Abstract
Molecular shape defines function in both biological and material settings and, as such, chemists have developed an ever-increasing descriptive vernacular to describe these shapes. Non-canonical atropisomers—i.e., shape-defined molecules that are formally topologically trivial, but only interconvertible by complex, non-physical multibond torsions—form a unique subset of atropisomers that differ from both canonical atropisomers (e.g., binaphthyls) and topoisomers (i.e., molecules that have identical connectivity, but non-identical molecular graphs). Small molecules, in contrast to biomacromolecules, are not expected to exhibit such ambiguous shapes. Herein, we present the discovery through total synthesis that the peptidic alkaloid tryptorubin A can be one of two non-canonical atropisomers. A synthetic strategy was subsequently devised that drives the first atropospecific synthesis of a non-canonical atrop-defined small molecule.
One Sentence Summary
Atypical atropisomerism is observed in the natural product tryptorubin A, and a synthetic strategy to achieve atropospecificity is devised.
In 1894, Emil Fischer proposed a lock and key analogy for how biological molecules interact to carry out biological functions, and the three-dimensional shapes of molecules have been a major focus of biological chemistry ever since (1). Since then, the structure of small molecules has been assumed to be defined solely by atomic connectivity and point or axial chirality. For example, the steroid hormones all have the same basic carbon skeleton: a rigid assembly of four rings fused one to another, and their different biological roles depend on the modifications to the periphery of this basic skeleton. In contrast, large molecules like proteins can reversibly self-organize into well-defined three-dimensional structures, and the rules governing this ability are increasingly well-understood (2). This structural feature of biological macromolecules encodes for many of the functions that form the basis of life (1). For example, hydrogen-bonding, hydrophobic, arene-π, and solvation interactions drive proteins to fold into specific tertiary structures that render them operable (2). Molecular shapes (i.e., tertiary structures) for most macromolecules are derived, but fundamentally separate, from atomic connectivity; that is, many proteins can be folded and unfolded without the breaking or forming covalent bonds (3).
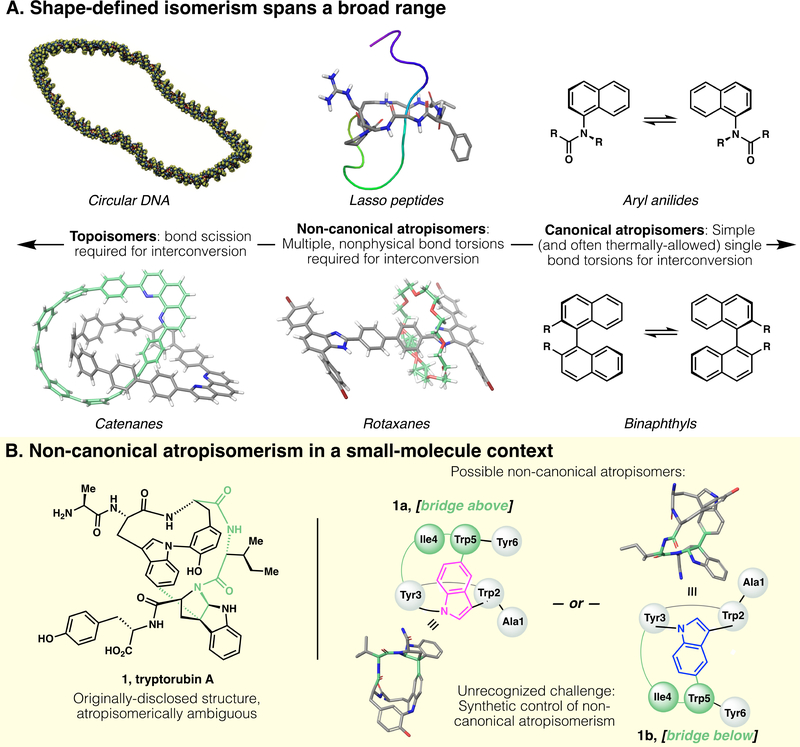
For certain macromolecules, however, shape is directly tied to atomic connectivity rather than conformational changes (Fig. 1A, left): In the case of cyclic DNA, for example, the wound and unwound topologies are interconvertible only by the scission and reformation of phosphate linkages (8). Likewise, molecular catenanes have been synthesized with defined topology (6). Such non-superimposable and non-interconvertible topologies are called topoisomers, a term defined as molecules that have identical connectivity, but non-identical molecular graphs; that is, molecular pairs that are non-interconvertible without the breaking and reformation of chemical bonds (9).
Fig. 1.
(A) Shape-based isomerism in synthetic and natural products spans a broad range. At one end (left), defined topology encodes topoisomers. At the other end (right) canonical atropisomerism is defined by simple axial differences (i.e., torsion of a single bond). Under the broad umbrella of atropisomerism, but distinct from more canonical examples, are (center) non-canonical atropisomers that are formally topologically trivial, but whose interconversion requires complex multibond rotations and unphysical torsions. Historically, this area has been occupied only by macromolecules; in this work, we disclose a small molecule natural product that presents this type of non-canonical atropisomerism. Structures obtained from PDB and/or CCDC database: circular DNA, reproduced from ref (4); lasso peptide, PDB 5TJ1 (5); catenane, CCDC #1835146 (6); rotaxane, CCDC #1576710 (7). (B) Left, originally-proposed structure of tryptorubin A; Right, two non-canonical atropisomers are possible within the limits of the originally-proposed 2D structure. Note: 3D structures of 1a and 1b are computed, not crystallographic, and their terminal residues are truncated for clarity.
This type of defined topoisomerism is conspicuously absent from small-molecule natural products. A distinct, if seemingly analogous, isomerism in a small-molecule context is atropisomerism; i.e., shape isomerism through hindered bond rotation. Canonically, atropisomerism involves a single torsionally-hindered bond that bestows axial chirality; hindered biaryls (Figure 1A, right) represent a prototypical example.
In contrast to both canonical, singly-axially-chiral atropisomerism, and topoisomerism, exist a variety of shape-defined molecules theoretically interconvertible by bond rotation, but categorically distinct from canonical atropisomers because of the multiple and non-physical bond torsions required for their interconversion. Many mechanically interlocked molecules (MIMs) fit into this middle ground; for example, both rotaxanes(7) and lasso peptides(5) (Figure 1A, middle) are topologically trivial and should formally be considered atropisomers with their unthreaded counterparts, but are clearly categorically distinct from simple prototypical examples of atropisomerism. (For another compelling case of non-canonical atropisomerism, see ref. 10) In a physical (rather than theoretical) sense, most members of the lasso peptide class of natural products can be interconverted from unthreaded to threaded shapes only by breakage and repair of the peptide backbone. It is worth noting that the terms ‘topology’ and ‘topoisomer’ are often erroneously used in the literature to refer to these shape-defined (but topologically trivial) structures, when non-canonical atropisomerism is a more accurate vernacular.
Shapes defined by features beyond atomic connectivity and point chirality—both topoisomerism and non-canonical atropisomerism—are not a mere structural curiosities: The shape of DNA completely drives its function (1), and an entire highly-conserved class of enzymes (topoisomerases) have evolved to control DNA’s tertiary structure (11). Similarly, the superior proteolytic stability of lasso peptides is attributed to their threaded shape (12).
Whereas non-canonical atropisomerism is prevalent in macromolecular examples, in the realm of small-molecule natural product chemistry, it is generally not even considered: Point chirality, E/Z alkene geometry, and more typical biaryl atropisomerism are all prevalent definers of structure in secondary metabolites, but structures with ambiguous molecular shape are conspicuously absent. Indeed, upon isolation of novel small-molecule natural products, considerations of shape may never even be part of the structural assignment process.
Tryptorubin A (1, Figure 1B) is a small-molecule peptidic indole alkaloid containing a highly-strained bridging bis(macrocycle) (13). Upon isolation, potential atropisomerism was not considered. Therefore, no explicit shape was initially assigned; the 2D rendering in the isolation disclosure (see Fig. 1 in ref. 13, as well as this report’s Fig. 1B, left) was drawn in a shape-ambiguous manner, and the 3D rendering (see Figure 2A in ref. 13) was arbitrarily drawn with the “bridge below” shape (i.e., equivalent to compound 1b in Fig. 1B, right).
Herein, we report the discovery that tryptorubin A (1), based on chirality and connectivity alone, could theoretically present as two possible non-canonical atropisomers. We describe an atroposelective synthesis of atrop-tryptorubin A (1b), the discovery of its atypical atropisomerism, and a hypothesis-driven atropospecific strategy that led to the synthesis of the natural product (1a) and its unambiguous atropisomeric assignment. Additionally, new genomic details on the biogenesis of 1a are reported; these data suggest a biosynthetic pathway involving ribosomal peptide synthesis followed by atroposelective post-translational modification.
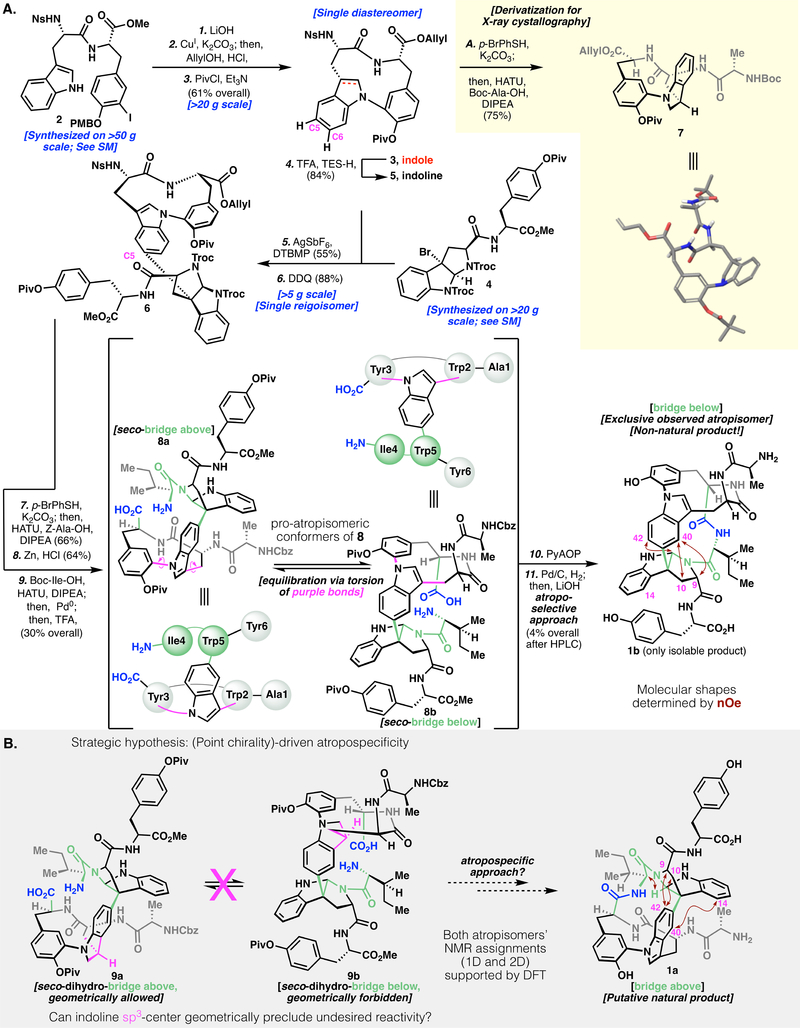
We were interested in pursuing the total synthesis of tryptorubin A for its sheer structural complexity. At the outset of synthetic work, atropisomerism was not considered, and the synthesis of 1 was pursued according to Figure 2A: Protected dipeptide 2 was prepared on multi-decagram scale (see Supplementary Materials for details). In an effort to cyclize the first macrocycle, we subjected 2 to Ullmann-Goldberg-Buchwald-Ma conditions(14) (i.e., CuI, diamine ligand, and carbonate base). This reaction resulted in substantial epimerization adjacent to the methyl ester; 2 was thus first saponified to lower its α-acidity. The Ullmann cyclization proceeded smoothly on the free acid without α-epimerization. Cyclization was possible at concentrations as high as 40 mM (>20 g scale) without substantial loss of material to dimeric or oligomeric species.
Fig. 2.
(A) Synthetic route to atrop-tryptorubin A (1b). (B) Synthetic hypothesis to use point chirality to drive an atropospecific synthesis of tryptorubin A. Definitions: SM, Supplementary Materials; HATU, hexafluorophosphate azabenzotriazole tetramethyl uronium; DDQ, 2,3-dichloro-5,6-dicyanobenzoquinone; PyAOP, (7-Azabenzotriazol-1-yloxy) tripyrrolidinophosphonium hexafluorophosphate; nOe, nuclear Overhauser effect; DFT, density functional theory.
Subsequent protecting group manipulation yielded protected indole scaffold 3, which was subjected to Movassaghi’s silver-mediated alkylation (15–16) to establish the key non-proteogenic C-C bond. Reaction of 3 with alkyl bromide 4 resulted in low yields and complex mixtures of regioisomers at C5 and C6. We hypothesized that reducing 3 to its indoline congener (5) would both increase the overall nucleophilicity of the arene and also inhibit C6-based reactivity. This tactic was realized by reduction of 3 with TFA/Et3SiH, followed by an alkylation/oxidation sequence, affording the desired product (6) as a single regioisomer in moderate yield. In parallel to this work, homologation of indoline 5 with an additional alanine unit (to form 7) allowed for unambiguous stereochemical assignment by X-ray crystallography.
Subsequent elongation of the peptide chain by appending Ala and Ile residues, followed by deprotection, gave seco-amino acid 8. At this juncture, characterization by NMR spectroscopy became challenging (even at high temperature), presumably due to cis/trans amide isomerization of the tertiary pyrroloindolinyl amide, various rotameric populations, and conformational equilibrium between 8a and 8b. Nonetheless, 8 appeared as a single sharp HPLC peak and exhibited a high-resolution mass spectrum (HRMS) consistent with the postulated structure. After extensive experimentation (see Supplementary Materials), this structure could be cyclized in low yield to a bis(macrocycle). Global deprotection yielded 1b, with HRMS data indicating the same molecular formula as the natural isolate (1). Unfortunately, the NMR data (1H, 13C, HMBC, HSQC, ROESY) and liquid chromatography (LC) retention of 1b were distinct from the natural product (1) (vide infra and see Supplementary Materials for details).
With these contrasts in spectral data in mind, we began to consider possible explanations for the structural discrepancy between 1 and 1b. The possibilities of stereochemical (e.g., a D-amino acid) or regiochemical misassignments (e.g., alternate regiochemistry in the indole-pyrroloindoline C-C bond) in the natural and/or synthetic products were considered. After exhaustive review of natural 1 and synthetic 1b’s respective spectral data as well as a separate total synthesis of C26-epimeric species epi-8 (See Supplementary Materials for this additional synthesis), we confirmed that natural 1 and synthetic 1b had the same connectivity and point-stereochemistry (See Supplementary Materials for details). It was only upon careful analysis of the two compounds’ ROESY spectra that a key insight was discovered: Although the natural product (1a, Fig. 2B) showed strong nuclear Overhauser effect correlations from H9 and H10 to H42, the analogous H9 and H10 protons in synthetic (1b, Fig. 2A) compound’s ROESY spectrum showed correlations to H40. This key geometric constraint, combined with additional spectral evidence (1b and 1a in Figs. 2A and 2B; see Supplementary Materials for additional details and full skeletal numbering system), illuminated our understanding that even within the limits of identical connectivity and stereochemistry, 1 could potentially exist as two non-canonical atropisomers (“bridge above,” 1a; or “bridge below,” 1b). Subsequent DFT-based NMR prediction validated our assignment of the synthesized product as 1b, and supported our hypothesis that the natural product was the other atropisomer (1a). (For a compelling example of use of these predictions to validate structural assignments, see ref. 17.) In addition to computational NMR prediction, computation of the respective atropisomers’ polar and hydrophobic surface area showed that the unnatural atropisomer (1b) had substantially less polar surface area, consistent with its increased retention relative to 1a on reverse-phase LC (See Supplementary Materials for details).
Retrospectively, we reasoned that seco compound 8 likely exists as two interconverting pro-atropisomeric conformers (8a and 8b, Fig. 2A), and that the “bridge below” conformer (8b) has superior kinetic facility for cyclization, leading exclusively to 1b as the observed cyclization product. We hypothesized that by geometrically locking the cyclization precursor into the “bridge above” conformation, we could achieve inversion of atroposelectivity. Combining this hypothesis with crystallographic evidence of the geometry of indoline 7, we recognized that in a substrate such as indoline 9, the point chirality at indoline (Fig. 2B, purple methine) would geometrically preclude the “bridge below” conformer (9b); indeed, geometric limitations of 9 would render the cyclization atropospecific for the “bridge above” atropisomer 1a (resulting from cyclization of 9a). Such a strategy is reminiscent of methods to control more canonical atroposelectivity by point-to-axial chirality transfer (18).
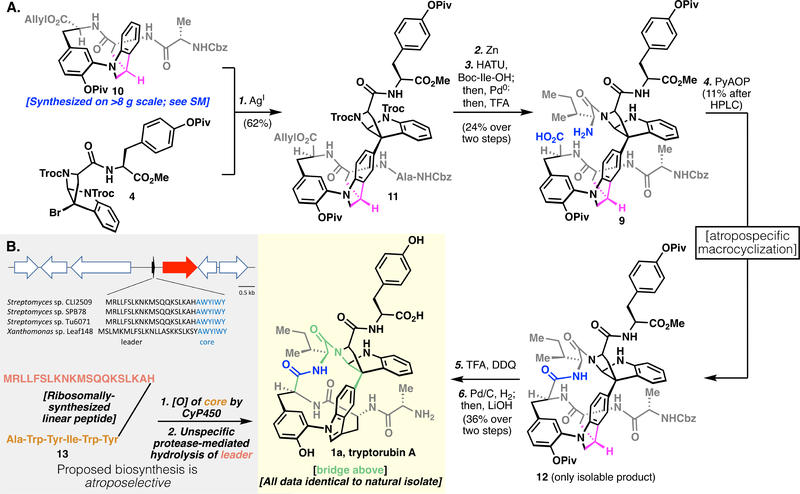
Figure 3A describes our successful execution of the atropospecific strategy laid out in Figure 2B, and the subsequent total synthesis of the natural isomer of tryptorubin A (1a). We prepared indoline analog 10 in multigram quantities, and a similar Friedel-Crafts sequence as previously described yielded pentamer 11. We subjected this pentamer to a peptide homologation sequence to give the key seco-amino acid (9), with the indoline oxidation state geometrically enforcing a “bridge above” conformation. Subsequent cyclization gave 12 as a single isomer. However, simple dehydrogenation of indoline 12 proved challenging. Attempts at metal-catalyzed dehydrogenation (i.e., Pd/C) gave only recovered starting material, and use of cerium ammonium nitrate (CAN) resulted in complete decomposition of the substrate. Although in the acyclic system (vide supra), dichlorodicyanobenzoquinone (DDQ) had given the desired reactivity, in the present case, we observed unselective oxidation of the pyrroloindoline aminal to an amidine, as well as the desired indoline-to-indole oxidation. We reasoned that this overoxidation pathway was due to the excessively electron rich, unprotected anilinic nitrogen. Transient protection of this basic nitrogen with a proton from solvent-quantity TFA allowed for the desired monodehydrogenation.
Fig. 3.
(A) Atropospecific synthesis of tryptorubin A (1a). (B) Above, a RiPP codon that encodes for tryptorubin A’s linear peptide sequence; below, proposed biosynthetic pathway to 1a.
Global deprotection of the oxidized material gave 1a, whose spectral data matched closely with the natural product (See Supplementary Materials). In light of the complex shape-driven assignment of 1b and 1a, we tried to obtain solid-state structures in several ways, including classical small molecule X-ray crystallographic methods, high-throughput protein crystallography approaches (19), and small molecule MicroED(20–21). Unfortunately, no solid-state structure could be determined, and our structural assignment remains reliant on solution-phase NMR. Although we observed slight deviations in 1H NMR resonances, these types of deviations are common for natural products rich in potential hydrogen-bonding groups, and the 13C NMR and all 2D (HSQC, HMBC, ROESY) correlations were near-identical. Co-injection of authentic natural product and synthetic material to LC resulted in identical retention as well as a single species in 1H NMR. We thus assign tryptorubin A as having the “bridge above” geometry of 1a.
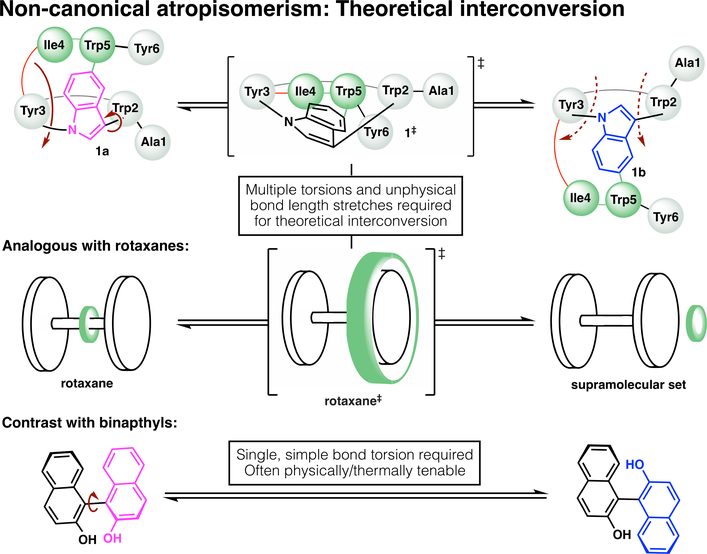
Isomers 1a and 1b do not interconvert at elevated temperature (See Supplementary Materials for details), further showcasing the (likely infinitely) high barrier to interconversion in this atypical example of atropisomerism. Geometrically, interconversion of the atropisomers would require one macrocycle to pass through the other (Fig. 4); it is unsurprising that this is thermally unfeasible, as it would appear based on first principles to be physically impossible without the breakage and reformation of covalent bonds. One might naively assign these compounds as topoisomers due to this physical limitation; however, non-canonical atropisomerism is a more accurate descriptor, as the word topoisomerism is defined without respect to such limitations. In other words, if one could hypothetically stretch bonds beyond the limits of covalency, 1a and 1b would be interconvertible, and thus they are topologically trivial. This contrasts with a topologically-defined catenane, for example, where even with infinite (non-physical) bond stretching, the rings could not be disentangled.
Fig. 4.
Graphical thought experiment considering putative interconversion of tryptorubin (1a) and its non-canonical atropisomer (1b). Theoretically, interconversion would require an unphysical inside-out flipping of the molecule, in which one macrocycle passed through the other (above). This is analogous to atropisomeric inversion of a rotaxane (center), which would require unphysical stretching of the ring (green) over the dumbbell. Such non-canonical atropisomers should be contrasted with prototypical atropisomers like binaphthol (below), which can interconvert through simple bond torsion.
The discovery of tryptorubin A’s geometric isomerism in the total synthesis effort prompted a re-examination of its biosynthesis. The original bioinformatic analysis identified 18 biosynthetic gene clusters, none of which could be confidently predicted to encode the biosynthesis of tryptorubin A (13). The most plausible candidate was a modular non-ribosomal peptide synthethase (NRPS) by which the hexapeptide chain would be assembled sequentially by dedicated enzymes. However, the selectivity of the module-encoded adenylation domains did not convincingly match the tryptorubin A peptide sequence and additional genes involved in the biosynthesis of amino acids that are not incorporated into tryptorubin A were present in the direct vicinity (22–23). We decided to evaluate other possible biosynthetic origins and thus considered the possibility that tryptorubin A is a ribosomally synthesized and post-translationally modified peptide (RiPP) that is missed by conventional bioinformatic analysis tools because of its small size, lack of homology to characterized ribosomal peptides, and the presence of non-canonical tailoring genes involved in carbon-carbon bond formation. Lack of robust genetic tools to modify genes in the native producer limits the degree to which bioinformatics predictions can be checked against experimental outcomes, but we tested our hypothesis with bioinformatics and analytical tools.
Screening the translated Streptomyces sp. CLI2509 genome sequence for the tryptorubin core peptide sequence (AWYIWY) resulted in a single hit. Close inspection of the unannotated region revealed a ribosomal binding site followed by a transcriptional start site, a putative RiPP precursor gene encoding a 20 amino acid leader, a core peptide, and a stop codon downstream of a transcriptional start site and a ribosomal binding site (Fig. 3B and S17). This sequence is followed by a gene encoding a cytochrome P450 enzyme that is likely involved in the formation of the non-proteogenic carbon-carbon and carbon-nitrogen bridges. Although cytochrome P450 enzymes that catalyze carbon-carbon bond formation in ribosomal peptides have not been reported (24), analogous carbon-carbon linkages between the aromatic residues in the non-ribosomal peptide vancomycin have been shown to be installed by cytochrome P450 enzymes (25–28). A specific protease in the same vicinity is missing, so an unspecific protease may cleave the cyclized tryptorubin. The lack of a dedicated protease would also explain the presence of tryptorubin B, a congener of tryptorubin A that differs by a terminal alanine residue (See Supplementary Materials). The same gene tandem was identified in the tryptorubin producer Streptomyces sp. SPB78 as well as a variety of other Streptomyces and more distantly related bacteria (See Supplementary Materials, Fig. S18 and S19). Comparison of this likely tryptorubin locus with other putative producers of tryptorubin-like peptides suggests that the precursor gene and the cytochrome P450 are the only genes conserved in all putative tryptorubin-like BGCs and hence may be sufficient for the biosynthesis of tryptorubin-like metabolites (See Supplementary Materials, Fig. S19). Since we were not able to knock out the putative trp BGC in the native producer, we subjected to metabolic analysis Xanthomonas sp. Leaf 148, which does not share any other peptide biosynthetic gene cluster with the reported tryptorubin producers, other than the candidate trp locus. We detected by LC-MS analysis a metabolite with the same mass (m/z of 826.356 [M+H]+) and similar fragmentation patterns, as revealed by molecular network analysis, with tryptorubin B (29) (See Supplementary Materials, Fig. S21). This preliminary work suggests that the tryptorubins are natural products produced by a RiPP biosynthetic pathway, and that a new family of cytochrome P450s is likely responsible for the unusual carbon-carbon and carbon-nitrogen bridges embedded in their structures.
Despite the extensive vernacular to describe regio-, stereo-, and atropisomers, the nuances of molecular shape can be lost within the realm of small molecule natural product chemistry. Although most practicing synthetic chemists are intimately familiar with the canonical examples of biaryl atropisomerism, the dramatically more complex examples of atropisomerism in polycyclic and mechanically-interlocked molecules often remain under-examined. Indeed, the possibility of non-canonical atropisomerism was initially missed during both the isolation and synthesis of tryptorubin A. We present this case as a cautionary tale in structural definition, a demonstration of the power of transferring point chirality to molecular shape, and a reminder that small-molecule organic chemists can greatly benefit from the deep understanding of three-dimensional structure known in the biomacromolecular and supramolecular literature.
Supplementary Material
Acknowledgments
We thank L. Pasternack and D.-H. Huang (Scripps Research Institute) for assistance with NMR spectroscopy; A. Rheingold, M. Gembicky, C. Moore, and J.B. Bailey (University of California, San Diego) for assistance with X-ray crystallography; J. Nowick and M. Wierzbicki (University of California, Irvine) for attempts at high-throughput protein crystallography screening; H.M. Nelson, L.J. Kim, and C. Jones (University of California, Los Angeles) for attempts at elucidating structures by MicroED; A.L. Vagstad (ETH Zurich) for assistance in identifying the trp BGC; J. Vorholt and M. Bortfeld-Miller (ETH Zurich) for providing Xanthomonas sp. Leaf148; J.S. Chen and B.B. Sanchez (Scripps Research Institute) for assistance with HPLC purification and HRMS; and T.P. Wyche and E. Mevers (Harvard University) for useful discussions.
Funding: Financial support for this work was provided by NIH (GM-118176 for P.S.B.; R01AT009874 for J.C.). S.H.R. gratefully acknowledges an NSF GRFP (#2017237151) and a Donald and Delia Baxter fellowship. A.S.W. gratefully acknowledges an NIH post-doctoral fellowship (F32GM128267). E.J.N.H. gratefully acknowledges funding by the Swiss National Science Foundation (Postdoctoral Mobility Fellowship).
Footnotes
Competing interests: The authors declare no competing interests.
Data availability: Detailed experimental and analytical procedures and full spectral data are available in the Supplementary Materials.
References and Notes
- 1.Remus D, Beall EL, Botchan MR EMBO J, 2004, 23, 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halling PJ. Proteins: structures and molecular properties, second ed., in: Creighton TE, Freeman WH (Eds.), New York, 1992. ISBN: 0–7167–7030-X [Google Scholar]
- 3.Onuchic JN, Wolynes PG Curr. Op. Struct. Bio. 2004, 14, 70. [DOI] [PubMed] [Google Scholar]
- 4.Pennisi E Science 2017, 356, 996. [DOI] [PubMed] [Google Scholar]
- 5.Zong C, Wu MJ, Qin JZ, Link AJ J. Am. Chem. Soc, 2017, 139, 10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan YY, Chen D, Huang ZA, et al. Nat. Commun, 2018, 9, 3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu K, Baggi G, Loeb SJ Nat. Chem, 2018, 10, 625. [DOI] [PubMed] [Google Scholar]
- 8.Roca J Trends Biochem. Sci, 1995, 20, 156. [DOI] [PubMed] [Google Scholar]
- 9.Frisch HL, Wasserman E, J. Am. Chem. Soc, 1961, 83, 3789. [Google Scholar]
- 10.Canfield PJ et al. Nature Chem, 2018, 10, 615. [DOI] [PubMed] [Google Scholar]
- 11.Withoff S, De Jong S, Devries EG, Mulder NH Anticancer Res, 1996, 16, 1867. [PubMed] [Google Scholar]
- 12.Hegemann JD, Zimmermann M, Xie X, Marahiel MA J. Am. Chem. Soc, 2013, 135, 210. [DOI] [PubMed] [Google Scholar]
- 13.Wyche TP, Ruzzini AC, Schwab L, Currie CR, Clardy JJ Am. Chem. Soc, 2017, 139, 12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunz K, Scholz U, Ganzer D Synlett 2003, 15, 2428. [Google Scholar]
- 15.Kim J; Movassaghi MJ Am. Chem. Soc, 2011, 133, 14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer N; Movassaghi M Chem. Sci, 2012, 3, 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rychnovsky SD Org. Lett, 2006, 13, 2895. [DOI] [PubMed] [Google Scholar]
- 18.Burns NZ, Krylova IN, Hannoush RN, Baran PS J. Am. Chem. Soc, 2009, 131, 9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer RK, Nowick JS Isr. J. Chem, 2015, 55, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones CG, Martynowycz MW, Hattne J, et al. ACS Cent. Sci, 2018, 4, 1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruene T, Wennmacher JTC, Zaubitzer C, et al. Angew. Chem., Int. Ed, 2018, 57, 16313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blin K, Shaw S, Steinke K et al. Nucleic Acids Res, 2019, 47, W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skinnider MA, Merwin NJ, Johnston CW, Magarvey NA Nucleic Acids Res, 2017, 45, W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greule A, Stok JE, De Voss JJ, Cryle MJ Nat. Prod. Rep, 2018, 35, 757. [DOI] [PubMed] [Google Scholar]
- 25.Haadatsch B, Butz D, Schmiederer T, et al. Chem. Biol, 2007, 14, 1078. [DOI] [PubMed] [Google Scholar]
- 26.Bischoff D, Bister B, Bertazzo M et al. Chembiochem, 2005, 6, 267. [DOI] [PubMed] [Google Scholar]
- 27.Bischoff D, Pelzer S, Holtzel A, et al. Angew. Chem., Int. Ed, 2001, 40, 1693. [DOI] [PubMed] [Google Scholar]
- 28.Bischoff D, Pelzer S, Bister B, et al. Angew. Chem., Int. Ed, 2001, 40, 4688. [DOI] [PubMed] [Google Scholar]
- 29.Wang M, Carver JJ, Phelan VV et al. Nat. Biotechnol, 2016, 34, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He C, Stratton TP; Baran PS J. Am. Chem. Soc, 2019, 141, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Commerical CuI was purified via Soxhlet washing with refluxing THF for 24 h, then stored in an Argon-filled glovebox. [Google Scholar]
- 32.Chiarello J and Joullie MM Syn. Comm, 1988, 18, 2211. [Google Scholar]
- 33.It should be noted for data in this table, and the following ROESY spectral data table, that the natural material’s NMR was acquired at Harvard, while NMR of synthetic material was acquired at Scripps. Slight differences in the respective experimental pulse sequences may explain slight differences in observed correlations.
- 34.Comparison of crystal structures of intermediates with the C26-epi configuration, vs. the natural C26 configuration (i.e., 7 vs. epi-SI-21; Fig. S9), helped us form a mechanistic hypothesis for the epi scaffold’s reticence to cyclization. Specifically, for the epi scaffold to lactamize, the newly-formed ring would have to be supra- facial to the pendant alanine (represented by in epi-SI-21 as a benzamide by analogy), and this steric clash strongly disfavors cyclization. In contrast, the natural stereochemistry allows the macrolactam to be antara-facial to the alanine, which is more sterically feasible.
- 35.Gaussian 16, Revision A.03, Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Petersson GA; Nakatsuji H; Li X; Caricato M; Marenich AV; Bloino J; Janesko BG; Gomperts R; Mennucci B; Hratchian HP; Ortiz JV; Izmaylov AF; Sonnenberg JL; Williams-Young D; Ding F; Lipparini F; Egidi F; Goings J; Peng B; Petrone A; Henderson T; Ranasinghe D; Zakrzewski VG; Gao J; Rega N; Zheng G; Liang W; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y; Kitao O; Nakai H; Vreven T; Throssell K; Montgomery JA Jr.; Peralta JE; Ogliaro F; Bearpark MJ; Heyd JJ; Brothers EN; Kudin KN; Staroverov VN; Keith TA; Kobayashi R; Normand J; Raghavachari K; Rendell AP; Burant JC; Iyengar SS; Tomasi J; Cossi M; Millam JM; Klene M; Adamo C; Cammi R; Ochterski JW; Martin RL; Morokuma K; Farkas O; Foresman JB; Fox DJ Gaussian, Inc., Wallingford CT, 2016. [Google Scholar]
- 36.Konstantinov IA, Broadbelt LJ, Regression formulas for density functional theory calculated 1H and 13C NMR chemical shifts in toluene-d8. J Phys Chem A 115, 12364–12372 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Blin K et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47, W81–W87, doi: 10.1093/nar/gkz310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skinnider MA, Merwin NJ, Johnston CW & Magarvey NA PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res 45, W49–W54, doi: 10.1093/nar/gkx320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letunic I & Bork P Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259, doi: 10.1093/nar/gkz239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang MX et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol 34, 828–837, doi: 10.1038/nbt.3597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.