Abstract
Innate immune sensors can recognize when host cells are irrevocably compromised by pathogens, and in response can trigger programmed cell death (pyroptosis, apoptosis, and necroptosis). Innate sensors can directly bind microbial ligands; for example, NAIP/NLRC4 detects flagellin/rod/needle, whereas caspase-11 detects lipopolysaccharide. Other sensors are guards that monitor normal function of cellular proteins; for instance, pyrin monitors Rho GTPases, whereas caspase-8 and receptor-interacting protein kinase (RIPK)3 guards RIPK1 transcriptional signaling. Some proteins that need to be guarded can be duplicated as decoy domains, as seen in the integrated decoy domains within NLRP1 that watch for microbial attack. Here, we discuss the evolutionary battle between pathogens and host innate immune sensors/guards, illustrated by the Red Queen hypothesis. We discuss in depth four pathogens, and how they either fail in this evolutionary race (Chromobacterium violaceum, Burkholderia thailandensis), or how the evolutionary race generates increasingly complex virulence factors and host innate immune signaling pathways (Yersinia species, and enteropathogenic Escherichia coli [EPEC]).
The innate immune system can combat intracellular bacteria by inducing programmed cell death, which eliminates the infected cell niche. Killing host cells can also be useful in the innate immune response in cases in which a cell has been irrevocably reprogrammed to act in the benefit of the pathogen. Programmed cell death can be initiated by a variety of interconnected pathways, resulting in pyroptosis, apoptosis, or necroptosis. There are notable recent advances in our understanding of how programmed cell death either restricts bacterial pathogens, or fails to do so as certain pathogens have evolved to evade this defense. The evolutionary race between pathogen virulence and host defense is vital in determining whether a bacterial pathogen can cause disease, or is readily cleared.
Direct sensors, guards, and decoys initiate specific forms of programmed cell death, including pyroptosis, necroptosis, and apoptosis. Pyroptosis and necroptosis result in membrane rupture that releases soluble cytosolic contents to the extracellular space, whereas apoptosis converts a cell into apoptotic bodies that retain cellular contents within membranes. Cell death may lead to enhanced clearance of the pathogen; however, inappropriate or excessive cell death can be extremely detrimental to the host. These tightly regulated cell death mechanisms are often triggered by the activation of specific caspase proteases. Pyroptosis results from activation of caspase-1 or murine caspase-11/human caspase-4, -5 (Jorgensen et al. 2017). Apoptosis ensues when apoptotic initiators (caspase-8, -9) activate apoptotic effectors (caspase-3, -6, -7) (Taylor et al. 2008). Lastly, necroptosis occurs when RIPK3 phosphorylates the pseudokinase MLKL (Dondelinger et al. 2016a; Vanden Berghe et al. 2016). Many of these signaling pathways use death fold family domains to drive homotypic interactions, including death domains (DDs), death effector domains (DEDs), caspase activation and recruitment domains (CARDs), and pyrin domains (PYDs) (Weber and Vincenz 2001).
RED QUEEN HYPOTHESIS
The evolutionary biologist Leigh Van Valen proposed that each species must constantly evolve to avoid extinction in the face of competitors who are also constantly evolving. To illustrate this race, Van Valen drew upon the imagery in Lewis Carol's book Through the Looking Glass, in which the protagonist, Alice, engages in a footrace with the Red Queen. Alice soon finds that they have been running, but have stayed in the same place. The Red Queen informs Alice “Now, here, you see, it takes all the running you can do, to keep in the same place.” Van Valen's Red Queen hypothesis proposes that organisms must constantly evolve to maintain their place in an environment where competitors are also constantly evolving (Van Valen 1973).
The Red Queen hypothesis also applies to host–pathogen interactions; as hosts evolve defenses against infection, pathogens must evolve virulence factors to overcome those defenses. This constant evolutionary race by both competitors ensures that neither the host nor pathogen go extinct. In the perpetual drive to outrun each other, hosts evolve increasingly complex defense mechanisms while, simultaneously, pathogens evolve equally complex virulence factors. Typically, pathogens are able to keep up with the evolutionary challenge in the race with the innate immune system. However, most pathogens lose the race against the adaptive immune system, which eventually resolves the infection, but not before the pathogen transmits to a new host. Thus, the host (which we herein personify as Alice) constantly evolves new immunologic defenses. Meanwhile, pathogens (which we herein personify as Red Queens), must constantly evolve novel virulence factors.
INNATE IMMUNE SENSORS: DIRECT SENSORS, GUARDS, AND DECOYS
Programmed cell death can be initiated by a variety of sensors in the innate immune system. Notable among these are the nucleotide-binding domain, leucine-rich repeat (NLR) superfamily of cytosolic sensors, which cause cell death in eukaryotes ranging from plants to animals. Jones, Vance, and Dangl proposed that NLRs fall into three categories: direct sensors, guards, and decoys (Jones et al. 2016). These concepts not only apply to NLRs, but are also broadly applicable to many pathways in the innate immune system (Fig. 1).
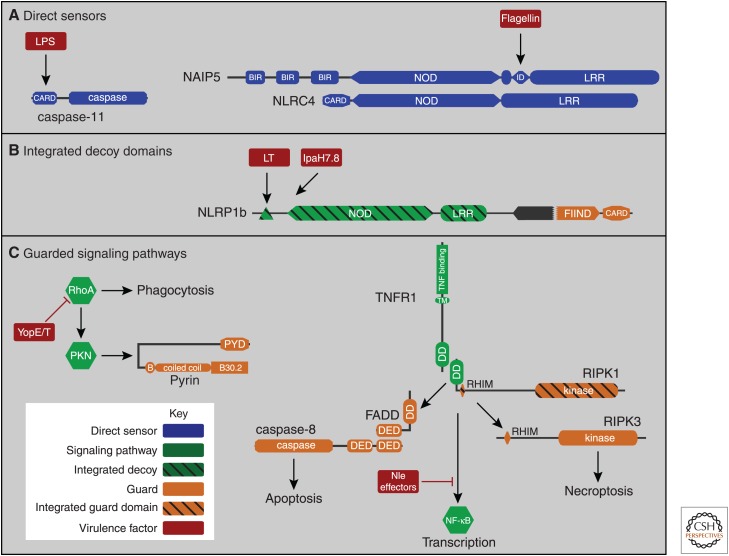
Figure 1.
Innate immune surveillance: direct sensors, integrated decoy domains, and guarded signaling pathways. (A) Direct sensors include both caspase-11 and NAIP5/NLRC4. Caspase-11 activates when it directly binds to bacterial lipopolysaccharide (LPS) via its caspase activation and recruitment domain (CARD). NAIP5 directly binds bacterial flagellin; the NAIP5-flagellin complex then oligomerizes with NLRC4 molecules to form a caspase-1 activating inflammasome. (B) Integrated decoy domains are similar to domains of other signaling pathways, acting as lures for bacterial virulence factors. Once the decoy domain is attacked, this triggers signaling by the innate immune guard domains within the same protein. For example, the anthrax lethal toxin (LT) intends to target host mitogen-activated protein kinase (MAPK) but also cleaves the amino-terminal integrated decoy domain of NLRP1. IpaH7.8 is an E3 ubiquitinase that presumably intends to target another host protein, but also targets the integrated decoy domains of NLRP1 for degradation. Either of these attack events cause the NLRP1 signaling guard domains (the FIIND-CARD fragment) to form an active inflammasome. (C) Guards can watch a protein or a specific signaling pathway. Pyrin monitors RhoA signaling to protein kinase (PKN). When the Yersinia effectors YopE and T attack RhoA, pyrin detects the loss of PKN activation and in response forms an active inflammasome. Integrated guard domains can also be added within normal signaling pathways; for example, receptor-interacting protein kinase (RIPK)1 has two guard functions that survey the tumor necrosis factor (TNF) transcriptional response, one of which uses an integrated guard domain. When bacterial virulence factors, such as Nle effectors, attack the TNF transcriptional response, the first guard pathway is triggered by exposed death domains (DDs) within RIPK1, which are normally occupied by interacting with the TNF receptor signaling complex. When these DDs are abnormally exposed, they are detected by the DD containing guard adaptor Fas-associated death domain protein (FADD), which in turn signals to the guard caspase-8 to initiate apoptosis. Additionally, RIPK1 also encodes a RHIM and a kinase domain that can be considered integrated guard domains. These domains activate RIPK3, but only when both the transcriptional response and the apoptosis guard pathways are inhibited.
“Direct sensors” bind microbial ligands. For example, TLR4 and its coreceptor MD2 directly bind lipopolysaccharide (LPS) in the extracellular space, triggering a transcriptional response. In the cytosol, caspase-11 is the direct sensor for LPS, triggering pyroptotic cell death. Similarly, extracellular flagellin detection by TLR5 drives a transcriptional response, whereas cytosolic flagellin detection by NAIP/NLRC4 triggers pyroptosis.
“Guards” monitor a cellular protein(s) for evidence of attack by virulence factors, and activate innate immune signaling in response. Guards can directly bind to the guarded protein, but we propose they may also monitor the enzymatic activity of the guarded protein. If the guarded protein is functioning normally, guards do not respond. However, if a guarded protein is attacked by a pathogen virulence factor, then the guard responds by activating and inducing a new response, often programmed cell death. Note that guards do not prevent the guarded protein from being attacked; rather they sound the alarm when such an attack occurs. More generally applied, the guard concept encompasses sensors that monitor whether signaling pathways are functioning normally. In this context, pyrin is a guard for Rho GTPases, and caspase-8/RIPK3 are guards for the tumor necrosis factor (TNF) signaling pathway. Extending the guard concept further, a guard function may be added to the protein that needs to be guarded within the same open reading frame (an “integrated guard”). Integrated guard domains are added to normal signaling proteins and are not strictly required for their normal function. The kinase domain of RIPK1 may be an example of an integrated guard domain whereby RIPK1 monitors its own scaffolding function.
“Decoys” are duplicates of the protein that needs to be monitored—the guard watches the decoy. Decoys act like lures, tempting virulence factors to attack them, triggering cell death in response. The decoy and guard can also be combined into a single protein; “integrated decoy domains” are decoys that are encoded in the same open reading frame as the guard domains. NLRP1 may be an example of an integrated decoy that monitors for attack on other NLR proteins.
PYROPTOSIS
Caspase-1 is activated by a variety of inflammasome sensors that act as direct sensors (NAIP/NLRC4, AIM2), guards (pyrin), or decoys (NLRP1), or whose mode of sensing remains to be elucidated (NLRP3). Inflammasomes can signal directly to caspase-1 if they contain a CARD (NLRC4, NLRP1), or indirectly via the ASC adaptor if they signal via a PYD (AIM2, pyrin, NLRP3). In contrast, caspase-11 is a combined direct sensor and protease that activates itself when it binds to LPS.
Either activated caspase-1 or caspase-11 can independently cleave and activate gasdermin D (Kayagaki et al. 2015; Shi et al. 2015) (for a gasdermin review, see Kovacs and Miao 2017). Cleavage of gasdermin D separates the amino-terminal pore-forming domain from the carboxy-terminal regulatory domain. Twenty-seven gasdermin pore-forming domains oligomerize to form an 18-nm pore that is large enough to allow the egress of all small molecules as well as small proteins (Ruan et al. 2018). In addition to gasdermin D, caspase-1 (but not caspase-11[Ramirez et al. 2018]) also cleaves IL-1β (4.5 nm) and IL-18 (5.0 nm), which will easily pass through the gasdermin pore. Sodium, and thereby water, enter the cell, increasing the cytosolic turgor pressure until the membrane ruptures. We define this rupture event as pyroptosis.
Pyroptosis is often described as lytic programmed cell death, which often conjures an image of a fully dispersed cell. Indeed, the membrane rupture is large enough to allow all soluble cytosolic contents to immediately escape from the cell. However, the torn plasma membrane remains otherwise intact, and retains the organelles, cytoskeleton, nucleus, and intracellular bacteria. These bacteria remain viable, but trapped within the remains of the pyroptotic cell. We termed the corpse of the pyroptotic cell as a pore-induced intracellular trap (PIT); as apoptosis converts cells into apoptotic bodies, pyroptosis converts cells into PITs. PITs simultaneously attract neutrophils, which efferocytose both the PIT and its trapped bacteria (Jorgensen et al. 2016a,b). The neutrophil then generates reactive oxygen species and kills the detained pathogen (Miao et al. 2010a).
NAIP–NLRC4 Defending against Chromobacterium violaceum
Perhaps the best understood inflammasome is NAIP/NLRC4, which detects the activity of bacterial type III secretion systems (T3SS). T3SS are syringe-like mechanisms that inject effector proteins into the cytosol of host cells. These effector proteins reprogram the host cell to the benefit of the pathogen. However, T3SS also aberrantly translocates flagellin, rod, and needle proteins into the host cytosol. Flagellin binds to mouse NAIP5 or NAIP6, T3SS rod proteins bind to NAIP2, whereas the T3SS needle binds to NAIP1 (Vance 2015). The structural basis for detection and activation have recently been delineated (Hu et al. 2015; Zhang et al. 2015; Tenthorey et al. 2017; Yang et al. 2018). For example, a single flagellin protein molecule binds to a single NAIP5, triggering a conformational change that exposes a polymerization interface on NAIP5. This interface recruits NLRC4, causing a similar conformational change that recruits another NLRC4, and so on until 10 NLRC4s have been oligomerized. This creates a hub-like structure composed of one NAIP and 10 NLRC4 proteins that is termed an inflammasome. The CARD domains of the 10 NLRC4 proteins are clustered in the middle of the inflammasome hub, and initiate caspase-1 polymerization. Thus, it is possible that a single molecule of flagellin, rod, or needle protein can result in pyroptosis. This seems to be an incredibly decisive system to destroy cells that have been compromised by T3SS injection.
One would expect that the existence of NLRC4 would make it very difficult for bacterial pathogens to use a T3SS. Yet, approximately half of all Gram-negative bacterial pathogens use T3SS. The Red Queen's race may have selected for pathogens that uniformly evade or inhibit NLRC4. For example, flagellin repression strategies are common and at least one case of an NLRC4-evasive T3SS has been described (Miao et al. 2010a,b). Remarkably, engineering bacteria to reverse flagellin repression generates stains that are eliminated by NLRC4 with exquisite sensitivity.
At least one bacterium, C. violaceum, appears to have no ability to evade NLRC4. C. violaceum is a ubiquitous environmental pathogen that encodes a T3SS, but whose natural host is unknown (Batista and da Silva Neto 2017). It infects people with significant immunologic defects, primarily those with chronic granulomatous disease (Batista and da Silva Neto 2017). Inflammasome responses clear C. violaceum infection in mouse models. Pyroptosis is required for splenic clearance, in which macrophages are most likely infected. In contrast, in the liver, inflammasome-driven IL-18 primes a natural killer (NK) cell response in which perforin-mediated cytotoxicity clears the hepatocyte niche, presumably by triggering apoptosis. Just as normal people are resistant to C. violaceum infection, wild-type (WT) mice resist high dose challenge (1,000,000 CFUs). However, Nlrc4−/− mice succumb to low-dose challenge (100 CFUs) (Maltez et al. 2015). We estimate that this represents a >50,000-fold change in the effective 100% lethal dose (LD100; Table 1) when comparing WT to inflammasome-deficient mice (Maltez and Miao 2016). The remarkable change in the effective lethal dose is a phenotype that is nearly unheard of in the inflammasome literature, equaled only by B. thailandensis (discussed in the next section).
Table 1.
Inflammasome and necroptosis survival studies
| Pathogen | Mutation | Dose | Route | Time to death (median days) | % Survival | Δ lethal dosea | Micro-biota ctrl | Background | KO used in paper to get results shown in Time and Survival columns (and other KO with similar results) | References | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | KO | WT | KO | |||||||||
| BACTERIA versus INFLAMMASOMES | ||||||||||||
| Acinetobacter baumanii | 5 × 108 | in | 1 | 2 | 88% | 25% | >2 | ns | C57BL/6 | Casp11−/− | Wang et al. 2017a | |
| Bacillus anthracis Ames 35—vegetative | 105 | sc | ∞ | 4 | 100% | 0% | >5 | ns | C57BL/6NTac | “WT” are Nlrp1S/S, KO are Nlrp1R/R | Moayeri et al. 2010 | |
| —vegetative | 105 | iv | ∞ | 4 | 100% | 0% | >5 | ns | C57BL/6NTac | “WT” are Nlrp1S/S, KO are Nlrp1R/R | Moayeri et al. 2010 | |
| —spores | 107 | sc | ∞ | 4.5 | 100% | 20% | >2 | ns | C57BL/6NTac | “WT” are Nlrp1S/S, KO are Nlrp1R/R | Moayeri et al. 2010 | |
| —spores | 2 × 107 | sc | ∞ | 1 | 100% | 0% | >5 | L | BALB/c | All Nlrp1S/S; Casp1/11+/+ or Casp1/11−/− | Moayeri et al. 2010 | |
| Bacillus anthracis Ames—spores | 4 × 102 | ip | 3 | 3 | 50% | 25% | >1 | L | C57BL/6 | “WT” are Nlrp1b129Tg; KO are no Tg | Terra et al. 2010 | |
| Bacillus anthracis Sterne—spores | 2.5 × 107 | ip | ∞ | 4 | 100% | 0% | >5 | L | C57BL/6 | “WT” are Nlrp1b129Tg; KO are no Tg | Terra et al. 2010 | |
| Bacillus anthracis Stern—spores | 106 | ip | ∞ | 3 | 100% | 0% | >5 | ns | C3H/HeJCr | Casp1/11−/− | Kang et al. 2008 | |
| Burkholderia cepacia | 106 | ip | ∞ | ∞ | 100% | 100% | 1 | N | C57BL/6 | Casp1/11−/− | Maltez et al. 2015 | |
| Burkholderia cenocepacia | 2 × 108 | ip | ∞ | 4 | 100% | 17% | >2 | ns | C57BL/6 | Mefv−/− | Aubert et al. 2016 | |
| Burkholderia pseudomallei | 102 | in | 4 | 4 | 65% | 0% | >5 | ns | C57BL/6 | Casp1/11−/− (Nlrc4−/−, Asc−/−) | Ceballos-Olvera et al. 2011 | |
| 25 | in | ∞ | 4 | 100% | 0% | >5 | ns | C57BL/6 | Casp1/11−/− (Nlrc4−/−, Asc−/−) | Ceballos-Olvera et al. 2011 | ||
| 102 | in | 4 | 5 | 65% | 20% | >1 | ns | C57BL/6 | Nlrp3−/− | Ceballos-Olvera et al. 2011 | ||
| 102 | in | ∞ | 2–3 | 100% | 0% | >5 | ns | C57BL/6 | Casp1/11−/− | Breitbach et al. 2009 | ||
| 102 | in | ∞ | 3.5 | 100% | 0% | >5 | N | C57BL/6 | Casp1/11−/− (Casp11−/−) | Aachoui et al. 2013 | ||
| Burkholderia thailandensis | 2 × 107 | ip | ∞ | 16 | 100% | 13% | >20 | N | C57BL/6 | Casp1/11−/− | Aachoui et al. 2013, 2015 | |
| (unpassaged) | 2 × 106 | ip | ∞ | 14 | 100% | 20% | >20 | N | C57BL/6 | Casp1/11−/− | Aachoui et al. 2013 | |
| 2 × 107 | ip | ∞ | ∞ | 100% | 100% | 1 | N | C57BL/6 | Nlrc4−/− | Aachoui et al. 2013 | ||
| 2 × 106 | ip | ∞ | ∞ | 100% | 100% | 1 | N | C57BL/6 | Nlrc4−/−Asc−/− | Aachoui et al. 2013 | ||
| 104 | in | ∞ | 5 | 100% | 0% | >5 | N | C57BL/6 | Casp1/11−/− | Aachoui et al. 2013 | ||
| 104 | in | ∞ | ∞ | 100% | 100% | 1 | N | C57BL/6 | Nlrc4−/−Asc−/− (Nlrc4−/−, Asc−/−, Nlrp3−/−) | Aachoui et al. 2013 | ||
| Burkholderia thailandensis | 2 × 107 | ip | ∞ | 1 | 100% | 0% | >1,000,000 | N | C57BL/6 | Casp1/11−/− (Casp11−/−) | Aachoui et al. 2013 | |
| (one mouse passage) | 2 × 107 | ip | ∞ | 1 | 100% | 0% | >1,000,000 | N | C57BL/6 | Casp1/11−/− (Casp11−/−) | Aachoui et al. 2015; Maltez et al. 2015 | |
| 106 | ip | ∞ | 2 | 100% | 0% | >1,000,000 | N | C57BL/6 | Casp1/11−/− | Aachoui et al. 2015 | ||
| 105 | ip | ∞ | 2 | 100% | 0% | >1,000,000 | N | C57BL/6 | Casp1/11−/− | Aachoui et al. 2015 | ||
| 104 | ip | ∞ | 2 | 100% | 0% | >1,000,000 | N | C57BL/6 | Casp1/11−/− (Casp11−/−) | Aachoui et al. 2015; Maltez et al. 2015 | ||
| 103 | ip | ∞ | 3 | 100% | 0% | >1,000,000 | N | C57BL/6 | Casp1/11−/− | Aachoui et al. 2015 | ||
| 102 | ip | ∞ | 3 | 100% | 0% | >1,000,000 | N | C57BL/6 | Casp1/11−/− (Casp11−/−) | Aachoui et al. 2015 | ||
| 2 × 107 | ip | ∞ | 2 | 100% | 0% | >5 | N | C57BL/6 | Nlrc4−/−Asc−/− (Nlrc4−/−Nlrp3−/−) | Aachoui et al. 2015 | ||
| 104 | ip | ∞ | ∞ | 100% | 100% | >5 | N | C57BL/6 | Nlrc4−/−Asc−/− | Aachoui et al. 2015 | ||
| 102 | ip | ∞ | ∞ | 100% | 100% | >5 | N | C57BL/6 | Nlrc4−/−Asc−/− | Aachoui et al. 2015 | ||
| 2 × 107 | ip | ∞ | ∞ | 100% | 100% | 1 | N | C57BL/6 | Nlrc4−/− (Nlrp3−/−,Nlrc3−/−, Nlrc5−/−,Nlrp12−/−) | Aachoui et al. 2015 | ||
| Chromobacterium violaceum | 106 | ip | ∞ | n.d. | 100% | n.d. | >50,000 | N | C57BL/6 | Casp1/11−/− (Nlrc4−/−, Nlrc4−/−Asc−/−) | Maltez et al. 2015 | |
| 104 | ip | ∞ | 3 | 100% | 0% | >50,000 | N | C57BL/6 | Casp1/11−/− (Nlrc4−/−, Nlrc4−/−Asc−/−) | Maltez et al. 2015 | ||
| 102 | ip | ∞ | 4 | 100% | 0% | >50,000 | N | C57BL/6 | Casp1/11−/− (Nlrc4−/−, Nlrc4−/−Asc−/−) | Maltez et al. 2015 | ||
| Ehrlichia (Ixodes ovatus ehrlichia) | ∼103 | ip | 9 | 9 | 100% | 100% | 1 | ns | C57BL/6 | Casp1/11−/− (Nlrp3−/−) | Yang et al. 2015 | |
| Escherichia coli O18:K1:H7 | Range | ip | ns | ns | ns | ns | 4 | L | CeH/HeJ | Casp1/11−/− | Joshi et al. 2002 | |
| Francisella tularensis subsp. novicida | 1.5 × 105 | sc | 4 | 3 | 65% | 0% | >2 | L | C57BL/6 | Aim2−/− | Fernandes-Alnemri et al. 2010 | |
| 1.5 × 105 | sc | 6 | 4 | 30% | 0% | ″ | N, (L) | C57BL/6 | Casp1/11−/− (Asc−/−) | Mariathasan et al. 2005 | ||
| 1.5 × 105 | sc | 5 | 4 | 22% | 0% | <1 | L | C57BL/6 | Nlrc4−/− | Mariathasan et al. 2005 | ||
| 5 × 103 | sc | 3 | 2.5 | 75% | 0% | ″ | ns | ns | Casp1/11−/− | Meunier et al. 2015 | ||
| 7.5 × 104 | sc | 4 | 4 | 65% | 0% | ″ | ns | ns | Casp1/11−/− (Aim2−/−) | Man et al. 2015 | ||
| Francisella turlarensis subsp. Holartica | LVS | 2 × 104 | in | 7 | 7 | 0% | 0% | 1 | ns | C57BL/6 | Nlrp3−/− | Duffy et al. 2016 |
| Francisella philomiragia | 106 | ip | ∞ | ∞ | 100% | 100% | 1 | N | C57BL/6 | Casp1/11−/− | Maltez et al. 2015 | |
| Klebsiella pneumoniae | 7.4 × 104 | it | 2.1 | 1.9 | 15% | 0% | >1 | ns | C57BL/6 | Nlrp3−/− (Asc−/−) | Willingham et al. 2009 | |
| 7.4 × 104 | it | 2 | 2.5 | 0% | 0% | 1 | ns | C57BL/6 | Nlrc4−/− | Willingham et al. 2009 | ||
| 103 | in | 5 | 5 | 75% | 40% | >1 | ns | C57BL/6 | Nlrc4−/− | Cai et al. 2012 | ||
| 104 | in | 4 | 6 | 50% | 15% | >1 | ns | C57BL/6 | Nlrc4−/− | Cai et al. 2012 | ||
| Listeria monocytogenes | 106 | iv | 5 | 3–4 | 35%– 65% | 0% | >2 | ns | C57BL/6J | Casp1/11−/− | Tsuji et al. 2004 | |
| 105 | iv | 5 | 6 | 0% | 50% | <2 | N | C57BL/6 | Casp11−/− (Nlrp6−/−) | Hara et al. 2018 | ||
| Mycobacterium tuberculosis | 250–350 | in | 200 | 148 | 0% | 0% | 1 | ns | C57BL/6 | Asc−/− | Mcelvania-Tekippe et al. 2010 | |
| 250–350 | in | 170 | 170 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− (Nlrp3−/−, Nlrc4−/−) | Mcelvania-Tekippe et al. 2010 | ||
| 50–100 | in | 200 | 110 | 90% | 0% | >2 | ns | C57BL/6 | Casp1/11−/− (Asc−/−) | Mayer-Barber et al. 2010 | ||
| 106 | it | ∞ | 6 | 0% | 0% | >5 | L | C57BL/6 | Aim2−/− | Saiga et al. 2012 | ||
| Pseudomonas aeruginosa | 2 × 107 | in | 1.5 | 1.7 | 20% | 65% | <1 | ns | C57BL/6 | Nlrc4−/− | Faure et al. 2014 | |
| ΔpopBD | 108 | in | 2.5 | 1.5 | 44% | 0% | >1 | ns | C57BL/6 | Nlrc4−/− | Faure et al. 2014 | |
| 7 × 105 | it | 3 | ∞ | 90% | 100% | <1 | ns | C57BL/6 | Nlrc4−/− | Tolle et al. 2015 | ||
| 3 × 107 | in | ∞ | 4 | 100% | 0% | >5 | ns | C57BL/6 | Nlrc4−/− | Iannitti et al. 2016 | ||
| 3 × 107 | in | ∞ | ∞ | 100% | 100% | 1 | ns | C57BL/6 | Nlrp3−/− | Iannitti et al. 2016 | ||
| 3.5 × 105 | in | 2.0 | 1.8 | 12% | 4% | >1 | ns | C57BL/6 | Casp1/11−/− | Hughes et al. 2018 | ||
| 3.5 × 105 | in | 2.0 | 2.9 | 12% | 46% | <1 | ns | C57BL/6 | Asc−/− | Hughes et al. 2018 | ||
| Salmonella enterica sv. Typhimurium | 100 | ip | 4.5 | 5.5 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− | Monack et al. 2000 | |
| 105 | po | 9 | 6 | 0% | 0% | 1 | L | C57BL/6 | Casp1/11−/− (Asc−/−) | Lara-Tejero et al. 2006 | ||
| 105 | po | 9 | 9 | 0% | 0% | 1 | L | C57BL/6 | Nlrp3−/− | Lara-Tejero et al. 2006 | ||
| 106 | po | 19 | 11 | 30% | 20% | >1 | L | Nramp1+/+ | Casp1/11−/− | Lara-Tejero et al. 2006 | ||
| 108 | po | 8 | 5.5 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− | Raupach et al. 2006 | ||
| 2 × 1010 | po | ∞ | 20 | 100% | 22% | >2 | ns | 129Sv/J | Casp1/11−/− | Raupach et al. 2006 | ||
| 108 | ip | 3 | 8 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− | Raupach et al. 2006 | ||
| 1–5 × 105 | po+s | 6 | 6 | 0% | 0% | 1 | ns | C57BL/6 | Nlrc4−/− | Franchi et al. 2012 | ||
| 105 | ip | 4 | 5 | 0% | 0% | 1 | ns | C57BL/6 | Nlrc4−/− | Franchi et al. 2012 | ||
| 1–5 × 105 | po | 8 | 7 | 0% | 0% | 1 | ns | BALB/c | Nlrc4−/− | Franchi et al. 2012 | ||
| 1–5 × 105 | po+s | 6 | 4 | 0% | 0% | 1 | ns | BALB/c | Nlrc4−/− | Franchi et al. 2012 | ||
| 105 | ip | 4 | 5 | 0% | 0% | 1 | ns | BALB/c | Nlrc4−/− | Franchi et al. 2012 | ||
| 103 | ip | 6 | 5 | 0% | 0% | 1 | N | C57BL/6 | Nlrc4−/− (Naip2−/−, Naip5−/−) | Zhao et al. 2016 | ||
| Salmonella enterica sv. Typhimurium | FliCON | 102 | ip | ∞ | 7 | 100% | 0% | >5 | N | C57BL/6 | Nlrc4−/− | Miao et al. 2010a |
| FliCON | 103 | ip | ∞ | 6 | 100% | 0% | >5 | ns | C57BL/6 | Nlrc4−/− (Naip5−/−) | Zhao et al. 2016 | |
| FliCON | 103 | ip | ∞ | ∞ | 100% | 100% | 1 | ns | C57BL/6 | Naip2−/− | Zhao et al. 2016 | |
| PrgJON | 103 | ip | ∞ | 5 | 100% | 0% | >5 | ns | C57BL/6 | Nlrc4−/− (Naip2−/−) | Zhao et al. 2016 | |
| PrgJON | 103 | ip | ∞ | ∞ | 100% | 100% | 1 | ns | C57BL/6 | Naip5−/− | Zhao et al. 2016 | |
| Shigella flexneri | 2 × 108 | in | 0.8 | 1.9 | 75% | 20% | >2 | ns | C57BL/6 | Casp1/11−/− | Sansonetti et al. 2000 | |
| Staphylococcus aureus | 104 | ic | 0.8 | 1 | 60% | 25% | >1 | ns | C57BL/6 | Casp1/11−/− (Asc−/−, Aim2−/−) | Hanamsagar et al. 2014 | |
| 104 | ic | 5 | 5 | 95% | 70% | >1 | ns | C57BL/6 | Nlrp3−/− | Hanamsagar et al. 2014 | ||
| 108 | ro | 2 | 1 | 84% | 40% | >1 | ns | C57BL/6 | Casp1/11−/− | Kitur et al. 2016 | ||
| 2 × 108 | iv | 0.5 | 1.5 | 0% | 60% | >2 | N | C57BL/6 | Casp11−/− (Nlrp6−/−) | Hara et al. 2018 | ||
| Streptococcus agalactiae (group B) | 105 | ip | ∞ | 1 | 100% | 40% | >2 | ns | C57BL/6 | Casp1/11−/− (Asc−/−, Nlrp3−/−) | Costa et al. 2012 | |
| Streptococcus pneumoniae | 105 | in | 3 | 2.5 | 87% | 90% | 1 | ns | C57BL/6 | Casp1/11−/− | Albiger et al. 2007 | |
| 103 | in | 6 | 5 | 90% | 50% | >1 | ns | C57BL/6 | Nlrp3−/−, Aim2−/− | Rodriguez et al. 2019 | ||
| Vibrio vulnificus | 1.5 × 104 | ip | ∞ | 1 | 100% | 60% | >1 | ns | C57BL/6 | Casp1/11−/− (Asc−/−, Nlrp3−/−) | Toma et al. 2010 | |
| Yersinia pestis | 104 | in | 3 | 3 | 0% | 0% | 1 | N | C57BL/6 | Casp1/11−/− | Sivaraman et al. 2015 | |
| Yersinia pestis | ΔyopJM | 2 × 102 | sc | 7 | 6 | 72% | 25% | >1 | ns | C57BL/6 | Casp1/11−/− | Ratner et al. 2016 |
| Yersinia pestis | 101 | sc | 7 | 5 | 0% | 0% | 1 | ns | C57BL/6 | Nlrp12−/− | Vladimer et al. 2012 | |
| Yersinia pestis | pYtbLpxL | 5 × 102 | sc | ∞ | 8 | 100% | 20% | >2 | ns | C57BL/6 | Nlrp12−/− (Nlrp3−/−) | Vladimer et al. 2012 |
| Yersinia pseudotuberculosis | 103 | ip | 6 | 4 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− | LaRock and Cookson 2012 | |
| Yersinia pseudotuberculosis | ΔyopM | 103 | ip | ∞ | 6 | 100% | 0% | >5 | ns | C57BL/6 | Casp1/11−/− | LaRock and Cookson 2012 |
| Yersinia pseudotuberculosis | 2 × 103 | iv | 7 | 7 | 0% | 0% | 1 | ns | C57BL/6 | Mefv−/− | Chung et al. 2016 | |
| Yersinia pseudotuberculosis | ΔyopM | 2 × 103 | iv | 13 | 7 | 89% | 0% | >2 | ns | C57BL/6 | Mefv−/− | Chung et al. 2016 |
| Yersinia pseudotuberculosis | Comp. mutant | 1 × 109 | po | 5 | 5 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− | Zheng et al. 2012 |
| VIRUSES versus INFLAMMASOMES | ||||||||||||
| Encephalomyocarditis virus | 2xLD50 | ip | 5 | 5 | 10% | 15% | >1 | N | C57BL/6 | Casp1/11−/− | Rajan et al. 2011 | |
| Influenza A virus | 6 × 104 | in | 8 | 7 | 65% | 40% | >1 | ns | C57BL/6 | Casp1/11−/− (Asc−/−, Nlrp3−/−) | Allen et al. 2009 | |
| 6 × 104 | in | 8 | 7 | 68% | 71% | 1 | ns | C57BL/6 | Nlrc4−/− | Allen et al. 2009 | ||
| 8 × 103 | in | 11 | 10 | 65% | 35% | >1 | ns | C57BL/6 | Casp1/11−/−, (Nlrp3−/−) | Thomas et al. 2009 | ||
| 8 × 103 | in | 12 | 9 | 83% | 85 | 1 | ns | C57BL/6 | Nlrc4−/− | Thomas et al. 2009 | ||
| 101 | in | ∞ | 11 | 100% | 0% | >5 | ns | C57BL/6 | Casp1/11−/− (Asc−/−) | Ichinohe et al. 2009 | ||
| 101 | in | ∞ | ∞ | 100% | 100% | 1 | ns | C57BL/6 | Nlrp3−/− | Ichinohe et al. 2009 | ||
| 1.25 × 102 | in | ∞ | ∞ | 100% | 100% | 1 | ns | C57BL/6 | Nlrp3−/− (Aim2−/−) | Rodriguez et al. 2019 | ||
| Influenza A virus H7N9 | 5 × 104 | in | 9 | 9 | 18% | 64% | <1 | ns | C57BL/6 | Casp1/11−/− (Nlrp3−/−, Asc−/−) | Ren et al. 2017 | |
| Herpes simplex virus-1 | 1.5 × 106 | scr | 11 | 12.5 | 71% | 73% | 1 | ns | C57BL/6 | Casp1/11−/− | Milora et al. 2014 | |
| Murine hepatitis virus A59 | 5 × 103 | ic | 10 | 10 | 90% | 35% | >2 | ns | C57BL/6 | Casp1/11−/− | Zalinger et al. 2017 | |
| Murine hepatitis virus-3 | 102 | ip | 4 | 5 | 0% | 28% | <1 | ns | C57BL/6 | Casp1/11−/− (Nlrp3−/−) | Guo et al. 2015 | |
| Murine norovirus | 102 | po | 6 | 7 | 0% | 0% | 1 | C57BL/6 Stat1−/− | Nlrp3−/− (Gsdmd−/−) | Dubois et al. 2019 | ||
| Vaccinia | 3 × 106 | iv | 6 | 7 | 0% | 0% | 1 | ns | C57BL/6 | Casp1−/− (Asc−/−) | Wang et al. 2017b | |
| Vesicular stomatitis virus | 2 × 105 | in | 7 | 7 | 40% | 20% | >1 | N | C57BL/6 | Casp1/11−/− | Rajan et al. 2011 | |
| 2 × 107 | iv | 5 | 5 | 0% | 0% | 1 | ns | C57BL/6 | Casp1−/− (Asc−/−) | Wang et al. 2017b | ||
| West Nile virus | 102 | sc | 11 | 11 | 81% | 50% | >1 | ns | C57BL/6 | Casp1/11−/− (Nlrp3−/−) | Ramos et al. 2012 | |
| 102 | sc | 11 | 12 | 75% | 75% | 1 | ns | C57BL/6 | Nlrc4−/− | Ramos et al. 2012 | ||
| 102 | sc | 11 | 10 | 42% | 11% | >1 | ns | C57BL/6 | Asc−/− | Kumar et al. 2013 | ||
| 101 | sc | 11 | 11 | 89% | 58% | >1 | ns | C57BL/6 | Asc−/− | Kumar et al. 2013 | ||
| FUNGI versus INFLAMMASOMES | ||||||||||||
| Aspergillus fumigatus | 2 × 107 | in | ∞ | ∞ | 100% | 100% | 1 | ns | C57BL/6 | Aim2−/−Nlrp3−/− | Man et al. 2017 | |
| 2 × 107 | in | ∞ | ∞ | 100% | 100% | 1 | ns | C57BL/6 | Nlrp3−/−, (Nlrc4−/−) | Iannitti et al. 2016 | ||
| Aspergillus fumigatus (imm. suppressed) | 105 | in | 6 | 4 | 70% | 0% | >2 | ns | C57BL/6 | Casp1/11−/− (Aim2−/−Nlrp3−/−, Asc−/−) | Man et al. 2017 | |
| 105 | in | 6 | 5 | 70% | 50% | >1 | ns | C57BL/6 | Nlrp3−/− | Man et al. 2017 | ||
| 105 | in | 6 | 5.5 | 70% | 67% | >1 | ns | C57BL/6 | Aim2−/− | Man et al. 2017 | ||
| 5 × 105 | in | 5 | 4 | 43% | 0% | >1 | ns | C57BL/6 | Casp1−/− (Casp1/11−/−) | Karki et al. 2015 | ||
| 5 × 105 | in | 5 | 5 | 43% | 0% | >1 | ns | C57BL/6 | Casp11−/− | Karki et al. 2015 | ||
| Candida albicans | 105 | iv | 9 | 5 | 40% | 0% | >1 | ns | C57BL/6 | Nlrp3−/− | Gross et al. 2009 | |
| 2 × 105 | iv | 18 | 17 | 83% | 50% | >1 | ns | C57BL/6 | Casp1/11−/− (Asc−/−) | Van De Veerdonk et al. 2011 | ||
| 5 × 106 | ts | N/A | 3 | 100% | 60% | >1 | ns | C57BL/6 | Nlrc4−/− | Tomalka et al. 2011 | ||
| ns | ts | ∞ | 5 | 97% | 60% | >1 | ns | C57BL/6 | Casp1/11−/− (Asc−/−) | Hise et al. 2009 | ||
| Paracoccidioides brasiliensis | 2 × 106 | iv | 98 | 93 | 73% | 0% | >2 | ns | C57BL/6 | Casp1/11−/− | Ketelut-Carneiro et al. 2015 | |
| 2 × 106 | iv | 98 | 93 | 73% | 30% | >1 | ns | C57BL/6 | Asc−/− | Ketelut-Carneiro et al. 2015 | ||
| 106 | it | 85 | 50 | 54% | 0% | >2 | ns | C57BL/6 | Casp1/11−/− | Feriotti et al. 2017 | ||
| 106 | it | 85 | 120 | 54% | 0% | >2 | ns | C57BL/6 | Asc−/− (Nlrp3−/−) | Feriotti et al. 2017 | ||
| PARASITES versus INFLAMMASOMES | ||||||||||||
| Plasmodium berghei sporozoites | 10 | iv | 9 | 12 | 45% | 73% | <1 | ns | C57BL/6 | Nlrp3−/− | Dostert et al. 2009 | |
| Plasmodium berghei sporozoites | 104 | iv | 6.5 | 6.5 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− | Kordes et al. 2011 | |
| Plasmodium berghei iRBCs | 104 | iv | 6.5 | 6.5 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− | Kordes et al. 2011 | |
| Plasmodium berghei iRBCs | 106 | ip | 6 | 6 | 0% | 0% | 1 | ns | C57BL/6 | Casp1/11−/− (Asc−/−) | Reimer et al. 2010 | |
| 106 | ip | 6 | 8 | 0% | 0% | 1 | ns | C57BL/6 | Nlrp3−/− | Reimer et al. 2010 | ||
| Plasmodium chabaudi adami | 5 × 104 | ip | 11 | 12 | 0% | 0% | 1 | ns | C57BL/6 | Nlrp3−/− | Shio et al. 2009 | |
| Toxoplasma gondii tachyzoites | 104 | ip | 10 | 9 | 75% | 10% | >2 | ns | C57BL/6 | Casp1/11−/− (Asc−/−, Nlrp3−/−, Nlrp1abc−/−) | Gorfu et al. 2014 | |
| 103 | ip | 13 | 13 | 50% | 30% | >1 | ns | C57BL/6 | Asc−/− | Coutermarsh-Ott et al. 2016 | ||
| 103 | ip | 13 | ∞ | 50% | 100% | <2 | ns | C57BL/6 | Casp11−/− | Coutermarsh-Ott et al. 2016 | ||
| Trypanosoma cruzi trypomastigotes | 103 | ip | 28 | 22 | 90% | 0% | >2 | ns | C57BL/6 | Asc−/− | Silva et al. 2013 | |
| 103 | ip | 28 | 28 | 90% | 17% | >2 | ns | C57BL/6 | Casp1/11−/− | Silva et al. 2013 | ||
| 103 | ip | 28 | 28 | 90% | 67% | >1 | ns | C57BL/6 | Nlrp3−/− | Silva et al. 2013 | ||
| 103 | sc | 22 | 28 | 85% | 92% | 1 | ns | C57BL/6 | Casp1/11−/− (Nlrp3−/−) | Gonçalves et al. 2013 | ||
| 103 | ip | ∞ | 26 | 100% | 50% | >2 | ns | C57BL/6 | Casp1/11−/− | Paroli et al. 2018 | ||
| 103 | ip | ∞ | ∞ | 100% | 100% | 1 | ns | C57BL/6 | Nlrp3−/− | Paroli et al. 2018 | ||
| BACTERIA versus NECROPTOSIS | ||||||||||||
| Salmonella enterica sv. Typhimurium | 102 | iv | 11 | 10 | 0% | 0% | 1 | ns | C57BL/6 | Ripk3−/− | Robinson et al. 2012 | |
| 108 | po | 10 | 7 | 0% | 0% | 1 | ns | C57BL/6 | Mlkl−/− | Yu et al. 2018 | ||
| Staphylococcus aureus | 108 | ro | 2 | 4 | 84% | 88% | 1 | ns | C57BL/6 | Ripk3−/− | Kitur et al. 2016 | |
| 108 | ro | 2 | 3 | 84% | 44% | >1 | ns | C57BL/6 | Mlkl−/− | Kitur et al. 2016 | ||
| Yersinia pestis | pEcLpxL | 5 × 102 | sc | ∞ | 11 | 100% | 83% | >1 | ns | C57BL/6 | Ripk3−/− (BMT) | Weng et al. 2014 |
| 2 × 105 | fp | 5 | 7 | 36% | 70% | <1 | ns | C57BL/6 | Ripk1D138N/D138N | Arifuzzaman et al. 2018 | ||
| Yersinia pseudotuberculosis | 9 × 107 | po | >6 | >6 | 100% | 100% | 1 | ns | C57BL/6 | Ripk3−/− (BMT) | Philip et al. 2014 | |
| VIRUSES versus NECROPTOSIS | ||||||||||||
| Influenza A | 13 HAU | in | 11 | 13 | 78% | 62% | 1 | ns | C57BL/6 | Ripk3−/− | Rodrigue-Gervais et al. 2014 | |
| 4 × 103 | in | 11 | 11 | 71% | 38% | >1 | ns | C57BL/6 | Ripk3−/− | Nogusa et al. 2016 | ||
| 4 × 103 | in | 10 | 11 | 74% | 81% | 1 | ns | C57BL/6 | Mlkl−/− | Nogusa et al. 2016 | ||
| 103 | in | 8.5 | ∞ | 67% | 100% | <1 | ns | C57BL/6 | Zbp1−/− | Kuriakose et al. 2016 | ||
| 103 | in | ∞ | 10 | 100% | 22% | >2 | L | C57BL/6 | Zbp1−/− | Thapa et al. 2016 | ||
| Herpes simplex virus-1 | 2 × 107 | ip | ∞ | 6 | 100% | 58% | >2 | ns | ns | Ripk3−/− | Wang et al. 2014 | |
| 107 | iv | ∞ | 5 | 100% | 50% | >2 | ns | ns | Ripk3−/− | Huang et al. 2015 | ||
| Herpes simplex virus-1 | ΔICP6 | 107 | iv | 3.5 | 3.5 | 38% | 56% | 1 | ns | ns | Ripk3−/− | Huang et al. 2015 |
| Vaccinia | 2 × 106 | ip | 9 | 7 | 86% | 0% | >2 | ns | C57BL/6 | Ripk3−/− | Cho et al. 2009 | |
| West Nile virus | 102 | sc | 11 | 11 | 56% | 0% | >2 | ns | C57BL/6 | Ripk3−/− | Daniels et al. 2017 | |
| 102 | sc | 11 | 12 | 40% | 46% | 1 | ns | C57BL/6 | Mlkl−/− | Daniels et al. 2017 | ||
| 102 | sc | 11 | 11 | 56% | 0% | >2 | ns | C57BL/6 | Ripk1KD/KD | Daniels et al. 2017 | ||
| 101 | ic | 9 | 7 | 0% | 0% | 0 | ns | C57BL/6 | Ripk3−/− | Daniels et al. 2017 | ||
| Zika virus | 103 | ic | 7 | 6.5 | 73% | 0% | >2 | ns | C57BL/6N | Ripk3−/− | Daniels et al. 2019 | |
| 103 | ic | 7.5 | 7 | 60% | 0% | >2 | ns | C57BL/6J | Ripk1KD/KD | Daniels et al. 2019 | ||
| 103 | ic | 7.5 | 7.5 | 60% | 70% | 1 | ns | C57BL/6J | Mlkl−/− | Daniels et al. 2019 | ||
| 103 | ic | 6.5 | 7 | 82% | 7% | >2 | L | C57BL/6 | Ripk3fl/fl CamKIIα-Cre+ | Daniels et al. 2019 | ||
| 103 | ic | 7.5 | 8 | 80% | 20% | >2 | ns | C57BL/6J | Zbp1−/− | Daniels et al. 2019 | ||
The investigators intend to continue to update this table and thus would appreciate notification of omissions or errors. This table includes experiments performed with mice defective for necroptosis genes, but does not include apoptosis knockout (KO) mice such as Casp8−/− mice.
BMT, KO mice instead are wild-type (WT) mice that received knockout bone marrow; iv, intravenous; sc, subcutaneous; scr, scratch to flank skin; po, per oral; po+s, per oral pretreated with streptomycin; in, intranasal; ro, retroorbital intravenous; it, intratracheal; ic, intracranial; fp, footpad; ts, tongue scratch; L, littermate; CO; cohoused; N; not littermate and not cohoused; ns, not stated.
aEstimated change in lethality between WT and KO mice. A difference in survival percentages of (1) 0% change was estimated to be onefold increase in infectious dose (or changes that were not statistically significant); (2) 1%–49% was estimated to be greater than onefold increase in the infectious dose; (3) 50%–99% was estimated to be greater than twofold; and (4) >100% was estimated to be greater than fivefold. Where multiple doses were examined, the high dose was divided by the low dose and then multiplied by the aforementioned estimator. As an example, in Ceballos-Olvera et al. (2011), WT mice had 100% lethality at 200 CFU but many survived at 100 CFU and Casp1−/−Casp11−/− mice succumbed to 25 CFU; 200/25 = 8, and so the change in lethal dose is listed as greater than eightfold since Casp1−/−Casp11−/− mice may succumb to even lower doses. Note that many infectious models have only examined one dose, so the change in lethal dose may turn out to be much larger than listed in this table once additional doses are tested. A final caveat is the all these manuscripts use mice on the C57BL/6 background, which carries MX1 and Nramp1 mutations that cause susceptibility to viral and bacterial infection, respectively, it is difficult to detect increases in the lethal dose when the lethal dose is already very low in WT C57BL/6 mice.
Caspase-11 Defending against Burkholderia thailandensis
Caspase-11 is the most direct pathway to programmed cell death, in that it requires just two proteins: caspase-11 and gasdermin D. Caspase-11 itself serves both as a sensor and as a catalytic protease. The CARD domain of caspase-11 directly binds to LPS in the cytosol, which triggers CARD-oligomerization and activation of the protease (Hagar et al. 2013; Kayagaki et al. 2013; Shi et al. 2014). Active caspase-11 then cleaves gasdermin D and induces pyroptosis (Kayagaki et al. 2015; Shi et al. 2015).
By sensing cytosolic LPS, caspase-11 discriminates cytosol-invasive bacteria from vacuolar or extracellular bacteria (Aachoui et al. 2013). Caspase-11 efficiently detects B. thailandensis, a cytosol invasive bacterium, in vivo and is incredibly effective at clearing the bacterium. This defense pathway is so effective that a systemic challenge of 20,000,000 CFUs are cleared within 1 day. In contrast, Casp11−/− mice succumb following challenge with as few as 100 CFUs (Aachoui et al. 2015). This is the strongest in vivo effect for caspase-11 against any infectious challenge (Maltez and Miao 2016). We estimate a 1,000,000-fold change in the effective LD100 (Table 1). It is remarkable that the strongest phenotypes for caspase-1 and caspase-11 in defense against infection arise from two ubiquitous environmental soil microbes, which only infect humans under extraordinary settings of host compromise such as patients with chronic granulomatous disease or after near drowning (Macher et al. 1982; Glass et al. 2006). This suggests that caspase-1/11 provide near permanent evolutionary victories over ubiquitous environmental pathogens, which are running the Red Queen's race against other hosts that lack caspase-1/11. (This hypothesis is described in more depth in Maltez and Miao 2016 and Box 1 of Jorgensen et al. 2017.)
Some pathogens alter their LPS structure to evade detection by caspase-11 (Hagar et al. 2013; Kayagaki et al. 2013; Paciello et al. 2013; Yang et al. 2019). Additionally, some bacteria avoid recognition by caspase-11 by remaining within the vacuolar space (Aachoui et al. 2013). There are several examples of caspase-11 responding to pathogens that are typically considered to be vacuolar. In these cases, there must be either vacuolar leakage or rupture to introduce LPS into the cytosol. In such cases, caspase-11 can reduce vacuolar pathogen burdens (Lacey et al. 2018). Nevertheless, it is likely that caspase-11 evolved to combat cytosol-invasive pathogens.
LPS is an incredibly abundant ligand. Thus, LPS sensing risks detection of LPS that enters the cytosol aberrantly, as in endotoxic shock models (Hagar et al. 2013; Kayagaki et al. 2013). This risk is partially ameliorated by tight regulation of caspase-11; caspase-11 cannot be activated in the absence of interferon (IFN)-γ or type I IFN signaling (Broz et al. 2012; Rathinam et al. 2012; Aachoui et al. 2015).
The Evolutionary Race between Yersinia and the Pyrin Inflammasome
One of the most notable examples of the never-ending race between host and pathogen are pathogenic Yersinia spp. The causative agent of plague (Yersinia pestis) and the enteric pathogens (Yersinia pseudotuberculosis and Yersinia enterocolitica) all encode a T3SS enabling them to infect humans and other mammals. These T3SS inject effectors called Yersinia outer proteins (Yops) into the cytosol of host cells (Bliska et al. 2013). Given that Yersinia expresses a T3SS, it should be detected by the NLRC4 inflammasome. Additionally, the NLRP3 inflammasome may detect the YopB and YopD translocon proteins. However, YopK reportedly restricts both these detection events although through an unclear mechanism (Brodsky et al. 2010; Zwack et al. 2015). A more explicit example of Yersinia species running the Red Queen's race has recently been shown in Yersinia’s ability to evade the pyrin inflammasome.
During bacterial infection, neutrophils are the first immune cells recruited to the site of infection. Neutrophils phagocytose bacteria by activating Rho GTPases, such as RhoA, Rac1, and CDC42, which polymerize actin driving phagocytosis (Mao and Finnemann 2015). The primary virulence strategy of Yersinia spp. is to inhibit phagocytosis and replicate extracellularly (Ke et al. 2013). Once neutrophils arrive, they are the predominant cell type targeted by the Yersinia T3SS (Pechous et al. 2013). Yersinia prevents actin polymerization in part by using YopE and YopT. YopE facilitates GTPase hydrolysis, keeping RhoA in the inactive GDP-bound state, whereas YopT is a cysteine protease that cleaves the carboxyl terminus of Rho GTPases, releasing them from the membrane (Black and Bliska 2000; Shao et al. 2003). Thus, both YopE and YopT disable Rho GTPase activity to prevent phagocytosis (Grosdent et al. 2002). At this point, the pathogen is winning the evolutionary race (Fig. 2).
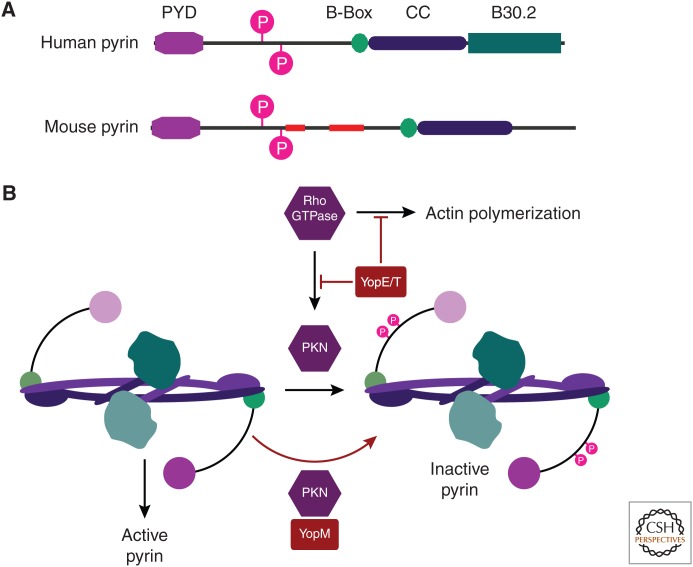
Figure 2.
The pyrin inflammasome guards Rho GTPases. Pyrin was the first gene described to encode the pyrin domain (PYD) that is also found in many other inflammasomes. PYD signals through the PYD-CARD adaptor protein ASC, and thereby to caspase-1. In addition to the PYD, pyrin is a member of the tripartite motif (TRIM) family, and thus contains a B-Box, a coiled coil (CC), and a B30.2 domain (Kawai and Akira 2011; Weinert et al. 2015). (A) Domain structure of human and mouse pyrin. Slim red boxes indicate regions that are present in the human but absent in the mouse, or vice versa. (B) Rho GTPases are activated in response to signals for immune cell motility and/or phagocytosis. As part of their effector functions, Rho GTPases activate actin polymerization as well as effector protein kinases (including PKN1 and PKN2). Pyrin (shown in dimeric form) guards these Rho GTPase signaling pathways by mechanisms that are only partially understood. Successful Rho GTPase signaling will activate PKN1/PKN2, which phosphorylate and thereby inactivate pyrin. Thus, pyrin acts as a checkpoint to verify Rho GTPase function. If phosphorylation fails, pyrin becomes an active inflammasome. Yersinia encodes YopE and YopT that prevent phagocytosis by attacking Rho GTPases, but at the cost of preventing pyrin phosphorylation. Another Yersinia effector, YopM, reattaches PKN1/PKN2 to pyrin, driving pyrin phosphorylation. Thus, Yersinia successfully blocks phagocytosis with YopE/YopT while simultaneously defusing the pyrin guard with YopM.
In an effort to combat antiphagocytic effectors, the host evolved a functional guard called pyrin (encoded by Mefv). Pyrin essentially monitors for normal biochemical function of the Rho GTPases; when the Rho GTPases are perturbed, pyrin activates caspase-1 (Xu et al. 2014). This guard function is accomplished not by direct interaction between pyrin and Rho GTPases, but via monitoring Rho effector protein kinases N1 and N2 (PKN1 and PKN2; also called PRK1/PRK2). Although the exact biochemical mechanism remains unclear, current literature suggests that RhoA activates PKN1/PKN2, activating their normal effector functions (Thumkeo et al. 2013) and also permitting PKN1/PKN2 to phosphorylate pyrin (Gao et al. 2016; Park et al. 2016). Thus, when RhoA is present and is capable of inducing phagocytosis, PKN1/PKN2 phosphorylate pyrin and repress inflammasome assembly. However, if RhoA is degraded or enzymatically modified by YopT and/or YopE, PKN1/PKN2 no longer phosphorylate pyrin. By an unclear mechanism, this lack of phosphorylation results in pyrin activation (Chung et al. 2016). Exactly how inflammasome activation at this point helps the host remains to be elucidated. Induction of pyroptosis should delete phagocytes that have been debilitated by the Yops. Additionally, the capase-1-driven release of IL-1β should recruit new neutrophils to the site of infection. In total, the pyrin inflammasome prevents the pathogen from creating a favorable environment for extracellular bacterial replication. Pyrin is also important for detection of a variety of pathogens that produce toxins or T3SS effectors that perturb Rho GTPases, indicating that pyrin is a general guard for actin cytoskeletal function (Xu et al. 2014; Aubert et al. 2016).
The evolutionary race was not finished; Yersinia spp. evolved an additional virulence factor in an effort to overthrow the host. Yersinia developed the effector YopM, which recruits PKN1/PKN2 to pyrin, driving pyrin phosphorylation even in the absence of RhoA activity (Mcdonald et al. 2003; Chung et al. 2016). Thus, WT mice are susceptible to WT Yersinia infection, but resistant to yopM mutants. Yersinia yopM mutants, on the other hand, are virulent in mice deficient in pyrin or caspase-1 (LaRock and Cookson 2012; Chung et al. 2016) (also see Table 1).
The basic function of YopM, to inhibit pyrin, is conserved among Yersinia spp. Nevertheless, the Red Queen's race appears to be ongoing as YopM is polymorphic between Yersinia isolates and species (Chung et al. 2016). Similarly, there are several key differences between mouse and human pyrin, suggesting some YopM variants may work effectively against the pyrin of one host, but fail against others (Fig. 2). This extreme evolutionary pressure may push the host into a precarious position. Autosomal recessive mutations in pyrin result in the most common autoinflammatory disease worldwide, familial Mediterranean fever (FMF) (Özen 2018). These mutant pyrin proteins are not phosphorylated to the same degree by PKN1/PKN2 (Park et al. 2016). Thus, pyrin is more easily activated, resulting in high IL-1β levels and reoccurring inflammation and fever, but only in the homozygous state (Park et al. 2016). FMF is highly prevalent in parts of the Mediterranean region, thus populations that are native to the area may have evolved this gain of function if it confers a selective advantage in the heterozygous state to a pathogen. Given the impact of plague throughout history, it is tempting to speculate that Y. pestis provided the selective pressure to induce expansion of pyrin mutations.
NLRP1—One NLR to Guard Them All
Although it was the first inflammasome identified (Martinon et al. 2002), how the NLRP1 inflammasome detects pathogens has only recently been elucidated. NLRP1 in humans and mice contains a unique domain structure among NLRs (Fig. 3). Like NLRC4, NLRP1 has a CARD that directly activates caspase-1, yet it is located on the carboxyl terminus rather than the amino terminus. NLRP1 is also unique in that it includes a carboxy-terminal function-to-find domain (FIIND) directly upstream of its CARD. The FIIND undergoes constitutive autoproteolysis, but remains noncovalently associated with the rest of NLRP1. This noncovalent association is required for activation of NLRP1 (D'Osualdo et al. 2011; Finger et al. 2012; Frew et al. 2012). Furthermore, the minimal active component of NLRP1 is actually the cleaved FIIND-CARD fragment alone; the NOD and LRR are surprisingly dispensable. Activation of this cleaved-FIIND-CARD inflammasome occurs only when the other domains of NLRP1 are degraded (Xu et al. 2018; Chui et al. 2019; Sandstrom et al. 2019). This amino-terminal degradation is thought to be induced, at least in part, by pathogen-mediated mechanisms (Frew et al. 2012).
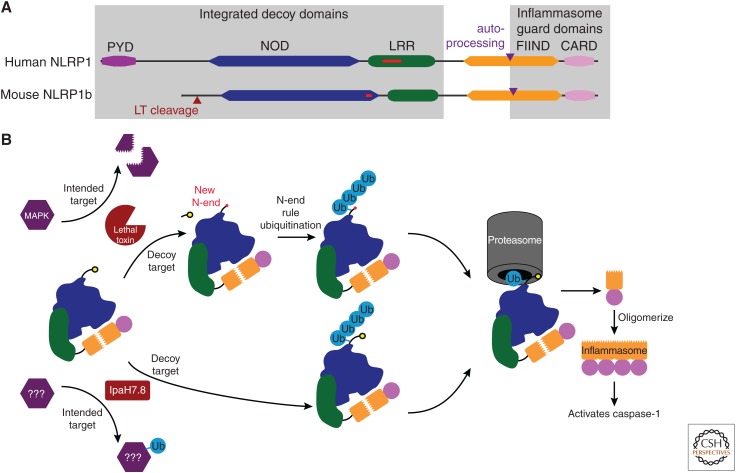
Figure 3.
The NLRP1 inflammasome has integrated decoy domains to detect virulence factor attack. (A) Diagram of the human NLRP1 protein, and one of the NLRP1 proteins in mice (NLRP1b). Domains are indicated by colored boxes. Slim red boxes indicate regions that are present in the human, but absent in the mouse protein, or vice versa. Red triangle indicates the lethal toxin cleavage site in mouse NLRP1b. Purple triangle indicates the autoprocessing site within the FIIND domain. (B) Schematic of murine NLRP1b pathways. In one mode of activation (top), anthrax lethal toxin intends to cleave MAP kinase kinases; however, a decoy sequence in NLRP1b is also cleaved by lethal toxin. This exposes a new amino terminus (New N-end) in NLRP1b, which is detected by the N-end rule ubiquitinases that attach a ubiquitin to a nearby lysine residue. In a second mode (bottom) a bacterial effector such as IpaH7.8 intends to attack and ubiquitinate a different cellular protein, but also inadvertently ubiquitinates the decoy domains of NLRP1b. Ubiquitinated NLRP1b is then degraded by the proteasome, but when the precleaved FIIND domain approaches the proteasome, the carboxy-terminal FIIND fragment and attached CARD domain dissociate and are therefore not degraded. This dissociation results from the FIIND-CARD domains not being covalently attached to the rest of the protein. The liberated FIIND-CARD then oligomerizes to form an inflammasome, clustering the CARD domains that activate caspase-1. The FIIND-CARD inflammasome is different from typical inflammasomes in which the NOD domain drives oligomerization and clustering of an amino-terminal CARD domain. (From Lacey and Miao 2019; adapted, with permission, from the authors.)
NLRP1 was first recognized as a sensor of anthrax lethal toxin (LT), a metalloprotease produced by Bacillus anthracis (Boyden and Dietrich 2006). LT cleaves MAP kinase kinases (MAPKKs) to prevent innate immune signaling (Turk 2007). The host response to this in certain mouse strains seems to be to integrate a decoy domain into NLRP1b, such that now LT cleaves NLRP1b in addition to its primary target. This cleavage event occurs at the amino terminus and activates NLRP1 (Levinsohn et al. 2012). Proteolysis of amino-terminal residues induces a process known as the N-end rule (Lucas and Ciulli 2017), wherein amino acid residues at the new amino terminus are modified by cellular E3 ubiquitin ligases and targeted for degradation by the proteasome. Thus, when LT cleaves NLRP1b, the amino-terminal portion of NLRP1 is ubiquitinated and degraded by the proteasome. The cleaved-FIIND-CARD disassociates from the rest of NLRP1 during this process because of its noncovalent association (Squires et al. 2007; Wickliffe et al. 2008; Xu et al. 2018; Chui et al. 2019; Sandstrom et al. 2019), and is then free to oligomerize and form potent inflammasomes (Xu et al. 2018; Chui et al. 2019; Sandstrom et al. 2019).
FIIND or similar domains are found in other innate immune genes, including CARD8 (present in humans but absent in mice) and PIDD, suggesting that activation by proteolysis may be useful for regulating other pathways (Tinel et al. 2007; D'Osualdo et al. 2011). Given this mechanism of NLRP1 activation, there needs to be a way to safely degrade old NLRP1, CARD8, and PIDD during cell homeostasis without activating caspase-1. In this regard, inhibitors of serine dipeptidases, Dpp, induce activation of NLRP1 and/or CARD8 without proteolytic cleavage, but the proteasome is still required (Okondo et al. 2017; 2018; Johnson et al. 2018; Zhong et al. 2018). Thus, Dpp may participate in controlled NLRP1 and/or CARD8 degradation during normal homeostasis, thereby reducing the risk of NLRP1 autoactivation.
Nonproteolytic pathogen-mediated mechanisms may also target NLRP1 for degradation in the proteasome. Shigella flexneri has a T3SS that secretes the E3 ubiquitin ligase, IpaH7.8 (Rohde et al. 2007; Singer et al. 2008; Zhu et al. 2008), which is detected by the NLRP1b inflammasome (Sandstrom et al. 2019). NLRP1 is ubiquitinated by IpaH7.8, which targets it for proteasomal degradation, and consequently releases the FIIND-CARD fragment that activates caspase-1. Ubiquitination and degradation of inhibitory proteins is also used in normal signaling pathways; for example, the NEMO/IKKα/IKKβ complex ubiquitinates IκB to drive its degradation, releasing nuclear factor (NF)-κB to translocate to the nucleus. In contrast, we speculate that NLRP1 acts as a decoy to detect ubiquitin attack on other NLRs. Thus, the NOD and LRR domains of NLRP1 would be defined as integrated decoy domains (Fig. 1). This idea would be supported if future research discovers that IpaH7.8 actually evolved to target a different NLR for degradation. The host may use these accessory integrated decoy domains to trick pathogens into targeting NLRP1 for degradation, creating a trip-wire that activates the inflammasome.
The number of NLRP1 genes and the domain structure of NLRP1 varies not only between species, but also within a species (Boyden and Dietrich 2006; D'Osualdo et al. 2011; Lilue et al. 2018). Unlike mouse macrophages, human macrophages exposed to IpaH7.8 do not activate caspase-1 (Muehlbauer et al. 2007). Thus, Shigella is winning the Red Queen's race against humans and losing against mice (Sharma et al. 2017). The versatility of NLRP1 is illustrated by the integration of an amino-terminal PYD in human NLRP1 (Moayeri et al. 2012), which perhaps evolved as an extra integrated decoy domain to lure virulence factors that attack PYDs. Pathogens that activate human NLRP1 have not yet been discovered, perhaps because NLRP1 eradicates them before disease development. If NLRs are the subject of attack by virulence factors, then NLRP1 could be considered the one NLR to guard them all.
Guarding Transcriptional Signaling with Apoptosis or Necroptosis
TNF is an important proinflammatory cytokine that is often targeted by bacterial virulence factors. Thus, it makes sense that TNF signaling is carefully guarded. TNF receptor signaling can have one of three outcomes: gene transcription, apoptosis, or necroptosis. The primary goal of TNF signaling is likely an NF-κB transcriptional response. When this transcriptional response is inhibited, guard functions detect the defective signaling pathway and, in response, trigger either apoptosis, pyroptosis, or necroptosis. These guard functions are likely the result of the ongoing evolutionary battle between host and pathogen, dating back to the dawn of primitive multicellular organisms (Quistad et al. 2014).
In the Red Queen's race, a hypothetical pathogen would attempt to inhibit the TNF transcriptional response with a virulence factor to dampen the host immune response. One key signaling point within the TNF signaling pathway is RIPK1, because it gets modified with polyubiquitin chains that recruit the TAK1 and IKK complexes needed to activate NF-κB (Dondelinger et al. 2016a). To counter this attack, the host has evolved guards for TNF transcriptional signaling mediated by the RIPK1 axis. Once the TNF guards detect that a virulence factor has intercepted the TNF to NF-κB pathway, the “interpretation” is that the cell has been irrevocably compromised. Therefore, the conservative response, erring on the side of assuming the worst, is to kill the cell.
The first pathway that evolved to guard RIPK1 was a branch to apoptosis (Lamkanfi et al. 2002; Dondelinger et al. 2016b). The DD of RIPK1 normally recruits it to the DD of TNF receptor 1 or the DD in the adaptor TRADD (Dondelinger et al. 2016a). When RIPK1 modification is perturbed, its DD becomes exposed and is detected by the guard protein Fas-associated death domain (FADD) protein (the caspase-8 adaptor composed of a DD and a DED). FADD binds to RIPK1 via DD–DD interactions, and then recruits caspase-8 to initiate apoptosis. Thus, FADD and caspase-8 are guards for RIPK1 (Fig. 1). This is mimicked in vitro through the addition of IAP antagonists, inhibitors of TAK1, or inhibitors of IKK, which inhibit the TNF transcriptional response and induce apoptosis (Dondelinger et al. 2016a). Although enacting apoptosis and killing a cell is a dramatic response, it allows the host to remove cells that have been irrevocably compromised. Thus, the host prevents the pathogen from hijacking cells and the pathogen is denied the ability to create its preferred environment.
Not only does caspase-8 guard RIPK1, but RIPK1 can conversely guard caspase-8. Many pathogens have evolved virulence factors to inhibit TNF transcriptional signaling, while simultaneously inhibiting caspase-8-mediated apoptosis (Kaiser et al. 2013). In response to this double attack, the host appears to have evolved another guard pathway attached to RIPK1, monitoring for abnormalities in caspase-8. Now, instead of RIPK1 triggering apoptosis, it instead induces a completely different form of programmed cell death termed necroptosis (Dondelinger et al. 2016a). Evolution selected for the addition of a RHIM domain and a kinase domain to RIPK1. The RIPK1 kinase domain autophosphorylates RIPK1 to promote RHIM–RHIM interactions that recruit RIPK3. This induces RIPK3 oligomerization and autophosphorylation. Then RIPK3 phosphorylates the pseudokinase MLKL, leading to cell lysis. In this pathway, the RHIM and kinase domains of RIPK1 can be considered integrated guard domains (Fig. 1) that signal to the guard protein RIPK3.
In summary, for a pathogen to prevent TNF (or the similarly guarded TLR3 and TLR4)-induced activation of NF-κB while also maintaining cell viability, the pathogen must run through multiple steps of evolution. The pathogen must simultaneously achieve its primary goal of blocking NF-κB, secondarily it must also block apoptosis, and finally necroptosis. This network of pathways and guards probably makes it incredibly difficult for pathogens to readily add NF-κB-inhibiting virulence factors to their repertoire.
EPEC TRIPLE ATTACKS TRANSCRIPTION, APOPTOSIS, AND NECROPTOSIS
Enteropathogenic E. coli (EPEC) is a human-specific pathogen that causes diarrhea and lives in close association with host cells (Fig. 4). It uses the locus of enterocyte effacement (LEE) T3SS to reprogram intestinal epithelial cells (IECs), permitting extracellular adherence of EPEC in the intestinal lumen. The translocated effectors cause the IEC microvilli to efface, and then induce the formation of a dense actin network, creating a pedestal on which EPEC closely adheres. The bacterium likely evolved this strategy to gain first access to oxygen and nutrients that diffuse across the IEC, gaining a replication advantage over luminal commensals (Lopez et al. 2016). EPEC must reprogram the IEC, while also preventing the IEC from noticing it is compromised. Despite the fact that the EPEC rod protein is detected by the NAIP/NLRC4 inflammasome, NLRC4 is inefficient at detecting the live bacteria (Miao et al. 2010b). Detection by NLRC4 in IECs would otherwise trigger immediate extrusion (Rauch et al. 2017), which would be devastating to the virulence strategy of EPEC. We therefore speculate that EPEC has an undiscovered strategy to evade NLRC4.
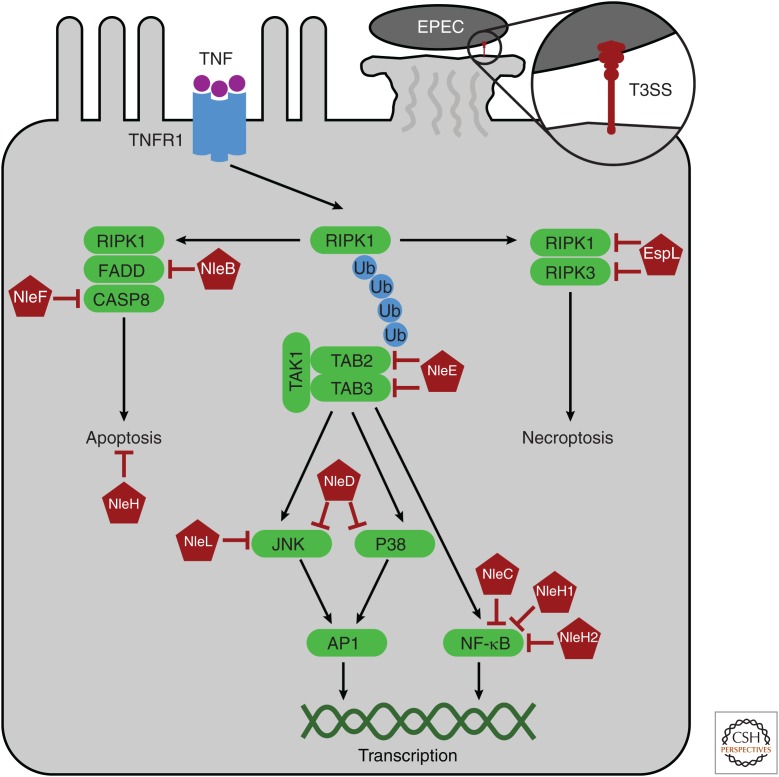
Figure 4.
Enteropathogenic Escherichia coli (EPEC) virulence factors target pathways for transcription, apoptosis, and necroptosis. Tumor necrosis factor (TNF) receptor signaling can trigger one of three responses. The cell will first try to initiate transcriptional signaling pathways. RIPK1 acts as a scaffold for polyubiquitin chains, on which assemble TAB2 and TAB3 in a complex with TAK1. Downstream from TAK1, JNK and p38 signal to the transcription factor AP1, and other complexes signal to nuclear factor (NF)-κB. Transcription can be blocked by EPEC effectors: NleE inhibits TAB2 and TAB3, NleD obstructs JNK and p38, and NleC and NleH1/2 block NF-κB. When transcription pathways are hindered, guards of RIPK1 initiate apoptosis. RIPK1 death domain (DD) becomes exposed allowing homotypic interactions with the DD of the adaptor FADD, which in turn recruits caspase-8 to initiate apoptosis. EPEC has additional virulence strategies to inhibit the apoptotic guard pathway: NleB and NleF attack FADD and caspase-8, respectively. Finally, the host cell has a third guard pathway in which RIPK1 uses its integrated guard RHIM and kinase domains to activate the guard, RIPK3. RIPK3 signaling then induces necroptosis of the host cell. Similar to transcription and apoptosis, EPEC has a virulence factor, EspL, to inhibit signaling to necroptosis.
There is mounting evidence that EPEC inhibits the transcriptional responses of the IEC to which it is attached, effectively shutting down the TNF signaling pathway (Pearson and Hartland 2014). EPEC attacks TAB2 and TAB3, which are adaptors for TAK1 downstream from RIPK1. EPEC accomplishes this by injecting the T3SS effector NleE, which methylates cysteines in the zinc finger domains of TAB2 and TAB3, thus inhibiting NF-κB responses (Zhang et al. 2011). EPEC also has two metalloproteases, NleC and NleD, which degrade key proteins in the transcriptional response. NleC degrades NF-κB by cleaving its p65 (RelA) subunit (Yen et al. 2010; Baruch et al. 2011; Mühlen et al. 2011; Pearson et al. 2011). Meanwhile, NleD degrades JNK and p38, which are activators of the AP1 transcription factor (Baruch et al. 2011). Another effector, NleL, also attacks JNK, but by ubiquitinating it, hence blocking JNK phosphorylation and activation (Sheng et al. 2017). Finally, NleH1 and NleH2 inhibit NF-κB-driven gene expression in part by NleH1, preventing IKKβ phosphorylation of RPS3, a specifier subunit of certain NF-κB complexes (Gao et al. 2009; Royan et al. 2010; Wan et al. 2011). The fact that EPEC translocates a plethora of T3SS effectors that redundantly attack NF-κB and AP1 indicate that preventing transcriptional responses is very important to its virulence strategy. However, attacks on these transcriptional signaling pathways theoretically should be detected by their guards.
EPEC must inhibit the caspase-8 apoptosis guard pathway to maintain its adherent niche on the IEC. The T3SS effector NleB attacks DD-containing proteins by adding an N-acetylglucosamine (called GlcNAcylation), with a preference for attacking the caspase-8 adaptor FADD (Li et al. 2013; Pearson et al. 2013; Scott et al. 2017). Thus, NleB prevents guard signaling from RIPK1 to caspase-8 and prevents apoptosis. Another effector, NleF, also attacks caspase-8 (as well as some other caspases) by direct binding to inhibit its catalytic activity (Blasche et al. 2013). In addition to attack on caspase-8 apoptotic guard function, EPEC also attacks other aspects of apoptosis—NleH binds Bax inhibitor-1 (BI-1) to prevent cell-intrinsic initiation of apoptosis (Hemrajani et al. 2010).
Lastly, EPEC also attacks the necroptotic guard pathway. EspL is a cysteine protease that cleaves the RHIM domains in RIPK1, RIPK3, and other RHIM-containing proteins, thereby preventing oligomerization and activation of RIPK3 (Pearson et al. 2017). This should prevent the IEC necroptosis guard pathway from activating in response to simultaneous attacks upon transcriptional signaling and apoptosis. The fact that EPEC delivers multiple effectors to prevent apoptosis and necroptosis illustrates that maintaining the viability of the IEC is important to the virulence strategy of EPEC, allowing it to replicate in its adherent niche.
The compendium of EPEC effectors that attack NF-κB signaling and simultaneously attack the two guard pathways to apoptosis and necroptosis are an excellent illustration of the Red Queen's race between pathogen and host. We expect that other bacterial pathogens that attack NF-κB signaling will similarly need to simultaneously block apoptosis and necroptosis. This should be particularly important for pathogens such as EPEC that replicate in intimate contact with the host cell it has reprogramed. Similarly, intracellular pathogens like Salmonella replicate inside a single host cell, and thus must also keep that host cell alive. S. Typhimurium attacks NF-κB signaling, which should trigger the apoptotic and necroptotic guard pathways. However, S. Typhimurium encodes SseK proteins that are similar to the EPEC apoptosis inhibitor NleB. Although homologs of the EPEC EspL necroptosis inhibitor are not present in the commonly used strains of S. Typhimurium, we predict that S. Typhimurium inhibits necroptosis by using undiscovered effectors. Yersinia species also attack transcriptional responses. For example, YopJ attacks TAK1, but at the cost of also triggering the apoptotic guard functions (Paquette et al. 2012). In contrast to EPEC and S. Typhimurium, the host cell does not need to remain viable for Yersinia to replicate. Thus, it remains to be determined whether Yersinia actually benefits from the apoptotic guard pathway triggered by its inhibition of the transcriptional response. Given that all pathogens are running the Red Queen's race against the host, it seems likely that many other pathogens will have virulence strategies that have equal complexity to those illustrated by the EPEC T3SS effectors.
CONCLUDING REMARKS
Evolution is limited by organism replication rates, thus it is easy to assume multicellular organisms are at a disadvantage as many have life span that are significantly longer than bacteria, which allows them less time to mutate and evolve between generations. The solution to being faced with a pathogen that step-for-step keeps up with the continuing evolution of the innate immune system was to generate an arm of the immune system that can evolve at a rate faster than bacterial evolution. The adaptive immune system accomplishes this feat—B cells and T cells evolve new antibody and T-cell receptors within one week. Ultimately, almost all infections in which pathogens are running the Red Queen's race against innate immunity are cleared by the adaptive immune system.
Footnotes
Editors: Kim Newton, James M. Murphy, and Edward A. Miao
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
- Aachoui Y, Leaf IA, Hagar JA, Fontana MF, Campos CG, Zak DE, Tan MH, Cotter PA, Vance RE, Aderem A, et al. 2013. Caspase-11 protects against bacteria that escape the vacuole. Science 339: 975–978. 10.1126/science.1230751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aachoui Y, Kajiwara Y, Leaf IA, Mao D, Ting JPY, Coers J, Aderem A, Buxbaum JD, Miao EA. 2015. Canonical inflammasomes drive IFN-γ to prime caspase-11 in defense against a cytosol-invasive bacterium. Cell Host Microbe 18: 320–332. 10.1016/j.chom.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiger B, Dahlberg S, Sandgren A, Wartha F, Beiter K, Katsuragi H, Akira S, Normark S, Henriques-Normark B. 2007. Toll-like receptor 9 acts at an early stage in host defence against pneumococcal infection. Cell Microbiol 9: 633–644. 10.1111/j.1462-5822.2006.00814.x [DOI] [PubMed] [Google Scholar]
- Allen IC. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30: 556–565. 10.1016/j.immuni.2009.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arifuzzaman M, Ang WXG, Choi HW, Nilles ML, St John AL, Abraham SN. 2018. Necroptosis of infiltrated macrophages drives Yersinia pestis dispersal within buboes. JCI Insight 3: 122188 10.1172/jci.insight.122188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert DF, Xu H, Yang J, Shi X, Gao W, Li L, Bisaro F, Chen S, Valvano MA, Shao F. 2016. A Burkholderia type VI effector deamidates Rho GTPases to activate the pyrin inflammasome and trigger inflammation. Cell Host Microbe 19: 664–674. 10.1016/j.chom.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Baruch K, Gur-Arie L, Nadler C, Koby S, Yerushalmi G, Ben-Neriah Y, Yogev O, Shaulian E, Guttman C, Zarivach R, et al. 2011. Metalloprotease type III effectors that specifically cleave JNK and NF-κB. EMBO J 30: 221–231. 10.1038/emboj.2010.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista JH, da Silva Neto JF. 2017. Chromobacterium violaceum pathogenicity: updates and insights from genome sequencing of novel Chromobacterium species. Front Microbiol 8: 2213 10.3389/fmicb.2017.02213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DS, Bliska JB. 2000. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol Microbiol 37: 515–527. 10.1046/j.1365-2958.2000.02021.x [DOI] [PubMed] [Google Scholar]
- Blasche S, Mörtl M, Steuber H, Siszler G, Nisa S, Schwarz F, Lavrik I, Gronewold TMA, Maskos K, Donnenberg MS, et al. 2013. The E. coli effector protein NleF is a caspase inhibitor. PLoS ONE 8: e58937 10.1371/journal.pone.0058937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska JB, Wang X, Viboud GI, Brodsky IE. 2013. Modulation of innate immune responses by Yersinia type III secretion system translocators and effectors. Cell Microbiol 15: 1622–1631. 10.1111/cmi.12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ED, Dietrich WF. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet 38: 240–244. 10.1038/ng1724 [DOI] [PubMed] [Google Scholar]
- Breitbach K, Sun GW, Köhler J, Eske K, Wongprompitak P, Tan G, Liu Y, Gan YH, Steinmetz I. 2009. Caspase-1 mediates resistance in murine melioidosis. 77: 1589–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky IE, Palm NW, Sadanand S, Ryndak MB, Sutterwala FS, Flavell RA, Bliska JB, Medzhitov R. 2010. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe 7: 376–387. 10.1016/j.chom.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. 2012. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490: 288–291. 10.1038/nature11419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Batra S, Wakamatsu N, Pacher P, Jeyaseelan S. 2012. NLRC4 inflammasome-mediated production of IL-1β modulates mucosal immunity in the lung against Gram-negative bacterial infection. J Immunol 188: 5623–5635. 10.4049/jimmunol.1200195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos-Olvera I, Sahoo M, Miller MA, Del Barrio L, Re F. 2011. Inflammasome-dependent pyroptosis and IL-18 protect against Burkholderia pseudomallei lung infection while IL-1β is deleterious. PLoS Pathog 7: e1002452 10.1371/journal.ppat.1002452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. 2009. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137: 1112–1123. 10.1016/j.cell.2009.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui AJ, Okondo MC, Rao SD, Gai K, Griswold AR, Johnson DC, Ball DP, Taabazuing CY, Orth EL, Vittimberga BA, et al. 2019. N-terminal degradation activates the NLRP1B inflammasome. Science 364: 82–85. 10.1126/science.aau1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung LK, Park YH, Zheng Y, Brodsky IE, Hearing P, Kastner DL, Chae JJ, Bliska JB. 2016. The Yersinia virulence factor YopM hijacks host kinases to inhibit type III effector-triggered activation of the pyrin inflammasome. Cell Host Microbe 20: 296–306. 10.1016/j.chom.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Gupta R, Signorino G, Malara A, Cardile F, Biondo C, Midiri A, Galbo R, Trieu-Cuot P, Papasergi S, et al. 2012. Activation of the NLRP3 inflammasome by group B Streptococci. J Immunol 188: 1953–1960. 10.4049/jimmunol.1102543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutermarsh-Ott SL, Doran JT, Campbell C, Williams TM, Lindsay DS, Allen IC. 2016. Caspase-11 modulates inflammation and attenuates Toxoplasma gondii pathogenesis. Mediators Inflamm 2016: 9848263 10.1155/2016/9848263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels BP, Snyder AG, Olsen TM, Orozco S, Oguin TH III, Tait SWG, Martinez J, Gale M Jr, Loo YM, Oberst A. 2017. RIPK3 restricts viral pathogenesis via cell death-independent neuroinflammation. Cell 169: 301–313.e11. 10.1016/j.cell.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels BP, Kofman SB, Smith JR, Norris GT, Snyder AG, Kolb JP, Gao X, Locasale JW, Martinez J, Gale M Jr, et al. 2019. The nucleotide sensor ZBP1 and kinase RIPK3 induce the enzyme IRG1 to promote an antiviral metabolic state in neurons. Immunity 50: 64–76.e4. 10.1016/j.immuni.2018.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y, Darding M, Bertrand MJM, Walczak H. 2016a. Poly-ubiquitination in TNFR1-mediated necroptosis. Cell Mol Life Sci 73: 2165–2176. 10.1007/s00018-016-2191-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y, Hulpiau P, Saeys Y, Bertrand MJM, Vandenabeele P. 2016b. An evolutionary perspective on the necroptotic pathway. Trends Cell Biol 26: 721–732. 10.1016/j.tcb.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Dostert C, Guarda G, Romero JF, Menu P, Gross O, Tardivel A, Suva ML, Stehle JC, Kopf M, Stamenkovic I, et al. 2009. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PLoS ONE 4: e6510 10.1371/journal.pone.0006510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Osualdo A, Weichenberger CX, Wagner RN, Godzik A, Wooley J, Reed JC. 2011. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS ONE 6: e27396 10.1371/journal.pone.0027396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois H, Sorgeloos F, Sarvestani ST, Martens L, Saeys Y, Mackenzie JM, Lamkanfi M, van Loo G, Goodfellow I, Wullaert A. 2019. Nlrp3 inflammasome activation and Gasdermin D-driven pyroptosis are immunopathogenic upon gastrointestinal norovirus infection. PLoS Pathog 15: e1007709 10.1371/journal.ppat.1007709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy EB, Periasamy S, Hunt D, Drake JR, Harton JA. 2016. FcγR mediates TLR2- and Syk-dependent NLRP3 inflammasome activation by inactivated Francisella tularensis LVS immune complexes. J Leukoc Biol 100: 1335–1347. 10.1189/jlb.2A1215-555RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E, Mear JB, Faure K, Normand S, Couturier-Maillard A, Grandjean T, Balloy V, Ryffel B, Dessein R, Chignard M, et al. 2014. Pseudomonas aeruginosa type-3 secretion system dampens host defense by exploiting the NLRC4-coupled inflammasome. Am J Respir Crit Care Med 189: 799–811. 10.1164/rccm.201307-1358OC [DOI] [PubMed] [Google Scholar]
- Feriotti C, de Araújo EF, Loures FV, da Costa TA, Galdino NAL, Zamboni DS, Calich VLG. 2017. NOD-like receptor P3 inflammasome controls protective Th1/Th17 immunity against pulmonary paracoccidioidomycosis. Front Immunol 8: 786 10.3389/fimmu.2017.00786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol 11: 385–393. 10.1038/ni.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger JN, Lich JD, Dare LC, Cook MN, Brown KK, Duraiswami C, Bertin J, Bertin JJ, Gough PJ. 2012. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J Biol Chem 287: 25030–25037. 10.1074/jbc.M112.378323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, Núñez G. 2012. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol 13: 449–456. 10.1038/ni.2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frew BC, Joag VR, Mogridge J. 2012. Proteolytic processing of nlrp1b is required for inflammasome activity. PLoS Pathog 8: e1002659 10.1371/journal.ppat.1002659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan F, Mateo K, Callegari E, Wang D, Deng W, Puente J, Li F, Chaussee MS, Finlay BB, et al. 2009. Bacterial effector binding to ribosomal protein S3 subverts NF-κB function. PLoS Pathog 5: e1000708 10.1371/journal.ppat.1000708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Yang J, Liu W, Wang Y, Shao F. 2016. Site-specific phosphorylation and microtubule dynamics control pyrin inflammasome activation. Proc Natl Acad Sci 113: E4857–E4866. 10.1073/pnas.1601700113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass MB, Gee JE, Steigerwalt AG, Cavuoti D, Barton T, Hardy RD, Godoy D, Spratt BG, Clark TA, Wilkins PP. 2006. Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J Clin Microbiol 44: 4601–4604. 10.1128/JCM.01585-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves VM, Matteucci KC, Buzzo CL, Miollo BH, Ferrante D, Torrecilhas AC, Rodrigues MM, Alvarez JM, Bortoluci KR. 2013. NLRP3 controls Trypanosoma cruzi infection through a caspase-1-dependent IL-1R-independent NO production. PLoS Negl Trop Dis 7: e2469 10.1371/journal.pntd.0002469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfu G, Cirelli KM, Melo MB, Mayer-Barber K, Crown D, Koller BH, Masters S, Sher A, Leppla SH, Moayeri M, et al. 2014. Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. mBio 5: e01117 10.1128/mBio.01117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdent N, Maridonneau-Parini I, Sory MP, Cornelis GR. 2002. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect Immun 70: 4165–4176. 10.1128/IAI.70.8.4165-4176.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross O, Poeck H, Bscheider M, Dostert C, Hannesschläger N, Endres S, Hartmann G, Tardivel A, Schweighoffer E, Tybulewicz V, et al. 2009. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature 459: 433–436. 10.1038/nature07965 [DOI] [PubMed] [Google Scholar]
- Guo S, Yang C, Diao B, Huang X, Jin M, Chen L, Yan W, Ning Q, Zheng L, Wu Y, et al. 2015. The NLRP3 inflammasome and IL-1β accelerate immunologically mediated pathology in experimental viral fulminant hepatitis. PLoS Pathog 11: e1005155 10.1371/journal.ppat.1005155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. 2013. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341: 1250–1253. 10.1126/science.1240988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R, Aldrich A, Kielian T. 2014. Critical role for the AIM2 inflammasome during acute CNS bacterial infection. J Neurochem 129: 704–711. 10.1111/jnc.12669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Seregin SS, Yang D, Fukase K, Chamaillard M, Alnemri ES, Inohara N, Chen GY, Núñez G. 2018. The NLRP6 inflammasome recognizes lipoteichoic acid and regulates Gram-positive pathogen infection. Cell 175: 1651–1664.e14. 10.1016/j.cell.2018.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemrajani C, Berger CN, Robinson KS, Marches O, Mousnier A, Frankel G. 2010. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc Natl Acad Sci 107: 3129–3134. 10.1073/pnas.0911609106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hise AG, Tomalka J, Ganesan S, Patel K, Hall BA, Brown GD, Fitzgerald KA. 2009. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe 5: 487–497. 10.1016/j.chom.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zhou Q, Zhang C, Fan S, Cheng W, Zhao Y, Shao F, Wang HW, Sui SF, Chai J. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science 350: 399–404. 10.1126/science.aac5489 [DOI] [PubMed] [Google Scholar]
- Huang Z, Wu SQ, Liang Y, Zhou X, Chen W, Li L, Wu J, Zhuang Q, Chen C, Li J, et al. 2015. RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe 17: 229–242. 10.1016/j.chom.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Knoten CA, Morris AR, Hauser AR. 2018. ASC acts in a caspase-1-independent manner to worsen acute pneumonia caused by Pseudomonas aeruginosa. J Med Microbiol 67: 1168–1180. 10.1099/jmm.0.000782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannitti RG, Napolioni V, Oikonomou V, De Luca A, Galosi C, Pariano M, Massi-Benedetti C, Borghi M, Puccetti M, Lucidi V, et al. 2016. IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis. Nat Commun 7: 10791 10.1038/ncomms10791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 206: 79–87. 10.1084/jem.20081667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Taabazuing CY, Okondo MC, Chui AJ, Rao SD, Brown FC, Reed C, Peguero E, de Stanchina E, Kentsis A, et al. 2018. DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat Med 24: 1151–1156. 10.1038/s41591-018-0082-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Vance RE, Dangl JL. 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395–aaf6395. 10.1126/science.aaf6395 [DOI] [PubMed] [Google Scholar]
- Jorgensen I, Lopez JP, Laufer SA, Miao EA. 2016a. IL-1β, IL-18, and eicosanoids promote neutrophil recruitment to pore-induced intracellular traps following pyroptosis. Eur J Immunol 46: 2761–2766. 10.1002/eji.201646647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Zhang Y, Krantz BA, Miao EA. 2016b. Pyroptosis triggers pore-induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J Exp Med 213: 2113–2128. 10.1084/jem.20151613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Rayamajhi M, Miao EA. 2017. Programmed cell death as a defence against infection. Nat Rev Immunol 17: 151–164. 10.1038/nri.2016.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi VD, Kalvakolanu DV, Hebel JR, Hasday JD, Cross AS. 2002. Role of caspase 1 in murine antibacterial host defenses and lethal endotoxemia. Infect Immun 70: 6896–6903. 10.1128/IAI.70.12.6896-6903.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Mocarski ES. 2013. Viral modulation of programmed necrosis. Curr Opin Virol 3: 296–306. 10.1016/j.coviro.2013.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TJ, Basu S, Zhang L, Thomas KE, Vogel SN, Baillie L, Cross AS. 2008. Bacillus anthracis spores and lethal toxin induce IL-1β via functionally distinct signaling pathways. Eur.J Immunol 38: 1574–1584. 10.1002/eji.200838141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, Man SM, Malireddi RKS, Gurung P, Vogel P, Lamkanfi M, Kanneganti TD. 2015. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe 17: 357–368. 10.1016/j.chom.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. 2011. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med 3: 513–527. 10.1002/emmm.201100160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, et al. 2013. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341: 1246–1249. 10.1126/science.1240248 [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526: 666–671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Ke Y, Chen Z, Yang R. 2013. Yersinia pestis: Mechanisms of entry into and resistance to the host cell. Front Cell Infect Microbiol 3: 106 10.3389/fcimb.2013.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelut-Carneiro N, Silva GK, Rocha FA, Milanezi CM, Cavalcanti-Neto FF, Zamboni DS, Silva JS. 2015. IL-18 triggered by the Nlrp3 inflammasome induces host innate resistance in a pulmonary model of fungal infection. J Immunol 194: 4507–4517. 10.4049/jimmunol.1402321 [DOI] [PubMed] [Google Scholar]
- Kitur K, Wachtel S, Brown A, Wickersham M, Paulino F, Peñaloza HF, Soong G, Bueno S, Parker D, Prince A. 2016. Necroptosis promotes Staphylococcus aureus clearance by inhibiting excessive inflammatory signaling. Cell Rep 16: 2219–2230. 10.1016/j.celrep.2016.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordes M, Matuschewski K, Hafalla JCR. 2011. Caspase-1 activation of interleukin-1β (IL-1β) and IL-18 is dispensable for induction of experimental cerebral malaria. Infect Immun 79: 3633–3641. 10.1128/IAI.05459-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs SB, Miao EA. 2017. Gasdermins: Effectors of pyroptosis. Trends Cell Biol 27: 673–684. 10.1016/j.tcb.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Roe K, Orillo B, Muruve DA, Nerurkar VR, Gale M Jr, Verma S. 2013. Inflammasome adaptor protein apoptosis-associated speck-like protein containing CARD (ASC) is critical for the immune response and survival in West Nile virus encephalitis. J Virol 87: 3655–3667. 10.1128/JVI.02667-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose T, Man SM, Malireddi RK, Karki R, Kesavardhana S, Place DE, Neale G, Vogel P, Kanneganti TD. 2016. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol 1: aag2045 10.1126/sciimmunol.aag2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey CA, Miao EA. 2019. NLRP1 - One NLR to guard them all. EMBO J 38: e102494 10.15252/embj.2019102494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey CA, Mitchell WJ, Dadelahi AS, Skyberg JA. 2018. Caspase-1 and caspase-11 mediate pyroptosis, inflammation, and control of Brucella joint infection. Infect Immun 86: e00361-18 10.1128/IAI.00361-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Declercq W, Kalai M, Saelens X, Vandenabeele P. 2002. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ 9: 358–361. 10.1038/sj.cdd.4400989 [DOI] [PubMed] [Google Scholar]
- Lara-Tejero M, Sutterwala FS, Ogura Y, Grant EP, Bertin J, Coyle AJ, Flavell RA, Galán JE. 2006. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med 203: 1407–1412. 10.1084/jem.20060206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock CN, Cookson BT. 2012. The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe 12: 799–805. 10.1016/j.chom.2012.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinsohn JL, Newman ZL, Hellmich KA, Fattah R, Getz MA, Liu S, Sastalla I, Leppla SH, Moayeri M. 2012. Anthrax lethal factor cleavage of nlrp1 is required for activation of the inflammasome. PLoS Pathog 8: e1002638 10.1371/journal.ppat.1002638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang L, Yao Q, Li L, Dong N, Rong J, Gao W, Ding X, Sun L, Chen X, et al. 2013. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature 501: 242–246. 10.1038/nature12436 [DOI] [PubMed] [Google Scholar]
- Lilue J, Doran AG, Fiddes IT, Abrudan M, Armstrong J, Bennett R, Chow W, Collins J, Collins S, Czechanski A, et al. 2018. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat Genet 50: 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CA, Miller BM, Rivera-Chávez F, Velazquez EM, Byndloss MX, Chávez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, Bäumler AJ. 2016. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353: 1249–1253. 10.1126/science.aag3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas X, Ciulli A. 2017. Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr Opin Struct Biol 44: 101–110. 10.1016/j.sbi.2016.12.015 [DOI] [PubMed] [Google Scholar]
- Macher AM, Casale TB, Fauci AS. 1982. Chronic granulomatous disease of childhood and Chromobacterium violaceum infections in the southeastern United States. Ann Intern Med 97: 51–55. 10.7326/0003-4819-97-1-51 [DOI] [PubMed] [Google Scholar]
- Maltez VI, Miao EA. 2016. Reassessing the evolutionary importance of inflammasomes. J Immunol 196: 956–962. 10.4049/jimmunol.1502060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltez VI, Tubbs AL, Cook KD, Aachoui Y, Falcone EL, Holland SM, Whitmire JK, Miao EA. 2015. Inflammasomes coordinate pyroptosis and natural killer cell cytotoxicity to clear infection by a ubiquitous environmental bacterium. Immunity 43: 987–997. 10.1016/j.immuni.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD. 2015. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol 16: 467–475. 10.1038/ni.3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Briard B, Burton A, Gingras S, Pelletier S, Kanneganti TD. 2017. Differential roles of caspase-1 and caspase-11 in infection and inflammation. Sci Rep 7: 45126 10.1038/srep45126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Finnemann SC. 2015. Regulation of phagocytosis by Rho GTPases. Small GTPases 6: 89–99. 10.4161/21541248.2014.989785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Dixit VM, Monack DM. 2005. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med 202: 1043–1049. 10.1084/jem.20050977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. 2002. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of pro-IL-β. Mol Cell 10: 417–426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- Mayer-Barber KD, Barber DL, Shenderov K, White SD, Wilson MS, Cheever A, Kugler D, Hieny S, Caspar P, Núñez G, et al. 2010. Caspase-1 independent IL-1β production is critical for host resistance to mycobacterium tuberculosis and does not require TLR signaling in vivo. J Immunol 184: 3326–3330. 10.4049/jimmunol.0904189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald C, Vacratsis PO, Bliska JB, Dixon JE. 2003. The Yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J Biol Chem 278: 18514–18523. 10.1074/jbc.M301226200 [DOI] [PubMed] [Google Scholar]
- Mcelvania-Tekippe E, Allen IC, Hulseberg PD, Sullivan JT, McCann JR, Sandor M, Braunstein M, Ting JP. 2010. Granuloma formation and host defense in chronic Mycobacterium tuberculosis infection requires PYCARD/ASC but not NLRP3 or caspase-1. PLoS ONE 5: e12320 10.1371/journal.pone.0012320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Rühl S, Dussurgey S, Dick MS, Kistner A, Rigard M, et al. 2015. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol 16: 476–484. 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. 2010a. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11: 1136–1142. 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. 2010b. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci 107: 3076–3080. 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milora KA, Miller SL, Sanmiguel JC, Jensen LE. 2014. Interleukin-1α released from HSV-1-infected keratinocytes acts as a functional alarmin in the skin. Nat Commun 5: 5230 10.1038/ncomms6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Crown D, Newman ZL, Okugawa S, Eckhaus M, Cataisson C, Liu S, Sastalla I, Leppla SH. 2010. Inflammasome sensor Nlrp1b-dependent resistance to anthrax is mediated by caspase-1, IL-1 signaling and neutrophil recruitment. PLoS Pathog 6: e1001222 10.1371/journal.ppat.1001222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Sastalla I, Leppla SH. 2012. Anthrax and the inflammasome. Microbes Infect 14: 392–400. 10.1016/j.micinf.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack DM, Hersh D, Ghori N, Bouley D, Zychlinsky A, Falkow S. 2000. Salmonella exploits caspase-1 to colonize Peyer's patches in a murine typhoid model. J Exp Med 192: 249–258. 10.1084/jem.192.2.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlbauer SM, Evering TH, Bonuccelli G, Squires RC, Ashton AW, Porcelli SA, Lisanti MP, Brojatsch J. 2007. Anthrax lethal toxin kills macrophages in a strain-specific manner by apoptosis or caspase-1-mediated necrosis. Cell Cycle 6: 758–766. 10.4161/cc.6.6.3991 [DOI] [PubMed] [Google Scholar]
- Mühlen S, Ruchaud-Sparagano M-H, Kenny B. 2011. Proteasome-independent degradation of canonical NF-κB complex components by the NleC protein of pathogenic Escherichia coli. J Biol Chem 286: 5100–5107. 10.1074/jbc.M110.172254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogusa S, Thapa RJ, Dillon CP, Liedmann S, Oguin TH III, Ingram JP, Rodriguez DA, Kosoff R, Sharma S, Sturm O, et al. 2016. RIPK3 activates parallel pathways of MLKL-driven necroptosis and FADD-mediated apoptosis to protect against influenza A virus. Cell Host Microbe 20: 13–24. 10.1016/j.chom.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okondo MC, Johnson DC, Sridharan R, Go EB, Chui AJ, Wang MS, Poplawski SE, Wu W, Liu Y, Lai JH, et al. 2017. DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat Chem Biol 13: 46–53. 10.1038/nchembio.2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okondo MC, Rao SD, Taabazuing CY, Chui AJ, Poplawski SE, Johnson DC, Bachovchin DA. 2018. Inhibition of Dpp8/9 activates the Nlrp1b inflammasome. Cell Chem Biol 25: 262–267.e5. 10.1016/j.chembiol.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özen S. 2018. Update on the epidemiology and disease outcome of familial Mediterranean fever. Best Pract Res Clin Rheumatol 32: 254–260. 10.1016/j.berh.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Paciello I, Silipo A, Lembo-Fazio L, Curcurù L, Zumsteg A, Noël G, Ciancarella V, Sturiale L, Molinaro A, Bernardini ML. 2013. Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proc Natl Acad Sci 110: E4345–E4354. 10.1073/pnas.1303641110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N, Conlon J, Sweet C, Rus F, Wilson L, Pereira A, Rosadini CV, Goutagny N, Weber ANR, Lane WS, et al. 2012. Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc Natl Acad Sci 109: 12710–12715. 10.1073/pnas.1008203109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YH, Wood G, Kastner DL, Chae JJ. 2016. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat Immunol 17: 914–921. 10.1038/ni.3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroli AF, Gonzalez PV, Díaz-Luján C, Onofrio LI, Arocena A, Cano RC, Carrera-Silva EA, Gea S. 2018. NLRP3 Inflammasome and caspase-1/11 pathway orchestrate different outcomes in the host protection against Trypanosoma cruzi acute infection. Front Immunol 9: 913 10.3389/fimmu.2018.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JS, Hartland EL. 2014. The inflammatory response during enterohemorrhagic Escherichia coli infection. Microbiol Spectr 2: EHEC–0012–2013 10.1128/microbiolspec.EHEC-0012-2013 [DOI] [PubMed] [Google Scholar]
- Pearson JS, Riedmaier P, Marches O, Frankel G, Hartland EL. 2011. A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-κB for degradation. Mol Microbiol 80: 219–230. 10.1111/j.1365-2958.2011.07568.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JS, Giogha C, Ong SY, Kennedy CL, Kelly M, Robinson KS, Lung TWF, Mansell A, Riedmaier P, Oates CVL, et al. 2013. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature 501: 247–251. 10.1038/nature12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JS, Giogha C, Mühlen S, Nachbur U, Pham CLL, Zhang Y, Hildebrand JM, Oates CV, Lung TWF, Ingle D, et al. 2017. EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat Microbiol 2: 16258 10.1038/nmicrobiol.2016.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechous RD, Sivaraman V, Price PA, Stasulli NM, Goldman WE. 2013. Early host cell targets of Yersinia pestis during primary pneumonic plague. PLoS Pathog 9: e1003679 10.1371/journal.ppat.1003679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip NH, Dillon CP, Snyder AG, Fitzgerald P, Wynosky-Dolfi MA, Zwack EE, Hu B, Fitzgerald L, Mauldin EA, Copenhaver AM, et al. 2014. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-κB and MAPK signaling. Proc Natl Acad Sci 111: 7385–7390. 10.1073/pnas.1403252111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quistad SD, Stotland A, Barott KL, Smurthwaite CA, Hilton BJ, Grasis JA, Wolkowicz R, Rohwer FL. 2014. Evolution of TNF-induced apoptosis reveals 550 My of functional conservation. Proc Natl Acad Sci 111: 9567–9572. 10.1073/pnas.1405912111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan JV, Rodriguez D, Miao EA, Aderem A. 2011. The NLRP3 inflammasome detects encephalomyocarditis virus and vesicular stomatitis virus infection. J Virol 85: 4167–4172. 10.1128/JVI.01687-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MLG, Poreba M, Snipas SJ, Groborz K, Drag M, Salvesen GS. 2018. Extensive peptide and natural protein substrate screens reveal that mouse caspase-11 has much narrower substrate specificity than caspase-1. J Biol Chem 293: 7058–7067. 10.1074/jbc.RA117.001329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos HJ, Lanteri MC, Blahnik G, Negash A, Suthar MS, Brassil MM, Sodhi K, Treuting PM, Busch MP, Norris PJ, et al. 2012. IL-1β signaling promotes CNS-intrinsic immune control of West Nile virus infection. PLoS Pathog 8: e1003039 10.1371/journal.ppat.1003039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VAK, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, Fitzgerald KA. 2012. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by Gram-negative bacteria. Cell 150: 606–619. 10.1016/j.cell.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner D, Orning MP, Starheim KK, Marty-Roix R, Proulx MK, Goguen JD, Lien E. 2016. Manipulation of interleukin-1β and interleukin-18 production by Yersinia pestis effectors YopJ and YopM and redundant impact on virulence. J Biol Chem 291: 9894–9905. 10.1074/jbc.M115.697698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I, Deets KA, Ji DX, Moltke Von J, Tenthorey JL, Lee AY, Philip NH, Ayres JS, Brodsky IE, Gronert K, et al. 2017. NAIP-NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity 46: 649–659. 10.1016/j.immuni.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raupach B, Peuschel SK, Monack DM, Zychlinsky A. 2006. Caspase-1-mediated activation of interleukin-1β (IL-1β) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect Immun 74: 4922–4926. 10.1128/IAI.00417-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer T, Shaw MH, Franchi L, Coban C, Ishii KJ, Akira S, Horii T, Rodriguez A, Núñez G. 2010. Experimental cerebral malaria progresses independently of the Nlrp3 inflammasome. Eur J Immunol 40: 764–769. 10.1002/eji.200939996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Wu S, Cai J, Yang Y, Ren X, Feng Y, Chen L, Qin B, Xu C, Yang H, et al. 2017. The H7N9 influenza A virus infection results in lethal inflammation in the mammalian host via the NLRP3-caspase-1 inflammasome. Sci Rep 7: 7625 10.1038/s41598-017-07384-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S. 2012. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol 13: 954–962. 10.1038/ni.2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue-Gervais IG, Labbé K, Dagenais M, Dupaul-Chicoine J, Champagne C, Morizot A, Skeldon A, Brincks EL, Vidal SM, Griffith TS, et al. 2014. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe 15: 23–35. 10.1016/j.chom.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Rodriguez AE, Bogart C, Gilbert CM, McCullers JA, Smith AM, Kanneganti TD, Lupfer CR. 2019. Enhanced IL-1β production is mediated by a TLR2-MYD88-NLRP3 signaling axis during coinfection with influenza A virus and Streptococcus pneumoniae. PLoS ONE 14: e0212236 10.1371/journal.pone.0212236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. 2007. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1: 77–83. 10.1016/j.chom.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Royan SV, Jones RM, Koutsouris A, Roxas JL, Falzari K, Weflen AW, Kim A, Bellmeyer A, Turner JR, Neish AS, et al. 2010. Enteropathogenic E. coli non-LEE encoded effectors NleH1 and NleH2 attenuate NF-κB activation. Mol Microbiol 78: 1232–1245. 10.1111/j.1365-2958.2010.07400.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Xia S, Liu X, Lieberman J, Wu H. 2018. Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557: 62–67. 10.1038/s41586-018-0058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga H, Kitada S, Shimada Y, Kamiyama N, Okuyama M, Makino M, Yamamoto M, Takeda K. 2012. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int Immunol 24: 637–644. 10.1093/intimm/dxs062 [DOI] [PubMed] [Google Scholar]
- Sandstrom A, Mitchell PS, Goers L, Mu EW, Lesser CF, Vance RE. 2019. Functional degradation: a mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364: eaau1330 10.1126/science.aau1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ, Phalipon A, Arondel J, Thirumalai K, Banerjee S, Akira S, Takeda K, Zychlinsky A. 2000. Caspase-1 activation of IL-1β and IL-18 are essential for Shigella flexneri–induced inflammation. Immunity 12: 581–590. 10.1016/S1074-7613(00)80209-5 [DOI] [PubMed] [Google Scholar]
- Scott NE, Giogha C, Pollock GL, Kennedy CL, Webb AI, Williamson NA, Pearson JS, Hartland EL. 2017. The bacterial arginine glycosyltransferase effector NleB preferentially modifies Fas-associated death domain protein (FADD). J Biol Chem 292: 17337–17350. 10.1074/jbc.M117.805036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Vacratsis PO, Bao Z, Bowers KE, Fierke CA, Dixon JE. 2003. Biochemical characterization of the Yersinia YopT protease: cleavage site and recognition elements in Rho GTPases. Proc Natl Acad Sci 100: 904–909. 10.1073/pnas.252770599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Yagnik B, Baksi R, Desai N, Padh H, Desai P. 2017. Shigellosis murine model established by intraperitoneal and intranasal route of administration: a comparative comprehension overview. Microbes Infect 19: 47–54. 10.1016/j.micinf.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Sheng X, You Q, Zhu H, Chang Z, Li Q, Wang H, Wang C, Wang H, Hui L, Du C, et al. 2017. Bacterial effector NleL promotes enterohemorrhagic E. coli-induced attaching and effacing lesions by ubiquitylating and inactivating JNK. PLoS Pathog 13: e1006534 10.1371/journal.ppat.1006534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. 2014. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514: 187–192. 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526: 660–665. 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- Shio MT, Eisenbarth SC, Savaria M, Vinet AF, Bellemare MJ, Harder KW, Sutterwala FS, Bohle DS, Descoteaux A, Flavell RA, et al. 2009. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog 5: e1000559 10.1371/journal.ppat.1000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GK, Costa RS, Silveira TN, Caetano BC, Horta CV, Gutierrez FR, Guedes PM, Andrade WA, De Niz M, Gazzinelli RT, et al. 2013. Apoptosis-associated speck-like protein containing a caspase recruitment domain inflammasomes mediate IL-1β response and host resistance to Trypanosoma cruzi infection. J Immunol 191: 3373–3383. 10.4049/jimmunol.1203293 [DOI] [PubMed] [Google Scholar]
- Singer AU, Rohde JR, Lam R, Skarina T, Kagan O, Dileo R, Chirgadze NY, Cuff ME, Joachimiak A, Tyers M, et al. 2008. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol 15: 1293–1301. 10.1038/nsmb.1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaraman V, Pechous RD, Stasulli NM, Miao EA, Goldman WE. 2015. Yersinia pestis activates both IL-1β and IL-1 receptor antagonist to modulate lung inflammation during pneumonic plague. PLoS Pathog 11: e1004688 10.1371/journal.ppat.1004688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires RC, Muehlbauer SM, Brojatsch J. 2007. Proteasomes control caspase-1 activation in anthrax lethal toxin-mediated cell killing. J Biol Chem 282: 34260–34267. 10.1074/jbc.M705687200 [DOI] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. 2008. Apoptosis: Controlled demolition at the cellular level. Nat Rev Mol Cell Biol 9: 231–241. 10.1038/nrm2312 [DOI] [PubMed] [Google Scholar]
- Tenthorey JL, Haloupek N, López-Blanco JR, Grob P, Adamson E, Hartenian E, Lind NA, Bourgeois NM, Chacón P, Nogales E, et al. 2017. The structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science 358: 888–893. 10.1126/science.aao1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra JK, Cote CK, France B, Jenkins AL, Bozue JA, Welkos SL, LeVine SM, Bradley KA. 2010. Cutting edge: resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. J Immunol 184: 17–20. 10.4049/jimmunol.0903114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa RJ, Ingram JP, Ragan KB, Nogusa S, Boyd DF, Benitez AA, Sridharan H, Kosoff R, Shubina M, Landsteiner VJ, et al. 2016. DAI senses influenza A virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe 20: 674–681. 10.1016/j.chom.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PG, Dash P, Aldridge JR, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, et al. 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30: 566–575. 10.1016/j.immuni.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumkeo D, Watanabe S, Narumiya S. 2013. Physiological roles of Rho and Rho effectors in mammals. Eur J Cell Biol 92: 303–315. 10.1016/j.ejcb.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, Quadroni M, Tschopp J. 2007. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-κB pathway. EMBO J 26: 197–208. 10.1038/sj.emboj.7601473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolle L, Yu FS, Kovach MA, Ballinger MN, Newstead MW, Zeng X, Nunez G, Standiford TJ. 2015. Redundant and cooperative interactions between TLR5 and NLRC4 in protective lung mucosal immunity against Pseudomonas aeruginosa. J Innate Immun 7: 177–186. 10.1159/000367790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma C, Higa N, Koizumi Y, Nakasone N, Ogura Y, McCoy AJ, Franchi L, Uematsu S, Sagara J, Taniguchi S, et al. 2010. Pathogenic Vibrio activate NLRP3 inflammasome via cytotoxins and TLR/nucleotide-binding oligomerization domain-mediated NF-κB signaling. J Immunol 184: 5287–5297. 10.4049/jimmunol.0903536 [DOI] [PubMed] [Google Scholar]
- Tomalka J, Ganesan S, Azodi E, Patel K, Majmudar P, Hall BA, Fitzgerald KA, Hise AG. 2011. A novel role for the NLRC4 inflammasome in mucosal defenses against the fungal pathogen Candida albicans. PLoS Pathog 7: e1002379 10.1371/journal.ppat.1002379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji NM, Tsutsui H, Seki E, Kuida K, Okamura H, Nakanishi K, Flavell RA. 2004. Roles of caspase-1 in Listeria infection in mice. Int Immunol 16: 335–343. 10.1093/intimm/dxh041 [DOI] [PubMed] [Google Scholar]
- Turk BE. 2007. Manipulation of host signalling pathways by anthrax toxins. Biochem J 402: 405–417. 10.1042/BJ20061891 [DOI] [PubMed] [Google Scholar]
- Vance RE. 2015. The NAIP/NLRC4 inflammasomes. Curr Opin Immunol 32: 84–89. 10.1016/j.coi.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe T, Hassannia B, Vandenabeele P. 2016. An outline of necrosome triggers. Cell Mol Life Sci 73: 2137–2152. 10.1007/s00018-016-2189-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Veerdonk FL, Joosten LA, Shaw PJ, Smeekens SP, Malireddi RK, van der Meer JW, Kullberg BJ, Netea MG, Kanneganti TD. 2011. The inflammasome drives protective Th1 and Th17 cellular responses in disseminated candidiasis. Eur J Immunol 41: 2260–2268. 10.1002/eji.201041226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. 1973. A new evolutionary law. Evol Theory 1: 1–30. [Google Scholar]
- Vladimer GI, Weng D, Paquette SW, Vanaja SK, Rathinam VA, Aune MH, Conlon JE, Burbage JJ, Proulx MK, Liu Q, et al. 2012. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37: 96–107. 10.1016/j.immuni.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, Lenardo MJ. 2011. IKKβ phosphorylation regulates RPS3 nuclear translocation and NF-κB function during infection with Escherichia coli strain O157:H7. Nat Immunol 12: 335–343. 10.1038/ni.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y, Liu S, Yu X, Li L, Shi C, He W, Li J, Xu L, Hu Z, et al. 2014. Direct activation of RIP3/MLKL-dependent necrosis by herpes simplex virus 1 (HSV-1) protein ICP6 triggers host antiviral defense. Proc Natl Acad Sci 111: 15438–15443. 10.1073/pnas.1412767111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Shao Y, Li S, Xin N, Ma T, Zhao C, Song M. 2017a. Caspase-11 plays a protective role in pulmonary Acinetobacter baumannii infection. Infect Immun 85: e00350 10.1128/IAI.00350-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ning X, Gao P, Wu S, Sha M, Lv M, Zhou X, Gao J, Fang R, Meng G, et al. 2017b. Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity 46: 393–404. 10.1016/j.immuni.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Weber CH, Vincenz C. 2001. The death domain superfamily: a tale of two interfaces? Trends Biochem Sci 26: 475–481. 10.1016/S0968-0004(01)01905-3 [DOI] [PubMed] [Google Scholar]
- Weinert C, Morger D, Djekic A, Grütter MG, Mittl PRE. 2015. Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: implications for the recognition of higher order oligomers. Sci Rep 5: 10819 10.1038/srep10819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng D, Marty-Roix R, Ganesan S, Proulx MK, Vladimer GI, Kaiser WJ, Mocarski ES, Pouliot K, Chan FK, Kelliher MA, et al. 2014. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proc Natl Acad Sci 111: 7391–7396. 10.1073/pnas.1403477111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Leppla SH, Moayeri M. 2008. Killing of macrophages by anthrax lethal toxin: involvement of the N-end rule pathway. Cell Microbiol 10: 1352–1362. 10.1111/j.1462-5822.2008.01131.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham SB, Allen IC, Bergstralh DT, Brickey WJ, Huang MT, Taxman DJ, Duncan JA, Ting JP. 2009. NLRP3 (NALP3, Cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J Immunol 183: 2008–2015. 10.4049/jimmunol.0900138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong Y-N, Peng X, Xi JJ, Chen S, et al. 2014. Innate immune sensing of bacterial modifications of Rho GTPases by the pyrin inflammasome. Nature 513: 237–241. 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- Xu H, Shi J, Gao H, Liu Y, Yang Z, Shao F, Dong N. 2019. The N-end rule ubiquitin ligase UBR2 mediates NLRP1B inflammasome activation by anthrax lethal toxin. EMBO J 38: e101996. 10.15252/embj.2019101996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Stevenson HL, Scott MJ, Ismail N. 2015. Type I interferon contributes to noncanonical inflammasome activation, mediates immunopathology, and impairs protective immunity during fatal infection with lipopolysaccharide-negative Ehrlichiae. Am J Pathol 185: 446–461. 10.1016/j.ajpath.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang F, Wang W, Lin G, Hu Z, Han Z, Qi Y, Zhang L, Wang J, Sui SF, et al. 2018. Structural basis for specific flagellin recognition by the NLR protein NAIP5. Cell Res 28: 35–47. 10.1038/cr.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Briones M, Chiou J, Lei L, Patton MJ, Ma L, McClarty G, Caldwell HD. 2019. Chlamydia trachomatis lipopolysaccharide evades the canonical and noncanonical inflammatory pathways to subvert innate immunity. mBio 10: e00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H, Ooka T, Iguchi A, Hayashi T, Sugimoto N, Tobe T. 2010. NleC, a type III secretion protease, compromises NF-κB activation by targeting p65/RelA. PLoS Pathog 6: e1001231 10.1371/journal.ppat.1001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SX, Chen W, Liu ZZ, Zhou FH, Yan SQ, Hu GQ, Qin XX, Zhang J, Ma K, Du CT, et al. 2018. Non-hematopoietic MLKL protects against Salmonella mucosal infection by enhancing inflammasome activation. Front Immunol 9: 119 10.3389/fimmu.2018.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalinger ZB, Elliott R, Weiss SR. 2017. Role of the inflammasome-related cytokines Il-1 and Il-18 during infection with murine coronavirus. J Neurovirol 23: 845–854. 10.1007/s13365-017-0574-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding X, Cui J, Xu H, Chen J, Gong YN, Hu L, Zhou Y, Ge J, Lu Q, et al. 2011. Cysteine methylation disrupts ubiquitin-chain sensing in NF-κB activation. Nature 481: 204–208. 10.1038/nature10690 [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen S, Ruan J, Wu J, Tong AB, Yin Q, Li Y, David L, Lu A, Wang WL, et al. 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350: 404–409. 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shi J, Shi X, Wang Y, Wang F, Shao F. 2016. Genetic functions of the NAIP family of inflammasome receptors for bacterial ligands in mice. J Exp Med 213: 647–656. 10.1084/jem.20160006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Lilo S, Mena P, Bliska JB. 2012. YopJ-induced caspase-1 activation in Yersinia-infected macrophages: independent of apoptosis, linked to necrosis, dispensable for innate host defense. PLoS ONE 7: e36019 10.1371/journal.pone.0036019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Robinson K, Teo DET, Tan KY, Lim C, Harapas CR, Yu C-H, Xie WH, Sobota RM, Au VB, et al. 2018. Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J Biol Chem 293: 18864–18878. 10.1074/jbc.RA118.004350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F. 2008. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol 15: 1302–1308. 10.1038/nsmb.1517 [DOI] [PubMed] [Google Scholar]
- Zwack EE, Snyder AG, Wynosky-Dolfi MA, Ruthel G, Philip NH, Marketon MM, Francis MS, Bliska JB, Brodsky IE. 2015. Inflammasome activation in response to the Yersinia type III secretion system requires hyperinjection of translocon proteins YopB and YopD. mBio 6: e02095 10.1128/mBio.02095-14 [DOI] [PMC free article] [PubMed] [Google Scholar]