Abstract
Apolipoprotein E (apoE) is associated with lipoproteins in the cerebrospinal fluid (CSF). APOE4 increases and APOE2 decreases the risk for Alzheimer disease (AD) compared to the risk associated with APOE3 . Because apoE4 is less efficient at cholesterol efflux than apoE2 or apoE3 in vitro , we hypothesized that APOE genotype may affect apoE particle size in vivo and that these size differences may be related to AD risk. We used nondenaturing gel electrophoresis to test for differences in the size of apoE complexes in human CSF samples of various APOE genotypes and created profiles of each sample to compare the patterns of apoE distribution. For middle-aged adults with no dementia, APOE 2.3 individuals had significantly larger apoE complexes than APOE 3.3 subjects, who had significantly larger apoE complexes than APOE 3.4 and APOE 4.4 individuals. Similarly, in an independent cohort of older adults, CSF apoE complexes of APOE4 -positive individuals were smaller than those of the APOE4 -negative individuals. Compared to individuals with no dementia, those with the mildest stages of dementia had similar sized CSF apoE complexes. These results identify a novel phenotypic difference in the size of CSF apoE complexes in middle age that correlate with the risk of AD later in life.
Keywords: Alzheimer disease, Cerebrospinal fluid, Clinical dementia rating, Cognitively normal, Lipoproteins, Risk factor
INTRODUCTION
Several apolipoproteins are found in association with lipids in the cerebrospinal fluid (CSF), including apolipoprotein E (apoE), apolipoprotein J, and apolipoprotein A–I ( 1 , 2 ). The most abundant apolipoprotein in the CSF, apoE, is a 34-kDa protein produced by astrocytes and secreted into the brain parenchyma ( 3 , 4 ). Newly synthesized astrocyte particles contain little lipid and are discoidal in shape, but by the time they reach the CSF, these particles have more spherical shapes and contain a cholesterol ester core ( 5 ). Nondenaturing gel electrophoresis has revealed that apoE lipoproteins secreted from astrocytes or present in the CSF ranged from 8 to 17 nm in diameter ( 6–8 ), which is comparable to the size of the HDL lipoproteins found in the plasma ( 9 ). However, not much is known about differences in the size of CSF lipoproteins among various groups of individuals.
In humans, apoE exists in 3 isoforms ( APOE2 , APOE3 , APOE4 ), which differ at amino acid residues 112 and 158. APOE4 is the strongest genetic risk factor for AD, while APOE2 strongly decreases a person’s risk compared to APOE3 ( 10 ). Because apoE2 is a more efficient acceptor of cholesterol efflux than apoE3 or apoE4, and because apoE3 can induce cholesterol efflux 3–4 times better than apoE4 ( 11 , 12 ), endogenous apoE4 lipoproteins might be less lipidated than apoE3 or apoE2 particles. In mouse brains, viral-mediated overexpression of apoE2 increased the average size of apoE lipoproteins, whereas overexpression of apoE4 decreased the average size of apoE lipoproteins ( 13 ). Here, we use nondenaturing gel electrophoresis to identify APOE genotypic differences in the size distribution of human CSF apoE that is complexed to other molecules both in middle aged and in older adults. We report that, in comparison to APOE 3.3 individuals, APOE 2.3 individuals have larger apoE complexes and individuals with an APOE4 allele have smaller apoE complexes. These findings suggest that the size distribution of apoE in CSF may serve as a biomarker correlating early measures with an individual’s risk of developing AD later in life.
MATERIALS AND METHODS
CSF Samples
Sixty-four anonymized human CSF samples and clinical information of the individuals were obtained from lumbar punctures at Washington University in St. Louis as part of a longitudinal study of CSF biomarkers in middle aged and preclinical AD individuals ( Table ) ( 14 ). Subjects had APOE genotypes of 2.3, 3.3, 3.4 or 4.4 with an average age of 54.5 years, and all had a Clinical Dementia Rating (CDR) of 0, indicating no cognitive impairment. Of these 64 young control CSF samples, [ 11 C]PiB PET imaging was conducted on 55 (86%); of those, 54 (98%) were scored as [ 11 C]PiB-negative. Another set of 65 CSF samples of an older cohort with an average age of 68.0 years was also obtained from Washington University ( Table ). These subjects had APOE genotypes of 3.3, 3.4, or 4.4 and had CDR scores of 0, 0.5, or 1 indicating no dementia, very mild dementia, or mild dementia. Of this older cohort, relatively fewer of the CDR 0 individuals had [ 11 C]PiB PET imaging (20/26, 74%), and a smaller proportion were scored as [ 11 C]PiB-negative (80%). All of the [ 11 C]PiB-positive CDR 0 individuals were APOE4.4 , as might be expected given the risk of amyloid accumulation in this population ( 15 ).
TABLE.
Characteristics of Individuals Contributing CSF Samples
| APOE genotype | CDR score | Average age (years±SD) | Male/female | Number of samples |
|---|---|---|---|---|
| 2.3 | 0 | 55.5±3.5 | 7/10 | 17 |
| 3.3 | 0 | 54.9±4.2 | 5/16 | 21 |
| 3.4 | 0 | 54.1±3.3 | 9/12 | 21 |
| 4.4 | 0 | 51.6±3.7 | 1/4 | 5 |
| 3.3 | 0 | 68.5±7.1 | 6/4 | 10 |
| 0.5 | 65.7±4.4 | 5/5 | 10 | |
| 1 | 72.8±6.4 | 4/1 | 5 | |
| 3.4 | 0 | 69.6±5.6 | 6/3 | 9 |
| 0.5 | 68.0±2.8 | 6/4 | 10 | |
| 1 | 68.5±10.1 | 3/2 | 5 | |
| 4.4 | 0 | 69.7±6.1 | 4/3 | 7 |
| 0.5 | 63.8±10.3 | 3/2 | 5 | |
| 1 | 69.5±10.3 | 2/2 | 4 |
Native Gel Electrophoresis
About 1 μL of CSF was added to 2.5 μL of NativePage 4× loading buffer and diluted with phosphate-buffered saline (PBS) to a total volume of 10 μL. Proteins were separated on Native Page 4–16% Bis–Tris gels (Invitrogen, Billerica, MA) at 125 V for 2 hours and transferred to polyvinylidene fluoride membrane (Millipore Billerica, MA) at 100 V for 2 hours. NativeMark Unstained Protein Standard (Invitrogen) was used to estimate protein complex size. After transfer, membranes were treated with boiling PBS 2 times for 5 minutes each and then 10% formic acid for 7 minutes. Membranes were then blocked overnight in 10% nonfat milk at 4 °C and subsequently incubated overnight at 4 °C with goat anti-apoE antibody (1:1000) (Calbiochem, San Diego, CA, Catalog #178479). Blots were washed with PBS-0.5% Tween-20 4 times for 7 minutes each and incubated with peroxidase-conjugated AffiniPure Rabbit Anti-Goat antibody (1:2500) (Jackson ImmunoResearch, West Grove, PA). Membranes were developed with SuperSignal West Dura (Thermo Scientific, Waltham, MA) and imaged via Amersham Imager 600. Nondenaturing polyacrylamide gel electrophoresis provides a relatively convenient method that allows separation of a tight range of particle sizes compared to other techniques such as density gradient electrophoresis, high performance liquid chromatography (either using gel filtration or anion exchange columns) ( 16 ) or dynamic light scattering ( 17 ). Nondenaturing polyacrylamide gel electrophoresis is particularly useful because it does not require isolation of fractions through ultracentrifugation or column separation, which have reduced ability to separate particles of subtle size differences ( 16 ). This approach has previously been used to study apoE complexes in brain samples ( 8 , 13 , 18 ).
Statistical Analysis
Plot profiles for each CSF sample were generated using ImageJ and normalized using GraphPad Prism 4 so that the darkest point of each lane was designated at 100%, defined as the point of “maximum apoE”. The x -axis of the plot profiles corresponds to the distance from the 1000-kDa molecular weight marker, where the 1000-kDa band is 0 and 450-kDa band is 0.83 in ( Fig. 1 ). The plot profiles were then averaged by APOE genotype and normalized so that the position of maximum apoE was equal to 100%. In addition to the average plot profiles, the area under the curve of the plot profile of each individual sample was summed and the distance from the 1000 kDa was calculated for the largest 25%, 50% and 75% of the total apoE. Individual data points for the positions at maximum, 25%, 50%, and 75% apoE were statistically analyzed with one-way ANOVA’s using Graph Pad Prism 4. Tukey’s multiple comparison test with p < 0.05 significance (*p < 0.05, **p < 0.01, ***p < 0.001) was used for post hoc analyses. For comparisons of 2 groups, a Student t -test was conducted. Averages are reported as ± SD.
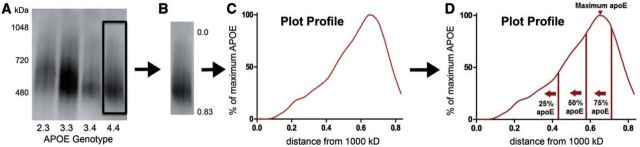
FIGURE 1.
Quantification of NativePage gel electrophoresis assay. (A) Nondenatured proteins from human CSF samples were separated using NativePage gel electrophoresis and immunostained for apoE. (B) A plot profile of each sample was generated starting from the 1000-kDa marker (0.0) and ending below the apoE staining under the 480-kDa marker (0.83). (C) Each plot profile was normalized so that the darkest apoE staining was the point of maximum apoE. (D) An example plot profile of the different measurements used in the data analysis.
RESULTS
We were interested in studying the effects of APOE genotype on CSF apoE complexes before the onset of AD pathology in order to identify biomarkers that correlated with an individual’s risk of developing AD. Sixty-four human CSF samples from nondemented individuals with APOE genotypes of 2.3 ( n = 17), 3.3 ( n = 21), 3.4 ( n = 21), or 4.4 ( n = 5) were analyzed ( Table ).
Using NativePage nondenaturing gel electrophoresis of these CSF samples, most of the apoE was in complexes between 1000 and 400 kDa in size ( Fig. 1A ), with <5% apoE at its molecular weight of 34 kDa (data not shown). The largest majority of the apoE runs between the 720- and 480-kDa molecular weight markers, corresponding to complexes between ∼17 and 12 nm in size ( 19 , 20 ). This size distribution of apoE complexes was consistent with patterns seen from apoE isolated from mouse brain ( 18 ) and human CSF ( 7 , 9 ). In order to compare the distributions of apoE complex sizes, we used the Plot Profile feature on ImageJ to generate a graph for each sample based on the intensity of the apoE bands ( Fig. 1B, C ). The x -axis of each plot spans from 1000 to ∼450 kDa ( Fig. 1B ); positions closer to 0.0 correspond to larger complexes, and positions closer to 0.83 correspond to smaller complexes. Because we wanted to assess patterns in the apoE complexes and not relative levels of apoE in the CSF samples, the plot profile data was then normalized so that the darkest point of each lane was 100%, or the point of “maximum apoE” ( Fig. 1C, D ). For the 64 CSF samples analyzed, the average maximum apoE was 0.55 ± 0.08, corresponding to complexes ∼500–600 kDa in size. From the plot profile, the area under each curve was summed, and the x -axis position of the largest quartile, median, and smallest quartile of apoE complexes were identified. We refer to these points as the largest 25%, 50% and 75% of the total apoE ( Fig. 1D ).
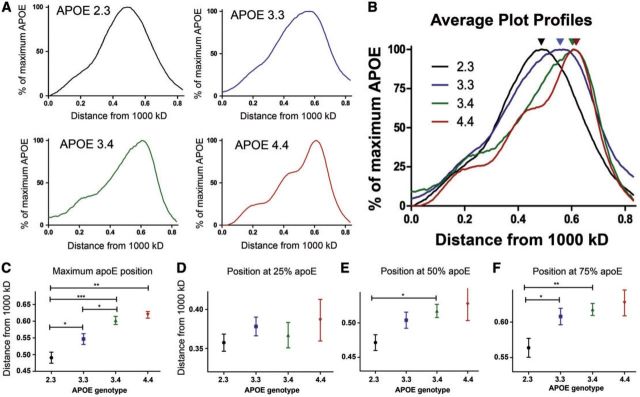
The plot profiles of each CSF sample were grouped by APOE genotype and averaged ( Fig. 2A ). Individuals with the APOE 2.3 genotype had a broad spread of apoE complexes between 720 and 480 kDa. Those with an APOE 3.3 genotype had a similar broad range of apoE complexes, with slightly smaller sizes. Individuals with an APOE4 allele (3.4 or 4.4 genotype) had the smallest complexes, with the average complexes <500 kDa. Qualitatively, each APOE genotype seemed to have a unique pattern of apoE that corresponded to differences in the size of complexes. The APOE 2.3 and APOE 3.3 groups had 1 broad peak while the APOE 3.4 s and APOE 4.4 s had a narrower main peak with several small peaks of larger sizes. The differences in the distribution of the apoE complexes are best seen when the average plot profiles are overlaid, particularly in the position of the maximum apoE indicated by the arrowheads ( Fig. 2B ). The APOE 2.3 graphs were shifted to the left, while the APOE4 genotypes were more shifted to the right compared to the APOE 3.3 genotype.
FIGURE 2.
The size of apoE complexes varies by APOE genotype in a middle aged cohort. Sixty-four CSF samples were analyzed and grouped by APOE genotype ( APOE 2.3 , n = 17; APOE 3.3 , n = 21; APOE 3.4 , n = 21; APOE 4.4 , n = 5). The positions at maximum, 25%, 50%, and 75% apoE were found from the plot profile of each sample. (A) The normalized plot profile of each sample was averaged by genotype. (B) An overlaid image of the average plot profiles for each APOE genotype. Arrows indicate the position of maximum apoE. (C–F) The x values for the point at (C) maximum apoE, (D) 25% apoE, (E) 50% apoE, and (F) 75% apoE were averaged and analyzed using a one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001).
We then quantified the position at maximum apoE for each individual sample and averaged these numbers by APOE genotype. Individuals with an APOE 2.3 genotype had an average position of maximum apoE at 0.49 ± 0.07, which corresponds to an average apoE complex of ∼600–650 kDa. APOE 3.3 individuals had an average position of maximum apoE at 0.55 ± 0.07, which corresponds to apoE complexes between ∼500 and 600 kDa. The APOE 3.4 and 4.4 individuals had average positions of maximum apoE at 0.60 ± 0.06 and 0.62 ± 0.02, respectively, corresponding to complexes of ∼500 kDa ( Fig. 2C ). The APOE 2.3 complexes were significantly larger than each of the APOE 3.3 , 3.4 and 4.4 complexes (p < 0.05, p < 0.001, p < 0.001, respectively), and APOE 3.3 complexes were significantly larger than the APOE 3.4 complexes (p < 0.05). Thus, CSF complexes from APOE 2.3 individuals were ∼100 kDa larger on an average than complexes from APOE 3.4 and APOE 4.4 individuals, a statistically significant difference in size.
Next, we tested whether there were differences in distribution patterns of the subpopulations of apoE-containing complexes by examining the positions of the largest 25%, 50% or 75% apoE. We determined the area under the curve of each of the average plot profiles, and found the positions of the largest 25%, 50%, and 75% of the total apoE ( Fig. 1D ). We did not detect a significant difference in the position at 25% apoE by APOE genotype ( Fig. 2D ), indicating no difference in the relative levels of the largest apoE complexes. There were significant differences in the relative levels between APOE 2.3 and APOE 3.4 CSF samples in the positions at 50% and 75% apoE ( Fig. 2E, F ). There was also a significant difference in the size of the largest 75% of apoE complexes between the APOE 2.3 and the APOE 3.3 genotypes ( Fig. 2F ). These data demonstrate that the main differences in CSF complex sizes are in the smaller complexes, which were most abundant in individuals carrying the APOE4 allele. These results show that the size of CSF apoE-containing complexes increase with an APOE2 allele and decrease with an APOE4 allele in cognitively normal individuals in their 50s.
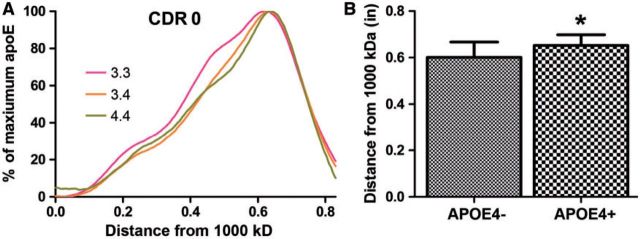
After analyzing this cohort of individuals, we asked whether the same pattern of apoE complex distribution by APOE genotype existed in an older cohort of control individuals or in a cohort demonstrating cognitive decline. This experiment tested whether the correlation between APOE genotype and size of apoE complex was maintained, strengthened, or lost with the onset of clinical disease to determine whether smaller complexes correlated with APOE4 through the earliest stages of AD. We examined CSF samples from 65 individuals with an average age of 68.0 years who had CDR scores of 0, 0.5 or 1, and APOE genotypes of 3.3, 3.4, or 4.4 ( Table ). For this set, we did not have CSF from individuals of the APOE 2.3 genotype. Again, we used nondenaturing gel electrophoresis and developed plot profiles to quantify the distribution of apoE-containing complexes. In the older cohort of cognitively normal individuals (CDR 0), we observed the same pattern of apoE complex sizes as in the younger cohort ( Fig. 3A ). The APOE 3.3 individuals had an average position of maximum apoE at 0.60 ± 0.07, corresponding to complexes ∼500 kDa in size, while APOE 3.4 and 4.4 individuals had an average position of maximum apoE at 0.64 ± 0.05 and 0.67 ± 0.04, respectively, which are both <480 kDa in size. Overall, the average migration of apoE complexes for all 3 APOE genotypes was slightly lower in the older cohort compared to the younger cohort ( Fig. 2C ), suggesting an effect of aging on the decreasing the size of CSF apoE complexes.
FIGURE 3.
The sizes of apoE complexes vary by APOE genotype in an older cohort without dementia. Twenty-six CSF samples were analyzed as in Figure 2 , and grouped by APOE genotype ( APOE 3.3 , n = 10; APOE 3.4 , n = 9; APOE 4.4 , n = 7). (A) Plot profiles from individuals with CDR scores of 0 were overlaid by APOE genotype. (B) The position of maximum apoE for APOE4 -positive individuals was significantly different from APOE4 -negative individuals (Student t -test, *p < 0.05).
There was a significant difference between the size of apoE-containing complexes from CDR 0 individuals who were APOE4 -negative vs. those who were APOE4 -positive ( Fig. 3B , p < 0.05). These data reproduced the finding from the younger cohort that apoE4 was part of smaller CSF complexes ( Fig. 2C ). There were no significant differences in the distribution of the largest 25%, 50%, or 75% of the complexes (data not shown).
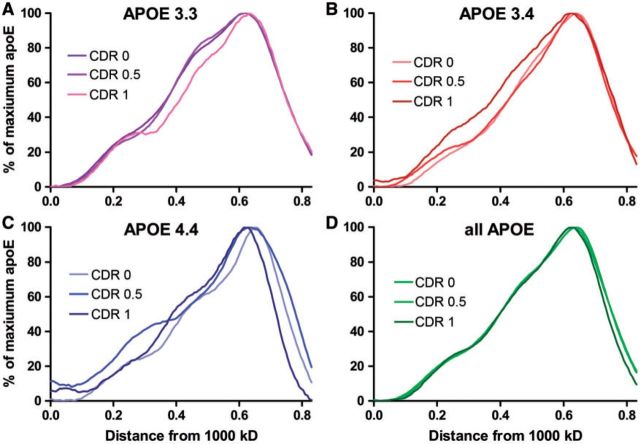
Next, we tested whether the early stages of dementia also affected the size distribution of apoE complexes. Within each of the APOE genotypes there were no significant differences in the position of maximum apoE between individuals with CDR scores of 0, 0.5, and 1 ( Fig. 4A–C ). Similarly, when we combined all of the APOE genotypes together and took the average plot profile by CDR score, there was no significant difference in the distribution of CSF apoE complexes at different stages of disease progression ( Fig. 4D ). Because there was no effect of CDR score on CSF apoE complex distribution, we compared CSF apoE complexes from individuals of the 3 APOE genotypes, combining the CDR scores. As with the CDR 0 subgroup, the position of maximum apoE of the APOE4 -positive individuals was significantly different from the APOE4 -negative individuals, with the APOE4 group showing smaller complexes (p < 0.03) (data not shown). Thus, although APOE genotype reproducibly affected the size of CSF apoE complexes, the stages of dementia did not have an effect.
FIGURE 4.
The sizes of apoE complexes in the older cohort do not differ by stage of dementia. (A–C)APOE 3.3(A) , 3.4(B) , and 4.4(C) samples ( Table ) were separated by CDR score and the average plot profiles were overlaid by APOE genotype. (D) All APOE genotypes were combined and analyzed by CDR scores 0, 0.5, and 1. No significant differences were observed among these grouped samples.
DISCUSSION
We report here APOE genotype-specific patterns of CSF apoE complex distribution in independent cohorts of middle aged and older adults, based on nondenaturing gel electrophoresis. In the middle-aged cohort, CSF apoE complexes from those with an APOE2 allele were significantly larger than those from APOE 3.3 individuals, who had significantly larger apoE complexes than those with an APOE4 allele. A similar pattern was also seen in a cohort over a decade older, which included APOE 3.3 , 3.4 , and 4.4 individuals. Patterns within individual APOE genotypes were not affected by the development or progression of dementia, as judged by CDR scores. We hypothesize that differences in the composition of these apoE-containing complexes may be responsible for differences in risk for AD. Indeed, these results suggest that more apoE4 is associated with smaller complexes in the CNS, and thus may be deficient in clearing lipid debris or delivering lipid to CNS cells, compared to apoE2 and apoE3.
Lipid metabolism is important in the context of neuronal injury and neurodegeneration. ApoE is upregulated following neuronal injury ( 21 , 22 ) and plays a role in cell survival and protection from neuronal injury and stress ( 23–25 ). APOE 4.4 AD patients have less neuronal reparative capacity compared to APOE 3.4 patients, who have less reparative capacity compared to APOE 3.3 patients ( 26 ). In mice, APOE4 was associated with impaired neuronal plasticity after entorhinal cortex injury compared to APOE3 ( 27 ), as well as with poorer outcomes after traumatic brain injury ( 28 ) and more cognitive deficits after experimental autoimmune encephalomyelitis ( 29 ). CNS apoE lipoproteins can clear lipids from dead or injured cells and deliver those lipids to neurons and astrocytes ( 30 ). Smaller CSF lipoprotein particles could be indicative of less lipid clearance from some locations and less lipid delivery to cells for membrane synthesis or other cellular functions. This hypothesis is consistent with other data that shows that apoE3 lipoproteins can enhance neurite outgrowth significantly better than apoE4 lipoproteins ( 31 , 32 ).
Lipid efflux via the ATP-binding cassette A1 (ABCA1) transporter is particularly important in AD. ABCA1 facilitates cholesterol efflux to apoE ( 33 ), and apoE4 is less efficient at this efflux in cell culture compared to apoE2 or apoE3 (11, 12). In mouse models, deletion of the ABCA1 gene decreases levels of apoE and increases deposition of Aβ in the amyloid plaques ( 34 , 35 ). Similarly, in humans, mutations in the ABCA1 gene are associated with increased risk of AD and decreased Aβ in the CSF, which is a core biomarker of AD ( 36 ), and the efficiency of ABCA1-mediated cholesterol efflux in the CSF is impaired in patients with mild cognitive impairment or AD ( 37 ). Activating expression of ABCA1 through the RXR agonist bexarotene has been explored as a therapeutic target because of its ability to decrease levels of Aβ plaques and improve cognition in mouse models of AD ( 38 ). Overexpression of ABCA1 may increase Aβ clearance in the brain ( 39 ) because increased lipidation of apoE facilitates proteolytic degradation of Aβ ( 40 ). Our data only address the size of apoE-containing complexes, which may represent forms of apoE on single or multiple lipoproteins, or apoE complexed to other molecules. However, these data support the hypothesis that the impaired formation of CNS lipoproteins may be an important risk factor of AD and that measuring lipidation levels might provide insight to an individual’s risk of AD.
These results highlight the importance of identifying differences between people at high and low risk of developing AD and of identifying biomarkers for high risk individuals. Currently, there are several well-accepted biomarkers of AD based on amyloid PET imaging and CSF protein levels. Decreased CSF levels of Aβ42 are associated with a greater risk of AD ( 41 , 42 ), and increased CSF levels of phosphorylated tau and total tau are seen in early stages of AD ( 43 , 44 ). These biomarkers based on AD neuropathological changes may help define the progression from β-amyloid and tau accumulation to clinical manifestations of AD ( 45 ). We tested whether the size of apoE complexes affected the transition from control to the earliest stages of dementia, but found that apoE complex size did not affect disease progression at this stage. Identifying biomarkers related to genetic risk of AD (specifically APOE ) early in life is a related but independent goal, potentially allowing early interventions to prevent disease. Focus on CSF lipoproteins is supported by data for the potential usefulness of measures of the CSF apolipoproteins apoE ( 46 ) and apoJ (clusterin) ( 47 ). Detailed examination of the protein and lipid composition of CSF lipoproteins may help in establishing new biomarkers correlating with the risk of amyloid accumulation prior to AD neuropathological changes.
Our observation that APOE genotype significantly affects the size of apoE complexes in 2 independent cohorts supports the hypothesis that there is a structural difference in CNS apoE isoform complexes in control individuals. A more quantitative analysis of the structure and composition of CSF apoE lipoproteins in the future would help evaluate whether their characteristics could be useful as biomarkers of AD risk. Moreover, if there are elements of apoE lipoproteins that are useful as preclinical biomarkers, then altering their characteristics through drugs or lifestyle changes might modify a person’s risk for AD before the onset of neuropathological alterations associated with AD.
This work was supported by NIH P01 AG030128. Mariam Gachechiladze was supported by a Neale-Oppenheimer Fellowship from the Department of Biology at Georgetown University. Cerebrospinal fluid samples were obtained from the Alzheimer’s Disease Research Center, Washington University in St. Louis, P50AG005681, P01AG003991 and PO1AG026276.
Conflict of interest: The authors have no duality or conflicts of interest to declare.
REFERENCES
- 1. Koch S, Donarski N, Goetze K , et al. . Characterization of four lipoprotein classes in human cerebrospinal fluid . J Lipid Res 2001. ; 42 : 1143 – 51 [PubMed] [Google Scholar]
- 2. Roheim PS, Carey M, Forte T , et al. . Apolipoproteins in human cerebrospinal fluid . Proc Natl Acad Sci USA 1979. ; 76 : 4646 – 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boyles JK, Pitas RE, Wilson E , et al. . Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system . J Clin Invest 1985. ; 76 : 1501 – 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pitas RE, Boyles JK, Lee SH , et al. . Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins . Biochim Biophys Acta 1987. ; 917 : 148 – 61 [DOI] [PubMed] [Google Scholar]
- 5. LaDu MJ, Gilligan SM, Lukens JR , et al. . Nascent astrocyte particles differ from lipoproteins in CSF . J Neurochem 1998. ; 70 : 2070 – 81 [DOI] [PubMed] [Google Scholar]
- 6. DeMattos RB, Brendza RP, Heuser JE , et al. . Purification and characterization of astrocyte-secreted apolipoprotein E and J-containing lipoproteins from wild-type and human apoE transgenic mice . Neurochem Int 2001. ; 39 : 415 – 25 [DOI] [PubMed] [Google Scholar]
- 7. Holtzman DM, Bales KR, Wu S , et al. . Expression of human apolipoprotein E reduces amyloid-beta deposition in a mouse model of Alzheimer’s disease . J Clin Invest 1999. ; 103 : R15 – 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ulrich JD, Burchett JM, Restivo JL , et al. . In vivo measurement of apolipoprotein E from the brain interstitial fluid using microdialysis . Mol Neurodegener 2013. ; 8 : 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fagan AM, Holtzman DM, Munson G , et al. . Unique lipoproteins secreted by primary astrocytes from wild type, apoE (-/-), and human apoE transgenic mice . J Biol Chem 1999. ; 274 : 30001 – 7 [DOI] [PubMed] [Google Scholar]
- 10. Liu CC, Kanekiyo T, Xu H , et al. . Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy . Nat Rev Neurol 2013. ; 9 : 106 – 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Michikawa M, Fan QW, Isobe I , et al. . Apolipoprotein E exhibits isoform-specific promotion of lipid efflux from astrocytes and neurons in culture . J Neurochem 2000. ; 74 : 1008 – 16 [DOI] [PubMed] [Google Scholar]
- 12. Minagawa H, Gong JS, Jung CG , et al. . Mechanism underlying apolipoprotein E (ApoE) isoform-dependent lipid efflux from neural cells in culture . J Neurosci Res 2009. ; 87 : 2498 – 508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu J, Liu CC, Chen XF , et al. . Opposing effects of viral mediated brain expression of apolipoprotein E2 (apoE2) and apoE4 on apoE lipidation and Abeta metabolism in apoE4-targeted replacement mice . Mol Neurodegener 2015. ; 10 : 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sutphen CL, Jasielec MS, Shah AR , et al. . Longitudinal cerebrospinal fluid biomarker changes in preclinical Alzheimer disease during middle age . JAMA Neurol 2015. ; 72 : 1029 – 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jansen WJ, Ossenkoppele R, Knol DL , et al. . Prevalence of cerebral amyloid pathology in persons without dementia: A meta-analysis . Jama 2015. ; 313 : 1924 – 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hafiane A, Genest J. High density lipoproteins: Measurement techniques and potential biomarkers of cardiovascular risk . BBA Clin 2015. ; 3 : 175 – 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakurai T, Trirongjitmoah S, Nishibata Y , et al. . Measurement of lipoprotein particle sizes using dynamic light scattering . Ann Clin Biochem 2010. ; 47 : 476 – 81 [DOI] [PubMed] [Google Scholar]
- 18. Boehm-Cagan A, Michaelson DM. Reversal of apoE4-driven brain pathology and behavioral deficits by bexarotene . J Neurosci 2014. ; 34 : 7293 – 301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rainwater DL, Moore PH, Jr., Shelledy WR , et al. . Characterization of a composite gradient gel for the electrophoretic separation of lipoproteins . J Lipid Res 1997. ; 38 : 1261 – 6 [PubMed] [Google Scholar]
- 20. Wittig I, Beckhaus T, Wumaier Z , et al. . Mass estimation of native proteins by blue native electrophoresis: Principles and practical hints . Mol Cell Proteomics 2010. ; 9 : 2149 – 61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ignatius MJ, Gebicke-Harter PJ, Skene JH , et al. . Expression of apolipoprotein E during nerve degeneration and regeneration . Proc Natl Acad Sci U S A 1986. ; 83 : 1125 – 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Snipes GJ, McGuire CB, Norden JJ , et al. . Nerve injury stimulates the secretion of apolipoprotein E by nonneuronal cells . Proc Natl Acad Sci U S A 1986. ; 83 : 1130 – 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitagawa K, Matsumoto M, Hori M , et al. . Neuroprotective effect of apolipoprotein E against ischemia . Ann N Y Acad Sci 2002. ; 977 : 468 – 75 [DOI] [PubMed] [Google Scholar]
- 24. Lee Y, Aono M, Laskowitz D , et al. . Apolipoprotein E protects against oxidative stress in mixed neuronal-glial cell cultures by reducing glutamate toxicity . Neurochem Int 2004. ; 44 : 107 – 18 [DOI] [PubMed] [Google Scholar]
- 25. Zhou S, Wu H, Zeng C , et al. . Apolipoprotein E protects astrocytes from hypoxia and glutamate-induced apoptosis . FEBS Lett 2013. ; 587 : 254 – 8 [DOI] [PubMed] [Google Scholar]
- 26. Arendt T, Schindler C, Bruckner MK , et al. . Plastic neuronal remodeling is impaired in patients with Alzheimer’s disease carrying apolipoprotein epsilon 4 allele . J Neurosci 1997. ; 17 : 516 – 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. White F, Nicoll JA, Roses AD , et al. . Impaired neuronal plasticity in transgenic mice expressing human apolipoprotein E4 compared to E3 in a model of entorhinal cortex lesion . Neurobiol Dis 2001. ; 8 : 611 – 25 [DOI] [PubMed] [Google Scholar]
- 28. Mannix RC, Zhang J, Park J , et al. . Age-dependent effect of apolipoprotein E4 on functional outcome after controlled cortical impact in mice . J Cereb Blood Flow Metab 2011. ; 31 : 351 – 61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tu JL, Zhao CB, Vollmer T , et al. . APOE 4 polymorphism results in early cognitive deficits in an EAE model . Biochem Biophys Res Commun 2009. ; 384 : 466 – 70 [DOI] [PubMed] [Google Scholar]
- 30. Rebeck GW, Alonzo NC, Berezovska O , et al. . Structure and functions of human cerebrospinal fluid lipoproteins from individuals of different APOE genotypes . Exp Neurol 1998. ; 149 : 175 – 82 [DOI] [PubMed] [Google Scholar]
- 31. Fagan AM, Bu G, Sun Y , et al. . Apolipoprotein E-containing high density lipoprotein promotes neurite outgrowth and is a ligand for the low density lipoprotein receptor-related protein . J Biol Chem 1996. ; 271 : 30121 – 5 [DOI] [PubMed] [Google Scholar]
- 32. Nathan BP, Bellosta S, Sanan DA , et al. . Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro . Science 1994. ; 264 : 850 – 2 [DOI] [PubMed] [Google Scholar]
- 33. Hirsch-Reinshagen V, Zhou S, Burgess BL , et al. . Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain . J Biol Chem 2004. ; 279 : 41197 – 207 [DOI] [PubMed] [Google Scholar]
- 34. Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice . J Biol Chem 2005. ; 280 : 43224 – 35 [DOI] [PubMed] [Google Scholar]
- 35. Wahrle SE, Jiang H, Parsadanian M , et al. . Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease . J Biol Chem 2005. ; 280 : 43236 – 42 [DOI] [PubMed] [Google Scholar]
- 36. Reynolds CA, Hong MG, Eriksson UK , et al. . A survey of ABCA1 sequence variation confirms association with dementia . Hum Mutat 2009. ; 30 : 1348 – 54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yassine HN, Feng Q, Chiang J , et al. . ABCA1-mediated cholesterol efflux capacity to cerebrospinal fluid is reduced in patients with mild cognitive impairment and Alzheimer’s disease . J Am Heart Assoc 2016. ; 5 : 1 – 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cramer PE, Cirrito JR, Wesson DW , et al. . ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models . Science 2012. ; 335 : 1503 – 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wahrle SE, Jiang H, Parsadanian M , et al. . Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease . J Clin Invest 2008. ; 118 : 671 – 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang Q, Lee CY, Mandrekar S , et al. . ApoE promotes the proteolytic degradation of Abeta . Neuron 2008. ; 58 : 681 – 93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fagan AM, Mintun MA, Mach RH , et al. . Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans . Ann Neurol 2006. ; 59 : 512 – 9 [DOI] [PubMed] [Google Scholar]
- 42. Grimmer T, Riemenschneider M, Forstl H , et al. . Beta amyloid in Alzheimer’s disease: Increased deposition in brain is reflected in reduced concentration in cerebrospinal fluid . Biol Psychiatry 2009. ; 65 : 927 – 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blennow K, Hampel H. CSF markers for incipient Alzheimer’s disease . Lancet Neurol 2003. ; 2 : 605 – 13 [DOI] [PubMed] [Google Scholar]
- 44. Blennow K, Hampel H, Weiner M , et al. . Cerebrospinal fluid and plasma biomarkers in Alzheimer disease . Nat Rev Neurol 2010. ; 6 : 131 – 44 [DOI] [PubMed] [Google Scholar]
- 45. Bateman RJ, Xiong C, Benzinger TL , et al. . Clinical and biomarker changes in dominantly inherited Alzheimer’s disease . N Engl J Med 2012. ; 367 : 795 – 804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cruchaga C, Kauwe JS, Nowotny P , et al. . Cerebrospinal fluid APOE levels: An endophenotype for genetic studies for Alzheimer’s disease . Hum Mol Genet 2012. ; 21 : 4558 – 71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deming Y, Xia J, Cai Y , et al. . A potential endophenotype for Alzheimer’s disease: Cerebrospinal fluid clusterin . Neurobiol Aging 2016. ; 37 : 208 e1 – 9 [DOI] [PMC free article] [PubMed] [Google Scholar]