Abstract
Tissue‐resident macrophages (MΦTR) originate from at least two distinct waves of erythro‐myeloid progenitors (EMP) arising in the yolk sac (YS) at E7.5 and E8.5 with the latter going through a liver monocyte intermediate. The relative potential of these precursors in determining development and functional capacity of MΦTR remains unclear. Here, we studied development of alveolar macrophages (AM) after single and competitive transplantation of different precursors from YS, fetal liver, and fetal lung into neonatal Csf2ra −/− mice, which lack endogenous AM. Fetal monocytes, promoted by Myb, outcompeted primitive MΦ (pMΦ) in empty AM niches and preferentially developed to mature AM, which is associated with enhanced mitochondrial respiratory and glycolytic capacity and repression of the transcription factors c‐Maf and MafB. Interestingly, AM derived from pMΦ failed to efficiently clear alveolar proteinosis and protect from fatal lung failure following influenza virus infection. Thus, our data demonstrate superior developmental and functional capacity of fetal monocytes over pMΦ in AM development and underlying mechanisms explaining replacement of pMΦ in fetal tissues.
Keywords: alveolar macrophages, fetal monocytes, metabolism, primitive macrophages
Subject Categories: Immunology, Metabolism
Transplantation studies aiming to identify the origin of tissue alveolar macrophages implicate a contribution of fetal monocytes with higher mitochondrial bioenergetic and glycolytic capacity than primitive macrophages.

Introduction
Tissue‐resident macrophages (MΦTR) in adults are multifunctional and extremely heterogeneous cells. They are present in almost all tissues and contribute to both host defense and local homeostasis (Davies et al, 2013; Ginhoux et al, 2016; Perdiguero & Geissmann, 2016). MΦTR arise from several sequential waves of embryonic precursors (Ginhoux & Guilliams, 2016; Palis, 2016; Perdiguero & Geissmann, 2016). In a first wave elicited in the extra‐embryonic yolk sac (YS) at embryonic day 7.5 (E7.25), primitive erythro‐myeloid progenitors (EMP) are generated and differentiated into primitive macrophages (pMΦ) at E9.0 before seeding all fetal tissues, where they appear as a population of F4/80hiCD11blo cells (Mucenski et al, 1991; Schulz et al, 2012; Hoeffel et al, 2015). YS‐derived pMΦ represent the primary source of microglia and generate a fraction of Langerhans cells, whereas the progenitors of most other MΦTR emerge subsequently, suggesting a distinct origin (Ginhoux et al, 2010; Hoeffel et al, 2012, 2015; Epelman et al, 2014; Gomez Perdiguero et al, 2015). Indeed, a second transient definitive wave of EMP has been suggested to arise in the hemogenic endothelium of the YS at E8.25, which colonize the fetal liver starting at E9.5 and there differentiate into fetal monocytes (Hoeffel et al, 2015; McGrath et al, 2015a). Whether or not EMPs are Myb‐dependent has been discussed controversially (Schulz et al, 2012; Hoeffel et al, 2015). Myb‐dependent hematopoietic stem cells (HSC) emerge in the aorta‐gonad‐mesonephros (AGM) around E10.5 and seed the fetal liver between E11.5 and E12.5. It is possible that HSC also could give rise to monocytes in the fetal liver then contribute to the MΦTR pool (Hoeffel & Ginhoux, 2015; Hoeffel et al, 2015; McGrath et al, 2015a,b). Apart from the brain, which is populated by pMΦ‐derived microglia, fetal liver monocytes seed the large majority of tissues starting from E13.5, where they expand and differentiate into MΦTR, thereby replacing pMΦ (Hoeffel et al, 2015). A fate‐mapping approach exploiting the expression of S100a4 provided further evidence for the hypothesis that, apart from microglia, most embryonic‐derived MΦ arise from fetal liver monocytes (Hoeffel et al, 2015). Moreover, adoptive cell transfer studies revealed that fetal liver monocytes have an advantage in colonizing the alveolar niche compared with pMΦ (Guilliams et al, 2013; Schneider et al, 2014b; van de Laar et al, 2016), although pMΦ can give rise to AM under some circumstances (van de Laar et al, 2016).
The underlying mechanism(s) for a potential advantage of fetal liver monocytes over pMΦ in MΦTR development remains unclear. Moreover, a key question in the field is whether pMΦ‐derived MΦTR and fetal monocyte‐derived MΦTR have different functional capacities in homeostasis and after challenge.
Alveolar macrophages (AM) comprise a subset of MΦTR localized in the terminal sacs of the lung airways, the alveoli, where they engulf inhaled particulate matter and microorganisms. Besides defense against pathogens, they have a vital function in supporting air exchange through the uptake and catabolism of surfactant produced constantly by type 2 alveolar epithelial cells, and by the removal of cellular debris accumulating in the alveoli (Kopf et al, 2015). The absence or dysfunction of AM in mice and humans results in the development of pulmonary alveolar proteinosis and accumulation of dead cells, which can result in lung failure and death during respiratory viral infection. GM‐CSF is essential for AM development, where it induces the transcription factor PPARγ in fetal lung monocytes. Indeed, the absence of the GM‐CSF receptor or PPARγ abrogates perinatal development of AM, which can be completely restored by transfer of wild‐type (WT) fetal lung monocytes (Guilliams et al, 2013; Schneider et al, 2014b).
In the present study, we have transferred EMP (from YS at E10.5), pMΦ, and fetal monocytes (both from liver and lung) either separately or in a competitive setting into Csf2ra −/− or Csf2rb −/− neonates in order to directly compare their capacities to restore development and function of mature AM. Our results revealed that pMΦ are strikingly impaired compared with fetal monocytes in AM development. pMΦ‐derived AM fail to fully replenish the empty niches, efficiently clear alveolar proteinosis, and protect mice from morbidity and lung failure following influenza infection. The residual developmental capacity of pMΦ waned with migration from YS to liver and with days of gestation. Mechanistically, we found that both EMP and fetal monocytes expressed Myb but repressed expression of c‐Maf/MafB in contrast to pMΦ. Furthermore, fetal monocytes showed increased mitochondrial respiratory and glycolytic capacity, which together explains increased proliferative and developmental capacity.
Results
Migration of transient definitive EMP from YS to fetal liver and accompanied differentiation to fetal liver monocytes enhances the developmental potential of MΦTR
We previously reported that transfer of CD11bintF4/80lo fetal lung monocytes to neonatal Csf2rb −/− recipient mice completely restored AM development (Schneider et al, 2014b). Analysis of the transfer efficiency showed that 5–10% of the cells put on the nostrils of Csf2rb −/− neonatal recipients reached the lung 1 h later (Fig EV1) and that as little as 5,000 transferred precursors (equaling 250–500 cells reaching the lung) could fully restore AM development in Csf2rb −/− mice. Harnessing the power of this cell transfer model, we wanted to compare the capacity of different embryonic macrophage precursors to differentiate into AM.
Figure EV1. Intranasal transfer efficiency.
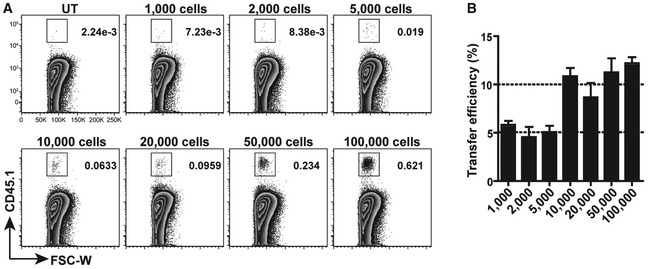
-
ARepresentative flow cytometric analysis of donor cells in neonatal lung from Csf2rb −/− recipients 1 h after being i.n. transferred with different numbers of AM from CD45.1+ adult WT mice and un‐transferred (UT) Csf2rb −/−. Pre‐gated viable singlets.
-
BIntranasal transfer efficiency after transfer of cells as described in (A) to neonatal Csf2rb −/− recipients (n = 3 mice each group).
Late EMP derived from the hemogenic endothelium of the YS seed the fetal liver at E9.5 and give rise to fetal monocytes around E12.5 (Lin et al, 2014; Hoeffel & Ginhoux, 2015). To assess the potential of YS‐derived late EMP to differentiate into AM, we sorted viable CD45loC‐kit+F4/80−CD11bloMHCII−CD11c− EMP (Fig EV2A) from the YS at E10.5 (CD45.1). A separated staining further confirmed that CD45lo EMP population are CX3CR1− and CD45hi population are CX3CR1+ pMΦ (Mass et al, 2016; Fig EV2B). A small number (i.e., 15,000 cells) of EMP were transferred to Csf2ra −/− neonates (CD45.2). Eight weeks later, EMP had expanded considerably (≥ 128‐fold) (Fig 1A–C) and developed into mature AM characterized as CD11chiF4/80+SiglecFhiCD11blo cells comparable to AM of untreated WT mice (Fig 1A). However, AM numbers in the BAL and lung of Csf2ra −/− mice transplanted with EMP were lower compared with unmanipulated WT mice (Fig 1C). EMP‐derived AM in Csf2ra −/− recipients prevented pulmonary alveolar proteinosis (PAP) in the BAL (Fig 1D), indicating that they developed into functionally competent AM, although the removal of dead cells (efferocytosis) in the BAL was impaired (Fig 1E).
Figure EV2. Sorting strategy for fetal precursors.
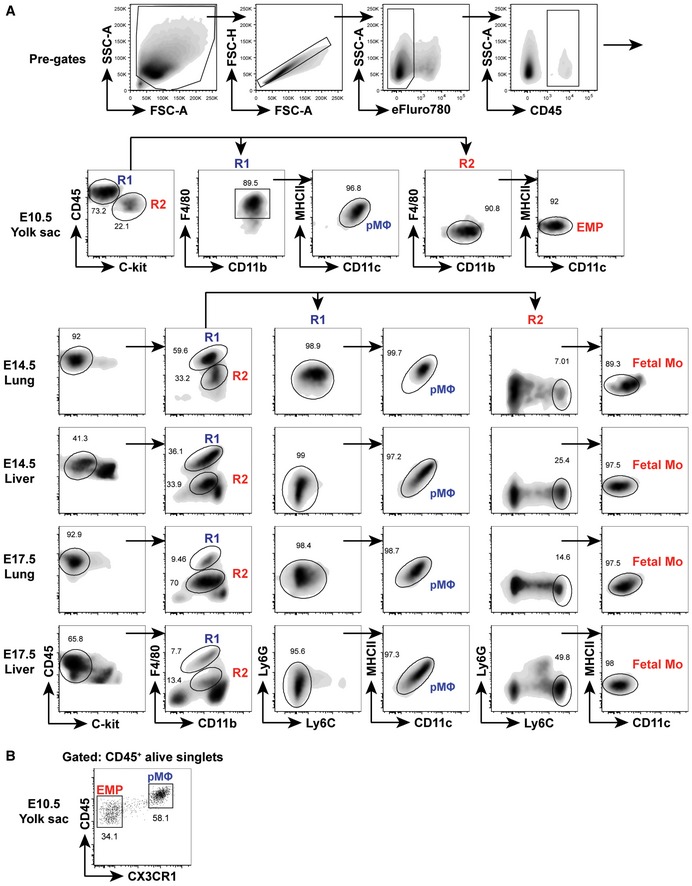
-
ASorting strategy for different fetal precursors which purified using flow cytometry. Doublets and debris were excluded using FSC and SSC. YS pMΦ were identified as viable CD45hiC‐kit−F4/80+CD11bhiMHCII−CD11c− cells. YS EMP were identified as viable CD45loC‐kit+F4/80−CD11bloMHCII−CD11c− cells. pMΦ from fetal lung and liver were identified as viable CD45+C‐kit−F4/80hiCD11bloLy6G−Ly6C−MHCII−CD11c− cells. Fetal Mo from lung and liver were identified as viable CD45+C‐kit−F4/80loCD11bintLy6G−Ly6ChiMHCII−CD11c− cells.
-
BRepresentative flow cytometric analysis of CD45 vs. CX3CR1 on E10.5 YS EMP and pMΦ.
Figure 1. Migration of transient definitive EMP from yolk sac to fetal liver and accompanied differentiation to fetal liver monocytes increases MΦTR developmental potential.
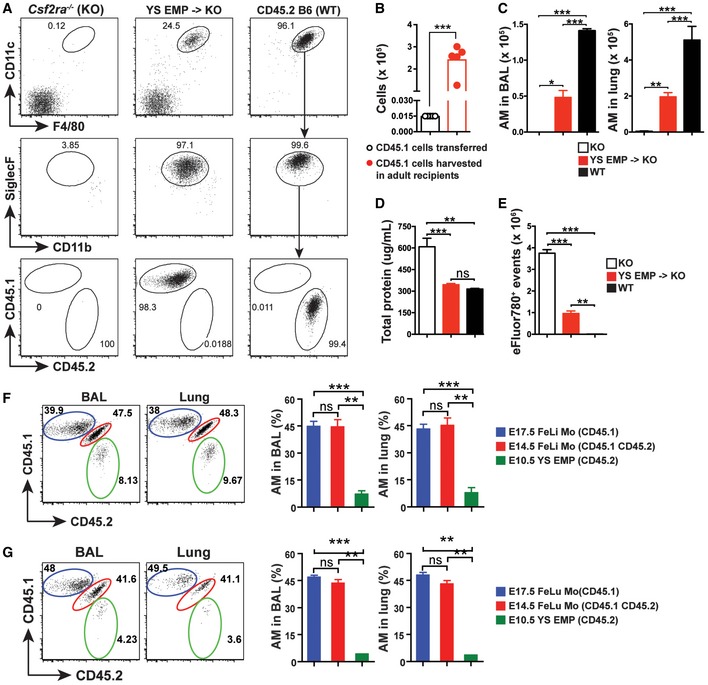
-
ARepresentative flow cytometric analysis of BAL from Csf2ra −/− recipients 8 weeks after neonatal i.n. transfer of YS EMP from CD45.1+ E10.5 embryos, 8‐week‐old un‐transferred Csf2ra −/−, and WT (CD45.2) mice. Pre‐gated viable CD45+ singlets.
-
BNumbers of EMP effectively transferred in neonates (white circle) and numbers of EMP‐derived AM (red circles) in BAL and lung of adult Csf2ra −/− recipients 8 weeks after transfer (n = 5 mice).
-
C–ENumbers of EMP‐derived AM in the BAL (left) and lung (right) (C) as well as total protein (D) and total eFluor780+ events (E) in the BAL of adult Csf2ra −/− recipients 8 weeks after transfer (n = 5 mice). Age‐matched Csf2ra −/− (KO) (n = 3 mice) and WT (n = 3 mice) were included as negative and positive controls.
-
F, GRepresentative flow cytometric analysis (left) and percentage of donor‐derived AM (right) in the BAL and lung from Csf2ra −/− recipients 8 weeks after neonatal transfer of 1:1:1 mixture with E17.5 fetal liver (FeLi) monocytes (Mo) (CD45.1+), E14.5 FeLi Mo (CD45.1+CD45.2+), and E10.5 YS EMP (CD45.2+) (F) or E17.5 fetal lung (FeLu) Mo (CD45.1+), E14.5 FeLu Mo (CD45.1+CD45.2+), and E10.5 YS EMP (CD45.2+) (G) (n = 3 mice).
Next, we compared the tissue macrophage differentiation capacity of E10.5 YS‐derived late EMP with fetal liver (FeLi) monocytes from E14.5 and E17.5 embryos in a competition experiment, by transfer of a 1:1:1 mixture. Monocytes were sorted as viable CD45+C‐kit−F4/80loCD11bintLy6G−Ly6ChiMHCII−CD11c− cells to avoid contamination by c‐kit+ late EMP and HSC precursors in the fetal liver (Fig EV2A). Analysis of BAL and lung of Csf2ra −/− recipients 8 weeks after transfer showed that around 90% of the mature AM were derived from FeLi monocytes, with equal proportions of E14.5 and E17.5 FeLi, while less than 10% came from late EMP (Fig 1F). Similar results were obtained by comparing E10.5 YS‐derived late EMP with E14.5 and E17.5 fetal lung (FeLu) monocytes (Fig 1G). These results demonstrate that YS‐derived late EMP can in principle develop into tissue macrophages (i.e., AM) without fetal liver transit. However, the liver stage enhances their capacity to expand. The data also show that liver and lung monocytes have comparable potential to develop into mature AM.
Superior capacity of fetal monocytes over pMΦ in AM development
Primitive MΦ (pMΦ) precursors can be found in the YS at E9.5, where they develop from E7.5 EMPs. They colonize the fetal liver and peripheral tissues beginning at E9.5 without passing through a monocyte stage and are replaced by late EMP‐derived monocytes between E13.5 and E17.5 in all tissues except the brain (Hoeffel & Ginhoux, 2015). We transferred a 1:1 mixture of E14.5 liver pMΦ (CD45.1) and monocytes (CD45.2) to Csf2ra −/− neonates to determine the competitive capacity of each precursor to reconstitute AM development. Eight weeks after transfer, mature AM in the recipients were derived predominantly from fetal monocytes (around 85%), while pMΦ contributed only 15% (Fig 2A and B). These data demonstrate a strikingly impaired capacity of pMΦ compared with fetal monocytes in development mature AM.
Figure 2. Superior capacity of fetal monocytes over pMΦ in AM development.
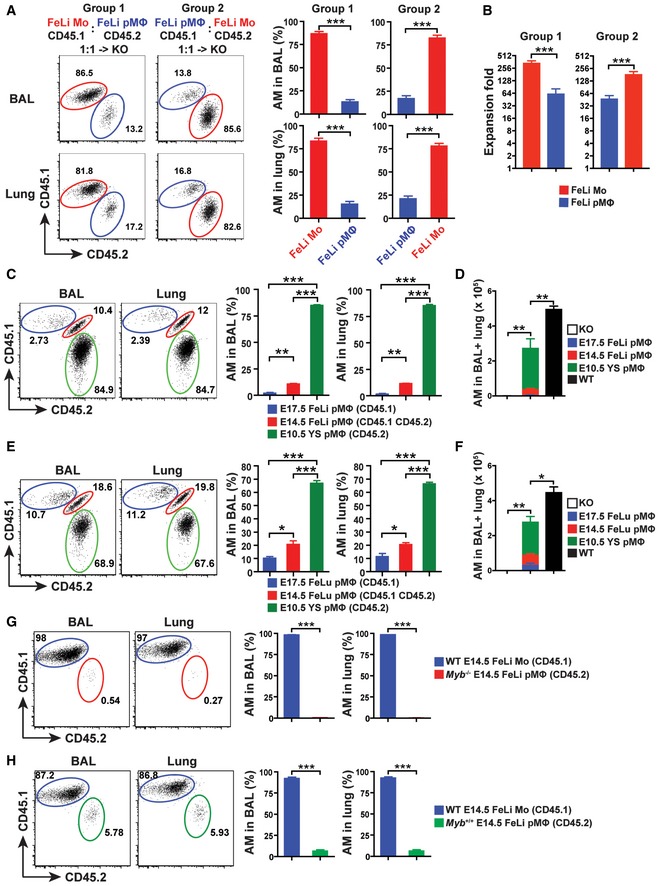
-
ARepresentative flow cytometric analysis (left) and percentage of donor‐derived AM (right) in the BAL (top) and lung (bottom) from Csf2ra −/− recipients 10 weeks after neonatal transfer of 1:1 mixture with CD45.1+ E14.5 FeLi Mo and CD45.2+ E14.5 FeLi pMΦ (Group 1) or CD45.1+ E14.5 FeLi pMΦ and CD45.2+ FeLi Mo (Group 2) (n = 3 mice).
-
BThe expansion fold of donor cells in Csf2ra −/− recipients 10 weeks after neonatal transfer of cells described above in (A) (n = 3 mice).
-
CRepresentative flow cytometric analysis (left) and percentage of donor‐derived AM (right) of BAL and lung from Csf2ra −/− recipients 8 weeks after neonatal transfer of 1:1:1 mixture with CD45.1+ E17.5 FeLi pMΦ, CD45.1+CD45.2+ E14.5 FeLi pMΦ, and CD45.2+ E10.5 YS pMΦ (n = 3 mice).
-
DTotal numbers of donor‐derived AM in the BAL and lung of adult Csf2ra −/− recipients 8 weeks after transfer of cells described above in (C) (n = 3 mice). Age‐matched Csf2ra −/− (KO) (n = 3 mice) and WT (n = 3 mice) were included as negative and positive controls.
-
ERepresentative flow cytometric analysis (left) and percentage of donor‐derived AM (right) of BAL and lung from Csf2ra −/− recipients 8 weeks after neonatal transfer of 1:1:1 mixture with CD45.1+ E17.5 FeLu pMΦ, CD45.1+CD45.2+ E14.5 FeLu pMΦ, and CD45.2+ E10.5 YS pMΦ (n = 3 mice).
-
FTotal numbers of donor‐derived AM in the BAL and lung of adult Csf2ra −/− recipients 8 weeks after transfer of cells described above in (E) (n = 3 mice). Age‐matched Csf2ra −/− (KO) (n = 3 mice) and WT (n = 3 mice) were included as negative and positive controls.
-
G, HRepresentative flow cytometric analysis (left) and percentage of donor‐derived AM (right) in the BAL and lung from Csf2ra −/− recipients 7 weeks after neonatal transfer of 1:1 mixture with CD45.1+ E14.5 FeLi Mo from WT embryos and CD45.2+ E14.5 FeLi pMΦ from Myb −/− (G) or Myb +/+ (H) embryos (n = 3 mice).
We next compared the tissue macrophage differentiation capacity of pMΦ from E10.5 YS with pMΦ derived from E14.5 and E17.5 fetal liver by transfer of a 1:1:1 mixture (including 5,000 cells each) of sorted populations into Csf2ra −/− neonates. We sorted pMΦ from E10.5 YS as viable CD45hiC‐kit−F4/80+CD11bhi MHCII−CD11c− cells and pMΦ from E14.5 and E17.5 fetal liver as viable CD45+C‐kit−F4/80hiCD11bloLy6G−Ly6C−MHCII−CD11c− cells (Fig EV2A). Analysis of reconstituted recipients showed that mature AM were predominantly derived from YS pMΦ (≥ 80%) followed by pMΦ from E14.5 liver (≥ 10%) and E17.5 liver (3%) (Fig 2C). Furthermore, similar results were obtained by comparing the AM maturation capacity of pMΦ from E10.5 YS with pMΦ from E14.5 and E17.5 fetal lung in a competitive reconstitution experiment (Fig 2E). Mature AM in reconstituted Csf2ra −/− recipients differentiated almost exclusively from YS pMΦ (70%) followed by pMΦ from E14.5 lung (20%) and E17.5 lung (10%) (Fig 2E). However, AM numbers in the BAL and lung of Csf2ra −/− mice transplanted with mixed pMΦ were lower compared with unmanipulated WT mice (Fig 2D and F), indicating that pMΦ fail to fully replenish the empty AM niches even in the absence of fetal monocytes. Thus, the residual developmental capacity of pMΦ in YS gradually wanes after their migration to liver and lung and with progressing gestation.
Myb expression in EMP and fetal liver monocytes promotes AM development
The pMΦ derived from the YS develop in a c‐Myb‐independent manner, in contrast to HSC (Schulz et al, 2012). The presence of F4/80hiCD11bint and the absence of F4/80intCD11bhi population in fetal tissues of Myb −/− mice have been proposed to demonstrate that pMΦ are the precursors of most MΦTR and that fetal CD11bhi cells are derived from HSC (Schulz et al, 2012). However, transient definitive EMP were recently shown to express Myb in the E9.5 YS and fetal liver (Hoeffel et al, 2015). We addressed the AM differentiation capacity of Myb‐deficient pMΦ from E14.5 fetal liver (and littermate control Myb +/+ pMΦ) in competition with WT FeLi monocytes (for sorting strategy see Fig EV3) by transferring respective populations in a 1:1 mixture into Csf2ra −/− neonates and analysis 7 weeks later (Fig 2G and H). Reconstituted mature AM were exclusively derived from FeLi monocytes (> 97%) with < 1% originating from Myb −/− pMΦ (Fig 2G), while control Myb +/+ pMΦ contributed around 5% in competition with WT FeLi monocytes (Fig 2H), consistent with the co‐transfer results shown in Fig 2A. Together, these data demonstrate that Myb expression in YS EMP and fetal liver monocytes promotes AM development.
Figure EV3. Sorting strategy for Myb KO and WT fetal precursors.
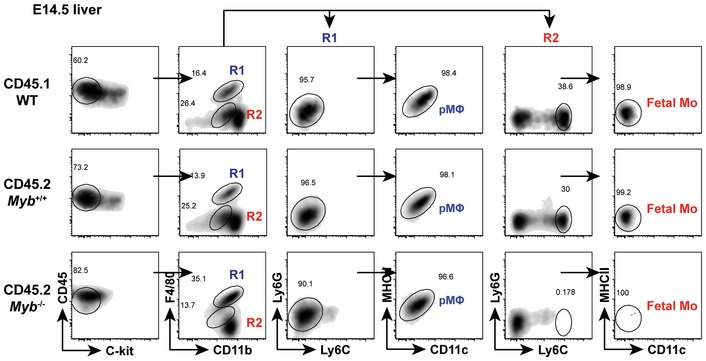
Sorting strategy for different fetal precursors which purified using flow cytometry. Doublets and debris were excluded using FSC and SSC. pMΦ were identified as viable CD45+C‐kit−F4/80hiCD11blo Ly6G−Ly6C−MHCII−CD11c− cells. Fetal Mo were identified as viable CD45+C‐kit−F4/80loCD11bintLy6G−Ly6ChiMHCII−CD11c− cells. There were no Mo in Myb −/− fetal liver.
pMΦ‐derived AM show impaired function compared with fetal monocyte‐derived AM
We have shown above that pMΦ precursors are almost completely outcompeted in the presence of fetal monocytes during AM development. To assess the potential of pMΦ in the absence of fetal monocytes, we transferred exclusively E14.5 liver pMΦ into Csf2ra −/− neonates and compared AM reconstitution with Csf2ra −/− recipients that received E14.5 liver monocytes. After 7 weeks, the number of FeLi monocyte‐derived AM in BAL and lung was slightly lower (reaching around 75%) than the AM number found in unmanipulated WT mice (Fig 3A and B). Interestingly, the AM reconstitution capacity of pMΦ precursors was reduced by > 60% compared with that of fetal monocyte precursors (Fig 3A and B). Full reconstitution of AM niches in Csf2ra −/− recipients by fetal monocytes is usually completed around 9–10 weeks after reconstitution (Appendix Fig S1). In contrast, even after 1 year, pMΦ‐derived AM in Csf2ra −/− recipients reached only around half of the number found in untreated WT (Fig 3C), indicating that their expansion capacity was exhausted already 7 weeks after transfer. Nevertheless, the pMΦ fare better alone than when in direct competition with fetal monocytes where they contributed around 15% to the pool of mature AM (Fig 2A). Csf2ra −/− recipients containing pMΦ‐derived AM showed mild PAP and a slight enrichment of dead cells. However, the BAL of Csf2ra −/− recipients with fetal monocyte‐derived AM looked as clean as the BAL of WT mice without any PAP or dead cell accumulation (Fig 3D–G). Thus, pMΦ‐derived AM show reduced function during homeostasis compared with fetal monocyte‐derived AM.
Figure 3. Function of pMΦ‐ and fetal monocyte‐derived AM during homeostasis.

-
ARepresentative flow cytometric analysis of BAL from Csf2ra −/− recipients 7 weeks after neonatal i.n. transfer of pMΦ or fetal Mo from CD45.1+ E14.5 fetal livers, 7‐week‐old un‐transferred Csf2ra −/−, WT (CD45.2) mice. Pre‐gated viable CD45+ singlets.
-
B, CNumbers of donor‐derived AM in the BAL (left) and lung (right) of adult Csf2ra −/− recipients 7 weeks (B) or 1 year (C) after neonatal transfer of cells described in (A) (n = 7 mice). Age‐matched Csf2ra −/− (KO) (n = 5 mice) and WT (n = 4 mice) were included as negative and positive controls.
-
D–GTotal protein and total eFluor780+ events in the BAL of adult Csf2ra −/− recipients 7 weeks (D, E) or 1 year (F, G) after neonatal transfer of cells described in (A) (n = 7 mice). Age‐matched Csf2ra −/− (KO) (n = 5 mice) and WT (n = 4 mice) were included as negative and positive controls.
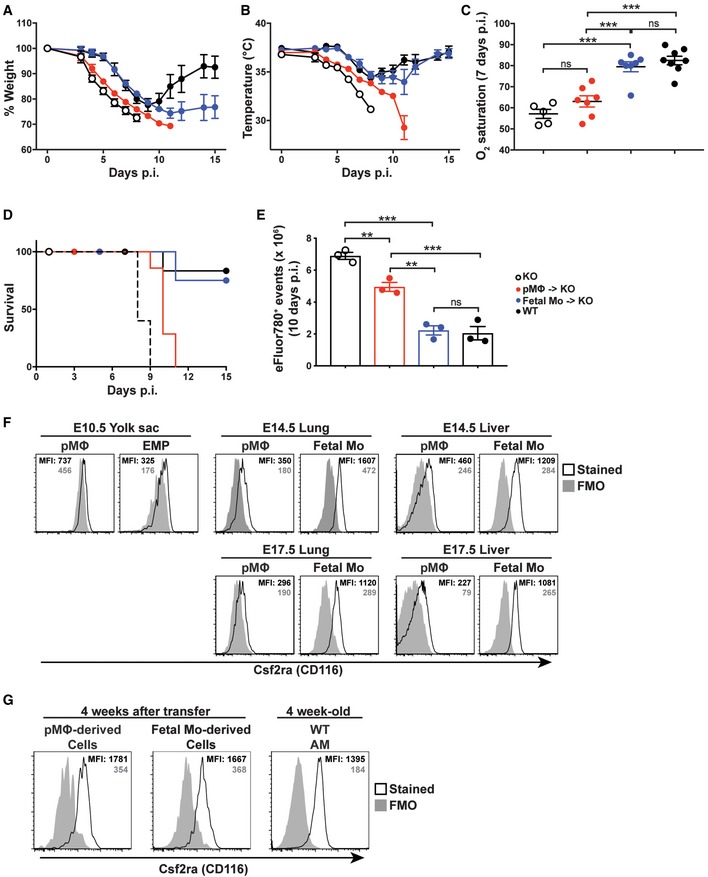
We have previously reported a critical role of AM for the prevention of lung failure and protection from morbidity and lethality following infection with influenza virus (Schneider et al, 2014a). To further compare the function of pMΦ‐ and fetal monocyte‐derived AM during infection, we infected such reconstituted Csf2ra −/− mice with influenza virus PR8. Expectedly, all Csf2ra −/− mice lacking AM succumbed to the infection due to lung failure (Fig 4A–E; Schneider et al, 2014b). The presence of pMΦ‐derived AM delayed the decline in body temperature (Fig 4B) and death only by 2 days without amelioration of O2 saturation (Fig 4C and D). In contrast, body temperature, O2 saturation, viability, and dead cell numbers in the BAL was comparable in infected WT and Csf2ra −/− recipients containing fetal monocyte‐derived AM (Fig 4B–E) indicating that fetal monocyte‐derived AM but not pMΦ‐derived AM protect from influenza‐induced morbidity and mortality. Together, these results show that pMΦ‐derived AM have impaired function in vivo compared with fetal monocyte‐derived AM.
Figure 4. Function of pMΦ‐ and fetal monocyte‐derived AM during infection.
-
A, BBody weight (A) and temperature (B) measurement of influenza virus (20 pfu of PR8 strain)‐infected Csf2ra −/− recipients 24 weeks after neonatal transfer of pMΦ or fetal Mo from E14.5 fetal livers (n = 7 mice from each transferred group), 24‐week‐old un‐transferred Csf2ra −/− (n = 5), and WT (n = 8).
-
COxygen (O2) saturation in influenza‐infected mice described in (A, B) 7 days post‐infection (p.i.) (n = 5 mice from Csf2ra −/− group; n = 7 mice from each transferred group; n = 8 from WT group).
-
DSurvival curve of influenza‐infected mice described in (A, B) (n = 5 mice from Csf2ra −/− group; n = 7 mice from each transferred group; n = 8 from WT group).
-
ETotal eFluor780+ events in the BAL of influenza‐infected mice described in (A, B) 10 days p.i. (n = 3 mice from each group).
-
FRepresentative flow cytometric analysis of Csf2ra (CD116) expression on different fetal precursors from WT embryos. The solid gray histogram represents the fluorescence minus one (FMO) controls, the open black histogram represents stained samples. The gray number represents mean fluorescence intensity (MFI) of FMO controls, the black number represents MFI of stained samples. E10.5 YS cells were pooled from 8 to 10 embryos, E14.5 lung cells were pooled from 3 to 4 embryos, and E14.5 liver cells were from individual embryo.
-
GRepresentative flow cytometric analysis of Csf2ra expressions on pMΦ‐ and fetal Mo‐derived AM in Csf2ra −/− recipients 4 weeks after transfer of pMΦ or fetal Mo from E14.5 fetal livers. AM from 4‐week‐old WT mice were included as controls.
Upregulation of c‐Myb and repression of Maf transcription factors in definitive EMP and fetal monocytes
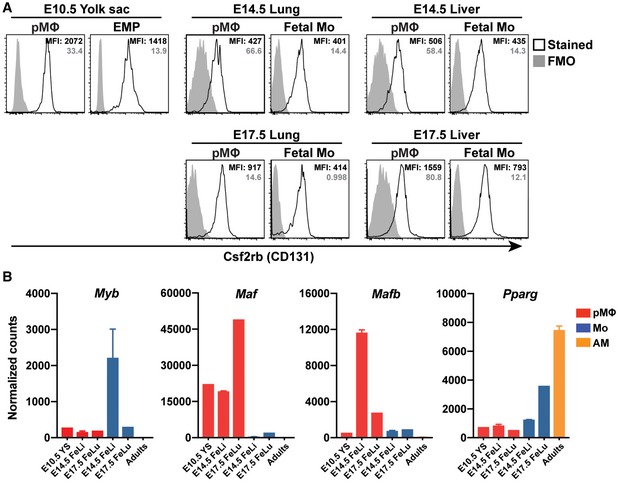
Previous studies have shown that the GM‐CSF/GM‐CSF receptor pathway is critical for the perinatal development of AM from fetal lung monocyte precursors but not from lung pMΦ (Guilliams et al, 2013; Schneider et al, 2014b). The GM‐CSF receptor is a heterodimer composed of the ligand‐specific CSF2RA chain (CD116) and the common CSF2RB chain (CD131), the latter being shared with the IL‐3 and IL‐5 receptors. Comparing CSF2RA and CSF2RB cell surface expression on different fetal precursors, we found that CSF2RB was comparably bright on YS‐derived EMP, as well as fetal monocytes and pMΦ (Fig EV4A). Interestingly, CSF2RA staining was bright on liver and lung monocyte precursors but dim on YS‐derived EMP and pMΦ from WT embryos prior to transfer into Csf2ra −/− recipients (Fig 4F). However, isolation of expanding fetal monocyte‐ and pMΦ‐derived AM 4 weeks after transplantation to the lungs of Csf2ra −/− recipients showed comparable levels of cell surface CSF2RA, indicating that it was upregulated on pMΦ after transplantation (Fig 4G). Since pMΦ precursors failed to fully replenish the AM pool of Csf2ra −/− recipients, even 1 year after transplantation, the difference in Csf2ra expression levels may contribute to but is certainly not the only reason for poor developmental capacity of pMΦ.
Figure EV4. Csf2rb expression on fetal precursors and the transcripts of self‐renewal and proliferation factors in fetal precursors.
-
ARepresentative flow cytometric analysis of Csf2rb (CD131) expressions on different fetal precursors from WT embryos. The solid gray histogram represents the FMO controls, and the open black histogram represents stained samples. The gray number represents MFI of FMO controls, the black number represents MFI of stained samples. E10.5 YS cells were pooled from 8 to 10 embryos, E14.5 lung cells were pooled from 3 to 4 embryos, and E14.5 liver cells were from individual embryos.
-
BRNA‐seq analysis of Myb, Maf, Mafb, and Pparg gene expression in different fetal precursors from WT embryos and mature AM from WT adult mice (n = 1–2 biological replicates each group).
We next compared the gene expression profiles of WT AM with AM derived from distinct fetal precursors 6 weeks after their transfer to Csf2ra −/− recipients (Fig 5A). Moreover, the transcriptome of the different precursors was determined. Principal component analysis (PCA) (Fig 5B) and clustering matrix (Fig 5C) based on all available genes showed that the analyzed precursors differ substantially, while mature AM derived from the distinct precursors had similar gene expression profiles, which cluster together with WT AM (van de Laar et al, 2016). By focusing the comparison on AM signature genes (Gautier et al, 2012), including AM signature‐up (Fig 5D), AM signature‐down genes (Fig 5E), and AM signature transcription factors (Fig 5F), we found that the transcriptomes of WT AM and AM derived from the distinct precursors looked alike. Together, these results indicate that in principle pMΦ have a genuine potential and inherent plasticity to develop to AM upon colonization of the AM niche.
Figure 5. Inverse expression of the transcription factors c‐Myb and c‐Maf/MafB in fetal monocytes and pMΦ.
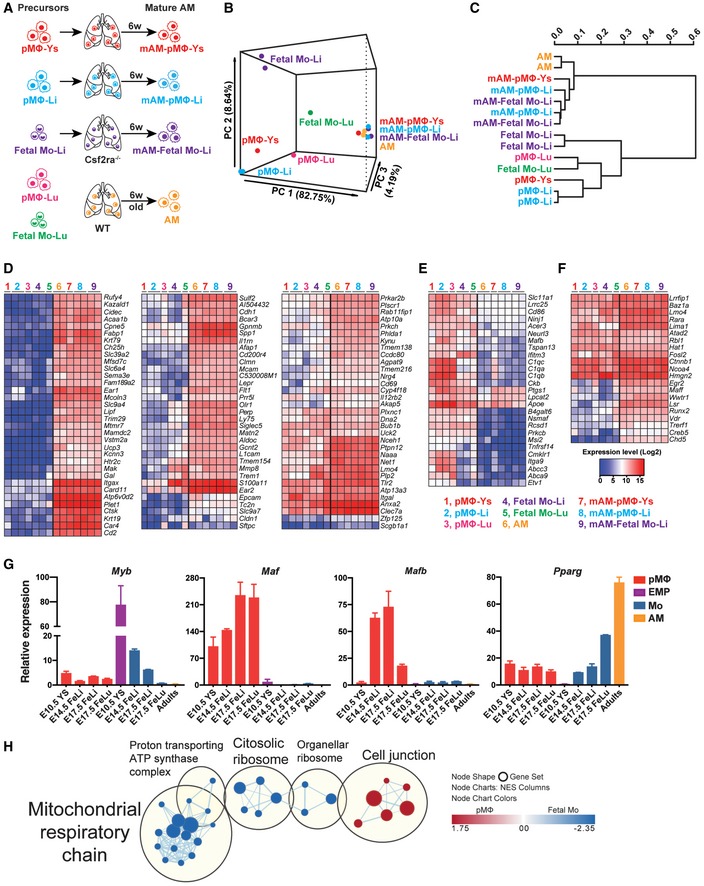
-
ASchematic of the samples for RNA‐seq analysis. pMΦ from E10.5 YS (pMΦ‐Ys), E14.5 liver (pMΦ‐Li), and E17.5 lung (pMΦ‐Lu) as well as fetal Mo form E14.5 liver (Fetal Mo‐Li) and E17.5 lung (Fetal Mo‐Lu) were sorted from WT embryos, respectively. pMΦ‐Ys, pMΦ‐Li, and Fetal Mo‐Li were i.n. transferred to neonatal Csf2ra −/− mice. Six weeks later, pMΦ‐Ys‐, pMΦ‐Li‐, and Fetal Mo‐Li‐derived mature AM (mAM‐pMΦ‐Ys, mAM‐pMΦ‐Li, mAM‐Fetal Mo‐Li) were sorted from the BAL of recipients, respectively. AM from 6‐week‐old WT were sorted as control.
-
B, CPrincipal component analysis (B) and clustering matrix (C) of the transcriptomes of precursors and mature AM described above in (A).
-
D–FHeatmap of AM signature‐up genes (D), AM signature‐down genes (E), and AM‐specific transcription factors (F) in the precursors and mature AM described above in (A).
-
GqPCR (n = 3 biological replicates) analysis of Myb, Maf, Mafb, and Pparg expression in different fetal precursors from WT embryos and mature AM from WT adult mice.
-
HGene Ontology (GO) term enrichment maps for Cellular Component (CC) of pMΦ and fetal Mo from E14.5 fetal liver. The maps show the top enriched gene networks with the FDR q value < 0.1. The line thickness connecting two gene set nodes corresponds to the number of genes overlapping between the two sets. Red nodes indicate gene sets enriched within genes strongly downregulated in fetal Mo. Blue nodes indicate gene sets enriched within genes strongly upregulated in fetal Mo compared with pMΦ.
We next determined expression of transcription factors known to control macrophage progenitor expansion and differentiation such as c‐Myb (Myb), c‐Maf (Maf), and MafB (Mafb) in sorted EMP and pMΦ from YS, fetal monocytes and pMΦ from fetal liver and lung at different days of gestation by qPCR (Fig 5G) and RNA‐seq (Fig EV4B). MafB activity is known to be involved in macrophage differentiation and in cooperation with c‐Maf inhibits stemness and self‐renewal of differentiated monocytes and macrophages (Aziz et al, 2009; Soucie et al, 2016). Interestingly, Maf expression and Mafb expression were completely repressed in EMP, fetal liver and lung monocytes, and mature AM. In contrast, expression was upregulated in fetal liver and lung pMΦ indicating differentiation and loss of proliferative capacity (Figs 5G and EV4B). c‐Myb is known to repress transcriptional activity of MafB. Expression of Myb was very high in E10.5 YS EMP and toned down in E14.5 fetal liver monocytes, while pMΦ from YS and fetal liver displayed very low levels (Figs 5G and EV4B; Hoeffel et al, 2015). These data support the concept that MafB/c‐Maf and c‐Myb antagonistically determine the capacity of progenitor expansion and differentiation. Pparg was selectively upregulated in E17.5 fetal lung monocytes compared with pMΦ and fetal liver monocytes (Fig 5G), confirming and extending a previous report demonstrating that GM‐CSF expression in the fetal lung instigates PPARγ and drives perinatal development of pre‐AM (Schneider et al, 2014b). The repression of Myb and upregulation of Pparg in E17.5 fetal lung (but not in liver) monocytes and mature AM (Figs 5G and EV4B) suggests that their expansion and tissue‐specific differentiation are driven by PPARγ rather than by the balance of c‐Myb and MafB that drives expansion and differentiation of definitive EMP and fetal liver monocytes.
Fetal monocytes display enhanced mitochondrial respiratory and glycolytic capacity
For an unbiased comparison of the phenotype of pMΦ and fetal monocytes, we performed gene set enrichment analysis (GSEA) on Gene Ontology (GO) for Cellular Component (CC) of their transcriptomes. Interestingly, we found that genes related to mitochondrial respiratory chain and ribosomal activity were highly enriched in fetal monocytes (Fig 5H), indicating that those are metabolically more active compared with pMΦ.
Accordingly, we assessed the bioenergetic profiles of pMΦ and fetal monocytes by measurement of the O2 consumption rate (OCR), an indicator of oxidative phosphorylation (OXPHOS) in a basal state and after the stimulation with oligomycin (to block ATP synthesis), FCCP (to uncouple ATP synthesis from the electron transport chain, ETC), and rotenone and antimycin A (to block complex I and III of the ETC, respectively) (Fig 6A; Gerencser et al, 2009; Nicholls et al, 2010). While basal respiration was comparable in pMΦ and fetal monocytes, the latter displayed higher maximal and spare respiratory capacity (SRC) (Fig 6A) (a measure for the efficiency of ETC‐mediated proton transport from the mitochondrial matrix to the intermembrane space upon uncoupling of ATP synthesis), which allows cell better survival in times of proliferation and energy demand. Moreover, measurement of aerobic glycolysis, an intrinsic metabolic feature of proliferative cells (Vander Heiden et al, 2009; Lunt & Vander Heiden, 2011), by the detection of lactic acid and extracellular acidification (ECAR) revealed an enhanced glycolytic reserve and glycolytic capacity in fetal liver and lung monocytes (Figs 6B and EV5A), which is in line with their increased expansion/differentiation ability in vivo (Figs 2A and 3). Furthermore, RNA sequencing and real‐time PCR results revealed increased expression of Eno1, Slc2a6, and Txn1, genes involved in glycolysis, glucose uptake, and cell proliferation, respectively, in fetal monocytes compared with pMΦ (Figs 6C and D, and EV5B). In contrast, Txnip, a negative regulator of glucose uptake and cell proliferation (Parikh et al, 2007; Muri et al, 2018), was increased in pMΦ (Figs 6C and D, and EV5B). Together, fetal monocytes possess increased mitochondrial bioenergetics and glycolytic capacity indicating increased survival and expansion capacities, which possibly explains their competitive advantage over pMΦ in empty niches allowing AM differentiation.
Figure 6. Increased metabolic fitness of fetal monocytes compared with pMΦ.
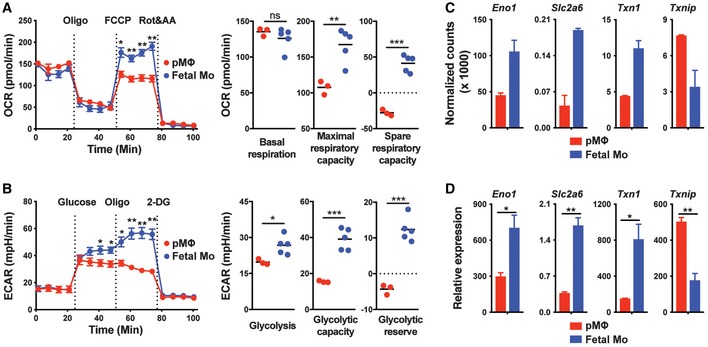
-
AOxygen consumption rate (OCR) of pMΦ and fetal Mo from E14.5 WT fetal liver at baseline and after treatment with oligomycin (Oligo), FCCP or rotenone (Rot), and antimycin A (AA).
-
BExtracellular acidification rate (ECAR) of pMΦ and fetal Mo from E14.5 WT fetal liver at baseline and after treatment with glucose, Oligo, or 2‐Deoxy‐D‐glucose (2‐DG).
-
C, DRNA‐seq (C, n = 2 biological replicates) and qPCR (D, n = 3 biological replicates) analysis of Eno1, Slc2a6, Txn1, and Txnip gene expression in pMΦ and fetal Mo from E14.5 WT fetal liver.
Figure EV5. Glycolytic ability of pMΦ and fetal Mo in fetal lung.
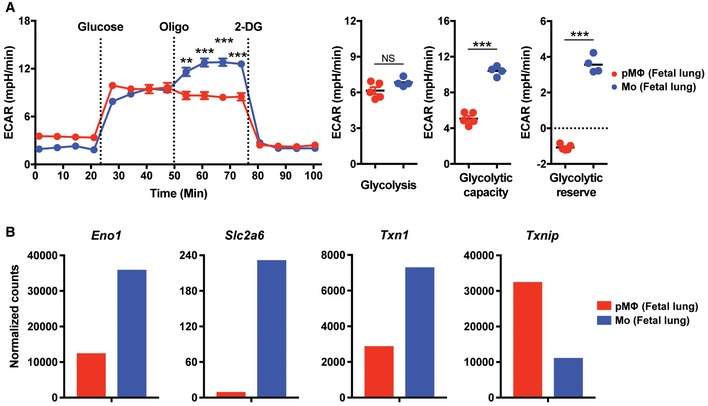
-
AExtracellular acidification rate (ECAR) of pMΦ and fetal Mo from E17.5 WT fetal lung at baseline and after treatment with glucose, Oligo, or 2‐Deoxy‐D‐glucose (2‐DG).
-
BRNA‐seq (n = 1) analysis of Eno1, Slc2a6, Txn1, and Txnip gene expression in pMΦ and fetal Mo from E17.5 WT fetal lung.
Discussion
Genetic fate mapping and parabiosis studies have convincingly demonstrated that most MΦTR develop from embryonic precursors. Several progenitors with macrophage potential arising at different days and sites of the embryo have been described but the relative contribution to the pool of mature self‐renewing macrophages in adult tissues still remains unclear. While genetic reporters enable tracking of putative precursors, they cannot accurately quantify or evaluate their functional long‐term output in vivo. In this study, by transferring different putative fetal macrophage precursors into the lungs of Csf2ra −/− neonates with empty AM niches, we were able to determine their developmental fate and definitive long‐term function as mature AM. In particular, when a mixture of two or even three different precursors was transferred into the same neonate recipient, we were able to directly compare their competitive fitness, analogous to the situation where precursors arising from overlapping developmental waves compete with each other. Such analysis provides evidence beyond a basal developmental potential. Indeed, while transfer of pMΦ harvested from fetal liver (E14.5) showed a basic potential to differentiate into AM, they were almost completely outcompeted in the presence of fetal liver (or lung) monocyte precursors, suggesting that the vast majority of mature AM in the lung is probably derived from fetal monocytes and not pMΦ.
From day E9.5 to E11.5, the YS contains the population of pMΦ that are derived from a primitive EMP (from a transient wave of primitive hematopoiesis), as well as late EMP from a wave of transient definitive hematopoiesis (Bertrand et al, 2005, 2007). The late EMP are thought to migrate to the fetal liver where they develop into monocytes prior to migration to the periphery and terminal differentiation into MΦTR (Hoeffel & Ginhoux, 2015; Palis, 2016). Consistently, we found that liver and lung monocytes outcompeted the late EMPs in a competitive setting, indicating that late EMPs require further differentiation signals from the liver. Interestingly, comparison of E10.5 pMΦ from YS with E14.5 and E17.5 pMΦ from fetal liver (or lung), in the absence of fetal monocytes, showed that the YS pMΦ possess a considerable advantage over E14.5 pMΦ from fetal liver (or lung) in AM development and that E17.5 pMΦ from liver (or lung) were almost completely outcompeted. This suggests a model where pMΦ from the YS effectively seed the fetal liver and all other tissues and are then successively replaced by monocytes due to exhaustion of the pMΦ and/or superior developmental fitness of the monocytes, consistent with the conclusions of a fate‐mapping approach (Hoeffel et al, 2015). A direct comparison of E10.5 pMΦ and late EMPs in the absence of fetal monocytes in a competitive setting remains to be done. The YS late pMΦ (here characterized as CD45hic‐kit−F4/80hiCD11bhi) may represent a heterogenous population that, among others, may also contain late EMP‐derived precursors. Unfortunately, the number of primitive (E7.5) EMP is too small to allow isolation and adoptive transfer experiments.
The transcription factor c‐Myb is essential for definitive but not primitive hematopoiesis. Consistently, primitive EMP do not express c‐Myb and embryos lacking the Myb gene show intact primitive erythropoiesis, allowing them to survive until E15.5 ± 1, after which they die due to the absence of definitive erythropoiesis and anemia (Mucenski et al, 1991; Sumner et al, 2000; Tober et al, 2008). Shortly before death, Myb‐deficient embryos contain normal numbers of F4/80hiCD11blo macrophages but no CD11bhi monocytes (and other HSC‐derived lineages) in every tissue analyzed, which has been suggested to demonstrate that tissue macrophages develop from primitive EMP, independent of c‐Myb (Schulz et al, 2012). Consistently, we found that transfer of E14.5 liver F4/80hi macrophages from c‐Myb −/− embryos can partially restore AM development in Csf2r‐deficient mice in the absence of fetal monocytes (data not shown). However, in competition with WT fetal liver monocytes, Myb‐deficient pMΦ completely failed to develop to AM indicating that Myb expression in definitive EMP and possibly fetal monocytes somehow promotes their developmental potential.
The ability of Myb‐deficient pMΦ precursors to develop into AM in the absence of fetal monocytes shows the enormous ability of empty niches to nurture second or third choice precursors in the absence of bona fide precursors to sustain development and function. Only the competitive transplantation of different precursors revealed the developmental capacity of the individual precursors.
During myelomonocytic differentiation, c‐Myb and MafB are known to interact in an antagonistic relationship and thereby balance progenitor expansion and differentiation. c‐Myb is upregulated and represses MafB in immature cells, while repression and upregulation of MafB results in inhibition of c‐Myb and differentiation of progenitors to macrophages (Hegde et al, 1999; Kelly et al, 2000; Lieu & Reddy, 2009). The c‐Myb/MafB antagonism appears to be regulated by SUMOylation of the transcription factors (Bies et al, 2002; Dahle et al, 2003; Tillmanns et al, 2007). Our data indicate that the balance of c‐Myb and Maf/MafB at least partially explains progenitor expansion capacity and macrophage differentiation capacities observed in the different precursors in YS, fetal liver, and lung. Expression of c‐Myb is very high in E10.5 YS definitive EMP and gradually wanes in fetal liver monocytes between E14.5 and E17.5, while it is poorly expressed in pMΦ. By contrast, Maf and MafB are highly expressed in pMΦ (except MafB in YS pMΦ), but are repressed (possibly by c‐Myb) in EMP and fetal monocytes suggesting that pMΦ have lost multi‐potent progenitor expansion potential and are undergoing differentiation. The absence of MafB in YS pMΦ may explain why they are better in reconstitution (in a competitive setting) of empty AM niches than pMΦ from fetal liver and lung. Importantly, the pMΦ from fetal liver and lung cannot be terminally differentiated Kupffer cells and AM, respectively, since transfer of mature AM but not fetal pMΦ can restore AM development in Csf2ra‐deficient mice. The absence of c‐Myb in fetal lung (but not liver) monocytes (and AM) are already committed pre‐AM, expansion of which is driven by an organ‐specific transcriptional program. Indeed, fetal monocytes in the lung but not in the liver express high levels of Pparg, similar to mature AM.
It should be noted that the population of fetal liver monocytes used for transfer consists of a mixture of monocytes derived from transient definitive (late) EMP and definitive fetal HSC arising in AGM regions at E10.5, which cannot be distinguished by surface markers. Fate‐mapping experiments using Flt3‐driven Cre recombinase, which labels progeny from definitive HSC, showed that around 15–20% of fetal liver monocytes and mature AM are derived from fetal HSC. It may be possible that this number is an underestimation due to a low expression of Flt3 and therefore inefficient labeling of fetal HSC. If HSC‐derived fetal monocytes have a proliferative advantage over EMP‐derived fetal monocytes, a considerable proportion of mature AM in adults may be derived from HSC‐derived fetal monocyte precursors, even if the precursor frequency is lower. It would be interesting to assess the development of fetal HSC‐derived and late EMP‐derived monocyte precursors in competition.
We have shown that the developmental capacity of fetal monocytes is at least 10‐fold greater compared with pMΦ precursors, raising questions about the underlying processes. GM‐CSF is required for perinatal development of AM by inducing PPARγ. We and others have shown that the GM‐CSF receptor alpha subunit (CSF2Rα) is more strongly expressed on fetal liver and lung monocytes compared with pMΦ precursors (Fig 4F), while the common signal‐transducing CSF2Rβ subunit, which is shared among receptors for GM‐CSF, IL‐5, and IL‐3, is similarly expressed on fetal monocytes and pMΦ (Fig EV4A; Schneider et al, 2014b; van de Laar et al, 2016), suggesting that GM‐CSF signaling is impaired in pMΦ. However, we found that CSF2Rα becomes upregulated on pMΦ soon after their transplantation to the lung of Csf2ra −/− mice. Nevertheless, pMΦ fail to fully replenish AM development within 12 months, indicating that GM‐CSFR signaling is unlikely to be the limiting factor for efficient AM differentiation of pMΦ precursors. However, we cannot fully exclude that the delayed upregulation of CSF2Rα on pMΦ contributes to a delay in reconstitution.
Instead, our data suggest that impaired glycolytic capacity limits the developmental capacity of pMΦ. Enhanced glycolytic flux and the pentose phosphate pathway support NADPH production and the biosynthesis of proteins, lipids, and ribonucleotides (O'Neill & Pearce, 2016; Van den Bossche et al, 2017). NADPH provides reducing equivalents to the thioredoxin (Trx) and glutaredoxin systems to prevent oxidative damage and allow DNA synthesis catalyzed by the ribonucleotide reductase (Lillig & Holmgren, 2007). Expression of thioredoxin‐1 (Txn1) and thioredoxin‐interacting protein (Txnip), an inhibitor of thioredoxin that is known to inhibit proliferation, is inversely expressed in proliferating and resting T cells (Muri et al, 2018). Similarly, Txn1 expression is increased and Txnip expression is decreased in E14.5 fetal monocytes compared with pMΦ, suggesting increased proliferation of fetal monocytes (Fig 6C and D). Upregulation of the hexose transmembrane transporter Slc2a6 (Glut6) and α‐enolase (Eno1) provides additional hints of the proliferative advantage of fetal monocytes. Eno1 catalyzes the conversion of 2‐phosphoglyceric acid to phosphoenolpyruvic acid in the glycolytic pathway. Eno1 upregulation has been related to aerobic glycolysis in cancer cells and associated with development of malignant tumors and a poorer prognosis (Fu et al, 2015).
It has been shown that endothelium‐specific plasmalemma vesicle‐associated protein (PLVAP) is required for the exit of fetal monocyte precursors from the liver and seeding of fetal tissues including the lung, while YS‐derived pMΦ migrate to fetal tissues in PLVAP‐independent manner (Rantakari et al, 2016). It should be noted that our preparation of the different precursors from YS and fetal liver and their transfer to the airways of neonates bypasses a potentially differential control of migration. We can also not completely exclude that the impaired capacity of pMΦ to reconstitute the AM pool after adoptive transfer to the airways is due to impaired migration and localization to the AM niche.
An important steady‐state function of AM is the removal and catabolism of pulmonary surfactant, secreted by type II alveolar epithelial cells into the alveoli. Csf2r‐deficient mice replenished with either fetal monocyte‐derived or pMΦ‐derived AM showed that the latter are slightly but significantly impaired in clearance of surfactant and dead cells, although they prevented development of pulmonary alveolar proteinosis (PAP) observed in AM‐deficient hosts. During a lytic respiratory viral infection causing massive death of epithelial cells, AM‐mediated efferocytosis of cellular debris becomes a vital function. Indeed, AM‐deficient mice succumb to influenza virus (IAV) infection due to a defect in gas exchange and insufficient peripheral oxygen saturation (Schneider et al, 2014b). We now found that pMΦ‐derived AM failed to prevent asphyxia and mortality following IAV infection, in contrast to fetal monocyte‐derived AM.
Taken together, our cell transfer studies demonstrate that fetal monocytes possess a high mitochondrial bioenergetics and glycolytic capacity allowing them to outcompete pMΦ in the development of AM. The results provide the rational and experimental evidence for a proposal based on genetic fate mapping indicating that a second wave of transient definitive EMP migrates from the YS to the fetal liver, where they develop into monocytes prior to entry into embryonal tissues (except the brain) where they can replace primitive EMP‐derived macrophages. The selective pressure for a second wave of fetal monocyte‐derived tissue macrophages can be explained by an increase in their functional capacity.
Materials and Methods
Mice
C57BL/6 CD45.2 and congenic CD45.1 mice were originally from the Jackson Laboratory. Csf2ra −/− mice were recently established in our laboratory (Schneider et al, 2017). Csf2rb −/− mice (Stanley et al, 1994) were originally provided by A. Dunn, Ludwig Institute for Cancer Research, Victoria, Australia, and backcrossed to C57BL/6 in our facility. Myb +/− mice were provided by J. Frampton at Birmingham University Medical School. All mice were housed and bred under specific pathogen‐free conditions in individually ventilated cages in a controlled day‐night cycle at the ETH Phenomics Facility and were used for experiments on 6–12 weeks (adults) unless otherwise stated. All animal experiments were performed according to the guidelines (Swiss Animal Protection Ordinance (TschV), Zurich) and Swiss animal protection law (TschG) and had been approved by the local animal ethics committee (cantonal veterinary office).
Time mating
Female C57BL/6 CD45.1, CD45.2, or Myb +/− mice were housed together with matching male mice overnight. The vaginal plug was checked on the next day morning that was designated as embryonic day 0.5 (E0.5).
Adoptive transfer
Neonatal (days 0–3 after birth) Csf2ra −/− recipient mice were transferred intranasally with sorted cells in 10 μl endotoxin‐free PBS. For single transfer experiments, 50,000 of fetal liver Mo and pMΦ, and 15,000 of YS EMP were transferred, respectively. For competitive transfer experiments, 10,000–25,000 cells in double‐transfer and 5,000 cells in triple‐transfer from each origin were mixed and transferred, respectively.
Viral infection
Mice were anesthetized and intratracheally (i.t.) infected with 20 pfu of influenza virus PR8 (A/Puerto Rico/34, H1N1) in 50 μl endotoxin‐free PBS. Temperature and weight of mice were monitored daily and animals were euthanized if they fulfilled severity criteria set out by institutional and cantonal guidelines.
Measurement of oxygen saturation
The MouseOx™ Pulse‐oximeter (Starr Life Sciences) was used to measure oxygen (O2) saturation in influenza‐infected mice on day 7 post‐infection. The depilatory agent was applied to the neck of mice 2–3 days prior to measurement to remove hair. Mice were sedated with 2.5 mg/kg intraperitoneal midazolam (Roche) 0.5–1 h before measurement. The sensor clip was placed on the neck, and O2 saturation was measured each second over 3–5 min per mouse. Data shown are the average value of each mouse.
Cell suspension preparation
Mice were euthanized by overdose (400 mg/kg body weight) of sodium pentobarbital by i.p. injection. For total protein quantification and cell detection, the lungs were washed three times with 0.4 ml of ice‐cold PBS containing 2 mM EDTA through an intratracheal cannula. BAL fluid was collected, and cells were harvested by centrifugation. Lungs were removed after perfusion with ice‐cold PBS from adult mice. YS, fetal liver, and fetal lung were removed at indicated time points, respectively. Organs were minced and then digested at 37°C in IMDM medium containing 2.0 mg/ml of type IV collagenase (Worthington), 0.125 mg/ml DNase I (Sigma) and 3% FCS for 45 min (lungs), 30 min (fetal lungs), 15 min (fetal livers), and 2 h (YS), respectively, and subsequently passed through a 70‐μm‐cell strainer. Ammonium‐chloride‐potassium (ACK) lysing buffer was used for erythrocyte lysis for all samples.
Flow cytometry and cell sorting
Multiparameter assessment and cell sorting were performed using LSRFortessa, BD FACS ARIA II, and ARIA III (BD Biosciences), and data were analyzed with FlowJo software (Treestar). After blocking the FcgIII/II receptors by incubation with homemade anti‐CD16/32 (2.4G2), single‐cell suspensions were incubated with the indicated fluorochrome‐conjugated or biotinylated monoclonal antibodies in FACS buffer (PBS containing 2% FCS and 2 mM EDTA) and then washed twice before detection. Monoclonal antibodies specific to mouse CD45 (30‐F11), CD11c (N418), F4/80 (BM8), CD11b (M1/70), SiglecF (E50‐2440, BD Biosciences), CD45.1 (A20), CD45.2 (104), C‐kit (2B8), Ly6C (HK1.4), CD131 (JORO50, BD Biosciences), CD116 (698423, R&D), Ly6G (1A8, BD Biosciences), MHC class II (M5/114.15.2, eBioscience), and CX3CR1 (SA011F11) were purchased from BioLegend unless otherwise stated. Dead cells were excluded using the live/dead marker eFluor780 (eBioscience).
Protein quantification
Total protein concentrations in BAL fluid were detected by Pierce BCA Protein Assay Kit according to the manufacturer's instructions (Thermo Scientific).
RNA analysis by real‐time quantitative PCR
Total RNA was extracted using RNAzol (Sigma‐Aldrich), followed by reverse transcription using GoScript Reverse Transcriptase (Promega) according to the manufacturer's instructions. Real‐time quantitative PCR (RT‐PCR) was performed using KAPA SYBR FAST (Sigma‐Aldrich) on an i‐Cycler (Bio‐Rad Laboratories) according to manufacturer's protocol. Expression was normalized to the housekeeping gene Tbp for mRNA expression and the value were calculated using the comparative threshold cycle method (2−ΔCt).
List of primers used:
Tbp Forward: 5′‐ TTGACCTAAAGACCATTGCACTTC ‐3′
Tbp Reverse: 5′‐ TTCTCATGATGACTGCAGCAAA ‐3′
Pparg Forward: 5′‐ GTGATGGAAGACCACTCGCATT ‐3′
Pparg Reverse: 5′‐ CCATGAGGGAGTTAGAAGGTTC ‐3′
Myb Forward: 5′‐ AGACCCCGACACAGCATCTA ‐3′
Myb Reverse: 5′‐ CAGCAGCCCATCGTAGTCAT ‐3′
Mafb Forward: 5′‐ ACTCCCTGTCCCTGCCATG ‐3′
Mafb Reverse: 5′‐ CGTCCTTCCTCCCTCTAGCT ‐3′
Maf Forward: 5′‐ GGATGGCTTCAGAACTGGCA ‐3′
Maf Reverse: 5′‐ AACATATTCCATGGCCAGGG ‐3′
Eno1 Forward: 5′‐ GCCCTAGAACTCCGAGACAA ‐3′
Eno1 Reverse: 5′‐ CAGAGCAGGCGCAATAGTTT ‐3′
Slc2a6 Forward: 5′‐ TGATACCTTCCCCGAGGTG ‐3′
Slc2a6 Reverse: 5′‐ TAGCCAGGAACACCCTTCTG ‐3′
Txn1 Forward: 5′‐ ATGACTGCCAGGATGTTGC ‐3′
Txn1 Reverse: 5′‐ CCTTGTTAGCACCGGAGAAC ‐3′
Txnip Forward: 5′‐ CCTGACCTAATGGCACCAG ‐3′
Txnip Reverse: 5′‐ AGGAATGAACATGCAGGAAAC ‐3′
RNA sequencing and gene set enrichment analysis
20,000–100,000 indicated cell populations were collected into TRIzol (Life Technologies), phase separation was achieved with the addition of chloroform (Sigma), and total RNA was precipitated from the aqueous layer with isopropanol (Sigma) using glycogen (Roche) as a carrier. RNA samples were sent to the Functional Genomics Center Zurich, where the RNA Sequencing was performed. The TruSeq RNA Stranded sample kit (Illumina) was used to construct the sequencing libraries. In brief, total RNA samples (100 ng) were poly (A) enriched and reverse‐transcribed into double‐stranded cDNA, and TruSeq adapters were then ligated to double‐stranded cDNA, then fragments containing TruSeq adapters on both ends were selectively enriched with PCR and subsequently sequenced on the Illumina HiSeq 2500 in single‐end mode for 150 cycles. The fragments were mapped to the ensemble mouse reference genome GRCm38 (version 25.06.2015) using the STAR aligner (Dobin et al, 2013). For normalization, the read counts were scaled with the use of the trimmed mean of M‐values (TMM) method proposed by Robinson and Oshlack (Robinson & Oshlack, 2010). Principal component analysis, clustering matrix, and heatmap were generated using R. Gene set enrichment analysis (GSEA) was performed with the GSEA v3.0 tool from Broad Institute (Mootha et al, 2003; Subramanian et al, 2005). Gene Ontology (GO) gene sets for mouse were downloaded from http://baderlab.org/GeneSets (version from 2018‐10‐01). Only gene sets consisting of more than three genes were analyzed, and the resulting enriched sets (with FDR q value < 0.1) were visualized with Cytoscape 3.5.0 (Shannon et al, 2003) with use of Enrichment Map plugin (Merico et al, 2010). Clustering of gene sets was performed with AutoAnnotate 1.2.
Seahorse extracellular flux analysis
FACS‐purified primitive MΦ and fetal monocytes from fetal liver were seeded at 200,000 cells per well in poly‐D‐lysine coated plates for 1 h. Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured in unbuffered RPMI 1640 with 2 mM l‐glutamine containing 11.11 mM or without glucose, using a 96‐well extracellular flux analyzer according to protocols from manufacturer (Seahorse Bioscience). Baseline levels and responses to the indicated compounds were determined. For mitochondrial stress analysis, 1.5 μM oligomycin, 1 μM FCCP, and 3 μM rotenone + 2 μg/ml antimycin A were provided (all from Seahorse Bioscience). For glycolysis stress analysis, 25 mM glucose (Sigma‐Aldrich), 1.5 μM oligomycin, and 45 mM 2′‐deoxy‐D‐glucose (Sigma‐Aldrich) were added. Primitive MΦ and fetal monocytes from fetal lung were seeded at 50,000 cells per well, and ECAR were measured using same protocol.
Statistical analysis
Prism software (GraphPad) was used for statistical analysis. The data were expressed as means and SEMs. Student's t‐test (unpaired), paired t‐test, and ANOVA (one‐way) were used. *P < 0.05, **P < 0.01, ***P < 0.001.
Author contributions
FL and MK designed the experiments; FL performed and analyzed the majority of experiments. KMO and LMP performed and analyzed specific experiments. CS and MK discussed data and provided conceptualization. FL and MK wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Peter Nielsen for discussion and editing of the manuscript, Lucia Balázová and Christian Wolfrum for support with the Seahorse instrument, Anette Schütz and Malgorzata Kisielow at the ETH Flow Cytometry Core Facility for cell sorting, and J. Frampton (Birmingham University Medical School) for c‐Myb +/− mice. We are grateful for research grants from SNF (310030_163443/1 and 310030B_182829).
The EMBO Journal (2020) 39: e103205
Data availability
The mouse RNA‐seq data first reported in this study are available at the Gene Expression Omnibus (GEO) repository under the accession number GSE140645 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140645).
References
- Aziz A, Soucie E, Sarrazin S, Sieweke MH (2009) MafB/c‐Maf deficiency enables self‐renewal of differentiated functional macrophages. Science 326: 867–871 [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Jalil A, Klaine M, Jung S, Cumano A, Godin I (2005) Three pathways to mature macrophages in the early mouse yolk sac. Blood 106: 3004–3011 [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D (2007) Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 134: 4147–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies J, Markus J, Wolff L (2002) Covalent attachment of the SUMO‐1 protein to the negative regulatory domain of the c‐Myb transcription factor modifies its stability and transactivation capacity. J Biol Chem 277: 8999–9009 [DOI] [PubMed] [Google Scholar]
- Dahle O, Andersen TO, Nordgard O, Matre V, Del Sal G, Gabrielsen OS (2003) Transactivation properties of c‐Myb are critically dependent on two SUMO‐1 acceptor sites that are conjugated in a PIASy enhanced manner. Eur J Biochem 270: 1338–1348 [DOI] [PubMed] [Google Scholar]
- Davies LC, Jenkins SJ, Allen JE, Taylor PR (2013) Tissue‐resident macrophages. Nat Immunol 14: 986–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT et al (2014) Embryonic and adult‐derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40: 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu QF, Liu Y, Fan Y, Hua SN, Qu HY, Dong SW, Li RL, Zhao MY, Zhen Y, Yu XL et al (2015) Alpha‐enolase promotes cell glycolysis, growth, migration, and invasion in non‐small cell lung cancer through FAK‐mediated PI3K/AKT pathway. J Hematol Oncol 8: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S et al (2012) Gene‐expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13: 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerencser AA, Neilson A, Choi SW, Edman U, Yadava N, Oh RJ, Ferrick DA, Nicholls DG, Brand MD (2009) Quantitative microplate‐based respirometry with correction for oxygen diffusion. Anal Chem 81: 6868–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER et al (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330: 841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Guilliams M (2016) Tissue‐resident macrophage ontogeny and homeostasis. Immunity 44: 439–449 [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK (2016) New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol 17: 34–40 [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F et al (2015) Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature 518: 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN (2013) Alveolar macrophages develop from fetal monocytes that differentiate into long‐lived cells in the first week of life via GM‐CSF. J Exp Med 210: 1977–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde SP, Zhao J, Ashmun RA, Shapiro LH (1999) c‐Maf induces monocytic differentiation and apoptosis in bipotent myeloid progenitors. Blood 94: 1578–1589 [PubMed] [Google Scholar]
- Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, Leboeuf M, Low D, Oller G, Almeida F et al (2012) Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac‐derived macrophages. J Exp Med 209: 1167–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC et al (2015) C‐Myb(+) erythro‐myeloid progenitor‐derived fetal monocytes give rise to adult tissue‐resident macrophages. Immunity 42: 665–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Ginhoux F (2015) Ontogeny of tissue‐resident macrophages. Front Immunol 6: 486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T (2000) MafB is an inducer of monocytic differentiation. EMBO J 19: 1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Schneider C, Nobs SP (2015) The development and function of lung‐resident macrophages and dendritic cells. Nat Immunol 16: 36–44 [DOI] [PubMed] [Google Scholar]
- van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, Hoffmann E, Beyaert R, Saeys Y, Lambrecht BN et al (2016) Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue‐resident macrophages. Immunity 44: 755–768 [DOI] [PubMed] [Google Scholar]
- Lieu YK, Reddy EP (2009) Conditional c‐myb knockout in adult hematopoietic stem cells leads to loss of self‐renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci U S A 106: 21689–21694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillig CH, Holmgren A (2007) Thioredoxin and related molecules–from biology to health and disease. Antioxid Redox Signal 9: 25–47 [DOI] [PubMed] [Google Scholar]
- Lin Y, Yoder MC, Yoshimoto M (2014) Lymphoid progenitor emergence in the murine embryo and yolk sac precedes stem cell detection. Stem Cells Dev 23: 1168–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG (2011) Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 27: 441–464 [DOI] [PubMed] [Google Scholar]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Gunther P, Crozet L, Jacome‐Galarza CE, Handler K, Klughammer J, Kobayashi Y et al (2016) Specification of tissue‐resident macrophages during organogenesis. Science 353: aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, Kingsley PD, Koniski AD, Palis J (2015a) Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep 11: 1892–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KE, Frame JM, Palis J (2015b) Early hematopoiesis and macrophage development. Semin Immunol 27: 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD (2010) Enrichment map: a network‐based method for gene‐set enrichment visualization and interpretation. PLoS ONE 5: e13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E et al (2003) PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273 [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ Jr, Potter SS (1991) A functional c‐myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65: 677–689 [DOI] [PubMed] [Google Scholar]
- Muri J, Heer S, Matsushita M, Pohlmeier L, Tortola L, Fuhrer T, Conrad M, Zamboni N, Kisielow J, Kopf M (2018) The thioredoxin‐1 system is essential for fueling DNA synthesis during T‐cell metabolic reprogramming and proliferation. Nat Commun 9: 1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls DG, Darley‐Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick DA (2010) Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp 46: 2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Pearce EJ (2016) Immunometabolism governs dendritic cell and macrophage function. J Exp Med 213: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J (2016) Interaction of the macrophage and primitive erythroid lineages in the mammalian embryo. Front Immunol 7: 669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ et al (2007) TXNIP regulates peripheral glucose metabolism in humans. PLoS Med 4: 868–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero EG, Geissmann F (2016) The development and maintenance of resident macrophages. Nat Immunol 17: 2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantakari P, Jappinen N, Lokka E, Mokkala E, Gerke H, Peuhu E, Ivaska J, Elima K, Auvinen K, Salmi M (2016) Fetal liver endothelium regulates the seeding of tissue‐resident macrophages. Nature 538: 392–396 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA‐seq data. Genome Biol 11: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Nobs SP, Heer AK, Kurrer M, Klinke G, van Rooijen N, Vogel J, Kopf M (2014a) Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog 10: e1004053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M (2014b) Induction of the nuclear receptor PPAR‐gamma by the cytokine GM‐CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol 15: 1026–1037 [DOI] [PubMed] [Google Scholar]
- Schneider C, Nobs SP, Heer AK, Hirsch E, Penninger J, Siggs OM, Kopf M (2017) Frontline Science: coincidental null mutation of Csf2ralpha in a colony of PI3Kgamma−/− mice causes alveolar macrophage deficiency and fatal respiratory viral infection. J Leukoc Biol 101: 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo‐Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW et al (2012) A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336: 86–90 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucie EL, Weng Z, Geirsdottir L, Molawi K, Maurizio J, Fenouil R, Mossadegh‐Keller N, Gimenez G, VanHille L, Beniazza M et al (2016) Lineage‐specific enhancers activate self‐renewal genes in macrophages and embryonic stem cells. Science 351: aad5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E, Lieschke GJ, Grail D, Metcalf D, Hodgson G, Gall JA, Maher DW, Cebon J, Sinickas V, Dunn AR (1994) Granulocyte/macrophage colony‐stimulating factor‐deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc Natl Acad Sci U S A 91: 5592–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al (2005) Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner R, Crawford A, Mucenski M, Frampton J (2000) Initiation of adult myelopoiesis can occur in the absence of c‐Myb whereas subsequent development is strictly dependent on the transcription factor. Oncogene 19: 3335–3342 [DOI] [PubMed] [Google Scholar]
- Tillmanns S, Otto C, Jaffray E, Du Roure C, Bakri Y, Vanhille L, Sarrazin S, Hay RT, Sieweke MH (2007) SUMO modification regulates MafB‐driven macrophage differentiation by enabling Myb‐dependent transcriptional repression. Mol Cell Biol 27: 5554–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tober J, McGrath KE, Palis J (2008) Primitive erythropoiesis and megakaryopoiesis in the yolk sac are independent of c‐myb. Blood 111: 2636–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bossche J, O'Neill LA, Menon D (2017) Macrophage immunometabolism: where are we (going)? Trends Immunol 38: 395–406 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Data Availability Statement
The mouse RNA‐seq data first reported in this study are available at the Gene Expression Omnibus (GEO) repository under the accession number GSE140645 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140645).


