO1 was essential for cell proliferation (8). shRNA knockdown of HBO1 in MCF7 cells (8) and HeLa cells (9) was reported to result in a dramatic (>2-fold) decrease in the percentage of BrdU-positive cells in the S phase of the cell cycle. In contrast, we observed only modest changes in HBO1-null 293T cells (16.8% decrease; P < 0.01), MCF7 cells (11.97% increase; not significant), and HeLa cells (17.4% decrease; P < 0.01) after the CRISPR-mediated complete loss of HBO1. Thus, it appears that HBO1 shRNA may have detrimental effects on cell cycle progression. Indeed, both studies utilized the same shRNA vectors and sequences (8, 9) and were hence potentially subject to the same off-target effects.
The initial discovery of HBO1 by Iizuka and Stillman (2) found HBO1 associated with ORC1 in 293 cells. However, only a small portion of the HBO1 protein associated with hORC1 in human cells (30). Nevertheless, this finding led to an initial focus on this association with the DNA prereplication complex, almost to the exclusion of the majority of HBO1 that is not associated with ORC1. It should also be noted that, aside from a role in DNA replication, the association between HBO1 and ORC1 may reflect a role in transcriptional regulation, as ORC proteins appear to be involved in transcriptional silencing in Saccharomyces cerevisiae (31–33). As observed by Iizuka and colleagues (6), we found normal levels of MCM6 in the chromatin-enriched fraction 0 h, 3 h, and 4 h after release from nocodazole but a pronounced decrease at 8 h. However, unlike the observations made by Iizuka and colleagues, in our experiments, this reduction in chromatin-associated MCM6 was not accompanied by a corresponding increase in the cytoplasmic fraction. Instead, we observed a reduction in MCM6 in the cytoplasmic, soluble nuclear, and chromatin-enriched fractions, suggesting that the lack of HBO1 affected the production of normal levels of protein in a more general way. Low levels of protein were observed for MCM6, ORC2, emerin, and tubulin but not histone H3 or RNA polymerase II (Pol II). It was previously suggested that HBO1 facilitates the efficient loading of replication-licensing proteins onto chromatin by acetylating histone H3 in the surrounds of replication initiation sites (34), and this is entirely consistent with our data. However, the continued exponential proliferation of human 293T, HeLa, and MCF7 cells in this study and of mouse embryonic fibroblasts (14) indicates that while HBO1 may aid in the production and chromatin association of replication-licensing proteins, DNA replication can occur in its absence at a near-normal rate.
We observed a marked loss of the cell adhesion capacity of HBO1-null 293T cells (97% of 293T cells did not adhere in a cell adhesion assay), which would have consequently reduced the potential of these cells to proliferate during culture. We suggest that the inability of HBO1-null 293T cells to attach efficiently contributed significantly to their reduced proliferation rates, rather than any underlying defect in DNA replication. Indeed, HBO1-null 293T, MCF7, and HeLa cells displayed robust DNA synthesis and were capable of long-term and sustained cell proliferation. Our data demonstrate that HBO1 is not essential for DNA replication or cell proliferation.
The work presented in this report is consistent with our previous findings that demonstrated a role for HBO1 in H3K14 acetylation in mice while being dispensable for H4 acetylation, cell proliferation, and DNA synthesis (14, 15). We proposed that HBO1-mediated H3K14ac was essential for regulating gene expression, particularly during cellular differentiation. Indeed, there have been subsequent reports confirming a role for HBO1 in H3K14 acetylation in vivo. As part of the HBO1-Brd1/Brpf2 complex, HBO1 has been reported to be essential for H3K14ac and the transcriptional activation of erythroid developmental regulator genes such as Gata1 but dispensable for cell proliferation (35). Interestingly, the use of two independent shRNAs to knock down Hbo1 in this study caused a loss of H3K14ac but not H3K9ac, H4K5ac, H4K8ac, H4K12ac, or H4K16ac (35). In addition, HBO1 H3K14ac activity as part of the HBO1-Brd1/Brpf2 complex has also been implicated in CD8 expression and thymocyte differentiation (15, 36). HBO1 was also reported to be required for H3K14ac at origins of replication found in the vicinity of transcription start sites (34). Furthermore, extensive biochemical characterizations of HBO1 protein complex members have demonstrated that HBO1 indeed acetylates histone H3 depending on the protein complex subunit composition (37, 38) and is essential for the transcriptional regulation of important cell cycle regulator genes such as p21 (39). These findings highlight the possibility that HBO1 primarily regulates gene expression by virtue of its H3K14ac activity and, hence, cell differentiation and proliferation, compared to being directly involved in the DNA replication process itself.
We propose that HBO1 and H3K14ac are important for the de novo activation of genes. Although we observed decreased proliferation and altered cell cycle profiles in HBO1-null 293T, MCF7, and HeLa cell types in this study, we did not observe increases in cell death or cessations of cell proliferation and DNA replication, as one would expect if HBO1 were indeed essential for DNA replication. Instead, we observed significant changes in cell morphology and cell adhesion properties in HBO1-null cells. HBO1 has been reported to be highly expressed in some cancer cell types (11), which may indicate that HBO1 is involved in key oncogenic processes, including cell adhesion, which is of particular importance during the metastatic spread of cancer cells. Transcriptomic analysis showed HBO1 is essential for regulating the expression of genes involved in fundamental processes during each passage interval, in particular cell adhesion (22 cell adhesion genes >5-fold downregulated). Interestingly, the expression of most genes, apart from those involved in cell adhesion, is unaffected by the loss of HBO1 and H3K14ac. This is consistent with our previous observation that transcriptionally active genes continue to be active in the absence of HBO1 but that HBO1 is essential for de novo gene activation, particularly during embryonic development (14). In contrast, cells with an established transcription profile, such as the immortalized cells and cancer cells examined here, are relatively insensitive to the loss of HBO1 and continue to express many of the genes required for their continued survival and proliferation.
In conclusion, we have shown that HBO1 is essential for H3K14 acetylation but not DNA replication or cell proliferation in 293T, MCF7, and HeLa cell lines. Our data suggest that HBO1 has important roles in regulating cell morphology and adhesion.
MATERIALS AND METHODS
Lentiviral vectors.
Our Dox-inducible sgRNA and constitutively expressed CRISPR/Cas9 lentiviral vectors were described in detail previously (19).
sgRNA design.
HBO1 sgRNA1 and sgRNA2 sequences were obtained using the MIT CRISPR Design tool (Zhang Lab, MIT, 2013; crispr.mit.edu). To clone HBO1 sgRNA1 and sgRNA2 into the inducible sgRNA lentiviral vector, 24-bp complementary oligonucleotides containing the sgRNA sequence and a 4-bp overhang (TCCC for the forward oligonucleotide and AAAC for the reverse oligonucleotide) were purchased (IDT, Australia). The sequence used for HBO1 sgRNA1 was ACCAGGTATCAAGCTCATAG. The sequence used for HBO1 sgRNA2 was TGTGCCGGCGGAGTATCGTT. The complementary oligonucleotides for sgRNA1 and sgRNA2 were each annealed, cloned into the sgRNA lentiviral vectors, and sequenced to confirm correct cloning.
Cell culture.
293T and HeLa cells were grown in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS; Gibco), 100 U/ml penicillin, 100 mg/ml streptomycin (Gibco), and 2 mM l-glutamine (Gibco). Cells were cultured at 37°C in 5% CO2. MCF7 cells were grown under the same conditions but with the addition of 10 μg/ml insulin.
Virus production and transduction of cell lines.
To produce lentiviral particles, 293T cells were grown on a 10-cm petri dish with the viral packaging constructs pMDL (5 μg), pRSV-rev (2.5 μg), and pVSV-G (3 μg) together with 10 μg of HBO1 sgRNA1 or sgRNA2 vector DNA and 10 μg of Cas9 vector DNA. X-tremeGENE 9 DNA transfection reagent (Roche) was used according to the manufacturer’s instructions for transient 293T cell transfections. The supernatant containing lentiviral particles was collected 48 h after transfection and passed through a 0.45-μm filter, and 1 ml of the viral supernatant was placed on 293T, HeLa, and MCF7 cells in a 12-well plate for 72 h at 37°C.
Doxycycline induction of sgRNA expression.
To induce sgRNA expression, cells were cultured in culture medium supplemented with 1 μg/ml doxycycline hyclate (Dox; Sigma) for 72 h, before changing Dox-supplemented culture medium to fresh medium. Cells were then expanded for another 7 days before they were used for either single-cell sorting or the assessment of parental cell lines. During this time in culture, no arrest in cell proliferation was observed. After the initial 72 h of Dox treatment, HeLa parental cell lines were exposed to Dox for 24 h at the start of each week until week 4 to obtain cells for additional histone acetylation analyses (Fig. 3D and E). These analyses served as an additional time point between week 1 parental cell line analyses (Fig. 3C and E) and single-cell-derived clonal cell line analyses (Fig. 3J and K). 293T and MCF7 parental cell lines were not repeatedly treated with Dox.
Fluorescence-activated cell sorting and next-generation sequencing of CRISPR/Cas9-mediated indels.
Cell lines successfully transduced with the HBO1 sgRNA vector and Cas9 were analyzed and sorted on an Aria W flow cytometer by gating on GFP- and mCherry-medium-to-high cells. After transduction, GFP- and mCherry-double-positive cells were enriched during the first cell sort. Cells were then exposed to Dox treatment before undergoing a second sort, again selecting GFP- and mCherry-double-positive cells for sorting into 96-well plates. A single cell was sorted into each well for the derivation of clonal cell lines. Non-Dox-treated parental cell lines were sorted in parallel to establish HBO1-expressing clonal cell lines for experimental controls. DNA was extracted from clonal cell lines and used for next-generation sequencing of CRISPR/Cas9-mediated indels using the Illumina MiSeq platform as described previously (19). Briefly, extracted DNA was used in a two-step PCR protocol to produce amplicon sequences with CRISPR/Cas9-mediated indels flanked with 8-bp indexing sequences. Each indexed pool of amplicons was diluted to 12 pM for sequencing on an Illumina MiSeq instrument with the 300-cycle kit according to the manufacturer’s instructions. HBO1 sgRNA off-target sites were determined using the CCTop CRISPR/Cas9 target online predictor tool (40) (http://crispr.cos.uni-heidelberg.de). The top 5 predicted off-target sites per sgRNA were assessed for CRISPR/Cas9-mediated indels. Full details of predicted off-target sites are listed in Tables S1 and S2 in the supplemental material. The actual frequencies of off-target mutations are listed in Table S3.
siRNA and transfection.
HBO1 siRNAMiotto (GGCUAAGCCAGAGUUCUCATT and the complementary sequence UGAGAACUCUGGCUUAGCCTG) was ordered from Ambion Thermo Fisher Scientific. HBO1 siRNA s253 was purchased from Ambion Thermo Fisher Scientific and is targeted to HBO1 (KAT7) exon 3 (s253 located at chromosome 17, 49,805,412 on build GRCh38; GGAUGCCCACUGUAUCAUATT and UAUGAUACAGUGGGCAUCCTG). Transfections were performed using the Lipofectamine RNAiMAX reagent in 2.3 μl/ml of culture medium, and the mixture was incubated for 30 min, essentially according to the manufacturer’s protocol (Thermo Fisher Scientific).
BrdU incorporation, cell cycle analysis, and cell proliferation assays.
BrdU/7-AAD cell cycle analyses were conducted using the APC BrdU flow kit (BD Pharmingen) according to the manufacturer’s instructions. Briefly, cells were incubated in medium supplemented with 10 μM BrdU for 45 min. Cells were then fixed, permeabilized, and treated with 300 μg/ml DNase for 1 h at 37°C to expose incorporated BrdU. Cells were stained with APC-conjugated anti-BrdU antibodies and a 7-AAD solution before flow cytometric analysis on an LSR II flow cytometer (BD Pharmingen). Cell debris and doublets were excluded using forward- and side-scatter parameters, and the remaining single cells were gated based on distinct cell populations analyzed by APC (BrdU) and 7-AAD intensities. For the long-term cell proliferation assays, cells were serially passaged approximately once a week for a total of 8 passages during approximately 8 weeks. At the start of each passage interval, 30,000 cells were seeded into a well of a 6-well plate and left to grow until subconfluent. Cells were then washed twice in a phosphate-buffered saline solution, trypsinized, and counted with a Countess automated cell counter with the exclusion of dead cells (Life Technologies), and 30,000 cells were reseeded for the next passage. Data are presented as either a cumulative proliferation curve (Fig. 1F) or individual passage intervals (Fig. S2).
Histone and HBO1 antibodies.
Antibodies used were raised against acetylated H4K12 (catalogue number AB1761; used at a 1:500 dilution), purchased from Abcam; H4K5 (catalogue number 07-327; 1:2,000), H4K8 (catalogue number 07-328; 1:2,000), and H4K16 (catalogue number 07-329; 1:2,000), purchased from Merck Millipore; pan-H4 (catalogue number 13919S; 1:2,000), acetylated H3K9 (catalogue number 9649; 1:2,000), and acetylated H3K14 (catalogue number 7627; 1:2,000), purchased from Cell Signaling; HBO1 (catalogue number 13751-1-AP; 1:1,000), purchased from Protein Tech; and actin (catalogue number sc-1616; 1:2,000), purchased from Santa Cruz.
Western immunoblotting.
Histone extraction and Western immunoblotting techniques were described previously (14). 293T and MCF7 parental cell lines were assessed for HBO1 protein levels and histone acetylation levels 1 week after Dox treatment. HeLa parental cell lines were assessed for HBO1 protein levels and histone acetylation levels 1 week and 4 weeks after Dox treatment. All cell lines were single-cell sorted 1 week after Dox treatment, expanded, and assessed for HBO1 protein levels and histone acetylation levels between 4 and 6 weeks after the initial Dox treatment.
Cell synchronization and subcellular fractionation.
HeLa cells were plated onto 10 15-cm tissue culture (TC)-treated dishes (2 dishes per time point) and grown for 2 days in DMEM containing 10% heat-inactivated fetal bovine serum (HI-FBS) and 1% penicillin-streptomycin. When the cell density reached 70% confluence, cells were synchronized by mitotic arrest through treatment with 50 ng/ml of nocodazole (Sigma-Aldrich) for 16 h. Mitotic cells were detached by manual shaking and gentle washing of medium over the plate, collected, and centrifuged at 200 × g for 3 min. The cells were washed with PBS twice, fresh growth medium was added, and cells were plated onto 10-cm dishes to resume progression through the cell cycle. Proteins from the cytoplasmic, soluble nuclear, and chromatin-bound cellular compartments were isolated using a subcellular protein fractionation kit for cultured cells (Thermo Scientific) according to the manufacturer’s instructions. This occurred 0, 2, 4, and 8 h after nocodazole release. The total protein concentration of each of the protein fractions was determined with the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific) according to the manufacturer’s instructions. Eight micrograms of total protein of each fraction was separated by SDS-PAGE, and proteins were blotted and probed with the following primary test antibodies: anti-MCM6 (1:2,000) (93-kDa rabbit monoclonal, catalogue number ab201683; Abcam), anti-ORC2 (1:3,000) (66-kDa rat monoclonal, clone 3G6, catalogue number ab22537; Abcam), anti-RNA polymerase II (1:2,000) (215-kDa mouse monoclonal CTD4H8, catalogue number 05-623; Millipore), anti-pan-histone H3 (1:30,000) (17-kDa rabbit monoclonal, catalogue number 05-928; Millipore), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:10,000) (rabbit monoclonal, clone 14C10; Cell Signaling catalogue number 2118S), anti-alpha tubulin (1:4,000) (50-kDa mouse monoclonal, catalogue number T5168; Sigma-Aldrich), and antiemerin (1:1,000) (29-kDa [observable, ∼32-kDa] rabbit monoclonal, catalogue number ab156871; Abcam). SYPRO ruby protein blot stain (catalogue number S11791; Invitrogen) was used as a loading control for the soluble nuclear fraction after the levels of four different nuclear proteins commonly used as nuclear loading controls were shown to be affected by the loss of HBO1.
RNA isolation, library preparation, sequencing, and analysis.
RNA was isolated from clonal 293T and HeLa cell lines with and without HBO1 deletion using an RNeasy microkit (Qiagen), and libraries were prepared using the TruSeq RNA library prep kit (Illumina). Within each cell line, there were 2 wild-type (WT) samples (sgRNA1 and sgRNA2, with no Cas9) and 4 HBO1 knockout (KO) (2 sgRNA1 and 2 sgRNA2, each with Cas9) samples. Libraries were sequenced on the Illumina NextSeq 500 machine to generate 80-bp paired-end reads for data analysis. Reads were aligned to the human genome, build hg38, using Rsubread align (1.30.5) (41). Approximately 89% of reads were successfully mapped for each sample. Reads overlapping each Entrez gene were summarized into counts using Rusbread’s featureCounts function and inbuilt RefSeq annotation. Differential expression analyses were then undertaken using the edgeR (42) and limma (43) software packages.
Lowly expressed genes were filtered from the data using edgeR’s filterByExpr function with default settings. Additionally, obsolete gene identifications were also removed. After filtering, 14,755 genes remained. TMM normalization (44) was then applied to all samples. Dispersions were estimated using edgeR’s estimateDisp function with robust set to true. Differential expression was then assessed between the KO groups and their respective WT samples as well as between the combined KO and WT groups. For this analysis, negative binomial generalized log-linear models were fit to the data, and differential expression was evaluated using likelihood ratio tests (44). P values were adjusted to control the false discovery rate (FDR) below 5% using the Benjamini-Hochberg method (45).
The barcode plot was generated using limma’s barcodeplot function, and the heat map was generated using the pheatmap software package. Gene ontology (GO) analyses were carried out using limma’s goana function.
Statistical analyses.
Statistical analyses were conducted using GraphPad Prism. Samples were compared using two-tailed unpaired t tests.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stephen Wilcox, Liz Milla, Liam O’Connor, and the WEHI Systems Biology and Personalized Medicine division for help with next-generation sequencing of CRISPR/Cas9-induced indels.
This work was supported by the Australian National Health and Medical Research Council under senior research fellowships 1003435 (T.T.) and 575512 (A.K.V.) and project grants 1029481, 103070, 1051078 (to T.T. and A.K.V.), and 1049720 (to M.J.H.) and by operational infrastructure grants from the Australian Federal Government (IRISS) and the Victorian State Government (OIS).
A.J.K., S.E., R.E.M., and L.T. carried out experiments. A.J.K., T.T., A.L.G., G.K.S., and A.K.V. analyzed data. T.T. conceived and initiated the project. A.K.V. and T.T. supervised the project. A.J.K., T.T., and A.K.V. wrote the manuscript. M.J.H. contributed ideas and reagents and interpreted results.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Voss AK, Thomas T. 2009. MYST family histone acetyltransferases take center stage in stem cells and development. Bioessays 31:1050–1061. doi: 10.1002/bies.200900051. [DOI] [PubMed] [Google Scholar]
- 2.Iizuka M, Stillman B. 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem 274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 3.Machida YJ, Hamlin JL, Dutta A. 2005. Right place, right time, and only once: replication initiation in metazoans. Cell 123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Burke TW, Cook JG, Asano M, Nevins JR. 2001. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem 276:15397–15408. doi: 10.1074/jbc.M011556200. [DOI] [PubMed] [Google Scholar]
- 5.Miotto B, Struhl K. 2008. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev 22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iizuka M, Matsui T, Takisawa H, Smith MM. 2006. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol 26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 8.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. 2006. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell 21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Wu ZQ, Liu X. 2008. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc Natl Acad Sci U S A 105:1919–1924. doi: 10.1073/pnas.0712063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miotto B, Struhl K. 2010. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iizuka M, Takahashi Y, Mizzen CA, Cook RG, Fujita M, Allis CD, Frierson HF Jr, Fukusato T, Smith MM. 2009. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene 436:108–114. doi: 10.1016/j.gene.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miotto B, Struhl K. 2011. JNK1 phosphorylation of Cdt1 inhibits recruitment of HBO1 histone acetylase and blocks replication licensing in response to stress. Mol Cell 44:62–71. doi: 10.1016/j.molcel.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, Vaziri C, Cote J, Panchenko MV. 2008. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem 283:28817–28826. doi: 10.1074/jbc.M801407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kueh AJ, Dixon MP, Voss AK, Thomas T. 2011. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol 31:845–860. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman DM, Voss AK, Thomas T, Allan RS. 2017. Essential role for the histone acetyltransferase KAT7 in T cell development, fitness, and survival. J Leukoc Biol 101:887–892. doi: 10.1189/jlb.1MA0816-338R. [DOI] [PubMed] [Google Scholar]
- 16.Baell JB, Leaver DJ, Hermans SJ, Kelly GL, Brennan MS, Downer NL, Nguyen N, Wichmann J, McRae HM, Yang Y, Cleary B, Lagiakos HR, Mieruszynski S, Pacini G, Vanyai HK, Bergamasco MI, May RE, Davey BK, Morgan KJ, Sealey AJ, Wang B, Zamudio N, Wilcox S, Garnham AL, Sheikh BN, Aubrey BJ, Doggett K, Chung MC, de Silva M, Bentley J, Pilling P, Hattarki M, Dolezal O, Dennis ML, Falk H, Ren B, Charman SA, White KL, Rautela J, Newbold A, Hawkins ED, Johnstone RW, Huntington ND, Peat TS, Heath JK, Strasser A, Parker MW, Smyth GK, Street IP, Monahan BJ, Voss AK, Thomas T. 2018. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560:253–257. doi: 10.1038/s41586-018-0387-5. [DOI] [PubMed] [Google Scholar]
- 17.MacPherson L, Anokye J, Yeung MM, Lam EYN, Chan YC, Weng CF, Yeh P, Knezevic K, Butler MS, Hoegl A, Chan KL, Burr ML, Gearing LJ, Willson T, Liu J, Choi J, Yang Y, Bilardi RA, Falk H, Nguyen N, Stupple P, Peat TS, Zhang M, de Silva M, Carrasco-Pozo C, Avery VM, Khoo PS, Dolezal O, Dennis ML, Nuttall S, Surjadi R, Newman J, Ren B, Leaver DJ, Sun Y, Aell JB, Dovey O, Vassiliou GS, Grebien F, Dawson SJ, Street IP, Monahan BJ, Burns CJ, Choudhary C, Blewitt ME, Voss AK, Thomas T, Dawson MD. HBO1 is required for the maintenance of leukaemia stem cells. Nature, in press. doi: 10.1038/s41586-019-1835-6. [DOI] [PubMed] [Google Scholar]
- 18.Miotto B, Struhl K. 2006. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol Cell Biol 26:5969–5982. doi: 10.1128/MCB.00696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, Wilcox S, Tai L, Strasser A, Herold MJ. 2015. An inducible lentiviral guide RNA platform enables the identification of tumor essential genes and novel tumor promoting mutations in vivo. Cell Rep 10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Martinez AO, Norwood TH, Prothero JW, Martin GM. 1978. Evidence for clonal attenuation of growth potential in HeLa cells. In Vitro 14:996–1002. doi: 10.1007/bf02616213. [DOI] [PubMed] [Google Scholar]
- 21.Hansen BK, Gupta R, Baldus L, Lyon D, Narita T, Lammers M, Choudhary C, Weinert BT. 2019. Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nat Commun 10:1055. doi: 10.1038/s41467-019-09024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas T, Dixon MP, Kueh AJ, Voss AK. 2008. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol 28:5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A. 2005. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voss AK, Collin C, Dixon MP, Thomas T. 2009. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev Cell 17:674–686. doi: 10.1016/j.devcel.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Sheikh BN, Phipson B, El-Saafin F, Vanyai HK, Downer NL, Bird MJ, Kueh AJ, May RE, Smyth GK, Voss AK, Thomas T. 2015. MOZ (MYST3, KAT6A) inhibits senescence via the INK4A-ARF pathway. Oncogene 34:5807–5820. doi: 10.1038/onc.2015.33. [DOI] [PubMed] [Google Scholar]
- 26.Jin Q, Yu L-R, Wang L, Zhang Z, Kasper LH, Lee J-E, Wang C, Brindle PK, Dent SYR, Ge K. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem 274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 28.Nagy Z, Tora L. 2007. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 29.Lee KK, Workman JL. 2007. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol 8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 30.Voss AK, Thomas T. 2018. Histone lysine and genomic targets of histone acetyltransferases in mammals. Bioessays 40:e1800078. doi: 10.1002/bies.201800078. [DOI] [PubMed] [Google Scholar]
- 31.Moazed D. 2011. Mechanisms for the inheritance of chromatin states. Cell 146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foss M, McNally FJ, Laurenson P, Rine J. 1993. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science 262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 33.Bell SP, Kobayashi R, Stillman B. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 34.Feng Y, Vlassis A, Roques C, Lalonde M-E, González-Aguilera C, Lambert J-P, Lee S-B, Zhao X, Alabert C, Johansen JV, Paquet E, Yang X-J, Gingras A-C, Côté J, Groth A. 2016. BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J 35:176–192. doi: 10.15252/embj.201591293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishima Y, Miyagi S, Saraya A, Negishi M, Endoh M, Endo TA, Toyoda T, Shinga J, Katsumoto T, Chiba T, Yamaguchi N, Kitabayashi I, Koseki H, Iwama A. 2011. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 118:2443–2453. doi: 10.1182/blood-2011-01-331892. [DOI] [PubMed] [Google Scholar]
- 36.Mishima Y, Wang C, Miyagi S, Saraya A, Hosokawa H, Mochizuki-Kashio M, Nakajima-Takagi Y, Koide S, Negishi M, Sashida G, Naito T, Ishikura T, Onodera A, Nakayama T, Tenen DG, Yamaguchi N, Koseki H, Taniuchi I, Iwama A. 2014. Histone acetylation mediated by Brd1 is crucial for Cd8 gene activation during early thymocyte development. Nat Commun 5:5872. doi: 10.1038/ncomms6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, Tan S, Yang XJ, Kutateladze TG, Cote J. 2013. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev 27:2009–2024. doi: 10.1101/gad.223396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Cote V, Yang XJ, Gozani O, Kutateladze TG, Cote J. 2009. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell 33:257–265. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avvakumov N, Lalonde ME, Saksouk N, Paquet E, Glass KC, Landry AJ, Doyon Y, Cayrou C, Robitaille GA, Richard DE, Yang XJ, Kutateladze TG, Cote J. 2012. Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol Cell Biol 32:689–703. doi: 10.1128/MCB.06455-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, Mateo JL. 2015. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao Y, Smyth GK, Shi W. 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.