Abstract
Purpose
To assess the performance of an automated myocardial T2 and extracellular volume (ECV) quantification method using transfer learning of a fully convolutional neural network (CNN) pretrained to segment the myocardium on T1 mapping images.
Materials and Methods
A single CNN previously trained and tested using 11 550 manually segmented native T1-weighted images was used to segment the myocardium for automated myocardial T2 and ECV quantification. Reference measurements from 1525 manually processed T2 maps and 1525 ECV maps (from 305 patients) were used to evaluate the performance of the pretrained network. Correlation coefficient (R) and Bland-Altman analysis were used to assess agreement between automated and reference values on per-patient, per-slice, and per-segment analyses. Furthermore, transfer learning effectiveness in the CNN was evaluated by comparing its performance to four CNNs trained using manually segmented T2-weighted and postcontrast T1-weighted images and initialized using random-weights or weights of the pretrained CNN.
Results
T2 and ECV measurements using the pretrained CNN strongly correlated with reference values in per-patient (T2: R = 0.88, 95% confidence interval [CI]: 0.85, 0.91; ECV: R = 0.91, 95% CI: 0.89, 0.93), per-slice (T2: R = 0.83, 95% CI: 0.81, 0.85; ECV: R = 0.84, 95% CI: 0.82, 0.86), and per-segment (T2: R = 0.75, 95% CI: 0.74, 0.77; ECV: R = 0.76, 95% CI: 0.75, 0.77) analyses. In Bland-Altman analysis, the automatic and reference values were in good agreement in per-patient (T2: 0.3 msec ± 2.9; ECV: −0.3% ± 1.7), per-slice (T2: 0.1 msec ± 4.6; ECV: −0.3% ± 2.5), and per-segment (T2: 0.0 msec ± 6.5; ECV: −0.4% ± 3.5) analyses. The performance of the pretrained network was comparable to networks refined or trained from scratch using additional manually segmented images.
Conclusion
Transfer learning extends the utility of pretrained CNN-based automated native T1 mapping analysis to T2 and ECV mapping without compromising performance.
Supplemental material is available for this article.
© RSNA, 2020
Summary
A single convolutional neural network, trained for myocardium segmentation on T1 maps, can be used to automate myocardial T2 and extracellular volume maps with comparable accuracy to that of manual assessment.
Key Points
■ Convolutional neural networks trained to segment the myocardium using cardiac native T1 mapping images can be used to automate myocardial T2 and extracellular volume (ECV) quantification without a need for retraining with separate T2 or ECV mapping images.
■ An automated myocardial tissue mapping platform using deep learning–based image segmentation has potential to facilitate the clinical utility and standardization of quantitative myocardial mapping.
Introduction
Cardiac MRI mapping allows noninvasive quantification of tissue alterations in the myocardium based on changes in T1 and T2 relaxation times and extracellular volume (ECV) (1–3). In myocardial mapping, a set of images is acquired with varying imaging parameters, such as varied inversion times and varied durations of T2 preparation pulses (TET2prep), to sample T1 or T2 relaxation curves (4–9). Image analysis of myocardial mapping typically involves three main steps: motion correction (10–13), curve fitting (4,7,14), and manual delineation of the myocardium for quantifying T1 or T2 values (15). Additional ECV map processing aligns native and postcontrast T1 maps prior to ECV calculations (16). This workflow is time-consuming, prone to subjective analysis errors, and burdens cardiac MRI readings, therefore creating an unmet clinical need for automating myocardial tissue mapping.
Deep learning models based on convolutional neural networks (CNNs) (17) have successfully been used to automate image analysis in different cardiac MRI applications, including segmentation of the left and right ventricles in cine sequences (18–20) and myocardial scarring by late gadolinium enhancement (21). Image segmentation requires a large dataset of manually segmented images to estimate the internal parameters of the network, referred to as weights (22). One of the main challenges of CNN training is the limited availability of manually segmented medical data for training (17). To mitigate this issue, transfer learning offers a way to adapt a trained network to different tasks (23) by transferring network-embedded knowledge to new applications. Transfer learning has been shown to perform better and provide more consistent tissue segmentation (24) and image retrieval (25) compared with networks trained from scratch.
Recently, a deep learning–based analysis platform was developed for automating analysis of native myocardial T1 mapping (26). T1 mapping data acquired by slice-interleaved myocardial T1 mapping sequence (STONE) (7) were used for training and validation. While similar network architectures could potentially automate other tissue parameters, such as ECV or T2 mapping, doing so would require a newly labeled dataset and a dedicated neural network for each individual parameter and sequence, which is not clinically feasible.
In this study, we sought to evaluate the performance of a pretrained deep neural network using native T1 mapping datasets to automate T2 and ECV measurements. Three neural networks with similar architectures were employed to analyze each parametric mapping sequence: (a) the native T1 pretrained network; (b) a refined native T1 pretrained network (trained using a small dataset of T2-weighted and postcontrast T1-weighted images); and (c) a new network trained from scratch using a small dataset of T2-weighted and postcontrast T1-weighted images. Network performance was evaluated with respect to corresponding reference measurements computed by manual analysis.
Materials and Methods
Processing Pipeline
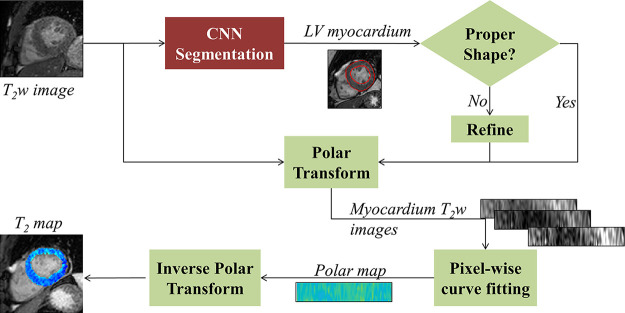
The proposed analysis pipeline (Fig 1) begins with automatic segmentation of the myocardium in each of the input of T1- or T2-weighted images. A CNN with a U-Net architecture (27) was used to segment the input images and produce a binary image containing a single object (ie, myocardium). If more than one object is segmented, only the largest object is maintained. To ensure that the segmented object is indeed the myocardium, two conditions are imposed on two automatically computed shape descriptors of the object: (a) Euler number and (b) eccentricity. The Euler number is the number of objects in an image subtracted by the number of holes in the objects; it equals 0 for a typical myocardium shape in the short axis. Eccentricity measures the aspect ratio of an ellipse (equals 0 for a circle and 1 for a line segment); in our dataset, the maximum eccentricity for a myocardium shape was 0.65 (26). An image was considered successfully segmented if it contained an object (ie, myocardium) with Euler number equal to 0 and eccentricity of less than 0.65. An unsuccessfully segmented object, termed Bu (ie, an incomplete annulus shape), was replaced by a successfully segmented object, termed Bs, of an image at the same slice location. Affine transformation was applied first to align Bs with Bu to compensate for breathing and cardiac motion (26). Refinement was not performed if Bs did not have a nonzero Euler number or eccentricity above 0.65, and subsequently Bu was excluded from analysis. The segmented myocardium on all weighted images of a given slice were transformed into polar coordinates for alignment (26). Pixelwise curve fitting was subsequently applied to the aligned images using three-parameter and two-parameter models for T2 mapping (14) and T1 mapping (7), respectively (see fitting method in Appendix E1 [supplement]). The ECV map was calculated as follows (16):
.
|
Figure 1a:
(a) Automated T2 mapping pipeline. A deep fully convolutional neural network (CNN) based on U-Net architecture is used for left ventricular (LV) myocardium segmentation followed by an automatic refinement step. The segmented myocardium is transformed into polar coordinates on a uniform grid to align the myocardial regions from different images. The T2 map is then fitted through the aligned myocardium. The resulting rectangular T2 map is finally transformed back to Cartesian coordinates using the stored correspondence between the Cartesian and polar coordinates of each myocardium point. (b) Automated extracellular volume (ECV) mapping pipeline. The CNN is used for LV myocardium segmentation followed by an automatic refinement step. The segmented myocardium is first transformed into polar coordinates, and the native and postcontrast T1 maps are then fitted through the aligned myocardium. Points located within 5 pixels of the segmented myocardial center of mass (ie, center of blood pool) are automatically selected. The average signal intensity of these points is calculated for all T1-weighted images followed by blood T1 fitting. The ECV map is calculated using the native and postcontrast myocardial T1 maps, blood T1 values, and the hematocrit. Last, the resulting rectangular ECV map is transformed back to Cartesian coordinates.
Figure 1b:
(a) Automated T2 mapping pipeline. A deep fully convolutional neural network (CNN) based on U-Net architecture is used for left ventricular (LV) myocardium segmentation followed by an automatic refinement step. The segmented myocardium is transformed into polar coordinates on a uniform grid to align the myocardial regions from different images. The T2 map is then fitted through the aligned myocardium. The resulting rectangular T2 map is finally transformed back to Cartesian coordinates using the stored correspondence between the Cartesian and polar coordinates of each myocardium point. (b) Automated extracellular volume (ECV) mapping pipeline. The CNN is used for LV myocardium segmentation followed by an automatic refinement step. The segmented myocardium is first transformed into polar coordinates, and the native and postcontrast T1 maps are then fitted through the aligned myocardium. Points located within 5 pixels of the segmented myocardial center of mass (ie, center of blood pool) are automatically selected. The average signal intensity of these points is calculated for all T1-weighted images followed by blood T1 fitting. The ECV map is calculated using the native and postcontrast myocardial T1 maps, blood T1 values, and the hematocrit. Last, the resulting rectangular ECV map is transformed back to Cartesian coordinates.
Blood T1 was automatically calculated from a circular region (radius = 5 pixels) located at the center of mass of the segmented myocardium. The resulting maps were inverse-transformed to Cartesian coordinates (26). To avoid errors from partial voluming, the myocardium maps were pruned to exclude boundary pixels using simple morphologic operations (26). Pixels with values outside typical ranges of myocardial T2 (30–100 msec) or ECV (10%–60%) were excluded from analysis. It should be noted that the CNN was only used for segmentation of the myocardium throughout the entire analysis pipeline.
Image Acquisition
Patients known to have or suspected of having cardiovascular disease referred for clinical cardiac MRI were prospectively recruited from 2016 to 2017. The institutional review board approved the study, written informed consent was obtained from each patient prior to examination, and patient data handling was Health Insurance Portability and Accountability Act compliant. A total of 305 patients (203 male patients and 102 female patients; mean age: 55 years ± 15 [standard deviation]) were recruited for myocardial T2 and ECV mapping (see acquisition parameters in Appendix E1 [supplement]).
Training and validation images.—T1- and T2-weighted images from 305 patients were randomly divided into (a) a network training subset (21 patients; 945 T2-weighted images and 1155 postcontrast T1-weighted images); (b) a validation subset (284 patients; 12 780 T2-weighted images and 15 620 postcontrast T1-weighted images); and (c) a network testing subset randomly selected from the validation subset (30 patients; 1650 postcontrast T1-weighted images and 1350 T2-weighted images). Network training and validation subsets were mutually exclusive. All images were standardized to 256 × 256 pixels using bicubic interpolation (28). Network training and testing images were manually segmented (Y.Z., 7 years of experience in cardiac MRI) to extract the endocardium and epicardium contours.
Reference measurements.—Reference T2 and ECV values were measured by an experienced reader (S.N., 8 years of experience in cardiac MRI) using an in-house tissue mapping analysis tool. First, T1- or T2-weighted images for each slice were registered using a nonrigid image registration algorithm (11) to compensate for in-plane motion. The ECV map was computed using Equation (1). Boundary pixels and image artifacts on T2 and ECV maps were manually excluded by the reader.
Evaluation of Transfer Learning Effectiveness
We employed a simple transfer learning approach to extend the utility of a single pretrained CNN network in all T2 and ECV mapping images. The network was pretrained from scratch and tested with 11 550 manually segmented native T1-weighted images (210 patients; publicly available at https://doi.org/10.7910/DVN/DHEUAV ). We refer to this network as 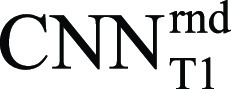 to indicate that it was trained using a native T1 image set and that its weights were initialized using standard Gaussian random (rnd) values (ie, from-scratch training).
to indicate that it was trained using a native T1 image set and that its weights were initialized using standard Gaussian random (rnd) values (ie, from-scratch training). 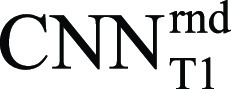 was used to automate T2-weighted mapping (Exp#1) and ECV mapping (Exp#4). We rigorously compared the performance of
was used to automate T2-weighted mapping (Exp#1) and ECV mapping (Exp#4). We rigorously compared the performance of 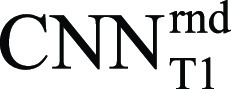 to four other networks (Table 1): (a)
to four other networks (Table 1): (a)
 , initialized using random weights and trained using 1155 postcontrast T1-weighted images; (b)
, initialized using random weights and trained using 1155 postcontrast T1-weighted images; (b)
 , initialized using random weights and trained using 945 T2-weighted images; (c)
, initialized using random weights and trained using 945 T2-weighted images; (c)
 , initialized using the weights of
, initialized using the weights of 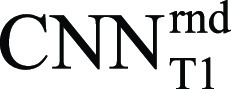 and refined using 1155 postcontrast T1-weighted images; and (d)
and refined using 1155 postcontrast T1-weighted images; and (d)
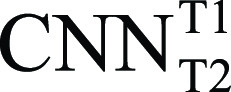 , initialized using the weights of
, initialized using the weights of 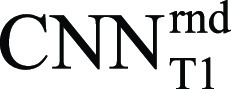 and refined using 945 T2-weighted images. Networks
and refined using 945 T2-weighted images. Networks 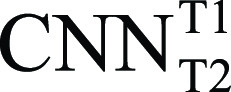 and
and  automated T2 mapping (Exp#2 and Exp#3), while
automated T2 mapping (Exp#2 and Exp#3), while  and
and  automated postcontrast T1 mapping (Exp#5 and Exp#6).
automated postcontrast T1 mapping (Exp#5 and Exp#6).
Table 1:
Performance Summary of Five Convolutional Neural Networks to Automate T2 and ECV Measurements

Data Analysis
Average myocardial T2 and ECV values were calculated from automatically reconstructed maps for each patient, slice, and segment. For per-patient analysis, measurements were defined as the mean T2 or ECV value of the myocardial pixels from all successfully analyzed slices. A 16-segment model for basal (six segments), midcavity (six segments), and apical (four segments) slices was used for per-segment analysis, and insertion points were manually selected. All other steps were performed using the automated analysis platform. We evaluated agreement between automated T2 and ECV values and corresponding reference values using linear regression and Pearson correlation and Bland-Altman analyses. Each network was evaluated according to per-patient and per-slice success rate, defined as percentage of successfully generated maps relative to the total number of patients and total number of slices, respectively. Mapping of a given slice was considered successful if the network successfully segmented the myocardium on at least six T2-weighted images (for T2 mapping), or on at least eight native and eight postcontrast T1-weighted images (for ECV mapping). Mapping of a given patient was considered successful if at least one apical, one midventricular, and one basal slice were successfully computed. The Wilcoxon signed-rank test was conducted to compare automatic and reference measurements. Statistical significance was set to P < .05. Additionally, we calculated the sensitivity and specificity of 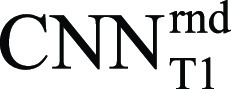 for identifying patients with myocardium mapping parameters above a cutoff threshold of 58 msec for T2 mapping and above 29% for ECV mapping. The threshold values were arbitrarily set to mean + 2 standard deviations of the parameter values reported in normal populations (29,30). Dice similarity coefficient was used to assess the CNN segmentation accuracy by measuring the overlap between the CNN-segmented and the manually segmented myocardium.
for identifying patients with myocardium mapping parameters above a cutoff threshold of 58 msec for T2 mapping and above 29% for ECV mapping. The threshold values were arbitrarily set to mean + 2 standard deviations of the parameter values reported in normal populations (29,30). Dice similarity coefficient was used to assess the CNN segmentation accuracy by measuring the overlap between the CNN-segmented and the manually segmented myocardium.
Results
The pretrained 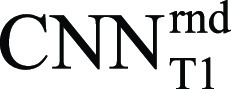 network delineated the epicardium and endocardium for different slices, inversion times, and TET2prep times (Fig 2). Representative T2 and ECV maps reconstructed using
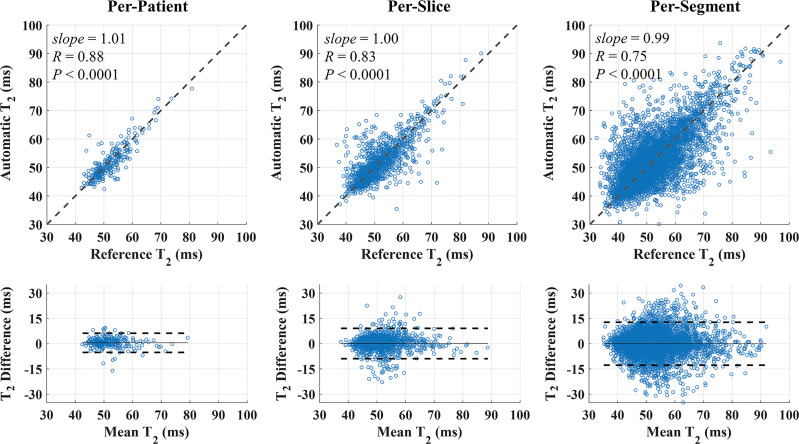
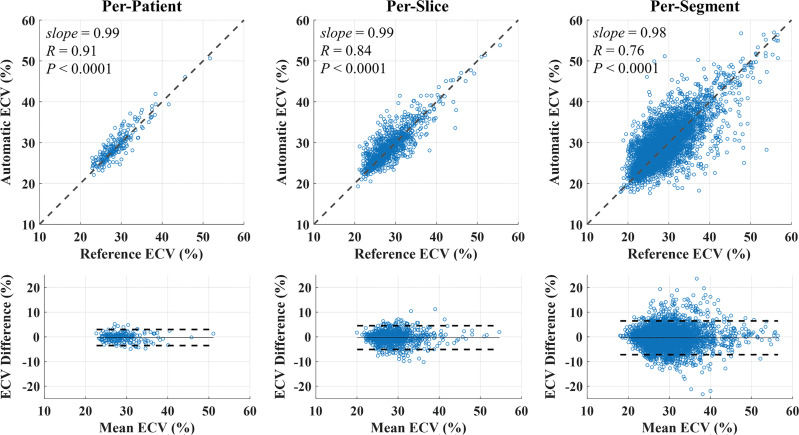
network delineated the epicardium and endocardium for different slices, inversion times, and TET2prep times (Fig 2). Representative T2 and ECV maps reconstructed using 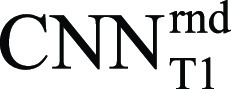 are shown (Fig 3). In Exp#1, the success rate for reconstructing T2 maps was 276 of 284 (97%) (per-patient) and 1329 of 1420 (94%) (per-slice) (Table 2). There were no significant differences between automatic and reference T2 measurements in per-patient (51.9 msec ± 5.5 vs 51.6 msec ± 5.7; P =.06), per-slice (51.7 msec ± 7.0 vs 51.6 msec ± 6.8; P =.08), and per-segment (51.5 msec ± 8.4 vs 51.6 msec ± 8.4; P =.38) analyses. There was a strong correlation between automated and reference T2 measurements in per-patient (R = 0.88, slope = 1.01, P <.0001), per-slice (1310 slices: R = 0.83, slope = 1.00, P <.0001), and per-segment (6899 segments: R = 0.75, slope = 0.99, P <.0001) analyses (Fig 4, top row). The automatic and reference T2 values were in good agreement in per-patient (0.3 msec ± 2.9, 95% confidence interval [CI]: −5.5 msec, 6.0 msec), per-slice (0.1 msec ± 4.6, 95% CI: −8.9 msec, 9.1 msec), and per-segment (0.0 msec ± 6.5, 95% CI: −12.8 msec, 12.7 msec) analyses (Fig 4, bottom row). Exp#4 had a success rate of 237 of 284 (83%) (per-patient) and 1111 of 1420 (78%) (per-slice) for reconstruction of ECV maps (Table 2). Automatic and reference ECV measurements were comparable in per-patient (28.7% ± 4.0 vs 29.0% ± 4.0; P =.06) and per-slice (28.7% ± 4.3 vs 29.0% ± 4.2; P =.05) analyses but not in per-segment analysis (28.% ± 5.0 vs 29.0% ± 5.0; P <.0001). The automatic ECV measurements showed a strong correlation with the reference ECV in per-patient (R = 0.91, slope = 0.99, P <.0001), per-slice (1101 slices: R = 0.84, slope = 0.99, P <.0001), and per-segment (6275 segments: R = 0.76, slope = 0.98, P <.0001) analyses (Fig 5, top row). Automatic and reference ECV values were in good agreement in per-patient (−0.3% ± 1.7, 95% CI: −3.5%, 3.0%), per-slice (−0.3% ± 2.5, 95% CI: −5.1%, 4.5%), and per-segment ECV (−0.4% ± 3.5, 95% CI: −7.2%, 6.5%) analyses (Fig 5, bottom row).
are shown (Fig 3). In Exp#1, the success rate for reconstructing T2 maps was 276 of 284 (97%) (per-patient) and 1329 of 1420 (94%) (per-slice) (Table 2). There were no significant differences between automatic and reference T2 measurements in per-patient (51.9 msec ± 5.5 vs 51.6 msec ± 5.7; P =.06), per-slice (51.7 msec ± 7.0 vs 51.6 msec ± 6.8; P =.08), and per-segment (51.5 msec ± 8.4 vs 51.6 msec ± 8.4; P =.38) analyses. There was a strong correlation between automated and reference T2 measurements in per-patient (R = 0.88, slope = 1.01, P <.0001), per-slice (1310 slices: R = 0.83, slope = 1.00, P <.0001), and per-segment (6899 segments: R = 0.75, slope = 0.99, P <.0001) analyses (Fig 4, top row). The automatic and reference T2 values were in good agreement in per-patient (0.3 msec ± 2.9, 95% confidence interval [CI]: −5.5 msec, 6.0 msec), per-slice (0.1 msec ± 4.6, 95% CI: −8.9 msec, 9.1 msec), and per-segment (0.0 msec ± 6.5, 95% CI: −12.8 msec, 12.7 msec) analyses (Fig 4, bottom row). Exp#4 had a success rate of 237 of 284 (83%) (per-patient) and 1111 of 1420 (78%) (per-slice) for reconstruction of ECV maps (Table 2). Automatic and reference ECV measurements were comparable in per-patient (28.7% ± 4.0 vs 29.0% ± 4.0; P =.06) and per-slice (28.7% ± 4.3 vs 29.0% ± 4.2; P =.05) analyses but not in per-segment analysis (28.% ± 5.0 vs 29.0% ± 5.0; P <.0001). The automatic ECV measurements showed a strong correlation with the reference ECV in per-patient (R = 0.91, slope = 0.99, P <.0001), per-slice (1101 slices: R = 0.84, slope = 0.99, P <.0001), and per-segment (6275 segments: R = 0.76, slope = 0.98, P <.0001) analyses (Fig 5, top row). Automatic and reference ECV values were in good agreement in per-patient (−0.3% ± 1.7, 95% CI: −3.5%, 3.0%), per-slice (−0.3% ± 2.5, 95% CI: −5.1%, 4.5%), and per-segment ECV (−0.4% ± 3.5, 95% CI: −7.2%, 6.5%) analyses (Fig 5, bottom row).
Figure 2:

Automated myocardial segmentation results by for T2 mapping with different TET2prep values and for native and postcontrast T1 mapping with different inversion times. CNN = convolutional neural network.
Figure 3:
Representative, A, T2 maps and, B, extracellular volume (ECV) maps reconstructed using the automated platform using . The subendocardial and subepicardial edges of the resulting myocardial maps were automatically pruned to exclude pixels in these areas from measurement calculation. The automated maps after pruning are also shown. CNN = convolutional neural network.
Table 2:
Evaluation Metrics for Automatic T2 and ECV Mapping by Network

Figure 4:
Scatterplots and Bland-Altman plots of automated (using ) and reference T2 values in per-patient, per-slice, and per-segment analyses. Automated T2 measurements showed a strong correlation with the reference T2 values in per-patient, per-slice, and per-segment analyses. Dashed lines in scatterplots represent the unity slope line. Automatic and reference T2 values were in good agreement in per-patient (0.3 msec 6 2.9, 95% confidence interval [CI]: −5.5 msec, 6.0 msec), per-slice (0.1 msec 6 4.6, 95% CI: −8.9 msec, 9.1 msec), and per-segment (0.0 msec 6 6.5, 95% CI: −12.8 msec, 12.7 msec) analyses. Solid and dashed lines in Bland-Altman plots represent the bias and 6 1.96 standard deviation limits, respectively. CNN = convolutional neural network.
Figure 5:
Scatterplots and Bland-Altman plots of automated (using ) and reference extracellular volume (ECV) values in per-patient, per-slice, and per-segment analyses. Automated ECV measurement showed a strong correlation with reference ECV values in per-patient, per-slice, and per-segment analyses. Automated and reference ECV values were in good agreement in per-patient (−0.3% 61.7, 95% confidence interval [CI]: −3.5%, 3.0%), per-slice (−0.3% 6 2.5, 95% CI: −5.1%, 4.5%), and per-segment ECV (−0.4% 6 3.5, 95% CI: −7.2%, 6.5%) analyses. Solid and dashed lines in Bland-Altman plots represent the bias and 6 1.96 standard deviation limits, respectively. CNN = convolutional neural network.
Table 2 summarizes the performance of various CNNs at automating T2 and ECV measurements. For T2 mapping, there was a strong correlation between the automated and reference T2 measurements among all three networks in per-patient (R > 0.88, P <.0001), per-slice (R > 0.83, P < .0001), and per-segment (R > 0.75, P < .0001) analyses. Addition of T2-weighted images to the network trained by native T1-weighted images did not significantly improve success rate (277 of 284 [98%] vs 276 of 284 [97%]) or segmentation accuracy (Dice similarity coefficient: 0.87 ± 0.07 vs 0.87 ± 0.11). However,  had a lower success rate than the other two pretrained networks
had a lower success rate than the other two pretrained networks 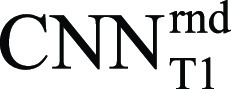 and
and  , presumably due to its smaller total number of training images (945 images for
, presumably due to its smaller total number of training images (945 images for  compared with 3465 and 4410 images for
compared with 3465 and 4410 images for 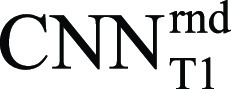 and
and 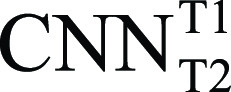 , respectively). These results suggest that a network pretrained with native T1 mapping data can be used to segment T2-weighted images without compromising its performance. For ECV mapping, addition of postcontrast T1-weighted images to the network trained with only native T1-weighted images resulted in 4% improvement in success rate (246 of 284 [87%] vs 237 of 284 [83%]) and 0.03 improvement in segmentation accuracy (0.87 ± 0.08 vs 0.84 ± 0.12). There was a strong correlation between automated and reference ECV measurements in all three CNNs in per-patient (R > 0.91, P < .0001), per-slice (R > 0.84, P < .0001), and per-segment (R > 0.76, P < .0001) analyses (Table 2).
, respectively). These results suggest that a network pretrained with native T1 mapping data can be used to segment T2-weighted images without compromising its performance. For ECV mapping, addition of postcontrast T1-weighted images to the network trained with only native T1-weighted images resulted in 4% improvement in success rate (246 of 284 [87%] vs 237 of 284 [83%]) and 0.03 improvement in segmentation accuracy (0.87 ± 0.08 vs 0.84 ± 0.12). There was a strong correlation between automated and reference ECV measurements in all three CNNs in per-patient (R > 0.91, P < .0001), per-slice (R > 0.84, P < .0001), and per-segment (R > 0.76, P < .0001) analyses (Table 2).
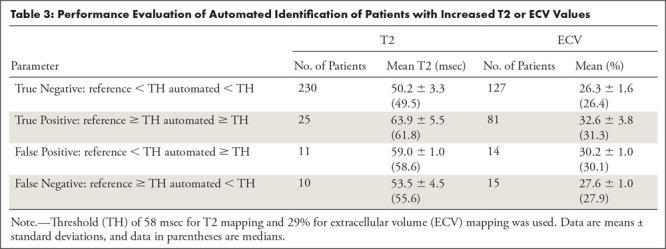
Table 3 summarizes the ability of 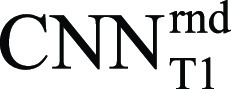 to identify patients with increased T2 (ie, T2 ≥ 58 msec) or ECV (ie, ECV ≥ 29%). The network’s sensitivity and specificity were 25 of 35 (71.4%) and 230 of 241 (95.4%) for T2 mapping, and 81 of 96 (84.4%) and (127 of 141) 90.1% for ECV mapping. Errors in patient identification occurred in patient images with parameter values close to the cutoff threshold; where the average values in the misidentified patients were less than 5 msec and less than 3% for T2 and ECV mapping, respectively.
to identify patients with increased T2 (ie, T2 ≥ 58 msec) or ECV (ie, ECV ≥ 29%). The network’s sensitivity and specificity were 25 of 35 (71.4%) and 230 of 241 (95.4%) for T2 mapping, and 81 of 96 (84.4%) and (127 of 141) 90.1% for ECV mapping. Errors in patient identification occurred in patient images with parameter values close to the cutoff threshold; where the average values in the misidentified patients were less than 5 msec and less than 3% for T2 and ECV mapping, respectively.
Table 3:
Performance Evaluation of Automated Identification of Patients with Increased T2 or ECV Values
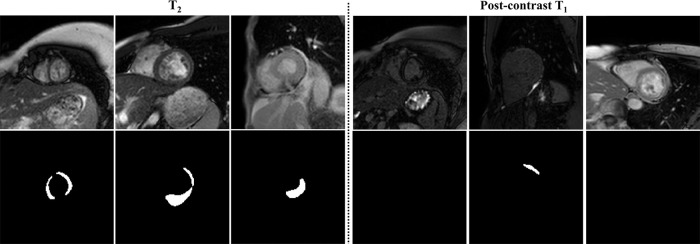
Although the network was capable of delineating the myocardium with high accuracy in the majority of cases, failure was more likely in images with low signal-to-noise ratio or low myocardium-blood contrast (eight in T2 mapping and 47 in ECV mapping) (Fig 6). Although CNN segmentation achieved good performance in per-slice and per-patient analyses, per-segment analysis showed relatively larger errors in estimating T2 and ECV values. The majority of the segments with large measurement errors was located in the apical slices, where the myocardium boundaries are often difficult to identify or had severe image artifacts.
Figure 6:
Representative failed cases of the segmentation for T2 mapping and postcontrast T1 mapping. CNN = convolutional neural network.
Discussion
Our study demonstrated the performance of transfer learning to automate T2 and ECV mapping analysis via a pretrained deep learning–based myocardial native T1 mapping analysis platform. The employed network yielded good performance, and the resulting T2 and ECV measurements agreed well with those from manual analyses. The employed transfer learning strategy proved effective, and networks with and without retraining showed comparable performance. Compared with transfer learning–based networks, networks trained from scratch achieved similar success rates for ECV mapping but showed lower success rates for T2 mapping. This may be due to the limited number of T2 mapping images in our training dataset, highlighting the value of transfer learning in the absence of large manually annotated datasets. In our study, from-scratch network training was performed using a manually segmented dataset of approximately 1000 images and employed two approaches to mitigate model overfitting: (a) a dropout layer (31) at the output of each network functional block (Figure E1 [supplement]) that randomly passed or blocked the processed data onto the next functional block, and (b) image augmentation to synthesize new training images by randomly translating, mirroring, and deforming training images. Although transfer learning produced superior network models, we expect the performance of from-scratch trained models to increase with the size of training set. However, we also note that, even with the availability of a larger manually segmented dataset, transfer learning can still be employed to allow efficient training of deeper models with a larger number of parameters.
It is also worth noting that we employed transfer learning mainly to mitigate the limitation of small annotated datasets of T2-weighted and postcontrast T1-weighted images. However, direct application of 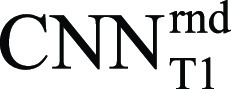 to T2 and ECV mapping has saved computational resources required for training of a new network. Further saving in computational resources might be needed on some platforms, which might be achieved through other approaches such as attention transfer and may require further investigation.
to T2 and ECV mapping has saved computational resources required for training of a new network. Further saving in computational resources might be needed on some platforms, which might be achieved through other approaches such as attention transfer and may require further investigation.
In our study, CNN was only used to segment the myocardium on T1- and T2-weighted images acquired for parametric mapping. In our pipeline, the CNN was predominantly used to segment the left ventricle, and aligned and/or segmented regions of interest were conventionally fitted to calculate T1 or T2 values. The CNN could alternatively be applied over the entire pipeline or used only to segment the resulting T1 or T2 maps; however, these alternatives were not investigated in our study. We chose to decouple the fitting and segmentation aspects of the myocardial T1 and T2 mapping processing pipeline to rigorously investigate the performance of segmentation in the pipeline. While we use a network trained to segment T1-weighted images, other networks trained for other segmentation tasks, such as cine or perfusion, could potentially be used and should be further investigated. Moreover, having motion correction and parametric fitting steps separated in the proposed analysis pipeline can be advantageous compared with previous methods (10,32) that combine both steps. For example, a separate step for parametric fitting allows flexibility in selecting the model type (eg, two-parameter or three-parameter) without a need to reperform the time-consuming motion correction step. Additionally, it allows exclusion of unsuccessfully segmented images from parametric fitting, thereby avoiding potential mapping errors.
Transfer learning–based segmentation of tissue mapping images is more challenging compared with segmentation in other sequences such as cine. Cine images acquired using balanced steady-state free precession sequences, for example, have high and temporally consistent myocardium-to-blood contrast ideal for efficient CNN training. Currently, most cardiac MRI analysis vendors offer automated cine segmentation for contouring endocardial and epicardial borders, and there are already several commercially available deep learning–based cine segmentation methods. On the other hand, to the best of our knowledge, there is no software available for automated analysis of T1, T2, or ECV mapping. Automated mapping analysis is challenged by the substantial variations in the myocardium-to-blood contrast caused by the different T1 or T2 weighting. At certain inversion or T2 preparation times, the myocardium-to-blood contrast is very low and usually impedes accurate myocardial segmentation. Also, there are currently numerous T1 and T2 mapping sequences that yield different contrasts. Different gadolinium-based contrast agents and acquisition times after contrast material administration also substantially impact the signal and contrast-to-noise ratio of postcontrast T1-weighted images. Learning image features from such diverse image contrast patterns requires large training datasets that might not be available, highlighting the benefit of transfer learning–based myocardium segmentation in tissue mapping images.
The current practice of quantifying T1, T2, or ECV varies across different centers. Septal measurements have been shown to have high reproducibility (33) and are often used in cardiac MRI tissue mapping (34,35). However, because diffuse fibrosis or inflammation could be regional (3,9), a septal measurement is insufficient. Myocardial tissue mapping sequences with whole heart coverage have been developed (7,9). However, manual analysis is time-consuming and operator dependent. The proposed automated analysis platform has the potential to facilitate adoption of whole-heart T1 or T2 mapping sequences for better characterization of tissue composition across different regions.
Our study had several limitations. All images were acquired using the same MRI scanner at 1.5 T. Because image quality varies with field strength and scanner type, further studies are warranted to evaluate the platform performance for myocardial tissue maps acquired at different field strengths and from different vendors. A preliminary experiment showed that the developed method may have the potential to automate T1 mapping using other sequences; eg, MOLLI (modified Look-Locker inversion recovery) (Appendix E1 [supplement]). However, a rigorous study with a large dataset is needed to evaluate the performance of generalizing transfer learning to a wider spectrum of mapping sequences. Also, myocardial alignment in our study was performed by forward and backward transformation from Cartesian to polar coordinates, which could introduce interpolation errors to the computed maps. Although interpolation errors may be suppressed during analysis through averaging of the myocardium maps, further investigation is needed to determine their impact on the computed maps compared with other sources of error such as imaging artifacts and noise. We used manual analysis by an experienced observer as the reference standard. However, there is still intraobserver variability in the manual measurements (29). For per-segment analysis, the selection of insertion points was manually selected in the current analysis platform.
In conclusion, our CNN-based automated analysis platform, trained on T1 mapping data, yielded T2 and ECV values in good agreement with manual measurements without the need to retrain the network with corresponding T2 and ECV mapping data. This platform has the potential to fully automate myocardial tissue mapping, allowing standardization of data analysis.
APPENDIX
SUPPLEMENTAL FIGURES
Acknowledgments
Acknowledgment
The authors wish to thank Jennifer Rodriguez for the editorial corrections.
Y.Z. and A.S.F. contributed equally to this work.
Current address: Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Current address: Digital Medicine & Translational Imaging Group, Pfizer, Cambridge, Mass
Supported by National Institutes of Health (R01HL129185 and R01HL129157) and American Heart Association (15EIA22710040). Y.Z. is funded by National Natural Science Foundation of China (61771463 and 81971611)
Disclosures of Conflicts of Interest: Y.Z. disclosed no relevant relationships. A.S.F. disclosed no relevant relationships. C.D. disclosed no relevant relationships. S.N. disclosed no relevant relationships. R.N. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: Institution receives royalties from Samsung, Philips, and Siemens. Other relationships: disclosed no relevant relationships.
Abbreviations:
- CI
- confidence interval
- CNN
- convolutional neural network
- ECV
- extracellular volume
- STONE
- slice-interleaved myocardial T1 mapping sequence
References
- 1.Taylor AJ, Salerno M, Dharmakumar R, Jerosch-Herold M. T1 Mapping: Basic Techniques and Clinical Applications. JACC Cardiovasc Imaging 2016;9(1):67–81. [DOI] [PubMed] [Google Scholar]
- 2.Haaf P, Garg P, Messroghli DR, Broadbent DA, Greenwood JP, Plein S. Cardiac T1 Mapping and Extracellular Volume (ECV) in clinical practice: a comprehensive review. J Cardiovasc Magn Reson 2016;18(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaert D, Thavendiranathan P, Giri S, et al. Direct T2 quantification of myocardial edema in acute ischemic injury. JACC Cardiovasc Imaging 2011;4(3):269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med 2004;52(1):141–146. [DOI] [PubMed] [Google Scholar]
- 5.Piechnik SK, Ferreira VM, Dall’Armellina E, et al. Shortened Modified Look-Locker Inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson 2010;12(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, Thompson RB. Saturation recovery single-shot acquisition (SASHA) for myocardial T(1) mapping. Magn Reson Med 2014;71(6):2082–2095. [DOI] [PubMed] [Google Scholar]
- 7.Wanderers S, Roujol S, Akçakaya M, Basha TA, Nezafat R. Free-breathing multislice native myocardial T1 mapping using the slice-interleaved T1 (STONE) sequence. Magn Reson Med 2015;74(1):115–124. [DOI] [PubMed] [Google Scholar]
- 8.Giri S, Chung YC, Merchant A, et al. T2 quantification for improved detection of myocardial edema. J Cardiovasc Magn Reson 2009;11(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato S, Nakamori S, Bellm S, et al. Myocardial Native T1 Time in Patients With Hypertrophic Cardiomyopathy. Am J Cardiol 2016;118(7):1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue H, Shah S, Greiser A, et al. Motion correction for myocardial T1 mapping using image registration with synthetic image estimation. Magn Reson Med 2012;67(6):1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roujol S, Foppa M, Weingärtner S, Manning WJ, Nezafat R. Adaptive registration of varying contrast-weighted images for improved tissue characterization (ARCTIC): application to T1 mapping. Magn Reson Med 2015;73(4):1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Rewaidy H, Nezafat M, Jang J, Nakamori S, Fahmy AS, Nezafat R. Nonrigid active shape model-based registration framework for motion correction of cardiac T1 mapping. Magn Reson Med 2018;80(2):780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roujol S, Basha TA, Weingärtner S, et al. Impact of motion correction on reproducibility and spatial variability of quantitative myocardial T2 mapping. J Cardiovasc Magn Reson 2015;17(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akçakaya M, Basha TA, Weingärtner S, Roujol S, Berg S, Nezafat R. Improved quantitative myocardial T2 mapping: Impact of the fitting model. Magn Reson Med 2015;74(1):93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon JC, Messroghli DR, Kellman P, et al. Myocardial T1 mapping and extracellular volume quantification: a Society for Cardiovascular Magnetic Resonance (SCMR) and CMR Working Group of the European Society of Cardiology consensus statement. J Cardiovasc Magn Reson 2013;15(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellman P, Wilson JR, Xue H, Ugander M, Arai AE. Extracellular volume fraction mapping in the myocardium, part 1: evaluation of an automated method. J Cardiovasc Magn Reson 2012;14(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19(1):221–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngo TA, Lu Z, Carneiro G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal 2017;35:159–171. [DOI] [PubMed] [Google Scholar]
- 19.Avendi MR, Kheradvar A, Jafarkhani H. Automatic segmentation of the right ventricle from cardiac MRI using a learning-based approach. Magn Reson Med 2017;78(6):2439–2448. [DOI] [PubMed] [Google Scholar]
- 20.Bai W, Sinclair M, Tarroni G, et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson 2018;20(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahmy AS, Rausch J, Neisius U, et al. Automated Cardiac MR Scar Quantification in Hypertrophic Cardiomyopathy Using Deep Convolutional Neural Networks. JACC Cardiovasc Imaging 2018;11(12):1917–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 23.Torrey L, Shavlik J. Transfer Learning. In: Soria Olivas E, Martín Guerrero JD, Martinez-Sober M, Magdalena-Benedito JR, Serrano López AJ, eds. Handbook of Research on Machine Learning Applications. Hershey, Pa: IGI Global, 2009; 242–264. 10.4018/978-1-60566-766-9 [DOI] [Google Scholar]
- 24.Girshick R, Donahue J, Darrell T, Malik J. Region-Based Convolutional Networks for Accurate Object Detection and Segmentation. IEEE Trans Pattern Anal Mach Intell 2016;38(1):142–158. [DOI] [PubMed] [Google Scholar]
- 25.Margeta J, Criminisi A, Cabrera Lozoya R, Lee DC, Ayache N. Fine-tuned convolutional neural nets for cardiac MRI acquisition plane recognition. Comput Methods Biomech Biomed Eng Imaging Vis 2015;5(5):339–349. [Google Scholar]
- 26.Fahmy AS, El-Rewaidy H, Nezafat M, Nakamori S, Nezafat R. Automated analysis of cardiovascular magnetic resonance myocardial native T1 mapping images using fully convolutional neural networks. J Cardiovasc Magn Reson 2019;21(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. ArXiv: 1505.04597 [preprint]. https://arxiv.org/abs/1505.04597. Posted May 18, 2015. Accessed May 18, 2015.
- 28.Keys RG. Cubic Convolution Interpolation for Digital Image-Processing. IEEE T Acoust Speech. 1981;29(6):1153–1160. [Google Scholar]
- 29.Bellm S, Basha TA, Shah RV, et al. Reproducibility of myocardial T1 and T2 relaxation time measurement using slice-interleaved T1 and T2 mapping sequences. J Magn Reson Imaging 2016;44(5):1159–1167. [DOI] [PubMed] [Google Scholar]
- 30.Shah RV, Kato S, Roujol S, et al. Native Myocardial T1 as a Biomarker of Cardiac Structure in Non-Ischemic Cardiomyopathy. Am J Cardiol 2016;117(2):282–288. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014;15(1):1929–1958. https://dl.acm.org/citation.cfm?id=2670313. [Google Scholar]
- 32.Xue H, Greiser A, Zuehlsdorff S, et al. Phase-sensitive inversion recovery for myocardial T1 mapping with motion correction and parametric fitting. Magn Reson Med 2013;69(5):1408–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers T, Dabir D, Mahmoud I, et al. Standardization of T1 measurements with MOLLI in differentiation between health and disease--the ConSept study. J Cardiovasc Magn Reson 2013;15(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roujol S, Weingärtner S, Foppa M, et al. Accuracy, precision, and reproducibility of four T1 mapping sequences: a head-to-head comparison of MOLLI, ShMOLLI, SASHA, and SAPPHIRE. Radiology 2014;272(3):683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pica S, Sado DM, Maestrini V, et al. Reproducibility of native myocardial T1 mapping in the assessment of Fabry disease and its role in early detection of cardiac involvement by cardiovascular magnetic resonance. J Cardiovasc Magn Reson 2014;16(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.