Abstract
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer deaths worldwide. The high mortality rate in HCC is largely due to the difficulty of early detection. In this study, to improve patient outcomes, serum samples from 345 patients with HCC, 46 patients with chronic hepatitis (CH), 93 patients with liver cirrhosis (LC), and 1,033 healthy individuals were analyzed with microRNA (miRNA) microarrays. We investigated the diagnostic potential of circulating miRNAs in serum and developed a detection model of HCC, including early stage. A diagnostic model was constructed based on the expression levels of a combination of miRNAs in a discovery set. We selected 52 miRNAs that had altered expressions according to disease progression status, established the diagnostic model with a combination of eight miRNAs in the discovery set, and tested the model in a validation set. The diagnostic values for discriminating cancer from HCC at‐risk control samples were as follows: area under the curve, 0.99; sensitivity, 97.7%; specificity, 94.7%. With this model, 98% of stage I HCC cases were detected; these results were much better than those observed from conventional methods. Conclusion: Circulating miRNAs could serve as biomarkers for the accurate detection of HCC. Because the diagnostic accuracy was maintained even in stage I, this may represent an accurate detection method even for early stage HCC.
Abbreviations
- AFP
α‐fetoprotein
- AUC
area under the curve
- CH
chronic hepatitis
- CI
confidence interval
- DCP
des‐γ‐carboxy prothrombin
- HCC
hepatocellular carcinoma
- LC
liver cirrhosis
- miRNA/miR
microRNA
- NC
noncancer
- NCC
National Cancer Center
- NCCH
National Cancer Center Hospital
- NCGG
National Center for Geriatrics and Gerontology
- qRT‐PCR
quantitative reverse‐transcription polymerase chain reaction
- ROC
receiver operating characteristics
Hepatocellular carcinoma (HCC) is a primary malignancy of the liver and is frequently developed in patients with hepatitis B and hepatitis C infection and advanced liver fibrosis.1 The number of cases of HCC is expected to increase worldwide in the coming years, although the incident percentage is higher in Asia and Africa because of the high prevalence of hepatitis B and hepatitis C. According to a previous meta‐analysis, HCC surveillance is associated with curative treatment rates (odds ratio [OR], 2.24; 95% confidence interval [CI], 1.99‐2.52) and prolonged survival (OR, 1.90; 95% CI, 1.67‐2.17).2 For the diagnosis and surveillance of HCC, radiologic approaches, such as ultrasonography and computed tomography, are generally used; however, it is hard to detect a small lesion in the liver.3 Additionally, another method for HCC screening is the serologic test of tumor markers, such as α‐fetoprotein (AFP) and protein induced by des‐γ‐carboxy prothrombin (DCP). The problem with these methods is that the sensitivity and specificity of elevated levels of serum AFP and DCP are insufficient for the accurate detection of HCC.4, 5, 6 Therefore, a new diagnostic system could further promote the accuracy of early detection for HCC.
MicroRNAs (miRNAs) are small noncoding RNA products that posttranscriptionally modulate gene expression by decreasing target messenger RNA (mRNA) stability or repressing translational efficiency, thus associating with various biological processes, such as development, differentiation, apoptosis, and proliferation. The abnormal expression of miRNAs has been reported in a wide variety of cancers and results in tumor initiation and progression.7, 8, 9, 10 It was recently reported that miRNAs are stably detectable in body fluids, including blood saliva and urine; furthermore, at least some miRNAs are packaged into extracellular vesicles and play crucial roles in intercellular communication.11 Serum biomarkers are attractive targets for disease screening because less invasive procedures are used; findings have indicated that there are new methods for measuring miRNA levels in patient blood samples that could serve as a new diagnostic tool for cancer detection.12, 13, 14, 15
Several research groups have performed circulating miRNA profiling to detect HCC.16, 17, 18, 19 However, these molecules are considered insufficient for clinical applications, primarily due to the lack of large‐scale validation and inconsistencies among detection devices. To standardize platforms for the collection and detection of serum miRNAs, we launched a national project in Japan in 2014 titled “Development and Diagnostic Technology for Detection of miRNA in Body Fluids”. This project includes the comprehensive characterization of serum miRNA profiles of 13 types of human cancers, including HCC, in more than 40,000 patients, using the same platform and technology.14, 20, 21 In this study, we performed miRNA microarrays for serum samples from 345 patients with HCC, 46 patients with chronic hepatitis (CH), 93 patients with liver cirrhosis (LC), and 1,033 healthy individuals. In the discovery set, we identified 52 miRNA candidates that were associated with the progression of HCC. In the combination of mathematical approaches, we determined the optimal combination of miRNAs and established an eight‐miRNA panel that enables the detection of HCC with high accuracy.
Materials and Methods
Clinical Samples of HCC Cases
A total of 353 serum samples were obtained from patients who were referred to the National Cancer Center Hospital (NCCH) between 2008 and 2016; histologically diagnosed as having HCC, based on the Barcelona Clinic Liver Cancer staging system; and were registered in the National Cancer Center (NCC) Biobank. Serum samples were stored at 4°C for 1 week and then stored at –20°C until further use. Patients with HCC who underwent surgical operation, chemotherapy, or radiotherapy before serum collection were excluded. Clinical information of all samples was obtained by reference to the cancer registry of the NCC and medical records.
Clinical Samples of CH and LC Cases
A total of 46 serum samples of CH and 93 serum samples of LC were obtained from patients who were referred to Osaka City University between 2015 and 2016. Serum samples were stored at –80°C for 24 weeks and then stored at –80°C until further use. Clinical information of all samples was obtained by reference to the registry of Osaka City University and medical records.
Clinical Samples of Noncancer Controls
Noncancer controls were from the same biobank of HCC cases. A total of 343 serum samples were obtained from patients who were referred to the NCCH between 2008 and 2016, were diagnosed with no malignant diseases, and were registered in the NCC Biobank. Serum samples were stored at –20°C. These patients were enrolled as noncancer control 1. In addition, noncancer control samples were obtained from the National Center for Geriatrics and Gerontology (NCGG) Biobank (noncancer control 2) and the Yokohama Minoru Clinic (noncancer control 3). The inclusion criteria for these sample sets were no history of cancer and no hospitalization during the previous 3 months. Noncancer control 2 included 345 individuals whose serum samples were collected between 2012 and 2017 and stored in the NCGG Biobank at –80°C. The third set (noncancer control 3) included 345 patients from a clinical health check‐up and were stored at –80°C.
Ethics Approval and Consent to Participate
The study was approved by the NCCH Institutional Review Board (2015‐376, 2016‐249), Osaka City University Institutional Review Board (No.1358), and the Research Ethics Committee of Medical Corporation Shintokai Yokohama Minoru Clinic (6019‐18‐3772). Written informed consent was obtained from each participant.
miRNA Expression Arrays of Clinical Samples
Total RNA was extracted from 300 µL of serum, using the 3D‐Gene RNA extraction reagent (Toray Industries, Inc., Kanagawa, Japan) and purified with RNeasy 96 QIAcube HT Kit (Qiagen). Comprehensive miRNA expression analysis was performed using the 3D‐Gene miRNA Labeling kit and the 3D‐Gene Human miRNA Oligo Chip (Toray Industries, Inc.), which was designed to detect 2,588 miRNA sequences registered in miRBase, release 21 (http://www.mirbase.org/).22 For quality control of the microarray data, criteria for low‐quality results were coefficient of variation for negative control probes >0.15 and the number of flagged probes, identified by 3D‐Gene Scanner, >10. Samples meeting these criteria were excluded from further analyses. The presence of miRNA was determined based on a corresponding microarray signal of greater than the mean + 2 × SD of the negative control signals, from which the most and least intense signals were removed. After an miRNA was considered present, the mean signal of the negative controls (from which the top and bottom 5%, ranked by signal intensity, were removed) was subtracted from the miRNA signal. To normalize the signals among the microarrays tested, three preselected internal control miRNAs (miR‐149‐3p, miR‐2861, and miR‐4463) were used as described.14 When the signal value was negative (or undetected) after normalization, the value was replaced by 0.1 on a base‐2 logarithmic scale. All microarray data in the present study were obtained in accordance with the Minimum Information About a Microarray Experiment guidelines. Data sets analyzed in this study were submitted to the National Center for Biotechnology Information Gene Expression Omnibus database (accession number http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113740).
Statistical Analysis
Prior to statistical comparisons, samples were divided into discovery and validation sets based on computer‐generated random numbers. The discovery set was used to select miRNA markers and construct discriminant models, and the validation set was used to validate the discriminant models. Based on combinatorial optimization for multicandidate miRNAs, diagnostic indices were generated using Fisher’s linear discriminant analysis (refer to Supporting Material). Using leave‐one‐out cross‐validation in the discovery set, the best diagnostic index was ultimately selected. An index score ≥0 indicated the presence of HCC, and an index score <0 indicated the absence of HCC. The diagnostic sensitivity, specificity, accuracy, and area under the receiver operating characteristics (ROC) curve (AUC) were calculated for a diagnostic index in the validation sets.
Statistical analyses were performed using R version 3.1.2 (R Foundation for Statistical Computing, http://www.R-project.org), compute.es package version 0.2‐4, hash package version 2.2.6, MASS package version 7.3‐45, mutoss package version 0.1‐10, pROC package version 1.8, and IBM SPSS Statistics version 22 (IBM Japan, Tokyo, Japan). Unsupervised clustering and heatmap generation with sorted data sets, using Pearson’s correlation and Ward’s method for linkage analysis, were performed using Partek Genomics Suite 6.6. Principal component analysis was also performed using Partek Genomics Suite 6.6. The limit of statistical significance for all analyses was defined as a two‐sided P value of 0.05.
Results
Study Design
A total of 1,526 serum samples, including 353 HCC, 46 CH, 93 LC, and 1,033 normal control samples (noncancer 1, 2, and 3 from different hospitals), were collected and analyzed by miRNA microarray, yielding comprehensive miRNA expression profiles. After the exclusion of eight samples with low‐quality results from HCC samples, 1,518 samples remained for analysis. These samples were randomly separated into the discovery set (759 serum samples: 172 HCC, 64 CH + LC, and 523 noncancer) and validation set (759 serum samples: 173 HCC, 75 CH + LC, and 511 noncancer) for biomarker discovery. An overview of the study design for HCC screening is illustrated in Fig. 1A. Clinicopathologic characteristics were analyzed, and there was no significant difference between the two sets (Table 1).
Figure 1.
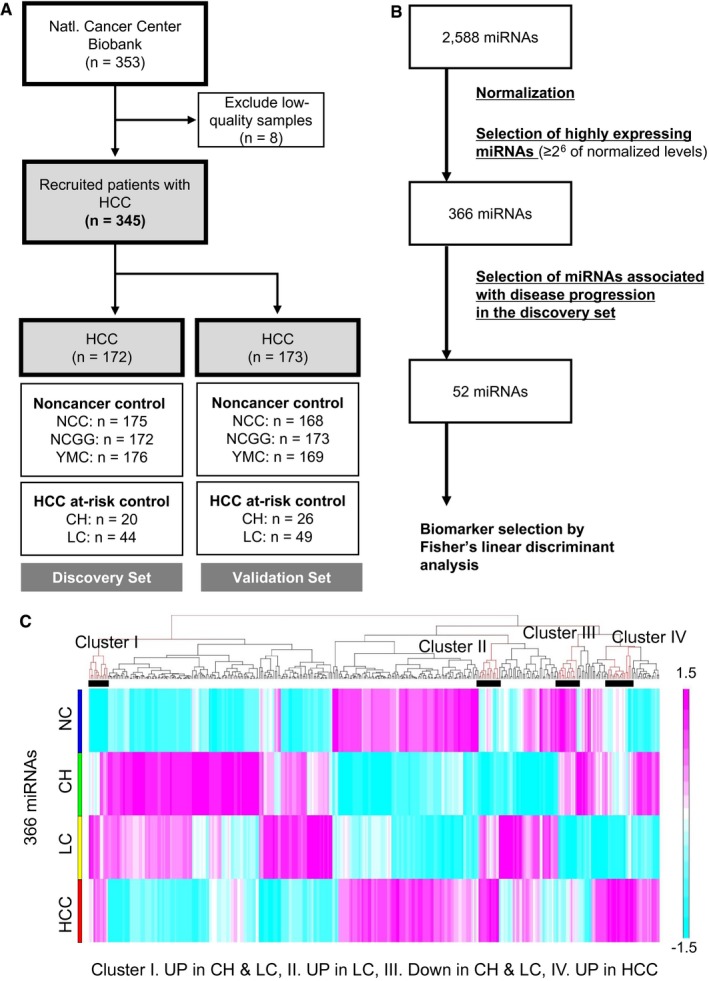
Flowchart of the process for the development of biomarkers of HCC. (A) A total of 353 serum samples were collected from patients with HCC, and eight samples were excluded due to their poor quality. A total of 1,033 serum samples were collected from the NCC, NCGG, and YMC as noncancer control samples. For HCC at‐risk controls, 46 CH and 93 LC samples were collected from Osaka City University. By randomly separating HCC samples and control samples into two groups, the discovery set and the validation set were prepared for the selection of miRNA biomarkers of HCC. (B) The selection process of miRNAs in this study. Based on the signal intensity of miRNAs (≥26 of normalized levels), highly expressing miRNAs were selected from 2,588 miRNAs. Among them, 52 miRNAs with expression patterns that were associated with disease progression were selected. (C) Heatmap showing average expression levels of each sample with 366 miRNAs; 52 miRNAs in clusters I, II, III, and IV were used for further analysis. Abbreviation: YMC, Yokohama Minoru Clinic.
Table 1.
Clinicopathologic Characteristics of Study Subjects
| Discovery | Validation | P Value | |
|---|---|---|---|
| Hepatocellular carcinoma | (n = 172) | (n = 173) | |
| Age, years | 67.6 ± 9.7 | 67.6 ± 8.8 | 0.97 |
| Sex | |||
| Men | 141 (82.0%) | 127 (73.4%) | 0.07 |
| Women | 31 (18.0%) | 46 (26.6%) | |
| Stage | |||
| I | 59 (34.3%) | 64 (37.0%) | 0.38 |
| II | 49 (28.5%) | 59 (34.1%) | |
| III | 43 (25.0%) | 37 (21.4%) | |
| IV | 13 (7.6%) | 6 (3.5%) | |
| X | 8 (4.7%) | 7 (4.0%) | |
| Child‐Pugh | |||
| A | 152 (88.4%) | 151 (87.3%) | 0.3 |
| B | 18 (10.5%) | 22 (12.7%) | |
| Unknown | 2 (1.2%) | 0 (0%) | |
| Virus | |||
| HBsAg+ | 27 (15.7%) | 30 (17.3%) | 0.71 |
| HCV Ab+ | 74 (43.0%) | 67 (38.7%) | |
| non‐B non‐C | 71 (41.3%) | 76 (43.9%) | |
| Tumor marker | |||
| AFP (ng/mL) | 7,435 ± 39,964 | 21,150 ± 138,510 | 0.21 |
| DCP (mAU/mL) | 10,162 ± 40,157 | 8,071 ± 28,113 | 0.58 |
| Noncancer control 1 | (n = 175) | (n = 168) | |
| Age | 62.9 ± 12.2 | 64.6 ± 10.0 | 0.16 |
| Sex | |||
| Men | 130 (74.3%) | 134 (79.8%) | 0.25 |
| Women | 45 (25.7%) | 34 (20.2%) | |
| Noncancer control 2 | (n = 172) | (n = 173) | |
| Age | 67.4 ± 9.9 | 67.5 ± 9.2 | 0.96 |
| Sex | |||
| Men | 126 (73.3%) | 139 (80.3%) | 0.13 |
| Women | 46 (26.7%) | 34 (19.7%) | |
| Noncancer control 3 | (n = 176) | (n = 169) | |
| Age | 65.2 ± 7.3 | 64.3 ± 8.4 | 0.28 |
| Sex | |||
| Men | 124 (70.5%) | 141 (83.4%) | 0.005 |
| Women | 52 (29.5%) | 28 (16.6%) | |
| Liver disease control | (n = 64) | (n = 75) | |
| Age | 70.1 ± 10.2 | 70.4 ± 9.0 | 0.82 |
| Sex | |||
| Men | 34 (53.1%) | 37 (49.3%) | 0.73 |
| Women | 30 (46.9%) | 38 (50.7%) | |
| Chronic hepatitis | 20 (31.3%) | 26 (34.7%) | 0.72 |
| Liver cirrhosis | 44 (68.8%) | 49 (65.3%) | |
| Virus | |||
| HBsAg+ | 0 | 0 | NA |
| HCV Ab+ | 64 (100%) | 75 (100%) | |
| non‐B non‐C | 0 | 0 | |
| Tumor marker | |||
| AFP (ng/mL) | 28.5 ± 66.3 | 28.0 ± 47.4 | 0.96 |
n represents total number of patients and respective percentage can be found in the parentheses (%). The values are shown as mean ± standard deviation.
Abbreviations: HBsAg+, hepatitis B surface antigen positive; HCV Ab+, hepatitis C virus antibody positive.
Selection of Serum miRNA Biomarker Candidates for HCC Screening
Using an miRNA microarray, expression levels of 2,588 miRNAs were comprehensively measured. To identify miRNA biomarker candidates, we focused on highly detected miRNAs (≥26 of normalized levels in signal intensity). The 366 miRNAs that were considered highly detected (Fig. 1B) were analyzed based on clinicopathologic characteristics and classification, i.e., sex, hepatitis status, stage, and Child‐Pugh score (Supporting Fig. S1); there was no clear segregation in miRNA expression profiles by these factors. These highly expressed miRNAs were applied for further selection in the discovery set. To select miRNAs with expressions that were associated with disease progression (i.e., sequentially increased or decreased during disease progress), we combined expression data of the samples from the same group into one and made a heatmap with 366 miRNAs (Fig1C; Supporting Table S1). Based on our criteria, we selected miRNAs in clusters I, II, III, and IV. For example, expression levels of miRNAs in clusters I and II gradually increased from noncancer samples to HCC samples. In this selection, 52 miRNAs passed the criteria (Fig. 1B,C; Supporting Table S2). With these 52 miRNAs, unsupervised hierarchical clustering analysis with a heatmap showed the separation of noncancer samples and HCC samples but not HCC at‐risk samples, such as CH and LC samples (Supporting Fig. S2).
Generation of Diagnostic Models for HCC Detection
To generate novel diagnostic models for the accurate discrimination between HCC and control samples in the discovery set, we used Fisher’s linear discriminant analysis. We designed comprehensive discriminants consisting of one to 10 miRNAs in the discovery set. Based on the optimal level of accuracy, the analysis identified a combination of eight miRNAs (miR‐320b, miR‐663a, miR‐4448, miR‐4651, miR‐4749‐5p, miR‐6724‐5p, miR‐6877‐5p, and miR‐6885‐5p) that provided the best discrimination in the discovery set: diagnostic index = (0.200683) × hsa‐miR‐320b + (1.83536) × hsa‐miR‐6724‐5p + (0.917188) × hsa‐miR‐6877‐5p + (−0.180267) × hsa‐miR‐4448 + (−1.06578) × hsa‐miR‐4749‐5p + (0.81661) × hsa‐miR‐663a + (−1.2972) × hsa‐miR‐4651 + (0.400051) × hsa‐miR‐6885‐5p − 17.7924 (Table 2). When we tested the combination for distinguishing HCC samples from noncancer controls or HCC at‐risk controls (Fig. 2A‐D), the discriminant showed high diagnostic power for noncancer controls (AUC, 1.0; Fig. 2B) and for HCC at‐risk controls (AUC, 0.97; Fig. 2D). Individual miRNA data for the discrimination of HCC are also shown (Fig. 2A,C).
Table 2.
Combination of miRNAs Identified in the Discovery Set
| miR_Number | Combination of miRNAs | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|---|
| 1 | (0.634497) × hsa‐miR‐320b − 3.67689 | 0.913 | 0.821 | 0.842 | 0.921 |
| 2 | (0.498834) × hsa‐miR‐320b + (1.82171) × hsa‐miR‐6724‐5p − 21.9504 | 0.971 | 0.920 | 0.932 | 0.975 |
| 3 | (0.502698) × hsa‐miR‐320b + (1.93218) × hsa‐miR‐6724‐5p + (−0.284752) × hsa‐miR‐3656 − 19.8097 | 0.983 | 0.917 | 0.932 | 0.977 |
| 4 | (0.426568) × hsa‐miR‐320b + (1.75338) × hsa‐miR‐6724‐5p + (0.68341) × hsa‐miR‐6877‐5p + (−0.161065) × hsa‐miR‐4448 − 25.068 | 0.959 | 0.961 | 0.961 | 0.984 |
| 5 | (0.448161) × hsa‐miR‐320b + (1.79821) × hsa‐miR‐6724‐5p + (0.627202) × hsa‐miR‐6877‐5p + (−0.116909) × hsa‐miR‐4448 + (−0.552478) × hsa‐miR‐6722‐3p − 20.6907 | 0.965 | 0.957 | 0.959 | 0.987 |
| 6 | (0.338274) × hsa‐miR‐320b + (2.12341) × hsa‐miR‐6724‐5p + (0.902937) × hsa‐miR‐6877‐5p + (−0.171105) × hsa‐miR‐3160‐5p + (−0.63432) × hsa‐miR‐4530 + (−0.644387) × hsa‐miR‐4749‐5p − 18.8571 | 0.959 | 0.974 | 0.971 | 0.989 |
| 7 | (0.214291) × hsa‐miR‐320b + (1.79304) × hsa‐miR‐6724‐5p + (0.961144) × hsa‐miR‐6877‐5p + (−0.205188) × hsa‐miR‐4448 + (−1.25349) × hsa‐miR‐4749‐5p + (1.06455) × hsa‐miR‐663a + (−1.0448) × hsa‐miR‐4651 − 17.0791 | 0.983 | 0.976 | 0.978 | 0.995 |
| 8 | (0.200683) × hsa‐miR‐320b + (1.83536) × hsa‐miR‐6724‐5p + (0.917188) × hsa‐miR‐6877‐5p + (−0.180267) × hsa‐miR‐4448 + (−1.06578) × hsa‐miR‐4749‐5p + (0.81661) × hsa‐miR‐663a + (−1.2972) × hsa‐miR‐4651 + (0.400051) × hsa‐miR‐6885‐5p − 17.7924 | 0.988 | 0.980 | 0.982 | 0.996 |
| 9 | (0.21609) × hsa‐miR‐320b + (1.84305) × hsa‐miR‐6724‐5p + (0.938715) × hsa‐miR‐6877‐5p + (−0.187943) × hsa‐miR‐4448 + (−1.04236) × hsa‐miR‐4749‐5p + (0.788289) × hsa‐miR‐663a + (−1.30622) × hsa‐miR‐4651 + (0.430173) × hsa‐miR‐6885‐5p + (−0.0931209) × hsa‐miR‐4731‐5p − 17.3143 | 1.000 | 0.969 | 0.976 | 0.995 |
| 10 | (0.181663) × hsa‐miR‐320b + (1.89463) × hsa‐miR‐6724‐5p + (0.907708) × hsa‐miR‐6877‐5p + (−0.202764) × hsa‐miR‐4448 + (−1.0582) × hsa‐miR‐4749‐5p + (0.751592) × hsa‐miR‐663a + (−1.25423) × hsa‐miR‐4651 + (0.382919) × hsa‐miR‐6885‐5p + (−0.174083) × hsa‐miR‐4530 + (0.0942364) × hsa‐miR‐4322 − 16.2484 | 1.000 | 0.971 | 0.978 | 0.996 |
Bold value indicates the final selection used to for the calculation of diagnostic value within this study.
Figure 2.
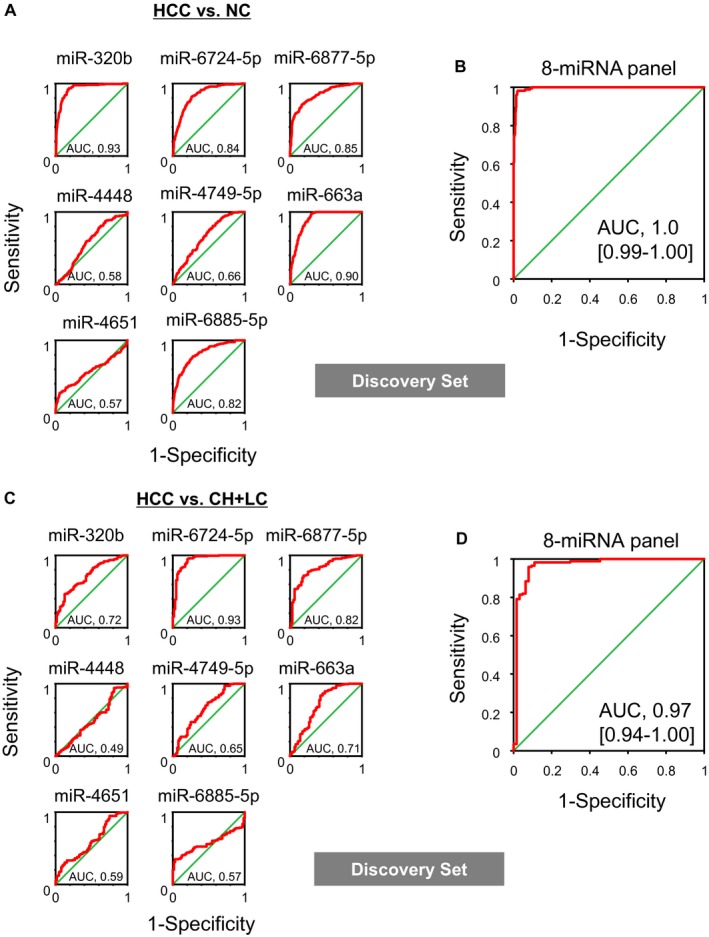
Discovering candidate miRNAs for HCC screening. (A,B) Discrimination of HCC samples from noncancer controls. In the discovery set, results of individual miRNAs and the eight‐miRNA panel are shown. (C,D) Discrimination of HCC samples from HCC at‐risk controls (CH and LC samples). In the discovery set, results of individual miRNAs and the eight‐miRNA panel are shown. AUC values are shown in all ROC curve analyses.
Expression Levels of Eight miRNAs in HCC, HCC At‐Risk, and Noncancer Samples
The analysis of serum levels of the eight miRNAs in HCC, HCC at‐risk controls, and noncancer controls revealed that averages of these miRNA levels were significantly higher in HCC cases, except for miR‐4448 and miR‐4651 (Fig. 3). The diagnostic performances of miR‐4448 and miR‐4651 were not high in either comparison with noncancer and HCC at‐risk controls (Fig. 2A,C), although most of the other miRNAs were individually effective to distinguish HCC cases from noncancer controls or HCC at‐risk controls (Fig. 2A,C). miR‐4448 and miR‐4651 alone had insufficient diagnostic performance; however, in combination with other miRNAs, they provided better diagnostic value (Table 2; Fig. 2B,D), leading to the best discrimination of HCC cases using an eight‐miRNA combination.
Figure 3.
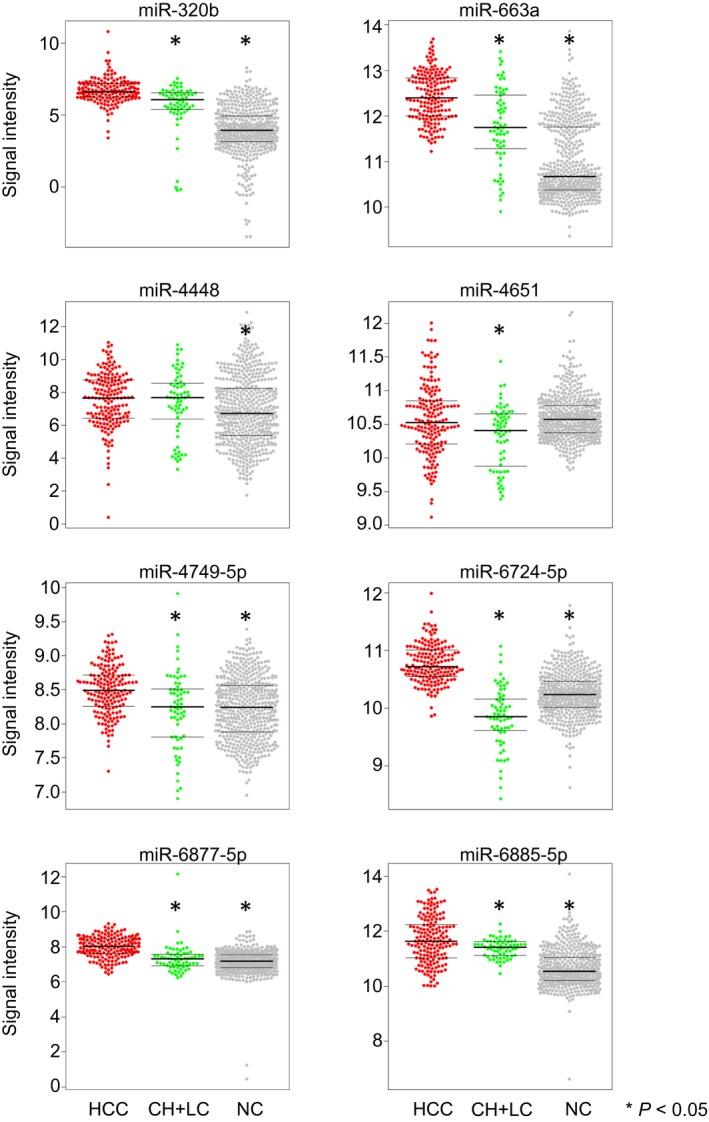
Selected eight‐miRNA expression patterns among samples. Dot plots for eight miRNAs (miR‐320b, miR‐663a, miR‐4448, miR‐4651, miR‐4749‐5p, miR‐6724‐5p, miR‐6877‐5p, and miR‐6885‐5p) in HCC, HCC at‐risk controls (CH and LC), and noncancer controls. Fold change, two‐sided Student t test; *P < 0.05. In each plot, first horizontal line represents 25th percentile, second line represents median and third line represents 75th percentile.
Verifying the Diagnostic Performance of an Eight‐miRNA Panel in the Validation Set
The diagnostic tool using an eight‐miRNA panel that was established from the discovery set was used to predict the probability of being diagnosed with HCC in the validation set (759 serum samples: 173 HCC, 75 HCC at‐risk controls [CH + LC], and 511 noncancer controls). Similar to the discovery set analysis, the predicted probability was used to construct the ROC curve. The AUC of the eight‐miRNA panel was 1.00 (95% CI, 0.99‐1.00; sensitivity, 97.7%; specificity, 98.4%; Fig. 4A) for the comparison with noncancer controls and 0.99 (95% CI, 0.98‐ 1.00; sensitivity, 97.7%; specificity, 94.7%; Fig. 4B) for the comparison with HCC at‐risk controls. When examining the stage I (HCC [hepatitis C virus positive] vs. CH+LC) samples, the conventional cancer marker AFP protein (>10 ng/mL) could not accurately distinguish HCC cases from HCC at‐risk controls (AUC, 0.65; sensitivity, 73.3%; specificity, 51.4%), although the eight‐miRNA panel provided a better result (AUC, 1.00; sensitivity, 100%; specificity, 94.7%) (Supporting Fig. S3). In addition, the analysis demonstrated that the performance of this eight‐miRNA panel was robustly validated and the specificities were nearly identical among the three noncancer control sets (collected from three different hospitals) as well as the HCC at‐risk controls (Fig. 4C).
Figure 4.
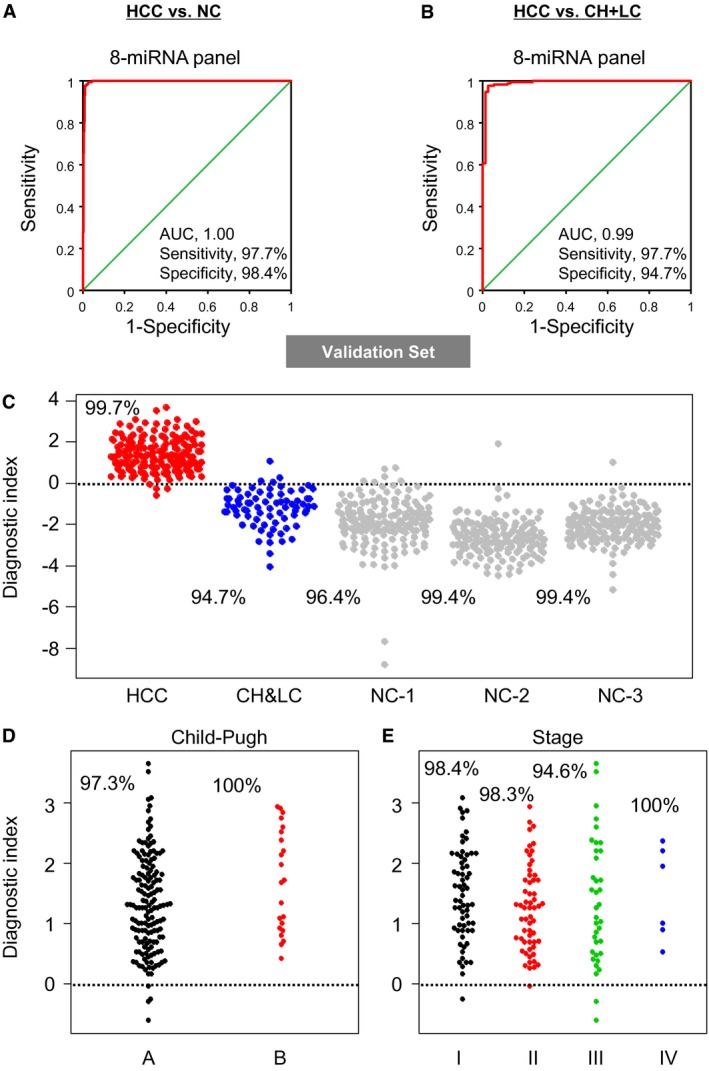
Diagnostic performance of the eight‐miRNA panel in the validation set. (A) ROC analysis for the discrimination of HCC from noncancer controls in the validation set. The AUC and 95% CI are shown in the plot. (B) ROC analysis for the discrimination of HCC from HCC at‐risk controls in the validation set. The AUC and 95% CI are shown in the plot. (C) Dot plot of diagnostic index of HCC, HCC at‐risk, and noncancer control samples. Noncancer samples are separately shown in the plot based on the hospitals where the samples were collected. (D) Dot plot of diagnostic index in HCC samples based on HCC stages. (E) Dot plot of diagnostic index in HCC samples based on Child‐Pugh score.
For the clinical significance of the eight‐miRNA panel for early detection of HCC, we compared the panel with conventional serologic approaches, i.e., serum levels of AFP and DCP (Table 3). In late‐stage HCC (stages III and IV), conventional AFP and DCP tests also showed high sensitivity, but in early stage HCC, only the eight‐miRNA panel maintained high sensitivity (stage I, 98% [63 out of 64] and stage II, 98% [58 out of 59]; Table 3; Fig. 4D). Based on the Child‐Pugh score, the analysis of the AFP test showed lower sensitivity in HCC cases with grade A, but the eight‐miRNA panel showed higher sensitivity in both grade A and grade B (Table 3; Fig. 4E). Because infection by hepatitis B and hepatitis C is one of the major causes of HCC, we examined the relationship of virus status and diagnostic value of the eight‐miRNA panel. The analysis clearly indicated that the effectiveness of the eight‐miRNA panel was not altered based on viral status (Table 3). These results suggest that the diagnostic model based on this combination of miRNAs is highly accurate in distinguishing patients with early stage HCC from noncancer sample sets.
Table 3.
Sensitivity for Each Subgroup of HCC in the Validation Set
| Stage | Child‐Pugh | Virus | |||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | A | B | HBV | HCV | nBnC | |
| n = 64 | n = 59 | n = 37 | n = 6 | n = 151 | n = 22 | n = 30 | n = 67 | n = 76 | |
| Eight‐miRNA panel | 63 | 58 | 35 | 6 | 147 | 22 | 30 | 65 | 74 |
| (>0) | 98% | 98% | 95% | 100% | 97% | 100% | 100% | 97% | 97% |
| AFP | 38 | 38 | 30 | 6 | 99 | 20 | 21 | 53 | 45 |
| (>10 ng/mL) | 59% | 64% | 81% | 100% | 66% | 91% | 70% | 79% | 59% |
| DCP | 26 | 39 | 32 | 6 | 98 | 9 | 20 | 38 | 49 |
| (>40 mAU/mL) | 40% | 66% | 87% | 100% | 65% | 41% | 67% | 57% | 65% |
Abbreviations: HBV, hepatitis B virus; HCV, hepatitis C virus; nBnC, non‐B non‐C.
Next, we investigated whether the eight‐miRNA panel could distinguish HCC from other solid cancers. The diagnostic index included 12 other types of solid tumors (n = 25 each) and 173 HCC samples and was calculated in the same manner. The eight‐miRNA panel could distinguish some solid tumors, such as breast cancer and prostate cancer, but was not as effective in distinguishing other types of cancers from HCC (Supporting Fig. S4). Thus, to obtain a better result, another discriminant model for separating HCC from other types of cancer could be required.
Discussion
Recently, comprehensive and integrative characterizations of HCC were reported and dysregulation of some miRNAs, such as miR‐122, a liver‐specific miRNA, were associated with poor survival in patients with HCC.23 In this study, we focused on circulating miRNA profiling in HCC and analyzed serum samples from 345 HCC, 139 HCC at‐risk controls, and 1,033 noncancer controls, using an miRNA microarray on a standardized platform (3D‐Gene).24 Similar studies have never performed a comprehensive analysis of all collected samples. Although miRNA biomarkers for HCC detection have been published using a smaller research cohort,25 we developed a large database of circulating miRNA expression in patients with HCC. After the selection of stringent criteria, 52 miRNAs passed as candidates for HCC screening. The diagnostic power of a single miRNA is unsatisfactory; however, the combination of eight miRNAs based on Fisher’s linear discriminant analysis provided a robust diagnostic tool. This study demonstrated that the eight‐miRNA panel comprising miR‐320b, miR‐663a, miR‐4448, miR‐4651, miR‐4749‐5p, miR‐6724‐5p, miR‐6877‐5p, and miR‐6885‐5p enabled the accurate early stage detection of HCC using patient serum samples. ROC analysis for the diagnostic value of the eight‐miRNA panel yielded an AUC of 0.99 with a sensitivity of 97.7% and specificity of 94.7% for discriminating HCC cases from HCC at‐risk controls in the independent validation set. Thus, we clearly showed that the eight‐miRNA panel is a valuable serum biomarker for HCC screening.
There are currently two major methods widely applied for HCC screening, imaging and serologic tests. The main issue with HCC screening is that when the HCC size is small or at an early stage, the diagnostic performance of these methods is insufficient to detect HCC. Historically, detection of AFP was clinically used as a general screening tool. However, it is well known that AFP levels in serum are elevated in CH or LC and have poor sensitivity in HCC detection; therefore, the diagnostic performance of AFP measurement is inadequate for HCC detection (Supporting Fig. S3). Compared with the conventional serum AFP and DCP tests, the diagnostic power of the eight‐miRNA panel is much higher, particularly at the early stage of HCC. Thus, efforts in our study to establish a less‐invasive approach with a serum miRNA combination have met with limited success.
Although it is not clear what type of cells express and secrete miRNAs into serum in patients with HCC (HCC cells or non‐HCC cells within or outside the tumor microenvironment) or the secretion and circulation mechanisms of miRNAs into serum, some miRNAs in the eight‐miRNA panel were shown to be deregulated in cancer. For example, miR‐663a expression levels were significantly decreased in HCC tissues compared with adjacent nontumor tissue and inhibited the proliferation and invasion of HCC cells by targeting high‐mobility group AT‐hook 2 (HMGA2).26 In addition, the expression levels of miR‐320b reportedly decrease in several types of cancer, such as colon cancer, and could act as a tumor suppressor miRNA by targeting c‐Myc.27 However, both miR‐320b and miR‐663a were highly detected in serum samples of HCC cases in this study. Although unclear, there might be molecular mechanisms through which cancer cells selectively secret these miRNAs into the blood. On the other hand, miR‐320b was reported as a cancer biomarker in the blood samples of patients with prostate cancer and lung squamous carcinoma28, 29; expression levels were associated with clinical parameters and diagnosis.28 Also, serum miR‐4651 levels were highly detected in patients with aflatoxin B1‐positive HCC, and higher levels of serum miR‐4651 were significantly correlated with poor prognosis.30 Investigating the expression levels of the eight‐miRNA panel between tissue and serum samples in patients with HCC would help to further clarify the physiological relevance of these miRNAs.
There are a few limitations in our study. First, we performed this analysis with retrospectively collected samples. Before microarray analysis, there were differences in the time interval of sample preparation, storage period, and storage temperature, which were not rigidly controlled at all institutes (refer to the Materials and Methods section). MiRNAs were considered to be more stable in serum than mRNA; however, this stability could affect the sample quality and miRNA expression levels.31, 32 To reduce sampling bias, we collected noncancer sample sets from three institutions and confirmed that diagnostic accuracy was not altered among the samples from the different institutes. When the HCC cohort was compared to the sample set of noncancer 1, which was stored under the same conditions as the HCC cohort, our diagnostic model successfully distinguished HCC samples from noncancer 1. For these reasons, we thought the difference in profiles was not generated from the storage condition or difference between facilities but from innate features of the samples. Furthermore, we have started a clinical prospective validation trial, and the prospective analysis with fresh blood samples will standardize the eight‐miRNA panel for HCC screening in the coming years. Second, there was no validation of the selected miRNA expression levels in the serum RNA samples by quantitative reverse‐transcription polymerase chain reaction (qRT‐PCR) or small RNA sequencing. One reason for this relates to the technical difficulty in specific primer design for all 2,588 miRNAs by qRT‐PCR. In addition, based on our experience, miRNA expression is not easily quantified by qRT‐PCR because some miRNA sequences are truncated in serum.33 However, considering the development of clinical applications for the eight‐miRNA panel based on the same microarray platform, it would not be necessary to verify the expression data by another modality. Third, we failed to show the physiological relevance of serum miRNAs in HCC. Serum miRNA profiling was altered based on disease status; however, the types of cells or tissues secreting these miRNAs into the blood and the function of circulating miRNAs remain unclear.
In summary, comprehensive analysis of serum miRNA profiles in 345 cases of HCC established a diagnosis system with an eight‐miRNA panel that is useful for early stage and AFP‐negative HCC and is particularly important for the surveillance of preclinical HCC in at‐risk groups. Liquid biopsy has recently gained the attention of both clinicians and researchers as a potentially powerful tool in a clinical setting.34 Of note, early stage detection of HCC by liquid biopsy improves the survival of patients with this disease. Liquid biopsy‐based miRNA profiling could be applicable as a combination with conventional serologic tests, such as AFP levels. In addition, serum miRNA profiling might be applicable as a future clinical practice for not only the prediction of prognosis of patients with HCC after surgical resection or systemic chemotherapy but also the judgment of HCC stage. The techniques for our method with the eight‐miRNA panel consisted of serum collection, RNA extraction, and microarray and are routinely used in clinical settings. Furthermore, data interpretation of miRNA expression profiles with algorithms is simple. Therefore, evaluation of an eight‐miRNA panel in clinical practice could be feasible and suitable for primary screening and surveillance of HCC.
Supporting information
Acknowledgment
We thank Tomomi Fukuda, Takumi Sonoda, Hiroko Tadokoro, Tatsuya Suzuki, Junpei Kawauchi, Satoshi Kondou, Makiko Ichikawa, and Kamakura Techno‐Science, Inc. for performing the microarray assays. We also thank Noriko Abe, Michiko Ohori, and Takumi Sonoda for collecting samples from the freezing room and Kazuki Sudo for independent confirmation of participant eligibility. Some of the samples and clinical information used in this study were obtained from the NCC Biobank, which is supported by National Cancer Center Research and Development Fund (29‐A‐1). We also thank the NCGG Biobank for providing the biological resources.
Supported by the Japan Agency for Medical Research and Development through a “Development of Diagnostic Technology for Detection of miRNA in Body Fluids” grant (number 16AE0101011H0003 to T.Oc.).
Potential conflict of interest: Dr. Kondo received grants from MSD, Pfizer, ASLAN, Takeda, Eli Lilly, and Boehringer Ingelheim. Dr. Okusaka received grants from Bristol Myers Squibb K.K. Dr. Takizawa is an employee of Toray Industries, Inc. The other authors have nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta‐analysis. PLoS Med 2014;11:e1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simmons O, Fetzer DT, Yokoo T, Marrero JA, Yopp A, Kono Y, et al. Predictors of adequate ultrasound quality for hepatocellular carcinoma surveillance in patients with cirrhosis. Aliment Pharmacol Ther 2017;45:169‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oka H, Tamori A, Kuroki T, Kobayashi K, Yamamoto S. Prospective study of alpha‐fetoprotein in cirrhotic patients monitored for development of hepatocellular carcinoma. Hepatology 1994;19:61‐66. [PubMed] [Google Scholar]
- 5. Marrero JA, Su GL, Wei W, Emick D, Conjeevaram HS, Fontana RJ, et al. Des‐gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in american patients. Hepatology 2003;37:1114‐1121. [DOI] [PubMed] [Google Scholar]
- 6. Seo SI, Kim HS, Kim WJ, Shin WG, Kim DJ, Kim KH, et al. Diagnostic value of PIVKA‐II and alpha‐fetoprotein in hepatitis B virus‐associated hepatocellular carcinoma. World J Gastroenterol 2015;21:3928‐3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857‐866. [DOI] [PubMed] [Google Scholar]
- 8. Esquela‐Kerscher A, Slack FJ. Oncomirs ‐ microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259‐269. [DOI] [PubMed] [Google Scholar]
- 9. Hironaka‐Mitsuhashi A, Matsuzaki J, Takahashi RU, Yoshida M, Nezu Y, Yamamoto Y, et al. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS One 2017;12:e0187638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuzaki J, Suzuki H, Tsugawa H, Watanabe M, Hossain S, Arai E, et al. Bile acids increase levels of microRNAs 221 and 222, leading to degradation of CDX2 during esophageal carcinogenesis. Gastroenterology 2013;145:1300‐1311. [DOI] [PubMed] [Google Scholar]
- 11. Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest 2016;126:1163‐1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamamoto Y, Kosaka N, Tanaka M, Koizumi F, Kanai Y, Mizutani T, et al. MicroRNA‐500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 2009;14:529‐538. [DOI] [PubMed] [Google Scholar]
- 13. Yokoi A, Yoshioka Y, Hirakawa A, Yamamoto Y, Ishikawa M, Ikeda SI, et al. A combination of circulating miRNAs for the early detection of ovarian cancer. Oncotarget 2017;8:89811‐89823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci 2016;107:326‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuzaki J, Ochiya T. Circulating microRNAs and extracellular vesicles as potential cancer biomarkers: a systematic review. Int J Clin Oncol 2017;22:413‐420. [DOI] [PubMed] [Google Scholar]
- 16. Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV‐positive hepatocarcinoma. Cancer Res 2010;70:9798‐9807. Erratum in: Cancer Res 2011;71:2022. [DOI] [PubMed] [Google Scholar]
- 17. Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus‐related hepatocellular carcinoma. J Clin Oncol 2011;29:4781‐4788. [DOI] [PubMed] [Google Scholar]
- 18. Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, et al. Circulating microRNA‐21 as a novel biomarker for hepatocellular carcinoma. J Hepatol 2012;56:167‐175. [DOI] [PubMed] [Google Scholar]
- 19. Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case‐control study. Lancet Oncol 2015;16:804‐815. [DOI] [PubMed] [Google Scholar]
- 20. Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka Y, Takahashi K, Shimizu H, et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun 2018;9:4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Usuba W, Urabe F, Yamamoto Y, Matsuzaki J, Sasaki H, Ichikawa M, Takizawa S, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci 2019;110:408‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kozomara A, Griffiths‐Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68‐D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cancer Genome Atlas Research Network . Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327‐1341.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sato F, Tsuchiya S, Terasawa K, Tsujimoto G. Intra‐platform repeatability and inter‐platform comparability of microRNA microarray technology. PLoS One 2009;4:e5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moshiri F, Salvi A, Gramantieri L, Sangiovanni A, Guerriero P, De Petro G, et al. Circulating miR‐106b‐3p, miR‐101‐3p and miR‐1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget 2018;9:15350‐15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang W, Li J, Guo X, Zhao Y, Yuan X. miR‐663a inhibits hepatocellular carcinoma cell proliferation and invasion by targeting HMGA2. Biomed Pharmacother 2016;81:431‐438. [DOI] [PubMed] [Google Scholar]
- 27. Wang H, Cao F, Li X, Miao H, E J, Xing J, Fu CG. miR‐320b suppresses cell proliferation by targeting c‐Myc in human colorectal cancer cells. BMC Cancer 2015;15:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lieb V, Weigelt K, Scheinost L, Fischer K, Greither T, Marcou M, et al. Serum levels of miR‐320 family members are associated with clinical parameters and diagnosis in prostate cancer patients. Oncotarget 2017;9:10402‐10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, et al. Evaluation of tumor‐derived exosomal miRNA as potential diagnostic biomarkers for early‐stage non‐small cell lung cancer using next‐generation sequencing. Clin Cancer Res 2017;23:5311‐5319. [DOI] [PubMed] [Google Scholar]
- 30. Wu XM, Xi ZF, Liao P, Huang HD, Huang XY, Wang C, et al. Diagnostic and prognostic potential of serum microRNA‐4651 for patients with hepatocellular carcinoma related to aflatoxin B1. Oncotarget 2017;8:81235‐81249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moreau MP, Bruse SE, David‐Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry 2011;69:188‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sourvinou IS, Markou A, Lianidou ES. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn 2013;15:827‐834. [DOI] [PubMed] [Google Scholar]
- 33. Juzenas S, Venkatesh G, Hübenthal M, Hoeppner MP, Du ZG, Paulsen M, et al. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res 2017;45:9290‐9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi‐analyte blood test. Science 2018;359:926‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


