Abstract
Tumor suppressor genes encode critical intracellular regulators, such as the retinoblastoma protein. They control processes including cell proliferation, cell survival, and responses to DNA damage and are frequently mutated in cancer. In this excerpt from his forthcoming book on the history of cancer research, Joe Lipsick looks back at the discovery of tumor suppressor genes, covering the early work on cell fusion by Henry Harris, Knudson's two-hit hypothesis, the genetic mapping studies that first identified the RB gene, and subsequent work on silencing.
HYBRID VIGOR?
In certain circumstances, cells can fuse to produce a hybrid cell. This happens at fertilization and during the normal embryonic development of skeletal muscle to produce multinucleated myotubes. Cell fusion can also occur in response to infection, when macrophages merge to become multinucleated giant cells. In the early 1960s, Georges Barski, Serge Sorieul, and Francine Cornefert observed spontaneous fusion during co-cultivation of cells from different subclones of a murine fibroblast cell line in culture. The hybrid cells were relatively stable and could be maintained for many passages. Remarkably, when cells with low cancer potential in vivo were fused to cells with high cancer potential, the stable hybrid cell line showed high cancer potential. Lawrence Scaletta and Boris Ephrussi then reported similar results when murine cancer cells were fused to normal murine fibroblasts. These results indicated that the cancer cell phenotype (and presumably the cancer cell genotype) acted in a dominant fashion. This was consistent with the oncogenic transformation of normal cells in culture by Rous sarcoma virus and murine polyoma virus that had recently been described.
Over the next few years, two technical advances greatly improved the efficiency of cell fusion in culture. First, John Littlefield developed an effective drug-selection scheme for producing hybrid cells. He selected one parental cell line for resistance to 8-azaguanine, a nucleotide analog. This resistance was due to a loss-of-function mutation in the HPRT gene. He selected the other parental cell line for resistance to 5-bromodeoxyuridine, another nucleotide analog. In this case, resistance was due to a loss-of-function mutation in the TK gene. In special growth medium containing hypoxanthine, thymidine, and aminopterin (an antifolate drug developed by Farber and Subbarow), neither parental cell could grow. Each lacked a component (either HPRT or TK) of the nucleotide salvage pathway that became essential when de novo nucleotide synthesis was blocked by aminopterin. However, a successful fusion of cells from the two different parental lines could survive in the selective growth medium. The TK− parental line was HPRT+; the HPRT− parental line was TK+. Because the normal wild-type (+) alleles of each gene were dominant, a successful fusion was phenotypically HPRT+, TK+. The ability to select for the hybrids and against both parental cell lines meant that he could select for hybrids even in cases in which one or both parental cell lines might otherwise outcompete the hybrid.
The second technical advance in cell fusion was based on the observation that some viruses caused the formation of multinucleated giant cells both in vivo and in cell culture. Productive viral infection was not required for giant cell formation, because irradiated virus preparations that were incapable of replication could still cause fusion. This separation of function was due to the much greater sensitivity of nucleic acids (the viral genome) than of proteins (including the fusogenic proteins on the surface of the virions) to ionizing radiation. Henry Harris and John Watkins showed that irradiated Sendai virus could be used to fuse cells from two different cell lines to create stable heterokaryons (hybrid cells containing nuclei from two different cell types). The ability to create heterokaryons at will became a powerful tool for cell biology. In a remarkable series of experiments, Harris showed that the highly condensed, transcriptionally inactive nuclei of chicken red blood cells could be reactivated by fusion to mitotically active cells.
Cell fusion also provided an important tool for investigating the genetic basis of inherited human metabolic defects. Cell lines from different patients could be fused to determine whether they had mutations in the same gene (no complementation, as seen by a lack of rescue of the metabolic defect) or in different genes (complementation, as seen by rescue of the metabolic defect). Unexpectedly, rodent–human hybrids preferentially lost human chromosomes, resulting in stable cell lines with only one or a few human chromosomes. These hybrid cells were very useful in mapping human genes to specific chromosomes. Furthermore, in 1975, Georges Köhler and César Milstein used this technology to develop hybridomas that secreted monoclonal antibodies. They fused a normal antibody-producing B lymphocyte with a nonproducing plasmacytoma cell line, and then selected a hybrid cell line that was able to survive in selective media (because of a wild-type HPRT gene from the normal B cells) and to proliferate continuously (because of cancer-causing mutations in the plasmacytoma, a tumor with a mature B cell phenotype). The resulting monoclonal antibodies became invaluable tools for the characterization of proteins, for the identification and purification of different cell types, and eventually for the treatment of some human cancers (see Lipsick 2019a).
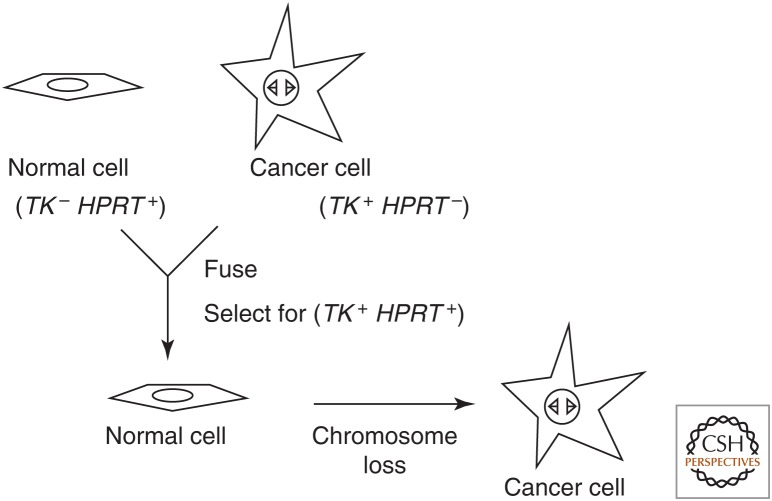
With these new tools in hand, Harris revisited the question of whether the oncogenic phenotype was indeed dominant in cell fusions. In contrast to the previous results of Barski and others, Harris reported in 1969 that the oncogenic phenotype was recessive in hybrids between normal cells and cancer cells of the laboratory mouse. These results remained controversial for some time. The apparent paradox was resolved when Harris together with George Klein showed that upon continued passage in culture or in animals, the hybrids of normal cells and cancer cells eventually exhibited on oncogenic phenotype (Fig. 1). Eric Stanbridge extended these results to human cells in 1976.
Figure 1.
Evidence for tumor suppressor genes from cell fusion experiments. Variations of this idealized experiment used senescing normal cells or nonadherent cancer cells without their own genetic markers.
By examining karyotypes (chromosome spreads from mitotic cells), they found that the oncogenic phenotype correlated with the loss of specific chromosomes from the hybrid cells. These results implied that normal cells contain genes that act in a dominant fashion within cells to prevent cancer. Such genes therefore became known as “tumor suppressors.” Additional experiments showed that tumor suppressor genes could be localized to specific regions of specific chromosomes. Despite considerable effort, attempts to clone the genes responsible based on their ability to suppress the oncogenic phenotype in cultured cells were not successful.
THE TWO-HIT HYPOTHESIS
Retinoblastoma, a malignant tumor of the eye, occurs in approximately 1 in 85,000 children between birth and 4 years of age. It can be diagnosed by leukocoria, a white instead of a normal red “reflex” of the eye that is seen upon routine examination. This cancer was described in detail in 1809 by James Waldrop, a Scottish surgeon, who proposed treatment by enucleation (removal of the eye). During the twentieth century, improved surgical techniques and radiation therapy permitted the survival of many affected patients. Follow-up of these patients presented an interesting puzzle.
Approximately two-thirds of the affected children presented with a solitary unilateral (in one eye only) tumor at the time of diagnosis. The remaining one-third presented with multiple bilateral tumors at the time of diagnosis. The children with bilateral tumors were, on average, much younger at the time of diagnosis and also had an increased incidence of other cancers, particularly osteosarcomas. As successfully treated children reached adulthood, and some eventually had children of their own, the resulting family histories further distinguished the two groups of patients. The children of parents with unilateral retinoblastoma usually did not develop tumors themselves. However, approximately one-half of the children of parents with bilateral retinoblastomas also developed bilateral retinoblastomas. Therefore, bilateral retinoblastoma was inherited in an autosomal dominant Mendelian fashion. An affected parent had two alleles of a putative retinoblastoma-susceptibility gene, one normal wild-type allele and one cancer-causing mutant allele. Thus, the chance of an individual offspring inheriting the cancer-causing allele from that parent was 50%.
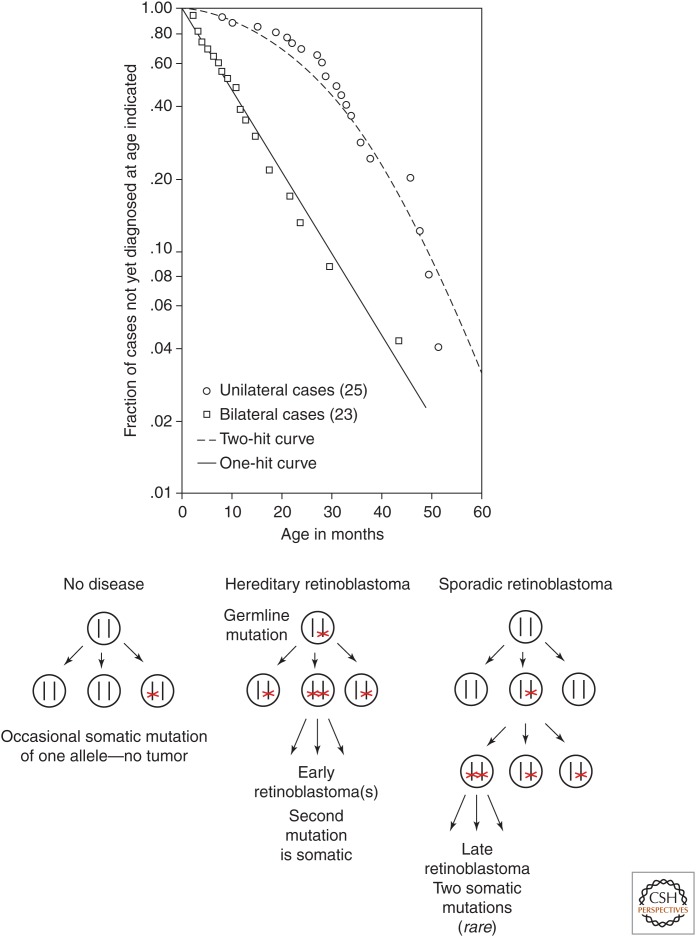
In 1971 Alfred Knudson performed a careful statistical analysis of data from retinoblastoma patients that included family histories and the number of tumors present in each patient. All the bilateral cases appeared to be hereditary, although 80% of these cases appeared to occur de novo (neither parent was affected). However, ∼15% of the unilateral cases were also hereditary. Furthermore, some individuals who had multiple children with bilateral tumors did not themselves have tumors. These individuals appeared to be “silent” carriers of the disease-causing allele. Together, these observations implied that inheritance of the cancer-causing allele was not by itself sufficient to cause retinoblastoma. Taking into account the statistical distribution of the number of tumors per patient in hereditary retinoblastoma (a mean of 3), the estimated number of retinal cells per eye (2 × 106), and the period of initial diagnosis (within 2 years of conception), Knudson developed a “two-hit” hypothesis consistent with the clinical observations (Fig. 2).
Figure 2.
Knudson's two-hit hypothesis. (Top) Semilogarithmic plot of fraction of retinoblastoma cases not yet diagnosed versus age of patient. (Bottom) Model of events in hereditary versus sporadic retinoblastoma. (Top, Reprinted from Knudson AG. 1971. Proc Natl Acad Sci 68: 820–823, Fig. 1, p. 823.)
The “two-hit” hypothesis proposed that for retinoblastoma to occur, two specific mutations must be present within the same cell. In the case of hereditary retinoblastoma, the first mutation is inherited and is present in every cell of the body. The probability that the second mutation occurs later within a somatic (nongermline) retinal cell had to be very low, because only a few tumors occurred per patient. However, the probability had to be high enough for one or a few of the ∼4 × 106 retinal cells to suffer a second mutation, given the nearly 100% tumor incidence in patients affected by hereditary retinoblastoma. In the case of sporadic (nonhereditary) retinoblastoma, the first somatic mutation needed to occur in a retinal cell or its embryonic progenitor. The second somatic mutation then had to occur within the mitotic descendants (cells that arise by subsequent divisions). If all somatic mutations were independent rare events, then the probability of two such mutations occurring in the same cell or its somatic progeny was much lower (“rare squared”).
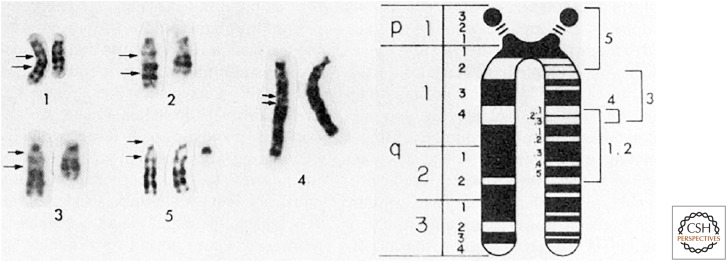
The two-hit hypothesis did not formally require mutation of both alleles of the same gene. It did not distinguish between dominant gain-of-function mutations in two different genes or recessive loss-of-function mutations in the same gene. However, subsequent cytogenetic observations argued strongly for loss-of-function mutations. Approximately 5% of patients with retinoblastoma had a cytogenetically visible constitutional (present in all cells) deletion within the long arm of chromosome 13, and nearly all of these patients had bilateral tumors. Although the extent of deletion varied from case to case, a relatively small region (13q14) was common to all such deletions (Fig. 3). In theory a deletion could cause an increase in the expression of an adjacent gene. However, the highly variable extent of the chromosome 13 deletions observed in patients with bilateral retinoblastoma implied that loss-of-function mutation was the most likely cause of the disease.
Figure 3.
Deletions of chromosome 13 in patients with (1–4) or without (5) retinoblastoma. Arrows to the left of each normal chromosome indicate deleted or duplicated segments. The smallest region of overlapping deletions in retinoblastoma was in 13q14. (Reprinted from Sparkes RS, et al. 1980. Science 208: 1042–1044, Fig. 1, p. 1043, with permission from AAAS.)
The statistical evidence for the two-hit hypothesis together with the cytogenetic evidence for loss-of-function mutation satisfyingly explained both hereditary and nonhereditary retinoblastoma. A mutant tumor-causing allele was recessive at the level of the individual cell, because both alleles had to be inactivated to cause cancer. However, the mutant allele appeared to be dominant at the level of the organism, because the chance of mutating the remaining wild-type allele in at least one developing retinal cell approached 100%. This model was reminiscent of what happened following the fusion of a cancer cell with a normal cell. The noncancerous phenotype initially “won” because the hybrid cell had a wild-type allele of a critical tumor suppressor gene. However, as the hybrid cells proliferated, the cancer phenotype eventually reappeared, because of the loss of the chromosomes that contained the wild-type alleles.
A LOST CAUSE
The identification of a putative retinoblastoma-susceptibility gene (RB) became a topic of intense interest. To pursue this goal, researchers needed a foothold near the 13q14 region that was deleted in many cases of retinoblastoma. In 1975, Walter Bodmer and his colleagues had mapped the human esterase D gene, which encodes an enzyme with two different alleles distinguishable by protein electrophoresis, to chromosome 13 by analysis of somatic cell hybrids. In 1980, Robert Sparkes and his colleagues further localized this gene to 13q14 by analyzing enzymatic activity in red blood cells and fibroblasts from patients with constitutional deletions of different portions of chromosome 13 (Fig. 3). However, mutation of this gene itself was unlikely to be a cause of retinoblastoma because most retinoblastomas expressed esterase D.
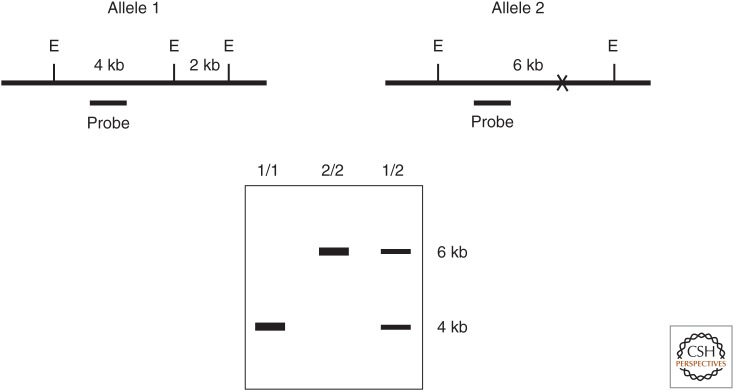
In 1980, David Botstein and his colleagues described a new general method for mapping human genes. Genetic linkage and the use of meiotic recombination to construct linkage maps had been described by Alfred Sturtevant in 1913 based on studies of visible mutant phenotypes in Drosophila. However, Botstein's new method did not rely on visible mutant phenotypes but instead utilized polymorphisms within the DNA itself. Genetics no longer depended on the expression of phenotype; rather the genotype itself was the phenotype. The initial description of the method used restriction fragment length polymorphism (RFLP) to provide markers for genetic mapping (Fig. 4).
Figure 4.
Genotyping using restriction fragment length polymorphism (RFLP). (Top) Cartoons of genomic DNA for two different alleles of the same locus. (E) EcoRI restriction enzyme recognition site, (X) absence of one EcoRI site in allele 2, (probe) radioactive DNA used for hybridization in the Southern blot below. (Bottom) Autoradiogram of a Southern blot of EcoRI-digested genomic DNA from individuals with the diploid genotypes indicated above each lane.
Restriction enzymes serve as an immune system in bacteria to resist infectious viruses. These enzymes recognize and then cleave very specific short sequences of DNA that differ for each enzyme. The host cell DNA is protected by specific methylation of these sequences that prevents recognition and cleavage by the restriction enzyme, whereas the unmethylated “foreign” viral DNA is digested and inactivated. Variations in DNA sequence from person to person (polymorphisms) can lead to differences in the recognition sites for these enzymes.
These variations could be detected by isolating and purifying DNA (often from a sample of a person's white blood cells), digesting it with a restriction enzyme, and then analyzing the length of the resulting DNA fragment by Southern blotting with a radioactive probe. The power of this approach was that it could identify phenotypically silent markers that vary among the human population. If such a marker was close to a particular gene of interest (e.g., a mutation causing a hereditary disease), the two are likely to remain linked during meiosis. Therefore, within an affected family, one allele of the marker would co-segregate with the disease. The closer the marker was to the gene of interest, the lower the frequency of recombination between them during meiosis. If a marker was sufficiently close to the gene of interest, it could be used as a probe to “walk” along a path of overlapping molecular clones to isolate the mutated gene of interest. Prior to the completion of the human genome project, this method was used to clone many genes mutated in hereditary diseases. As DNA sequencing became faster and less expensive, RFLP analysis was largely replaced by the direct analysis of all detectable single-nucleotide polymorphisms (SNPs), regardless of whether or not they alter restriction enzyme sites. This provided a greater number of more closely spaced markers for genetic mapping studies.
Webster Cavenee and Ray White at the University of Utah set out to map the putative RB gene by RFLP analysis in collaboration with several groups that had collected samples from retinoblastoma patients and their families. To make molecular probes specific for polymorphic regions of human chromosome 13, they constructed a recombinant bacteriophage library from genomic DNA isolated from a hamster–human somatic cell hybrid that contained human chromosomes 13, 12, and 6p. They identified phage that contained human DNA, and then isolated small segments of DNA from these phage and tested these probes against a panel of hamster–human somatic cell hybrids containing different complements of human chromosomes. Eventually, seven different DNA probes without repetitive sequences were identified that mapped to chromosome 13 and that also displayed RFLP in DNA samples from unrelated donors.
Cavanee and White used these probes in RFLP analysis of genomic DNA samples from normal cells and from retinoblastoma tumors from several affected individuals. In most cases in which normal cells showed heterozygosity in RFLP markers, there was a loss of heterozygosity (LOH) of at least one marker in the tumor tissue. This LOH of chromosome 13 markers occurred in both hereditary and nonhereditary cases of retinoblastoma. Loss of the normal allele of the retinoblastoma susceptibility gene (RB) and a closely linked RFLP marker, rather than a second independent mutation, was thus the most frequent “second hit” predicted by the two-hit hypothesis.
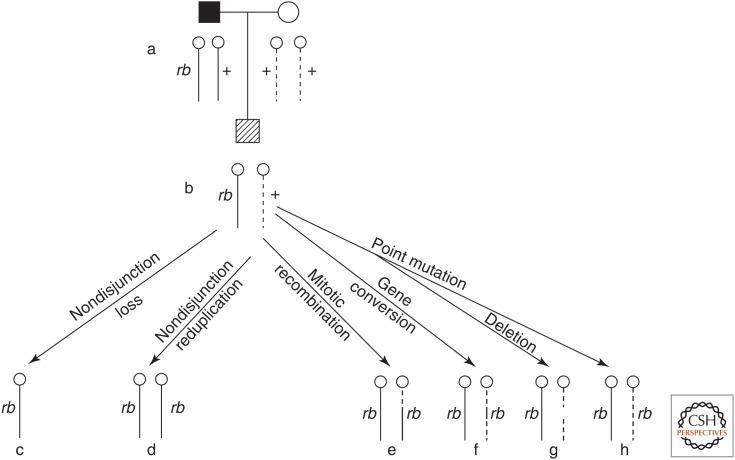
Analyses of RFLP markers, esterase D alleles, and karyotypes of normal versus tumor tissue indicated that a number of different mechanisms were responsible for the LOH observed in many tumors (Fig. 5). These mechanisms included nondisjunction (whole chromosome loss during cell division), mitotic recombination (crossing-over between homologous chromosomes), gene conversion (replacement of one allele by the other allele), and interstitial deletion at 13q14. Although LOH was observed in most cases of both hereditary and nonhereditary retinoblastoma, in some cases no LOH was observed in tumor tissue. Cavanee and White thought this was due either to a lack of an RFLP marker sufficiently close to the RB gene or a second independent mutation in the other RB allele.
Figure 5.
Mechanisms for generating somatic loss of heterozygosity (c–h) in retinoblasts of a heterozygous carrier (b) born to an affected father and unaffected mother (a). Solid lines represent paternal chromosomes; dashed lines represent maternal chromosomes. (Reprinted from Cavenee, et al. 1983. Nature 305: 779–784, Fig. 1, p. 780, by permission of Springer Nature.)
A SMOKING GUN
Several laboratories raced to clone the RB tumor suppressor gene. Thaddeus (Ted) Dryja's group made the breakthrough in 1986. He and his coworkers identified single-copy DNA probes whose targets on chromosome 13 were deleted in a number of retinoblastoma tumors. One group of adjacent probes hybridized to DNA sequences that were conserved in human and in mouse. This suggested that this segment of genomic DNA might include the coding region of a conserved gene. In collaboration with Stephen Friend and Robert Weinberg, they showed that probes for this region hybridized with an mRNA species present in many tissues, including normal retina, but not in several retinoblastomas. The mRNA was also absent in one osteosarcoma, a malignant bone tumor that sometimes occurs in patients with hereditary retinoblastoma. This provided strong circumstantial evidence that the gene was likely to be the long-sought RB tumor suppressor.
By the end of the following decade, the status of the RB gene had been analyzed in more than 350 cases of hereditary retinoblastoma. Visible cytogenetic abnormalities causing large deletions or translocations that included RB were present in ∼8% of the tumors. Tumors without gross cytogenetic abnormalities displayed a range of focal mutations. Single-nucleotide substitution mutants were found in ∼40% of the tumors; small insertions or deletions were found in ∼25% of the tumors; more complex mutations were found in ∼10% of the tumors. Strikingly, almost all of the focal mutations resulted in truncation or complete loss of the Rb protein because of nonsense mutations (premature stop codons), frameshift mutations, or aberrant mRNA splicing. Thus, there seemed to be a strong selection for the loss of Rb protein function during tumor initiation or growth. However, in ∼15% of tumors no RB mutations were identified. Nevertheless, many of the tumors without RB mutations displayed loss of heterozygosity at the RB locus. On the other hand, 35% of tumors with proven RB mutations did not show loss of heterozygosity at closely linked genetic markers. So, genetics alone did not seem to tell the whole story.
EPIGENETIC GENE SILENCING
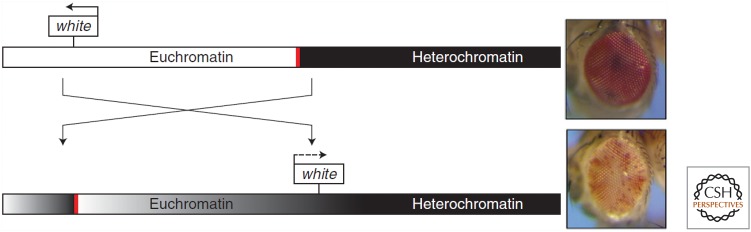
Among the first X-ray-induced mutations in Drosophila identified by Hermann Muller in the 1920s was a peculiar set of eye-color mutants called white-mottled. The eyes of the affected flies contained variable patches of red and white pigment. This phenotype was passed from parents to progeny, arguing against somatic mosaicism (mutations that occur in somatic cells and so are not inherited in the germline). Decades of work by many scientists revealed that in these flies, a wild-type allele of the white gene had been translocated from euchromatin (transcriptionally active open regions of chromosomes) to a position near heterochromatin (transcriptionally inactive condensed regions of chromosomes). In some cells the white gene remained transcriptionally active, whereas in other cells the heterochromatin spread into and silenced the white gene (Fig. 6). Both of these states appeared to be relatively stable and could be inherited during mitotic cell divisions, thereby explaining the variable patches of red (white gene expression) and white (white gene silencing) pigmentation within the eye. This phenomenon became known as position-effect variegation (PEV) because the eye pigment phenotype resulted from an altered location of the gene within the genome.
Figure 6.
Position effect variegation of the white gene in Drosophila. The upper chromosome and eye are wild-type. The lower chromosome has an inversion that placed the white gene next to constitutive heterochromatin (black bar), resulting in a mottled eye. The red bar indicates a boundary that normally prevents the spread of silencing. (Reprinted from Elgin SC, Reuter G. 2013. Cold Spring Harb Perspect Biol 5: a017780, doi:10.1101/cshperspect.a017780, Fig. 1A, p. 4, © Cold Spring Harbor Laboratory Press.)
PEV is an example of epigenetic regulation—there are two alternate states of expression that can be stably inherited through cell division, but this heritability is not directly determined by DNA sequence. In Drosophila and in most other eukaryotes, epigenetic silencing can involve reversible chemical modifications of histones, proteins around which DNA is wound in the cell nucleus. However, in many species, including humans, it may involve methylation of the DNA itself.
Nearly all DNA methylation in humans occurs on cytosine (C) residues in the context of a CpG dinucleotide. The “p” indicates that C and G are adjacent bases within the same strand and not paired bases on different strands of the double helix. In the 1970s, Arthur Riggs proposed that the main function of methylcytosine (meC) was to silence gene expression. He postulated the existence of “de novo” DNA methylases (enzymes that can infrequently place new methyl marks on DNA) and of “maintenance” DNA methylases (enyzmes that maintain existing methylation marks on new DNA strands during DNA replication).
DNA methylation was first mapped using restriction enzymes that cut unmethylated, but not methylated DNA. The strategy is similar to the RFLP method described above. However, in this case the methylation-sensitive restriction enzyme detects a change in DNA methylation rather than a change within the DNA sequence.
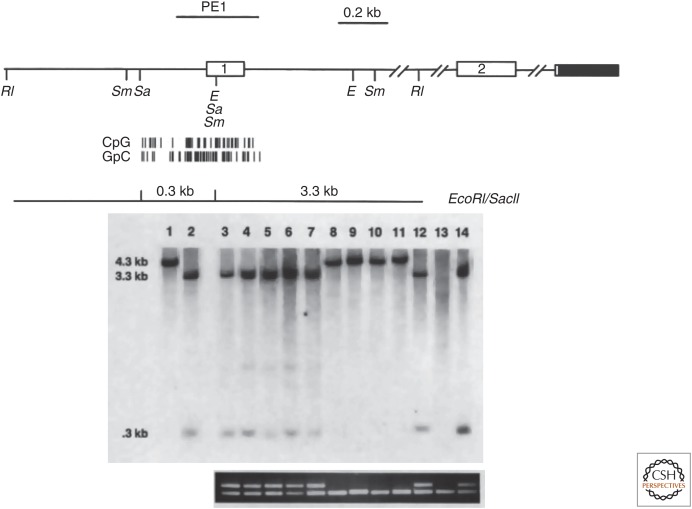
In 1989, Bernard Horsthemke's lab detected methylation of the RB gene, but not the adjacent esterase D gene in a unilateral sporadic retinoblastoma. This methylation was not present in normal lymphocytes from the same patient. Approximately 50% of the total RB DNA was methylated, which suggested allele-specific methylation. Two years later, Ted Dryja's lab published a similar analysis of 56 primary retinoblastoma tumors. They found that ∼10% of samples showed DNA methylation of the RB gene, all in patients with unilateral tumors. Furthermore, methylation correlated with the absence of RB gene expression. Another tumor suppressor gene in the same pathway, CDKN2A encoding p16INK4A, was later shown to be frequently methylated in a variety of human cancers not bearing alterations of RB itself (Fig. 7) (Lipsick 2019b).
Figure 7.
DNA methylation of the CDKN2A gene encoding p16INK4A in human lung cancer cell lines. (Top) Map of the locus showing restriction enzyme sites and the CpG island surrounding the first exon. (Middle) Southern blot with PE1 probe following digestion of genomic DNA with EcoRI (RI) and/or SacII (Sa). Lane 1 contains control DNA digested with EcoRI; lane 2 contains control DNA digested with EcoRI and SacII; lanes 3–14 contain DNAs from different lung cancer cell lines digested with EcoRI and SacII. Unmethylated DNA digested with EcoRI and SacII generates hybridizing fragments of 3.3 and 0.3 kb, whereas methylated DNA generates a hybridizing fragment of 4.0 kb because of the failure of SacII to cut at its recognition sites. (Bottom) RT-PCR detection of mRNA from the CDKN2A gene (top band) or a TP53 control gene (bottom band). Note the absence of CDKN2A mRNA in the cell lines in which the locus is methylated (lanes 8–11) or deleted (lane 13). (Adapted from Merlo et al. 1995. Nat Med 1: 686–682, Figs 1 and 2, pp. 687, 688, by permission of Springer Nature.)
A more comprehensive method for detecting genome-wide DNA methylation has largely replaced methylation-sensitive restriction enzymes. This method relies on the ability of bisulfite treatment of purified DNA to convert C to U, unless the C is methylated. Large-scale analysis of genome-wide DNA methylation patterns in different types of tumors is now possible with this strategy. Thus far, the patterns observed are rather complex, and it remains unclear whether most DNA methylation causes cancer or is instead a result of the general deregulation of gene expression that often occurs in cancer. However, there are some specific types of cancer in which mutation of genes encoding enzymes that regulate covalent modifications of histones and of DNA do act as drivers of oncogenesis (see Lipsick 2019c).
GENERALIZING THE RB STORY
The analysis of a number of other familial cancer syndromes (e.g., Wilm's tumor, neurofibromatosis, von Hippel–Lindau syndrome, and familial breast cancer) yielded findings similar to those initially discovered in retinoblastoma. In familial cases, a heterozygous germline mutation of a tumor suppressor gene acted in a dominant fashion in the family pedigree. In nonfamilial (sporadic) cases of the same tumor type, the same tumor suppressor gene was often mutated or epigenetically silenced in the tumor cells but not in normal cells from the same patient. In both cases, the tumor cells displayed LOH or epigenetic silencing of the normal allele of the same gene. These observations provided the basis for a general strategy for discovering new tumor suppressor genes that did not depend on familial cancers. The idea was to look for regions of the genome that displayed frequent deletion or LOH in cancer cells but not in normal cells from individual patients. If the same region was similarly affected in tumors from multiple unrelated patients, it might harbor a tumor suppressor gene. A number of different experimental strategies identified tumor-specific deletions and regions of LOH, including nucleic acid hybridization to a panel of probes for all human genes, single-nucleotide polymorphism screening, and, more recently, quantitative high-throughput DNA sequencing.
The work of many laboratories showed that proto-oncogenes were activated by dominant gain-of-function mutations, whereas tumor suppressor genes were inactivated by recessive loss-of-function mutations or epigenetic silencing. By 1990 tumor suppressor genes had gained equal billing with oncogenes as important players in cancer (Fig. 8). At that time, tumor suppressors were also known as “anti-oncogenes” or “recessive oncogenes,” usages that have faded over time. Furthermore, some tumor suppressor genes displayed “haploinsufficiency,” in that inactivation of only one allele caused cancer. This was first shown in 1998 by Christopher Kemp's laboratory, who created mice heterozygous for the CDKN1B gene encoding the p27Kip1 inhibitor of cyclin-dependent protein kinases.
Figure 8.
Poster and T-shirt design for the 6th Annual Meeting on Oncogenes. (© Jamie Simon.)
High-throughput sequencing of many human cancer cell genomes in the early twenty-first century led Bert Vogelstein's group to conclude that perhaps as few as 55 proto-oncogenes and 70 tumor suppressor genes accounted for most of the recurrently mutated drivers of oncogenesis in the most common types of lethal human cancer. However, an as yet unknown number of additional genes may act as drivers in rare types of cancer and, perhaps, as rare drivers in common types of cancer.
SUGGESTED READING
*Article is in this collection.
- Baylin S, Bestor TH. 2002. Altered methylation patterns in cancer cell genomes: Cause or consequence? Cancer Cell 1: 299–305. [DOI] [PubMed] [Google Scholar]
- Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL. 1983. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 305: 779–784. [DOI] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. 1986. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646. [DOI] [PubMed] [Google Scholar]
- Harris H. 1993. How tumour suppressor genes were discovered. FASEB J 7: 978–979. [DOI] [PubMed] [Google Scholar]
- Knudson AG. 2001. Two genetic hits (more or less) to cancer. Nat Rev Cancer 1: 157–162. [DOI] [PubMed] [Google Scholar]
- *.Lipsick J. 2019a. A history of cancer research: genetically targeted therapies. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lipsick J. 2019b. A history of cancer research: RB and cell cycle control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a035923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lipsick J. 2019c. A history of cancer research: epigenetics and differentiation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a035998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Toguchida J, Ohtani N, Yandell DW, Rapaport JM, Dryja TP. 1991. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am J Hum Genet 48: 880–888. [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. 2004. Principles of tumor suppression. Cell 116: 235–246. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]