Discovery of the role of bacterial RNase J1 in termination of transcription suggests common allosteric principles and mechanistic congruency of termination between bacteria and eukaryotes, in which an unrelated RNase Xrn2/Rat1 plays a similar role.
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; DNA Replication, Repair & Recombination; Microbiology, Virology & Host Pathogen Interaction
An RNase J1 role in Bacillus subtilis transcription termination supports the existence of common allosteric principles of termination mechanisms, but also suggests higher mechanistic diversity in bacteria than expected.

In its most simple conceptualization, RNA synthesis in bacteria begins at a defined DNA sequence recognized by RNA polymerase (RNAP) – the promoter – and ends at another defined DNA sequence, called terminator. Bacterial terminators are generally considered to be of two classes: intrinsic terminators do not require factors other than RNAP, or terminators dependent on Rho (a widely, albeit not universally, conserved RNA helicase [Fig 1]) (Porrua et al, 2016; Roberts, 2019). Promoters and terminators are often thought of as boundaries of transcription units. Many, if not most, transcription units in bacteria feature expansions of this simple architecture through multiplication and combination of different types of promoters and terminators/anti‐terminators. This combinatorial design of transcription units allows for adaptive regulation of gene expression. New work in The EMBO Journal by Krasny and colleagues (Šiková et al, 2020) further investigates how termination events, which occur when RNAP is unable to complete RNA synthesis and stalls/arrests before reaching the end of the transcription unit, are resolved.
Figure 1. Evolutionary distribution of the transcription factors Rho (blue), GreAB (orange), Mfd (green) and RNase J1 (purple) among Proteobacteria.
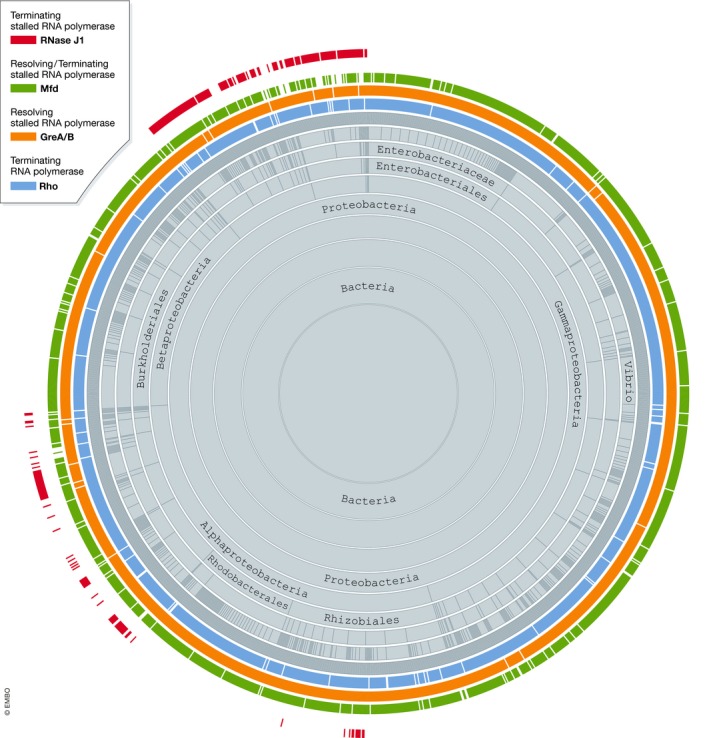
Figure generated using Aquerium (http://aquerium.zhulinlab.org/).
Bacterial RNAP is a highly processive mechano‐chemical machine that routinely traverses several kilobases of DNA template without dissociation, while synthesizing RNA molecules thousands of nucleotides in length. However, with yet‐to‐be rigorously quantified frequencies, the transcribing elongation complex (EC) encounters physical barriers that cause it to stall, such as DNA‐bound proteins or segments of damaged DNA. A large fraction of these stalled complexes retreat (backtrack), away from such roadblocks, making them the substrate for transcript cleavage factor(s) GreA(B) (Nudler, 2012; Abdelkareem et al, 2019). Reactivated through GreA(B)‐stimulated generation of the new 3′ in the nascent RNA, the stalled complexes can resume RNA synthesis. Complexes that fail to be reactivated in this fashion are often removed from the DNA template in an act of termination distinct from the intrinsic and Rho‐dependent termination mechanisms.
Mfd, a DNA translocase capable of binding RNAP in vivo and in vitro, is a transcription factor generally recognized to reactivate backtracked ECs or terminate permanently blocked ECs. Moving co‐directionally with transcribing RNAP as an ATP hydrolase/helicase, Mfd is perfectly suited to induce forward translocation of the EC (Le et al, 2018). In the absence of concomitant RNA synthesis, such translocation is thought to cause dissociation of RNA–DNA hybrid, and the entire EC. Although the basic mechanism of Mfd action is not in dispute, it is known that Mfd is present in the cell at a much lower concentration than RNAP (Schmidt et al, 2016) and that it lacks a discernible mechanism to distinguish between blocked, arrested, transiently paused and active ECs. Combined with its even more sporadic evolutionary conservation than that of Rho (Fig 1), these properties of Mfd suggest that additional factors involved in termination of stalled RNAPs may/must exist.
The report by Šiková et al (2020) identifies a novel mechanism of resolving/terminating stalled ECs by the action of RNase J1 in Bacillus subtilis, as a complementary or alternative pathway to Mfd‐mediated termination. The authors note that depletion of RNase J1 in vivo leads to a significant increase in RNAP density along some transcription units without a concomitant increase in the level of transcription, consistent with the accumulation of unresolved stalled ECs. Intriguingly, this identified role of RNase J1 is reminiscent of the RNase‐dependent mechanism of Pol II termination in eukaryotes, where Xrn2/Rat1 terminates ECs at the ends of the transcription units (Nagarajan et al, 2013; Porrua et al, 2016). Both RNase J1 and Xrn2/Rat1 possess 5′–3′ exoribonuclease activities; Xrn2/Rat1 function in termination depends on RNA cleavage by an external endoribonuclease (cleavage and polyadenylation specificity factor CPSF), whereas RNase J1 has an intrinsic endoribonuclease activity whose importance is yet‐to‐be rigorously studied. The apparent functional overlap between the two is remarkable: although the main role of Xrn2 is to stimulate termination at the ends of full‐length RNA transcripts, it might also function in eliminating stalled/arrested complexes, similarly to RNase J1 (Eaton et al, 2018).
Consistent with its role in terminating ECs, the authors found RNase J1 in B. subtilis co‐localizing largely with RNAP, as compared to elongation‐independent co‐localization with more widely distributed RNA. In vitro RNase J1 efficiently terminates stalled complexes of B. subtilis RNAP and less efficiently those of Escherichia coli. Heterologous 5′–3′ exonuclease Xrn1 is capable of doing the same, albeit with less efficiency, whereas 3′‐5′ RNase R failed to effect any detectable termination. These observations led Šiková et al (2020) to conclude that the 5′–3′ directionality of the terminating exoribonuclease is of absolute importance, with the efficiency of termination being further modulated by the structural differences in both the exonuclease and the RNAP. We agree with this assessment, although it is worth discussing potential caveats. In addition to structural differences, RNase J1 and Xrn1 also diverge in enzymatic activity: Xrn1 does not appear to exhibit similar intrinsic endoribonuclease function as RNase J1, whereas, unlike RNase J1, it acts to degrade single‐stranded DNA and to promote single‐/double‐stranded DNA strand exchange. Moreover, 5′–3′ exoribonuclease activity appears to be necessary but not sufficient for termination of stalled RNAP, as another 5′–3′ exoribonuclease, Xrn2, failed to terminate bacterial ECs.
Work studying the Pol II termination mechanism has led to the proposition that a CPSF‐induced conformational change in RNAP allosterically modulates its receptivity to subsequently recruited Rat1/Xrn2 (Baejen et al, 2017). We believe this to be true in most, if not all, termination events, where the common essential step is the allosteric transition from the processive EC to the dissociation‐prone pre‐termination complex (Epshtein et al, 2007, 2010). This transition enables vastly different molecular “devices”, such as RNA hairpins, exoribonucleases and RNA/DNA helicases to promptly disrupt the highly stable EC. Consequently, the differential action of various 5′–3′ exoribonucleases on B. subtilis and E. coli transcription complexes, reported by Šiková et al, is likely to reflect their respective propensities to induce such transition in cognate and non‐cognate RNAPs.
Overall, this discovery of the function of RNase J1 in transcription termination in B. subtilis reinforces the notion that the termination mechanisms are similar across evolution in terms of the fundamental allosteric principles. Yet, they are more diverse in bacteria than previously thought, calling for a renewed search for other potential termination factors.
The EMBO Journal (2020) 39: e104112
See also: https://doi.org/10.15252/embj.2019102500 (February 2020)
References
- Abdelkareem M, Saint‐Andre C, Takacs M, Papai G, Crucifix C, Guo X, Ortiz J, Weixlbaumer A (2019) Structural basis of transcription: RNA polymerase backtracking and its reactivation. Mol Cell 75: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baejen C, Andreani J, Torkler P, Battaglia S, Schwalb B, Lidschreiber M, Maier KC, Boltendahl A, Rus P, Esslinger S et al (2017) Genome‐wide analysis of RNA polymerase II termination at protein‐coding genes. Mol Cell 66: 38–49 [DOI] [PubMed] [Google Scholar]
- Eaton JD, Davidson L, Bauer DLV, Natsume T, Kanemaki MT, West S (2018) Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes Dev 32: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E (2007) An allosteric path to transcription termination. Mol Cell 28: 991–1001 [DOI] [PubMed] [Google Scholar]
- Epshtein V, Dutta D, Wade J, Nudler E (2010) An allosteric mechanism of Rho‐dependent transcription termination. Nature 463: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Yang Y, Tan C, Suhanovsky MM, Fulbright RM Jr, Inman JT, Li M, Lee J, Perelman S, Roberts JW et al (2018) Mfd dynamically regulates transcription via a release and catch‐up mechanism. Cell 172: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan VK, Jones CI, Newbury SF, Green PJ (2013) XRN 5′–>3′ exoribonucleases: structure, mechanisms and functions. Biochim Biophys Acta 1829: 590–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E (2012) RNA polymerase backtracking in gene regulation and genome instability. Cell 149: 1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Boudvillain M, Libri D (2016) Transcription termination: variations on common themes. Trends Genet 32: 508–522 [DOI] [PubMed] [Google Scholar]
- Roberts JW (2019) Mechanisms of bacterial transcription termination. J Mol Biol 431: 4030–4039 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Kochanowski K, Vedelaar S, Ahrne E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M (2016) The quantitative and condition‐dependent Escherichia coli proteome. Nat Biotechnol 34: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šiková M, Wiedermannová J, Převorovský M, Barvík I, Sudzinová P, Kofroňová O, Benada O, Šanderová H, Condon C, Krásný L (2020) The torpedo effect in Bacillus subtilis: RNase J1 resolves stalled transcription. EMBO J 39: e102500 [DOI] [PMC free article] [PubMed] [Google Scholar]


