Abstract
The tumor suppressor phosphatase and tension homolog (PTEN) is frequently mutated in human cancers, and it functions in multiple ways to safeguard cells from tumorigenesis. In the cytoplasm, PTEN antagonizes the PI3K/AKT pathway and suppresses cellular proliferation and survival. In the nucleus, PTEN is indispensable for the maintenance of genomic stability. In addition, PTEN loss leads to extensive changes in gene expression at the transcriptional level. The linker histone H1, generally considered as a transcriptional repressor, binds to the nucleosome to form a structure named the chromatosome. The dynamics between H1 and chromatin play an important role in determining gene expression. Here, we summarize the current understanding of roles of PTEN in controlling chromatin dynamics and global gene expression, which is crucial function of nuclear PTEN. We will also introduce the recent discovery of the PTEN family members and their functions.
It has been adequately indicated that the tumor suppressor phosphatase and tension homolog (PTEN) antagonizes the PI3K/AKT/mTOR signaling cascade in the cytoplasm, inhibiting cell proliferation and cell survival (Maehama and Dixon 1998; Stambolic et al. 1998). PTEN has long been thought to reside exclusively in the cytoplasm, and the first papers describing its presence in the nuclear compartment were in primary neurons and endothelial cells (Sano et al. 1999), as well as myoepithelial cells of normal breast ducts (Perren et al. 1999). In 2007, Shen et al. reported that PTEN is localized in nucleus and maintains chromosomal integrity (Shen et al. 2007). Since then, functions of nuclear PTEN have been greatly expanded to DNA- and chromatin-related events, including regulation of genomic stability, DNA replication, DNA repair, and cell-cycle progression (Planchon et al. 2008; Hou et al. 2017). However, molecular mechanisms of how PTEN is shuttled to the nucleus are still not fully understood. Because the PTEN protein lacks either a classical nuclear localization signal (NLS) or nuclear export signal (NES) motif, several mechanisms of its shuttling have been proposed, including simple diffusion (Liu et al. 2005), active shuttling by the RAN GTPase (Gil et al. 2006) or major vault protein (MVP) (Chung et al. 2005), phosphorylation-dependent shuttling (Planchon et al. 2008), and monoubiquitylation-dependent import (Trotman et al. 2007). Recently, SUMOylation has also been reported to control PTEN nuclear localization (Bassi et al. 2013). Nuclear PTEN plays an important role in tumor suppressing, and the absence of nuclear PTEN is associated with increased aggressiveness in cancers (Gimm et al. 2000; Baker 2007). Therefore, it is of great importance that we summarize the latest research progress on functions of nuclear PTEN, especially the interplay with histones on chromatin and gene expression resulting in biological and pathological activities.
DYNAMICS OF CHROMATIN STRUCTURE
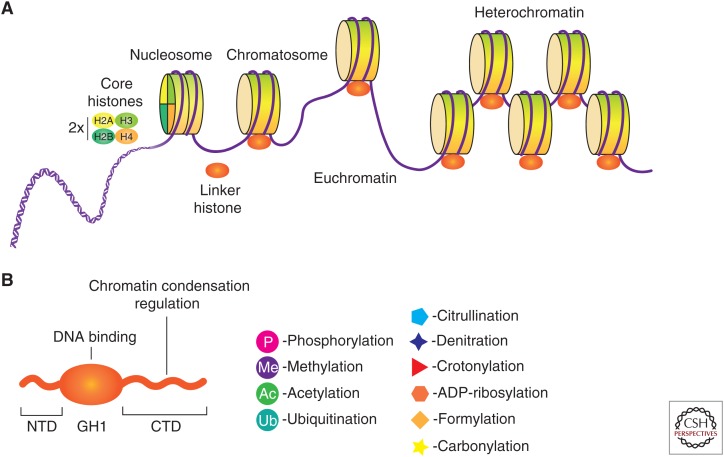
In eukaryotes, the 2 m of genomic DNA is compactly intertwined and bound to histone proteins, and together they form a fibrous architecture named “chromatin” (Kornberg 1974; Kornberg and Thomas 1974). The fundamental unit of chromatin is named the nucleosome, which consists of ∼147 bp of DNA wound around an octamer of core histones (two copies each of H2A, H2B, H3, and H4) and an additional variable length of linker DNA (Arents and Moudrianakis 1993; Luger et al. 1997). Further packaging of DNA involves a linker histone (H1) bound to the nucleosome with ∼10 bp of DNA at the entry/exit sites of the nucleosome core particle, generating a second-level structure called the chromatosome (Simpson 1978). Chromatin can be either densely packed in the form of heterochromatin, which is largely inaccessible to gene transcription and hence harboring inactive genes, or as open and accessible euchromatin, which contains numerous active genes (Fig. 1A). This dynamic, tightly controlled regulation of chromatin structure ensures timely, coordinated, and appropriate gene expression. Chromatin can be regulated by various means, including modifications of DNA (Schübeler 2015), modifications of histones (Tessarz and Kouzarides 2014), and architectural remodeling by protein complexes (Clapier and Cairns 2009). These mechanisms function individually and in concert to modulate genome-wide topology and gene expression, thereby regulating cell differentiation, cell division, and tissue and organism development.
Figure 1.
Multiple levels of chromatin assembly. (A) The overlong DNA is highly compacted through association with core histones to form nucleosomes, and then chromatosomes by histone H1 binding. Several aspects show a great impact on chromatin dynamics (interconversion of euchromatin and heterochromatin), such as modifications of DNA, modifications of histones, and other protein complexes that interact with chromatin. (B) Structural domains of the linker histone H1. Although the globular domain (GH1) is the only folded portion of the protein in solution, all three domains are subjected to multiple posttranslational modifications (PTMs). (NTD) amino-terminal domain, (CTD) carboxy-terminal domain.
ROLE OF THE LINKER HISTONE H1 IN GENE EXPRESSION AND CHROMATIN DYNAMICS
It is believed that the general role of core histones is to condense DNA and reduce its accessibility, resulting in the inhibition of transcription in vitro (Lorch et al. 1987). However, in vivo studies have revealed that the transcription regulation is largely dependent on the posttranslational modification (PTM) of core histones and have identified specific PTMs of core histones that are related to either transcription activation or repression (Eberharter and Becker 2002; Allis and Jenuwein 2016; Izzo and Schneider 2016). Proteins that add, recognize, and remove these PTMs have also been identified and termed writers, readers, and erasers, respectively (Torres and Fujimori 2015).
In contrast to the knowledge about core histones, much less is known about linker histones in regulation of chromatin structure and function. Histone H1 binds to nucleosomes and shows a dynamic binding affinity for chromatin (Bednar et al. 1998), which allows the modulation of chromatin architecture and gene transcription. Based on findings in Drosophila, Tetrahymena, and other model organisms, it is appreciated that histone H1 is generally a transcriptional repressor (Ura et al. 1997), and studies of its structure and PTMs have provided hints of how it achieves this function.
Histone H1 species consist of three domains: the central globular domain (GH1, ∼80 amino acids), the short amino-terminal domain (NTD, ∼35 amino acids), and the carboxy-terminal domain (CTD, ∼100 amino acids) (Fig. 1B; Allan et al. 1980). The GH1 domain is the only folded portion of the entire protein in free solution and is proposed to bind symmetrically to DNA on the dyad axis (Ramakrishnan et al. 1993). The NTD is disordered in free solution and does not greatly affect chromatin condensation but is subject to PTMs that play a regulatory role in H1 function (Allan et al. 1986). The CTD is also disordered in free solution and is assumed to play an important part in chromatin condensation (Hendzel et al. 2004).
The linker histone H1 undergoes various PTMs, including phosphorylation, acetylation, methylation, and ubiquitination. In addition, H1 can undergo various other PTMs, such as citrullination, formylation, denitration, ADP ribosylation, carbonylation, and crotonylation (Fig. 1B; Izzo and Schneider 2016). More than 150 histone-modifying proteins have been identified and their dysregulation can result in inappropriate activation or inactivation of genes (Valencia and Kadoch 2019).
As with core histones H2A and H3, there are also multiple variants of H1 histones in mammalian cells. Although linker histone subtypes are redundant proteins in knockout mice (Fan et al. 2003), suggesting no functional distinction between subtypes, different tissues are characterized by specific subtypes, and transitions in subtypes are observed during embryonic development (Terme et al. 2011; Li et al. 2012). H1.1 to H1.5 and the testis-specific H1t in humans are expressed in a replication-dependent (RD) manner, meaning that these H1 subtypes are synthesized only during S phase, whereas H1.0 and H1x are replication-independent and can be expressed in nonproliferating cells (Fyodorov et al. 2018). Some functional distinctions between the RD subtypes have been observed in gene expression regulation (Alami et al. 2003), and a specific contribution of H1 subtypes to other nuclear events, such as DNA methylation (Fan et al. 2005) and cell-cycle disruption (Sancho et al. 2008), have been shown.
Fluorescence recovery after photobleaching/fluorescence loss in photobleaching (FRAP/FLIP) experiments show that a large proportion of the H1 exchanges rapidly between sites on euchromatin with an average residence time of ∼4 min (Lever et al. 2000). This pattern is very different from that of core histones, for which almost no exchange is observed over the same period of time. Therefore, the conditions for generating a stable in vitro H1-containing chromatosome have not been achieved, and an understanding of how H1 is bound to the nucleosome and how chromatin structures are controlled to facilitate gene expression is greatly needed (Crane-Robinson 2016).
Multiple Localizations and Functions of PTEN Family Members
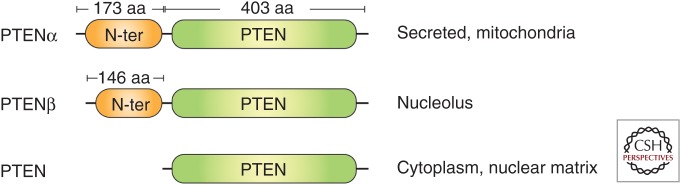
For more than 15 years since its discovery, PTEN was thought to be a nonredundant tumor suppressor gene with no variants or isoforms in humans (Keniry and Parsons 2008). Although it is well understood that germline mutations of PTEN induce cancer predisposition in various organs, PTEN dysfunction could also be an important factor in other diseases, such as diabetes and nervous system diseases (Worby and Dixon 2014). It has long been thought that PTEN is so multifunctional that this 403-amino acid (aa) protein plays a role almost everywhere. However, this notion was challenged when a new isoform of PTEN, PTEN-Long or PTENα, was independently discovered by two groups (Hopkins et al. 2013; Liang et al. 2014). In these studies, the new PTEN isoform was found to use a noncanonical translation initiation codon CUG to start translation on the canonical PTEN messenger RNA (mRNA), resulting in a 173-aa amino-terminal extended protein. These groups found that with the amino-terminal extended sequences, the localization of the protein was changed; it could be secreted from cells and reenter adjacent cells to suppress cell proliferation (Hopkins et al. 2013), or it could target mitochondria within cells and regulate energy metabolism (Liang et al. 2014). Soon after that discovery, in 2017, a third PTEN family member, PTENβ, was identified. Similar to PTENα, PTENβ used another noncanonical translational initiation codon AUU to start translation and resulted in a 146-aa amino-terminal extended protein that localized predominantly in the nucleolus and regulated pre–ribosomal RNA (rRNA) synthesis (Fig. 2; Liang et al. 2017).
Figure 2.
Structural and functional summary of the phosphatase and tension homolog (PTEN) family proteins. PTENα and PTENβ have a 173-aa and 146-aa extension, respectively, at the amino terminus of regular PTEN. The differences in amino terminus of the PTEN family decides each protein's subcellular location.
ROLE OF PTEN IN DNA REPLICATION AND REPAIR
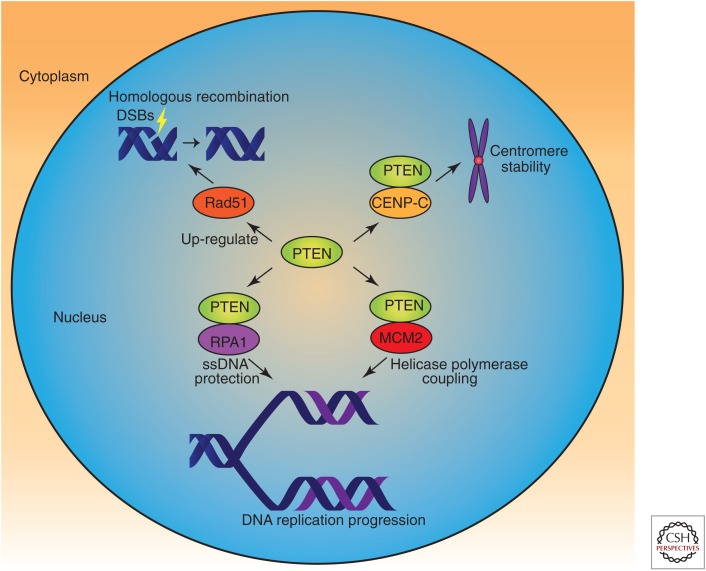
PTEN family members can be found in various locations within cells, and both PTEN and PTENβ show nuclear localization. However, the subnuclear localization of these two proteins is quite different. PTENβ is specifically localized in the nucleolus, whereas PTEN is mostly localized in the matrix. In the nucleus, PTEN is highly interactive with DNA-binding proteins and histones, and is crucial to the processes of DNA replication, DNA repair, and DNA transcription. In 2007, Shen and colleagues reported the intranuclear localization of PTEN and showed its colocalization with centromeres in mouse embryonic fibroblasts (MEFs). These researchers found that within the nucleus, PTEN physically interacts with centromere-specific binding protein C (CENP-C) to maintain chromosomal stability (Shen et al. 2007). PTEN is also involved in pathways of DNA double-strand break (DSB) repair, which is accomplished by homologous recombination (HR) induced by Rad51. PTEN also plays an indispensable role in DNA replication via association with many replication-related proteins, such as replication protein A1 (RPA1) on DNA replication forks (Wang et al. 2015) and minichromosome maintenance complex component 2 (MCM2) (Feng et al. 2015) on replication stress. Replication fork–localized PTEN can physically interact with Rad51 following replication stress for the recovery of stalled forks (Fig. 3; He et al. 2015). Therefore, PTEN guards the genome by controlling multiple processes of chromosome inheritance.
Figure 3.
Role of phosphatase and tension homolog (PTEN) in DNA replication and repair. Nuclear PTEN safeguards the genome in multiple ways. PTEN is involved in maintenance of centromere stability, DNA replication progression, and DNA repair via homologous recombination through interaction with or regulation of relative proteins, either dependent or independent of its protein kinase activity. (ssDNA) single-stranded DNA.
PTEN INTERACTS WITH HISTONE H1 AND CONTROLS CHROMATIN CONDENSATION
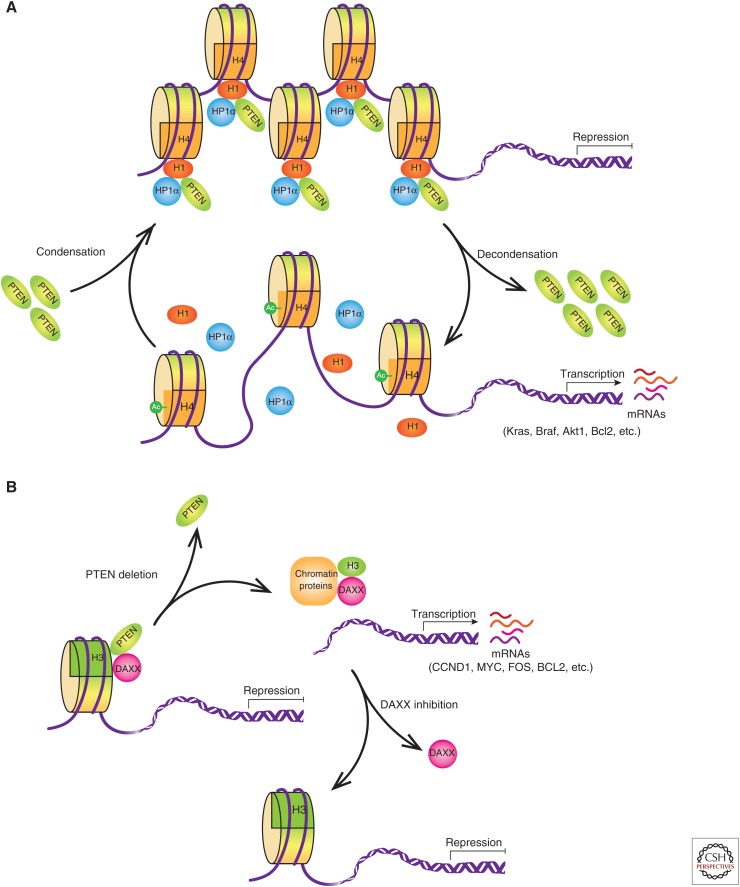
Several lines of evidence have shown that the PTEN status has an influence on global gene expression (Matsushima-Nishiu et al. 2001; Li et al. 2006), but the molecular mechanisms were not well understood. More recently, Chen et al. discovered an interplay between PTEN and the linker histone H1 as potential means of PTEN impact on gene expression (Chen et al. 2014). In this study, PTEN loss in MEFs leads to cumulative errors of heterochromatin protein 1α (HP1α) foci formation, indicating impaired structural organization of chromatin in the nucleus. Further investigation using FLAG-hemagglutinin (FH) tandem affinity purification with ectopic PTEN followed by SDS-PAGE and mass spectrometry analysis identified several subtypes of histone H1 as PTEN-interacting proteins and indicated that the C2 domain of PTEN and the CTD of H1 are responsible for the interaction. Because the CTD of histone H1 accounts for much of its binding affinity to chromatin, PTEN loss leads to histone H1 dissociation from chromatin and chromatin decondensation. Moreover, PTEN and histone H1 were found to act cooperatively to repress histone H4 acetylation at lysine 16, which is an epigenetic marker for chromatin activation. Microarray analysis of Pten-deficient MEFs revealed a number of up-regulated genes, including the oncogenes Kras, Braf, and Akt1; the survival factors Bcl2 and Nfkb2; the estrogen receptor Esr1; and the tumor invasion factor Cd44 (Fig. 4A). This mechanism appears to be a part of the cellular machinery that controls global gene expression profiles through regulating DNA accessibility and hierarchical levels of chromatin organization (Chen et al. 2014).
Figure 4.
Interplay between phosphatase and tension homolog (PTEN) and histones. (A) Interplay between PTEN, H1, and heterochromatin protein 1α (HP1α). PTEN forms a complex with H1 and HP1α on chromatin, which condenses the genome and represses oncogenic gene transcription. When PTEN is depleted in cells, H1 and HP1α fall off the chromatin, leading to acetylation of H4 at K16 and chromatin decondensation, which causes oncogenic gene transcription. (B) Interplay between PTEN and H3.3. PTEN interacts with H3.3 and its chaperone DAXX, and anchors the complex on the chromatin to repress oncogenic gene transcription. On PTEN deletion, the DAXX-H3.3 complex dissociates the chromatin, resulting in gene activation. DAXX inhibition suppresses tumor growth through global H3.3 distribution changes on chromatin leading to up-regulation of tumor suppressor genes and down-regulation of oncogenes. (mRNA) messenger RNA.
NUCLEAR PTEN BINDS TO HP1α AND MAINTAINS HETEROCHROMATIN STRUCTURE
In 2015, a study by Gong and colleagues revealed that PTEN could bind directly to heterochromatin and to the resident heterochromatin protein HP1α. In PTEN knockout in cells, there was a significant reduction in histone H3 trimethyl Lys 9 (H3K9me3) foci intensity, a hallmark of chromatin decondensation. Consistent with this, Gong and colleagues found, using quantitative real-time PCR (RT-qPCR), overexpression of satellite DNA following PTEN loss in both mouse and human cells (Gong et al. 2015). This research, together with Chen's study, reveals the existence of a complex regulatory network based on the relationship between PTEN, histones, and heterochromatin status, which is integral to the control of gene expression (Fig. 4A).
PTEN REGULATES GLIOBLASTOMA ONCOGENESIS THROUGH CHROMATIN-ASSOCIATED COMPLEX OF DAXX AND HISTONE H3.3
In addition to its association with the linker histone and HP1α, PTEN was recently shown to form a complex with the histone chaperone DAXX and the core histone H3.3 (Benitez et al. 2017). DAXX binds to the unfolded PTEN-hinge domain (amino acids 186–202), whereas PTEN binds to the histone-binding domain of DAXX. The DAXX-H3.3 interaction influences gene expression in several different ways (Puto and Reed 2008; Wethkamp and Klempnauer 2009; Wong et al. 2009; Goldberg et al. 2010). Using RT-qPCR and anti-H3.3 chromatin immunoprecipitation (ChIP) approaches, four genes with an inverse correlation between gene expression and H3.3 enrichment in PTEN-deficient cells compared with that in PTEN-WT cells were identified (CCND1, MYC, FOS, and BCL2) (Benitez et al. 2017), suggesting that PTEN represses oncogene expression by recruiting H3.3 to chromatin. Additionally, DAXX inhibition in glioblastoma patient-derived glioma neurosphere (GBM-PDX) cultures suppressed tumor growth and increases survival, specifically in a PTEN-deficient background (Benitez et al. 2017). Therefore, the PTEN-DAXX-H3.3 complex regulates gene transcription and can be a new therapeutic target to revert tumorigenesis caused by the loss of PTEN function in GBM (Fig. 4B).
CONCLUDING REMARKS
Nuclear PTEN maintains genomic stability through multiple mechanisms. PTEN-null cells show chromatin instability (CIN) and have many manifestations of CIN, including centromere breakage (Shen et al. 2007), spontaneous elevation of replication stress (Wang et al. 2015), failure of stalled fork recovery (Feng et al. 2015; He et al. 2015), checkpoint dysfunction throughout all phases of the cell cycle (Weng et al. 1999; Puc et al. 2005), genome-wide copy number alterations (Sun et al. 2014), and global chromatin decondensation (Chen et al. 2014; Gong et al. 2015). The interplay of PTEN with histone H1, HP1α, H3.3, and heterochromatin sheds light on how this powerful tumor suppressor influences expression of thousands of genes, as well as the extent of chromatin condensation. Despite this knowledge, much remains to be elucidated regarding the structural basis behind these interactions. Although the structure of the nucleosome core particle at atomic resolution was solved more than a decade ago (Richmond and Davey 2003), determination of the structures of the chromatin-containing linker histones has been very challenging. However, important progress has been made in resolving higher-order chromatin structures, including the crystal structure of the nucleosome-linker histone complex at near-atomic resolution (Zhou et al. 2015), the cryo-electron microscopic (cryo-EM) structure of the 30 nm chromatin fiber (Song et al. 2014), and the nuclear positioning of topologically associating domains (TADs) (Geeven et al. 2015), revealing detailed interactions between the linker histone and DNA in the nucleosome. The nucleosome forms a transcriptional barrier to RNA polymerase II (RNAPII) progression, resulting in transcriptional inhibition. A very recent study solved the cryo-EM structure of nucleosome-transcribing RNAPII elongation complexes (ECs), revealing that transcription elongation factors Elf1 and Spt4/5 reshape the downstream edge of the EC and intervene between RNAPII and the nucleosome. These mediators facilitate RNAPII progression through superhelical location SHL(-1) and suppress RNAPII pausing at SHL(-5), providing a platform for various chromatin activities (Ehara et al. 2019). In addition, “assay for transposase-accessible chromatin using sequencing” (ATAC-seq) has recently emerged as one of the most powerful techniques for studying genome-wide chromatin accessibility profiling (Buenrostro et al. 2013). By using this approach to study open chromatin under the regulation of PTEN and histones, we will better understand how PTEN regulates gene expression at the chromatin level. In summary, combining our increasing knowledge of the structure and principles of histone and nucleosome organization and techniques to evaluate whole genome-wide open chromatin regions, the specific role of PTEN in this elaborate and complex process will be revealed and understood.
ACKNOWLEDGMENTS
This work was supported by grants to Y.Y. including the National Key Research and Development Program of China (2016YFA0500302), the National Natural Science Foundation of China (81430056, 31420103905, 81874235, and 81621063), the Beijing Natural Science Foundation (7161007), and the Lam Chung Nin Foundation for Systems Biomedicine.
Footnotes
Editors: Charis Eng, Joanne Ngeow, and Vuk Stambolic
Additional Perspectives on The PTEN Family available at www.perspectivesinmedicine.org
REFERENCES
- Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin Q, Greally JM, Skoultchi AI, Bouhassira EE. 2003. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci 100: 5920–5925. 10.1073/pnas.0736105100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J, Hartman PG, Crane-Robinson C, Aviles FX. 1980. The structure of histone H1 and its location in chromatin. Nature 288: 675–679. 10.1038/288675a0 [DOI] [PubMed] [Google Scholar]
- Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. 1986. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol 187: 591–601. 10.1016/0022-2836(86)90337-2 [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17: 487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Arents G, Moudrianakis EN. 1993. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci 90: 10489–10493. 10.1073/pnas.90.22.10489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SJ. 2007. PTEN enters the nuclear age. Cell 128: 25–28. 10.1016/j.cell.2006.12.023 [DOI] [PubMed] [Google Scholar]
- Bassi C, Ho J, Srikumar T, Dowling RJ, Gorrini C, Miller SJ, Mak TW, Neel BG, Raught B, Stambolic V. 2013. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 341: 395–399. 10.1126/science.1236188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci 95: 14173–14178. 10.1073/pnas.95.24.14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez JA, Ma J, D'Antonio M, Boyer A, Camargo MF, Zanca C, Kelly S, Khodadadi-Jamayran A, Jameson NM, Andersen M, et al. 2017. PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nat Commun 8: 15223 10.1038/ncomms15223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Zhu M, Yang J, Liang H, He J, He S, Wang P, Kang X, McNutt MA, Yin Y, et al. 2014. PTEN interacts with histone H1 and controls chromatin condensation. Cell Rep 8: 2003–2014. 10.1016/j.celrep.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Ginn-Pease ME, Eng C. 2005. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) has nuclear localization signal-like sequences for nuclear import mediated by major vault protein. Cancer Res 65: 4108–4116. 10.1158/0008-5472.CAN-05-0124 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Crane-Robinson C. 2016. Linker histones: history and current perspectives. Biochim Biophys Acta 1859: 431–435. 10.1016/j.bbagrm.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. 2002. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 3: 224–229. 10.1093/embo-reports/kvf053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine SI. 2019. Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science 363: 744–747. 10.1126/science.aav8912 [DOI] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. 2003. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol 23: 4559–4572. 10.1128/MCB.23.13.4559-4572.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. 2005. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123: 1199–1212. 10.1016/j.cell.2005.10.028 [DOI] [PubMed] [Google Scholar]
- Feng J, Liang J, Li J, Li Y, Liang H, Zhao X, McNutt MA, Yin Y. 2015. PTEN controls the DNA replication process through MCM2 in response to replicative stress. Cell Rep 13: 1295–1303. 10.1016/j.celrep.2015.10.016 [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y. 2018. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol 19: 192–206. 10.1038/nrm.2017.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeven G, Zhu Y, Kim BJ, Bartholdy BA, Yang SM, Macfarlan TS, Gifford WD, Pfaff SL, Verstegen MJ, Pinto H, et al. 2015. Local compartment changes and regulatory landscape alterations in histone H1-depleted cells. Genome Biol 16: 289 10.1186/s13059-015-0857-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Andrés-Pons A, Fernández E, Valiente M, Torres J, Cervera J, Pulido R. 2006. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol Biol Cell 17: 4002–4013. 10.1091/mbc.e06-05-0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimm O, Perren A, Weng LP, Marsh DJ, Yeh JJ, Ziebold U, Gil E, Hinze R, Delbridge L, Lees JA, et al. 2000. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am J Pathol 156: 1693–1700. 10.1016/S0002-9440(10)65040-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691. 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Govan JM, Evans EB, Dai H, Wang E, Lee SW, Lin HK, Lazar AJ, Mills GB, Lin SY. 2015. Nuclear PTEN tumor-suppressor functions through maintaining heterochromatin structure. Cell Cycle 14: 2323–2332. 10.1080/15384101.2015.1044174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kang X, Yin Y, Chao KS, Shen WH. 2015. PTEN regulates DNA replication progression and stalled fork recovery. Nat Commun 6: 7620 10.1038/ncomms8620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Lever MA, Crawford E, Th'ng JP. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem 279: 20028–20034. 10.1074/jbc.M400070200 [DOI] [PubMed] [Google Scholar]
- Hopkins BD, Fine B, Steinbach N, Dendy M, Rapp Z, Shaw J, Pappas K, Yu JS, Hodakoski C, Mense S, et al. 2013. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 341: 399–402. 10.1126/science.1234907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou SQ, Ouyang M, Brandmaier A, Hao H, Shen WH. 2017. PTEN in the maintenance of genome integrity: from DNA replication to chromosome segregation. Bioessays 39 10.1002/bies.201700082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo A, Schneider R. 2016. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim Biophys Acta 1859: 486–495. 10.1016/j.bbagrm.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Keniry M, Parsons R. 2008. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene 27: 5477–5485. 10.1038/onc.2008.248 [DOI] [PubMed] [Google Scholar]
- Kornberg RD. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184: 868–871. 10.1126/science.184.4139.868 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Thomas JO. 1974. Chromatin structure: oligomers of the histones. Science 184: 865–868. 10.1126/science.184.4139.865 [DOI] [PubMed] [Google Scholar]
- Lever MA, Th'ng JP, Sun X, Hendzel MJ. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408: 873–876. 10.1038/35048603 [DOI] [PubMed] [Google Scholar]
- Li G, Hu Y, Huo Y, Liu M, Freeman D, Gao J, Liu X, Wu DC, Wu H. 2006. PTEN deletion leads to up-regulation of a secreted growth factor pleiotrophin. J Biol Chem 281: 10663–10668. 10.1074/jbc.M512509200 [DOI] [PubMed] [Google Scholar]
- Li JY, Patterson M, Mikkola HK, Lowry WE, Kurdistani SK. 2012. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet 8: e1002879 10.1371/journal.pgen.1002879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, He S, Yang J, Jia X, Wang P, Chen X, Zhang Z, Zou X, McNutt MA, Shen WH, et al. 2014. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab 19: 836–848. 10.1016/j.cmet.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Chen X, Yin Q, Ruan D, Zhao X, Zhang C, McNutt MA, Yin Y. 2017. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat Commun 8: 14771 10.1038/ncomms14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wagner S, Campbell RB, Nickerson JA, Schiffer CA, Ross AH. 2005. PTEN enters the nucleus by diffusion. J Cell Biochem 96: 221–234. 10.1002/jcb.20525 [DOI] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. 1987. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49: 203–210. 10.1016/0092-8674(87)90561-7 [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 273: 13375–13378. 10.1074/jbc.273.22.13375 [DOI] [PubMed] [Google Scholar]
- Matsushima-Nishiu M, Unoki M, Ono K, Tsunoda T, Minaguchi T, Kuramoto H, Nishida M, Satoh T, Tanaka T, Nakamura Y. 2001. Growth and gene expression profile analyses of endometrial cancer cells expressing exogenous PTEN. Cancer Res 61: 3741–3749. [PubMed] [Google Scholar]
- Perren A, Weng LP, Boag AH, Ziebold U, Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, Mutter GL, et al. 1999. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol 155: 1253–1260. 10.1016/S0002-9440(10)65227-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon SM, Waite KA, Eng C. 2008. The nuclear affairs of PTEN. J Cell Sci 121: 249–253. 10.1242/jcs.022459 [DOI] [PubMed] [Google Scholar]
- Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, Mansukhani M, Murty VV, Gaciong Z, Meek SE, et al. 2005. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell 7: 193–204. 10.1016/j.ccr.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Puto LA, Reed JC. 2008. Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev 22: 998–1010. 10.1101/gad.1632208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. 1993. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362: 219–223. 10.1038/362219a0 [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA. 2003. The structure of DNA in the nucleosome core. Nature 423: 145–150. 10.1038/nature01595 [DOI] [PubMed] [Google Scholar]
- Sancho M, Diani E, Beato M, Jordan A. 2008. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet 4: e1000227 10.1371/journal.pgen.1000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Lin H, Chen X, Langford LA, Koul D, Bondy ML, Hess KR, Myers JN, Hong YK, Yung WK, et al. 1999. Differential expression of MMAC/PTEN in glioblastoma multiforme: relationship to localization and prognosis. Cancer Res 59: 1820–1824. [PubMed] [Google Scholar]
- Schübeler D. 2015. Function and information content of DNA methylation. Nature 517: 321–326. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. 2007. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128: 157–170. 10.1016/j.cell.2006.11.042 [DOI] [PubMed] [Google Scholar]
- Simpson RT. 1978. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 17: 5524–5531. 10.1021/bi00618a030 [DOI] [PubMed] [Google Scholar]
- Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G. 2014. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344: 376–380. 10.1126/science.1251413 [DOI] [PubMed] [Google Scholar]
- Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. 1998. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95: 29–39. 10.1016/S0092-8674(00)81780-8 [DOI] [PubMed] [Google Scholar]
- Sun Z, Huang C, He J, Lamb KL, Kang X, Gu T, Shen WH, Yin Y. 2014. PTEN C-terminal deletion causes genomic instability and tumor development. Cell Rep 6: 844–854. 10.1016/j.celrep.2014.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terme JM, Sesé B, Millán-Ariño L, Mayor R, Izpisua Belmonte JC, Barrero MJ, Jordan A. 2011. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J Biol Chem 286: 35347–35357. 10.1074/jbc.M111.281923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Kouzarides T. 2014. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol 15: 703–708. 10.1038/nrm3890 [DOI] [PubMed] [Google Scholar]
- Torres IO, Fujimori DG. 2015. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol 35: 68–75. 10.1016/j.sbi.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, et al. 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128: 141–156. 10.1016/j.cell.2006.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe AP. 1997. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J 16: 2096–2107. 10.1093/emboj/16.8.2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia AM, Kadoch C. 2019. Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat Cell Biol 21: 152–161. 10.1038/s41556-018-0258-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li Y, Wang P, Liang H, Cui M, Zhu M, Guo L, Su Q, Sun Y, McNutt MA, et al. 2015. PTEN regulates RPA1 and protects DNA replication forks. Cell Res 25: 1189–1204. 10.1038/cr.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng LP, Smith WM, Dahia PL, Ziebold U, Gil E, Lees JA, Eng C. 1999. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res 59: 5808–5814. [PubMed] [Google Scholar]
- Wethkamp N, Klempnauer KH. 2009. Daxx is a transcriptional repressor of CCAAT/enhancer-binding protein β. J Biol Chem 284: 28783–28794. 10.1074/jbc.M109.041186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al. 2009. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res 19: 404–414. 10.1101/gr.084947.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. 2014. Pten. Annu Rev Biochem 83: 641–669. 10.1146/annurev-biochem-082411-113907 [DOI] [PubMed] [Google Scholar]
- Zhou BR, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. 2015. Structural mechanisms of nucleosome recognition by linker histones. Mol Cell 59: 628–638. 10.1016/j.molcel.2015.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]