Abstract
The aim of this study was to evaluate the feasibility of measuring ALK gene rearrangement in cell-free RNA (cf-RNA) of the supernatant from malignant pleural effusion (MPE). Supernatant, cell blocks, and matched sera samples were collected. Cf-RNA was isolated from the supernatant and sera, and cellular RNA was isolated from cell blocks. The ALK gene rearrangement in the cf-RNA was tested by the real-time polymerase chain reaction. Results showed that the concentration of cf-RNA was higher in the supernatant than in matched sera. ALK status concordance rates were 100% between the supernatant and cell blocks, while they were 0% between sera and cell blocks in ALK gene rearrangement cases. This suggests that using cf-RNA in MPE supernatant, but not in sera, could offer a reliable and robust surrogate strategy for the detection of ALK gene rearrangement.
1. Introduction
Lung cancer is the leading cause of cancer death among both men and women, of which non-small-cell lung cancer (NSCLC) represents approximately 85% of the total cases diagnosed [1]. Driver gene abnormality tests are the premise of targeted therapy for advanced NSCLC, particularly in the adenocarcinoma subtype [2]. Among these, EGFR gene mutations and ALK gene rearrangement are widely recognized alterations that respond to target agents in NSCLC [3]. Malignant pleural effusion (MPE) is a common manifestation in patient with advanced lung cancer. Most MPE contain tumor cells, which can be used for EGFR and ALK gene status tests [4–7].
Cell-free nucleic acids are extracellular nucleic acids found in cell-free plasma/serum and other biological fluids [8]. Detection of cell-free nucleic acids (DNA or RNA) using quantitative real-time PCR or quantitative real-time RT-PCR (qRT-PCR) methods have been suggested as a promising diagnostic tool for cancer detection [9]. A number of studies have reported that EGFR mutations can be detected in cell-free DNA (cf-DNA) of malignant pleural effusion rearrangement samples from lung cancer patients [10, 11]. Though researchers have showed that there was cell-free RNA (cf-RNA) in MPE [12], it is still not known whether ALK gene rearrangements can be detected by qRT-PCR using cf-RNA. Therefore, the subject of the present study was to explore the use of the newly developed method for the detection of ALK gene rearrangement in MPE.
2. Materials and Methods
2.1. Patients and Samples
A total of 168 MPE samples were collected and centrifuged. The tumor cell content in cell blocks was sufficient to perform the subsequent immunohistochemistry (IHC) and ALK gene rearrangement test. Twenty-three cases of cell-free supernatant and sera (matched with 12 ALK positive rearrangement and 11 ALK negative rearrangement cell blocks) were collected. The diagnosis of NSCLC was based on cytology via a combination of IHC staining results and clinical information. All of the samples were obtained from Guizhou Provincial People's Hospital during November 2015–June 2019. The age at the time of initial diagnosis and smoking status were obtained from the hospital medical records. The study was approval by the Ethics Committees of Guizhou Provincial People's Hospital.
2.2. RNA Extraction and ALK Gene Rearrangement Detection
The cell blocks were embedded by paraffin, and then cellular RNA was extracted by the use of an RNA FFPE Tissue Kit (AmoyDx, Xiamen, China) according to the manufacturer's protocols. The supernatant and sera were firstly centrifuged for 10 min at full speed (8000 g/min), and then cf-RNA was extracted by the use of a Circulate Nucleic Acids Kit (AmoyDx, Xiamen, China) according to the manufacturer's protocols. The quantity of isolated RNA was assessed by using a NanoDrop2000 spectrophotometer (Thermo, L.A., USA). The ALK gene rearrangement was detected by qRT-PCR using an ALK gene rearrangement Detection Kit (AmoyDx, Xiamen, China). The PCR reaction was performed on an Agilent Mx3000P QPCR instrument (Agilent Technologies, Santa Clara, CA). The following PCR procedure was used: an initial reverse transcription at 42°C for 5 min, denaturation at 95°C for 5 min, and then 10 annealing cycles at 95°C for 25 seconds, 64°C for 20 seconds, and 72°C for 20 seconds, followed by 36 extension cycles at 93°C for 25 seconds, 60°C for 35 seconds, and 72°C for 20 seconds to perform the data collection. The quantitative judgment was according to the gene rearrangement fluorescence signal. Assay reactions achieving threshold cycle (Ct) values of ≤35 cycles were considered as positive, otherwise as negative. Amplification of the control gene (beta-actin) in the assay demonstrates the presence of RNA.
2.3. Statistical Analysis
Data were analyzed using SPSS 13.0 software (SPSS Inc., Chicago, IL, USA). The chi-square test, Friedman test, and paired Student's t-test were performed to analyze variables, where appropriate. In addition, Kappa statistic and McNemar's test were performed to determine consistency between the supernatant, sera, and cell blocks. p < 0.05 was considered statistically significant.
3. Results
3.1. Patient Features
In total, 168 cases with MPE were eligible for analysis. The ALK gene rearrangement status was assessed by analysis of RNA extracted from cell blocks using the qRT-PCR method. The clinicopathological features of the patients are shown in Table 1. The current ALK positive rate was 7.1% (12/168) in patients with MPE. Further analysis found that the positive rate was significantly higher in patients with age under 60 years (14.1%) than in patients with age over 60 years (2.4%) (p=0.012; Table 1).
Table 1.
Clinicopathological features of patients with MPE (n = 168).
| Clinicopathological features | ALK positive rearrangement | ALK negative rearrangement | p value |
|---|---|---|---|
| Age (years) | |||
| <60 | 10 | 71 | p=0.012 |
| ≥60 | 2 | 85 | |
|
| |||
| Sex | |||
| Male | 4 | 83 | p=0.184 |
| Female | 8 | 73 | |
|
| |||
| Smoking | |||
| Never | 9 | 79 | p=0.104 |
| Ever | 3 | 77 | |
ALK, anaplastic lymphoma kinase; chi-square test.
3.2. Cf-RNA in Supernatant and Sera
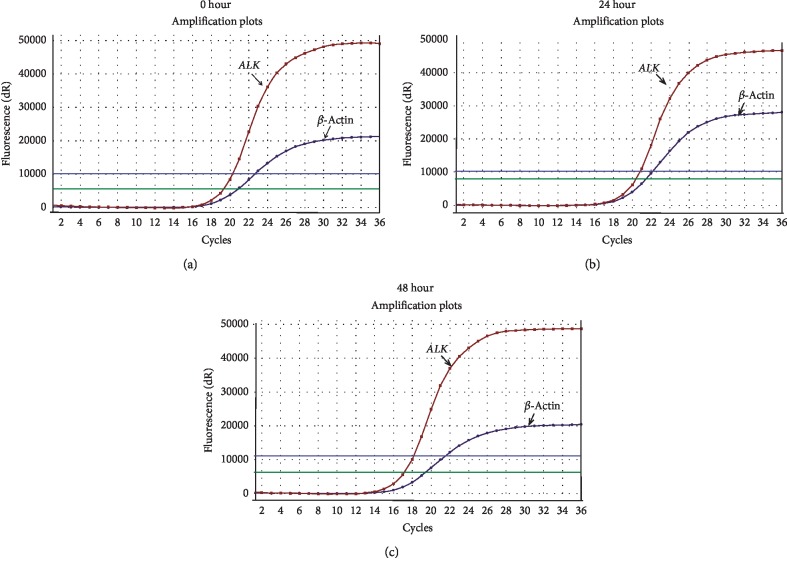
To determine whether delayed MPE supernatant processing would lead to the degradation of cf-RNA, 23 patients with MPE supernatant were collected for cf-RNA assay. The samples of MPE supernatant were placed in 4°C icebox at different time points (0, 24, and 48 hours) and then processed. Results showed that no significant difference was found for the concentrations of cf-RNA at three time points (p=0.632; Figure 1). We further detected the ALK gene rearrangement by qRT-PCR, nevertheless, there was no difference between the three time points too (Figure 2). We also tested the RNA concentration in the matched sera of patients with MPE (n = 23); however, the level of cf-RNA was very low in sera than in the supernatant (p < 0.0001; Figure 3).
Figure 1.
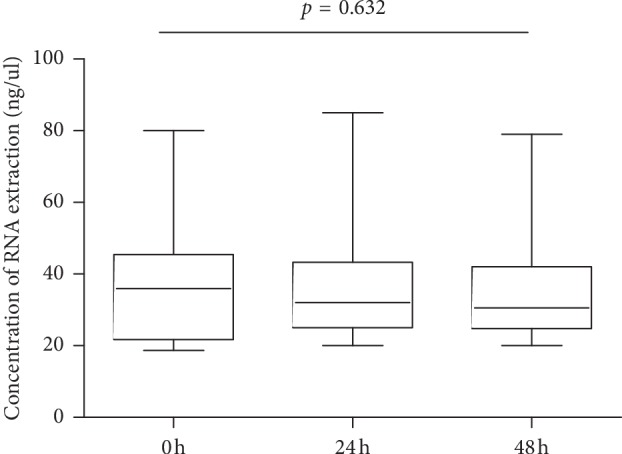
Concentration of cf-RNA extraction (ng/ul) in the MPE supernatant as determined by using a spectrometer is plotted on the y-axis. The processing time points are shown on the x-axis (0, 24, and 48 hours). Statistically significant differences were determined by using the Friedman test.
Figure 2.
Detection of ALK gene rearrangement at three processing time points in cf-RNA of the MPE supernatant. (a) Amplification curve of ALK rearrangement at 0 hour. (b) Amplification curve of ALK rearrangement at 24 hours. (c) Amplification curve of ALK rearrangement at 48 hours.
Figure 3.
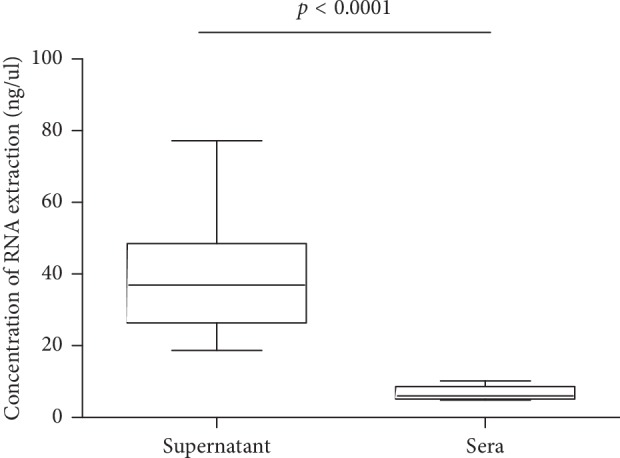
Concentration of cf-RNA extraction (ng/ul) in the MPE supernatant and matched sera as determined by using a spectrometer is plotted on the y-axis. Statistically significant differences were determined by using paired Student's t-test.
3.3. Comparison of Detection of ALK Gene Rearrangement between Sera, Supernatant, and Cell Blocks
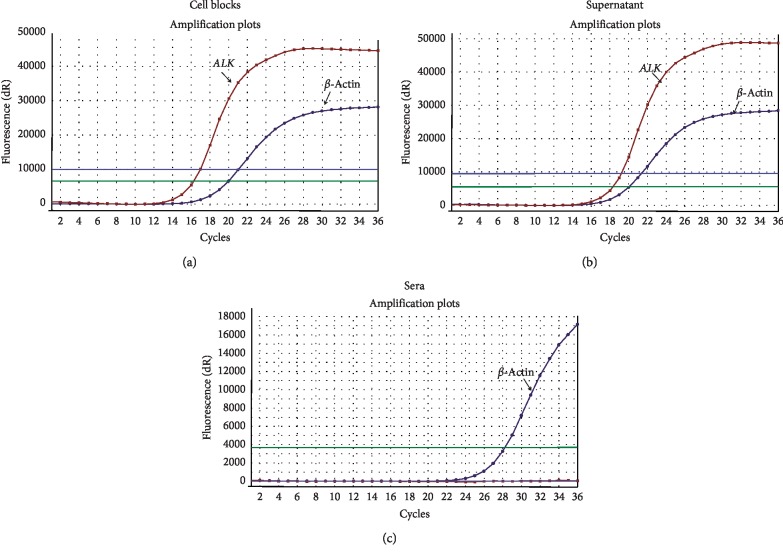
Of the 168 samples for which ALK gene rearrangement results were available, ALK-positive rearrangement of cell blocks was 7.1% (12/168) (Table 1). Cf-RNA of 23 cases of matched sera and supernatant was isolated (including 12 cases of ALK-positive rearrangement). Results showed that the detection efficiency of ALK gene rearrangement was completely consistent in the supernatant (cf-RNA) and cell blocks (cellular RNA) (McNemar's p=1.000; kappa = 1.000; Table 2; Figure 4). However, the detection of ALK gene rearrangement was wholly negative in all matched sera (McNemar's p < 0.001; kappa = 0.000; Table 2; Figure 4).
Table 2.
Comparison of ALK gene rearrangement results in supernatant, sera, and cell blocks.
| Cell blocks | Supernatant | Sera | ||
|---|---|---|---|---|
| + | − | + | − | |
| + | 12 | 0 | 0 | 12 |
| − | 0 | 11 | 0 | 11 |
| p value (McNemar's test) | =1.000 | ≤0.001 | ||
| Kappa value | =1.000 | =0.000 | ||
+, ALK positive rearrangement; −, ALK negative rearrangement.
Figure 4.
Detection of ALK gene rearrangement in cellular RNA of MPE cell blocks and cf-RNA of the MPE supernatant and sera. (a) Amplification curve of ALK rearrangement in cellular RNA of MPE cell blocks. (b) Amplification curve of ALK rearrangement in cf-RNA of the MPE supernatant. (c) Amplification curve of ALK rearrangement in cf-RNA of sera.
4. Discussion
MPE is a common manifestation of advanced NSCLC. For many patients, MPE can be used for driver gene status tests, including EGFR gene mutation and ALK gene rearrangement [4, 6, 7, 13]. Recently studies have showed that the cf-DNA of MPE could be used for the EGFR mutation test [10, 11]. However, whether the cf-RNA of MPE is appropriate for ALK gene rearrangement testing is not yet known and was therefore the subject of the current study.
We firstly detected the concentration of cf-RNA and ALK gene rearrangement in different processing time points (0, 24, and 48 hours), and the results showed that there was no difference between them (Table 1; Figure 2). That indicates that there will be plenty of time to process cf-RNA in clinical working practice for MPE samples. In addition, we found that the concentration of cf-RNA was very low in matched sera than in the supernatant (Figure 3). These findings suggest low level of cf-RNA burden in sera. Previous studies have showed that MPE cell blocks can be used for ALK gene rearrangement detection [2, 4]. Hence, in this study, we compared the mutation status in cf-RNA of sera and the supernatant to the mutation status in the cell blocks from the same patients. Our findings showed that the ALK status concordance rates were 100% between the supernatant and cell blocks, but 0% between sera and cell blocks in ALK gene rearrangement cases (Table 2; Figure 4). We speculated that very low level cf-RNA in sera maybe the cause of negative results, while high concentration of cf-RNA provide enough material to ensure the sensitivity testing ALK gene rearrangement status in the supernatant. So, we think that cf-RNA in MPE supernatant samples is one of the proper substrates for ALK gene rearrangement testing. In the future, cf-RNA and cf-DNA in the MPE supernatant would be an attractive alternative source supplying useful information about the mutation status of ALK, EGFR, and other genes.
In conclusion, this study explored a newly developed method for the detection of ALK gene rearrangement in MPE. Cf-RNA in the supernatant would be one potential substitute for molecular diagnostics.
Acknowledgments
This work was supported by the Guizhou Province Science and Technology Agency Fund (grant no. SY(2013)3002).
Contributor Information
Mingliang Chu, Email: chumingliang@foxmail.com.
Zhuxue Zhang, Email: zhzhxue0117@126.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors' Contributions
Mingliang Chu and Yanqiu Zhu contributed equally to this work.
References
- 1.Gridelli C., Rossi A., Carbone D. P., et al. Non-small-cell lung cancer. Nature Reviews Disease Primers. 2015;1(1) doi: 10.1038/nrdp.2015.9.15009 [DOI] [PubMed] [Google Scholar]
- 2.Wang Z., Wu X., Han X., et al. ALK gene expression status in pleural effusion predicts tumor responsiveness to crizotinib in Chinese patients with lung adenocarcinoma. Chinese Journal of Cancer Research. 2016;28(6):606–616. doi: 10.21147/j.issn.1000-9604.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo S. Y., Lam D. C. Oncogenic driver mutations in lung cancer. Translational Respiratory Medicine. 2013;1(1):p. 6. doi: 10.1186/2213-0802-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J., Yao H., Zhao J., et al. Cell block samples from malignant pleural effusion might be valid alternative samples for anaplastic lymphoma kinase detection in patients with advanced non-small-cell lung cancer. Histopathology. 2015;66(7):949–954. doi: 10.1111/his.12560. [DOI] [PubMed] [Google Scholar]
- 5.Lindeman N. I., Cagle P. T., Beasley M. B., et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the college of American pathologists, international association for the study of lung cancer, and association for molecular pathology. Journal of Thoracic Oncology. 2013;8(7):823–859. doi: 10.1097/jto.0b013e318290868f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akamatsu H., Koh Y., Kenmotsu H., et al. Multiplexed molecular profiling of lung cancer using pleural effusion. Journal of Thoracic Oncology. 2014;9(7):1048–1052. doi: 10.1097/jto.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 7.Agalioti T., Giannou A. D., Stathopoulos G. T. Pleural involvement in lung cancer. Journal of Thoracic Disease. 2015;7(6):1021–1030. doi: 10.3978/j.issn.2072-1439.2015.04.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischhacker M., Schmidt B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochimica et Biophysica Acta (BBA)—Reviews on Cancer. 2007;1775(1):181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Tzimagiorgis G., Michailidou E. Z., Kritis A., Markopoulos A. K., Kouidou S. Recovering circulating extracellular or cell-free RNA from bodily fluids. Cancer Epidemiology. 2011;35(6):580–589. doi: 10.1016/j.canep.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Shin S., Kim J., Kim Y., Cho S. M., Lee K. A. Assessment of real-time PCR method for detection of EGFR mutation using both supernatant and cell pellet of malignant pleural effusion samples from non-small-cell lung cancer patients. Clinical Chemistry and Laboratory Medicine (CCLM) 2017;55(12):1962–1969. doi: 10.1515/cclm-2016-0851. [DOI] [PubMed] [Google Scholar]
- 11.Lin J., Gu Y., Du R., Deng M., Lu Y., Ding Y. Detection of EGFR mutation in supernatant, cell pellets of pleural effusion and tumor tissues from non-small cell lung cancer patients by high resolution melting analysis and sequencing. International Journal of Clinical and Experimental Pathology. 2014;7(12):8813–8822. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T., Lv M., Shen S., et al. Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0043268.e43268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan L., Feng Y., Wan H., Shi G., Niu W. Clinicopathological and demographical characteristics of non-small cell lung cancer patients with ALK rearrangements: a systematic review and meta-analysis. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0100866.e100866 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.