Abstract
Protein arginine methyltransferase-5 (PRMT5) is overexpressed in aggressive B-cell non-Hodgkin's lymphomas, including mantle cell lymphoma and diffuse large B-cell lymphoma, and supports constitutive expression of CYCLIN D1 and c-MYC. Here, we combined ChIP analysis with next-generation sequencing to identify microRNA (miRNA) genes that are targeted by PRMT5 in aggressive lymphoma cell lines. We identified enrichment of histone 3 dimethylation at Arg-8 (H3(Me2)R8) in the promoter regions of miR33b, miR96, and miR503. PRMT5 knockdown de-repressed transcription of all three miRNAs, accompanied by loss of recruitment of epigenetic repressor complexes containing PRMT5 and either histone deacetylase 2 (HDAC2) or HDAC3, enhanced binding of co-activator complexes containing p300 or CREB-binding protein (CBP), and increased acetylation of specific histones, including H2BK12, H3K9, H3K14, and H4K8 at the miRNA promoters. Re-expression of individual miRNAs in B-cell lymphoma cells down-regulated expression of PRMT5, CYCLIN D1, and c-MYC, which are all predicted targets of these miRNAs, and reduced lymphoma cell survival. Luciferase reporter assays with WT and mutant 3′UTRs of CYCLIN D1 and c-MYC mRNAs revealed that binding sites for miR33b, miR96, and miR503 are critical for translational regulation of the transcripts of these two genes. Our findings link altered PRMT5 expression to transcriptional silencing of tumor-suppressing miRNAs in lymphoma cells and reinforce PRMT5's relevance for promoting lymphoma cell growth and survival.
Keywords: epigenetics, lymphoma, Myc (c-Myc), cyclin D1, microRNA (miRNA), post-translational modification, gene regulation, cancer, histone deacetylase (HDAC), PRMT5
Introduction
Tightly-regulated epigenetic modifications of chromatin structure affect recruitment of both repressor and activator transcription factors and remodeling complexes, which govern gene expression. Consequently, any disruption impacting the function or interaction of transcription factors with chromatin-remodeling enzymes can trigger cellular transformation (1–5).
Among the various modifications that can alter gene expression is methylation of arginine residues in histone tails catalyzed by protein arginine methyltransferases (PRMTs).4 PRMTs are classified either as type I, II, or III based on the type of methylation being catalyzed (6–8). All PRMTs carry out monomethylation at the ω-NH2 of arginine; however, they differ in their ability to add the second methyl group in either an asymmetric (type I) or symmetric (type II) fashion. Depending on the type of histone arginine methylation introduced, transcription can either be inhibited or induced (6, 7). PRMT5 is a type II arginine methyltransferase that utilizes S-adenosyl-l-methionine as a cofactor to catalyze symmetric dimethylation of arginine residues in histone proteins H3 arginine 8 (H3(Me2)R8) and H4(Me2)R3 (5–7). Our group and others have shown that PRMT5 functions as a chromatin remodeler by interacting with hSWI/SNF complexes to repress transcription of tumor suppressor genes such as suppressor of tumorigenicity 7 (ST7), nonmetastatic 23 (NM23), retinoblastoma-like 2 (RBL2), phosphotyrosine phosphatase receptor (PTPROt), and cell cycle regulator genes such as CYCLIN E1, P14ARF, and P16INK4a (9–13).
Both Mantle cell lymphoma (MCL) and diffuse large B-cell lymphoma (DLBCL) are aggressive subtypes of B-cell non-Hodgkin's lymphoma (NHL) with a broad spectrum of clinical, pathological, and biological features. Patients with relapsed/refractory MCL and DLBCL have an overall poor prognosis despite aggressive multimodal therapy (14, 15). Thus, identification of novel therapeutic targets and development of targeted treatment strategies remain a top priority for these patients. The genetic hallmark of MCL is the translocation t(11;14)(q13;q32), which leads to juxtaposition of the proto-oncogene CCND1 at 11q13 to the immunoglobulin heavy chain enhancer on chromosome 14q32 (16). This translocation leads to constitutive overexpression of CYCLIN D1 protein and cell cycle dysregulation through direct binding to CDK4/6 and phosphorylation of RB (17). Although the t(11;14) is very rare in DLBCL, CYCLIN D1 is overexpressed in a subset of DLBCL and associated with more aggressive behavior (18).
c-MYC is a transcription factor with oncogenic function that is overexpressed through a variety of mechanisms in many cancers, including a subset of DLBCL and MCL. c-MYC is an immediate early gene involved in promoting transition from the G0/G1 phase to the S phase, activating both directly and indirectly the expression of CCND2 and CDKs as well as many other genes required for onset of the S phase and down-regulation of cell cycle inhibitors (19). In addition, c-MYC up-regulates the oncogenic miR 17–92 cluster, but most microRNAs directly regulated by c-MYC have tumor suppressor function and are usually repressed (20).
We have previously shown that PRMT5 is overexpressed in MCL and DLBCL cell lines and primary lymphoma samples (10, 21). Our data show that PRMT5 knockdown with shRNA antagonizes CYCLIN D1–CDK4/6 signaling in MCL and DLBCL (21). In addition to the primary translocation event, other mechanisms that further increase CYCLIN D1 expression are frequently observed in MCL. These mechanisms include secondary chromosomal rearrangement at the 3′ end of the CCND1 locus or mutations in the 3′UTR that lead to expression of truncated CYCLIN D1 transcripts missing part of the 3′UTR (22, 23). These shorter transcripts, depleted of the destabilizing AU-rich elements and the binding sites for different microRNAs, have an extended half-life resulting in higher CYCLIN D1 protein levels, more aggressive disease phenotype, and poor clinical outcome (22, 23).
c-MYC overexpression can occur by translocation of t(8;14)(q24;q32), amplification, or dysfunction of the pathways regulating c-MYC expression (24, 25). c-MYC overexpression is associated with more aggressive disease behavior in patients with MCL and DLBCL (26–31). Prior work showed that c-MYC promotes malignant cell survival and proliferation through direct up-regulation of PRMT5 transcription (32). More recently, we have shown that PRMT5 overexpression is critical for initiation and maintenance of Epstein-Barr virus–induced B-cell transformation and that its inhibition using a first–in–class small-molecule PRMT5 inhibitor induces lymphoma cell death without affecting normal B-cell survival or viability (12, 21).
In this report, we shed light on the mechanisms by which PRMT5 regulates CYCLIN D1 and c-MYC expression in aggressive B-cell NHL. Screening experiments utilizing ChIP combined with next-generation sequencing (ChIP-Seq) analysis showed that PRMT5 overexpression is associated with enrichment of the H3(Me2)R8 epigenetic mark on promoter regions of over 9,000 genes in DLBCL cells, which was distinct from the enrichment observed in normal control B cells. We used these data sets to extract potential micro-RNAs (miRNAs) predicted to target the 3′-untranslated region (UTR) of CYCLIN D1 and c-MYC transcripts. Following experimental validation of the ChIP-Seq data, we identified three miRNAs (miR33b, miR96, and miR503) as direct targets of PRMT5-repressive complexes. PRMT5 inhibition triggered derepression of all three miRNAs and resulted in reduced expression of both CYCLIN D1 and c-MYC. Luciferase reporter analysis using 3′UTR of both WT and mutant CYCLIN D1 and c-MYC mRNAs showed that miR33b, miR96, and miR503 contribute differently to translational regulation of CYCLIN D1 and c-MYC. Furthermore, we show that re-expression of individual miRNAs reduces lymphoma cell survival and promotes cell death. These results demonstrate that PRMT5 epigenetically represses specific tumor suppressor miRNAs in B-cell lymphoma cells and provide additional rationale for targeting this enzyme in aggressive lymphomas.
Results
Genome-wide PRMT5 recruitment in B-cell lymphoma cells as determined by H3(Me2)R8 enrichment
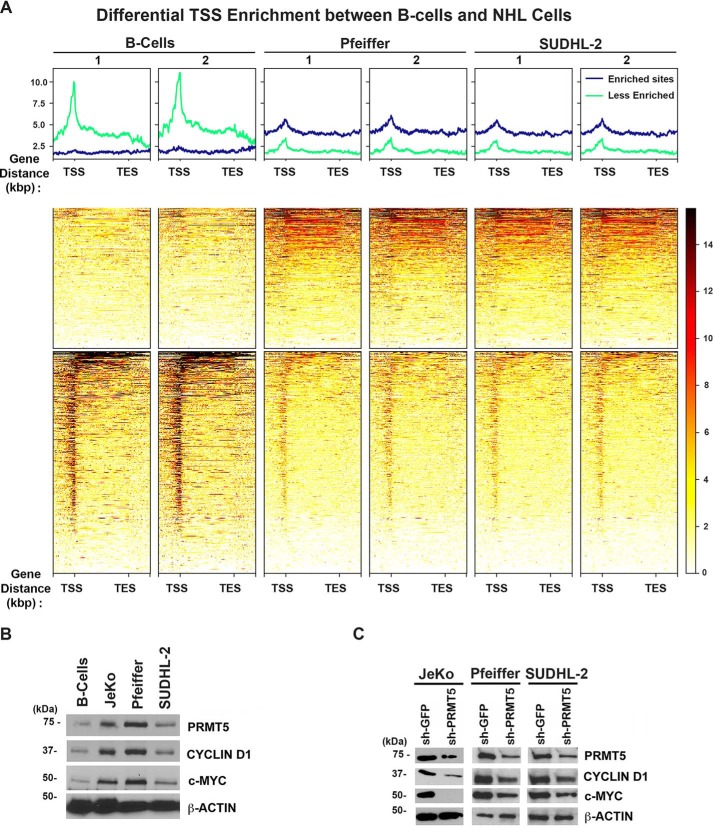
To identify genome-wide recruitment of PRMT5, cross-linked chromatin from tonsillar normal human B cells and two human DLBCL cell lines (Pfeiffer and SUDHL-2) was immunoprecipitated using a highly-specific anti-H3(Me2)R8 antibody (9, 10). We studied genome-wide recruitment of the H3(Me2)R8 PRMT5 epigenetic mark (enrichment sites) and differences between normal B cells and DLBCL cell lines. Comparison of PRMT5 enriched genomic sites in the promoter region of identified target genes showed that there was a significant difference between normal and transformed B cells (Fig. 1A). The heatmap of ChIP-Seq read densities in the promoter (−2.0 kbp upstream from the transcription start site (TSS)) and transcribed genomic regions (up to +2 kbp downstream of the transcription end site (TES)) ranked by decreasing combined occupancy signals showed that maximum PRMT5 occupancy is in the promoter region for both normal and transformed B cells (Fig. S1A). Genome-wide correlation of ChIP signals showed that Pfeiffer and SUDHL-2 cell lines have highly-similar PRMT5-binding profiles that were distinct from normal B cells with average correlation coefficient of 0.98 and 0.8, respectively (Fig. S1B).
Figure 1.
Differential H3(Me2)R8 recruitment in primary B cells compared with DLBCL cell lines. A, heatmap showing H3(Me2)R8 binding over the gene bodies of 504 and 322 genes with 2-fold or more and 2-fold or less H3(Me2)R8 load in B cells compared with both DLBCL cell lines, respectively. Heatmap depicts scaled gene bodies, including 2 kb upstream from TSS and 2 kb downstream from TES. B, RIPA extracts (20 μg) from control normal B cells or transformed B-cell lines JeKo-1, Pfeiffer, and SUDHL2 cells were analyzed by Western blotting using the indicated antibodies. Anti-β-ACTIN antibody was used as control to show equal loading. C, RIPA extracts (20 μg) were prepared from control sh-GFP or sh-PRMT5 JeKo-1, Pfeiffer, and SUDHL2 cells and analyzed by immunoblotting using the indicated antibodies. Anti-β-ACTIN antibody was included to show equal loading.
ChIP-Seq reads within 2 kbp upstream of the TSS and 2 kbp downstream of TES were quantified as total number of bases per million and reported as PRTM5 load at TSS (Table S1). We identified 504 TSS regions with a 2-fold or more load and 322 TSS regions with a 2-fold or less load in B cells compared with both DLBCL cell lines, respectively (Fig. 1A). Although the majority of H3(Me2)R8-enriched regions in B cells were located in TSS, selected enriched regions in DLBCL cell lines were located primarily in TES (Fig. 1A, bottom and middle panels, respectively). Top molecular functions of genes with differential PRMT5 load in B cells include transcription DNA-binding activity, transcription regulatory region sequence-specific DNA binding, and RNA polymerase II regulatory region sequence-specific DNA binding (Table S2).
We have previously reported that decreased expression of miR92b and miR96 promotes efficient PRMT5 translation, overexpression, and methylation of histone epigenetic marks in MCL (10). PRMT5 promotes MCL and DLBCL cell growth and proliferation through enhanced CYCLIN D1–CDK4/6 signaling (21). In light of these results and our ChIP-Seq data, we reasoned that there might be a common set of miRNAs targeted by PRMT5, which are directly involved in regulating prosurvival factors such as CYCLIN D1 and c-MYC.
To address this hypothesis, we examined expression of CYCLIN D1 and c-MYC in three different types of NHL lymphoma cell lines, including DLBCL cell lines (Pfeiffer and SUDHL-2) and an MCL cell line (JeKo-1) (Fig. 1B). Our findings show that both CYCLIN D1 and c-MYC are overexpressed in lymphoma cell lines compared with control normal B cells (Fig. 1B). In addition, PRMT5 genetic knockdown via short hairpin (sh)-RNA or pharmacological inhibition via a small molecule inhibitor (CMP5) leads to reduced CYCLIN D1 and c-MYC expression (Fig. 1C and Fig. S2) (12).
PRMT5 epigenetically represses miR33b, -96, and -503 and indirectly leads to enhanced CYCLIN D1 and c-MYC expression
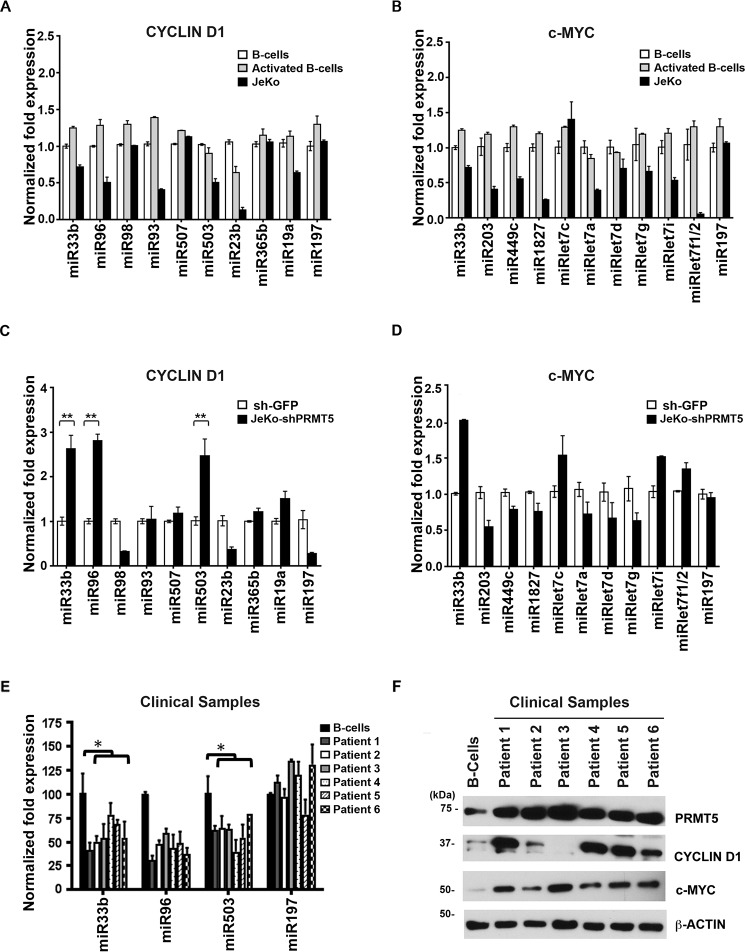
To determine whether there is a PRMT5-silenced set of miRNAs, which regulate expression of CYCLIN D1 and c-MYC, we analyzed the 3′UTRs of each transcript using TargetScan prediction algorithm (www.targetscan.org) for the presence of miRNA-binding sites. Furthermore, analysis of the ChIP-Seq list of target genes with enriched H3(Me2)R8 in their promoter region allowed us to identify miRNAs targeted by PRMT5 with average TSS load of 50 or more across the entire cellular genome. Comparison of the list of potential miRNAs binding to the 3′UTR of CYCLIN D1 and c-MYC with the list of miRNAs targeted by PRMT5 resulted in identification of a small set of candidate miRNAs for each gene (Table 1 and Fig. S3). To assess whether PRMT5 was involved in transcriptional silencing of the identified miRNAs, we measured their expression in both resting and activated normal B cells as well as in JeKo-1, Pfeiffer, and SUDHL-2 lymphoma cell lines (Fig. 2, A and B, and Fig. S4A). Real-time RT-PCR analysis showed that six of nine predicted CYCLIN D1–specific miRNAs exhibit 1.4–5-fold reduced expression in JeKo-1 cells compared with normal B cells (Fig. 2A). Similarly, 9 of 10 c-MYC-specific miRNAs showed 1.4–10-fold reduced expression in JeKo-1 (Fig. 2B). Similar CYCLIN D1–specific miRNA repression was seen in Pfeiffer and SUDHL-2 cell lines (Fig. S4A). As a control, we analyzed the levels of an unrelated miRNA, miR197, which is not a direct PRMT5 target as determined from the ChIP-Seq data, and we found that its expression was unaltered in normal B, JeKo-1, Pfeiffer, and SUDHL-2 cells. Comparison of miRNA levels between resting and activated B lymphocytes did not show any significant difference.
Table 1.
CYCLIN D1 and c-MYC miRNA
| miRNA | Fold enrichment H3R8 | Binding energy |
|---|---|---|
| CYCLIN D1 miRNA | ||
| miR33b | 74 | −21.19 |
| miR96 | 32 | −36.39 |
| miR98 | 11 | −24.27 |
| miR93 | 9.67 | −29.91 |
| miR503 | 8.5 | −40.39 |
| miR507 | 7.5 | −18.41 |
| miR23b | 7.2 | −21.89 |
| miR365b | 4.9 | −21.02 |
| miR19a | 1.59 | −27.23 |
| c-MYC miRNA | ||
| miR33b | 74 | −26.23 |
| miR203 | 14 | −31.3 |
| miR449c | 3.03 | −17.64 |
| miR1827 | 4.6 | −11.32 |
| miRLet7c | 4.69 | −21.54 |
| miRLet7a | 3.68 | −26.1 |
| miRLet7d | 3.96 | −30.32 |
| miRLet7g | 2.76 | −13.23 |
| miRLet7i | 4.48 | −19.21 |
| miRLet7f1 | 4.42 | −19.32 |
Figure 2.
PRMT5 represses transcription of CYCLIN D1- and c-MYC–specific miRNAs. A and B, expression profile of PRMT5 target miRNAs predicted to bind CYCLIN D1 (A) and c-MYC (B) 3′UTR in control normal B cells (resting and activated) versus JeKo-1 cells. Total RNA was isolated from each cell line, and small RNAs were enriched using spin columns per the manufacturer's protocol (Ambion, Inc.). Levels of individual miRNA were measured by real-time RT-PCR using specific primers and probes as described under “Experimental procedures.” miR197 was used as a control. C and D, expression of predicted CYCLIN D1- (C) and c-MYC (D)-specific miRNAs was analyzed in JeKo-1 cells that were infected with either sh-GFP or sh-PRMT5 using real-time RT-PCR as described in Fig. 2A. Data in each graph are represented as mean ± S.D. (**, p ≤ 0.0001). E, expression of CYCLIN D1 and c-MYC target miRNAs miR96, miR33b, and miR503 was analyzed in normal B cells versus patient sample cells using real-time RT-PCR as described in A. miR197 was used as a control. Data in each graph are represented as mean ± S.D. (*, p ≤ 0.005). F, RIPA extracts (20 μg) from control normal B cells or patient samples were analyzed by Western blotting using the indicated antibodies. Anti-β-ACTIN antibody was used as control to show equal loading.
PRMT5 knockdown in all three lymphoma cell lines resulted in de-repression of miR33b (p < 10−4), which is predicted to bind to both CYCLIN D1 and c-MYC 3′UTRs and miR96 (p < 10−4) and miR503 (p < 10−4), which are predicted to be CYCLIN D1–specific miRNAs (Fig. 2, C and D, and Fig. S4, B–F), confirming the direct role played by PRMT5 in miR33b, miR96, and miR503 transcriptional regulation. We also measured expression of CYCLIN D1 and c-MYC–specific miRNAs in primary MCL tumor cells (n = 6), and we found that expression of miR33b (p = 0.043), miR96 (p = 0.118), and miR503 (p = 0.0393) were reduced 1.8–2.3-fold compared with control normal B cells, demonstrating that expression of these three miRNAs is also suppressed in primary lymphoma cells (Fig. 2E). Reduced expression of CYCLIN D1 and c-MYC–specific miRNAs is in agreement with enhanced CYCLIN D1 and c-MYC protein expression in all primary tumors examined, except for patient 3 where there was no detectable CYCLIN D1 expression (Fig. 2F). Whereas our data convincingly show that miRs levels are significantly lower in malignant B cells compared with normal B cells, our results suggest that additional factors may play a role in regulating PRMT5, c-Myc, and Cyclin D1 expression in MCL.
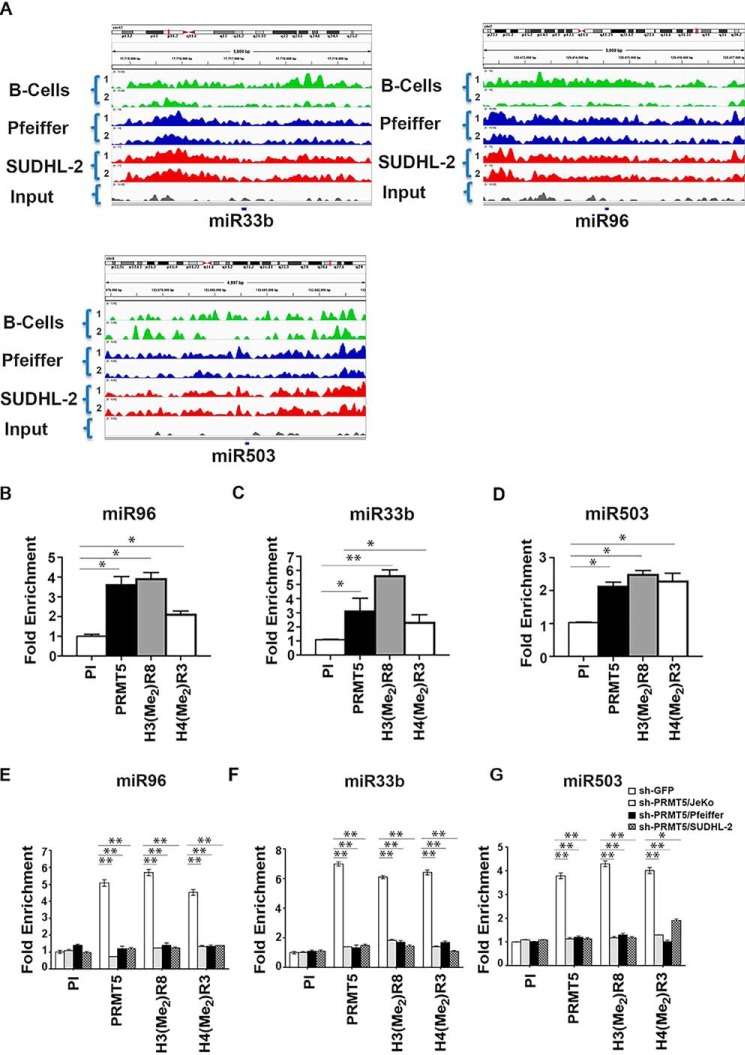
To confirm whether miR33b, miR96, and miR503 were direct PRMT5 targets, we analyzed H3(Me2)R8 methylation recruitment profiles obtained by ChIP-Seq data for promoter regions of each of the three miRNA (Fig. 3A). We found that there was enrichment of the PRMT5-induced H3(Me2)R8 methylation mark in the miR33b, miR96, and miR503 promoter regions in both normal and transformed B cells compared with input, and that there were clear qualitative differences between normal B and lymphoma cell lines. Consistent with the ChIP-Seq results, quantitative ChIP-PCR assays showed that PRMT5 and its epigenetic marks, H3(Me2)R8 and H4(Me2)R3, were enriched 2.5–4-fold (p = 2 × 10−4 for PRMT5 and for H3[Me2]R8, p = 5 × 10−4 for H4[Me2]R3) at the miR96 promoter (Fig. 3B), 2.5–6-fold (p = 10−3 for PRMT5, p = 10−4 for H3[Me2]R8, p = 2 × 10−3 for H4[Me2]R3) at the miR33b promoter in JeKo-1 cells (Fig. 3C), and 2–3-fold (p = 8 × 10−4 for PRMT5, p = 6 × 10−4 for H3[Me2]R8, p = 4 × 10−4 for H4[Me2]R3) at the miR503 promoter (Fig. 3D). The same experiment was performed in Pfeiffer and SUDHL2 with similar results (Fig. S5, A and B). PRMT5 knockdown with shRNA led to loss of recruitment of PRMT5 and associated epigenetic marks at the miR33b (Fig. 3E) (p < 10−4), miR96 (Fig. 3F) (p < 10−4), and miR503 (Fig. 3G) (p < 10−4 for PRMT5 and H3[Me2]R8, p = 9 × 10−4 for H4[Me2]R3) promoters, further supporting the direct role played by PRMT5 in regulating miR33b, miR96, and miR503 transcription.
Figure 3.
PRMT5 methylates promoter histones of target miRNAs. A, promoters of miR33b, miR96, and miR503 in B cells, Pfeiffer, and SUDHL-2 cells from ChIP-Seq libraries were analyzed using Integrated Genome Viewer software to show enrichment of H3(Me2)R8 compared with input. The numbers in braces represent two independent replicates for each cell line. B–D, cross-linked chromatin from JeKo-1 cells was immunoprecipitated using pre-immune- (PI), anti-PRMT5-, H3(Me2)R8-, or H4(Me2)R3-specific antisera, and promoter sequences of miR33b, miR96, and miR503 were detected using specific primers and probes. E–G, cross-linked chromatin from either sh-GFP- or sh-PRMT5-infected JeKo-1, Pfeiffer, and SUDHL-2 cells was immunoprecipitated using the indicated antibodies, and promoter sequences of miR33b, miR96, and miR503 were detected using specific primers and probes. sh-GFP bar is normalized for each cell line. All ChIP assays were carried out twice in triplicate, and fold enrichment with each antibody was calculated relative to the PI sample. Data in each graph are represented as mean ± S.D. (*, p ≤ 0.005; **, p ≤ 0.0001.)
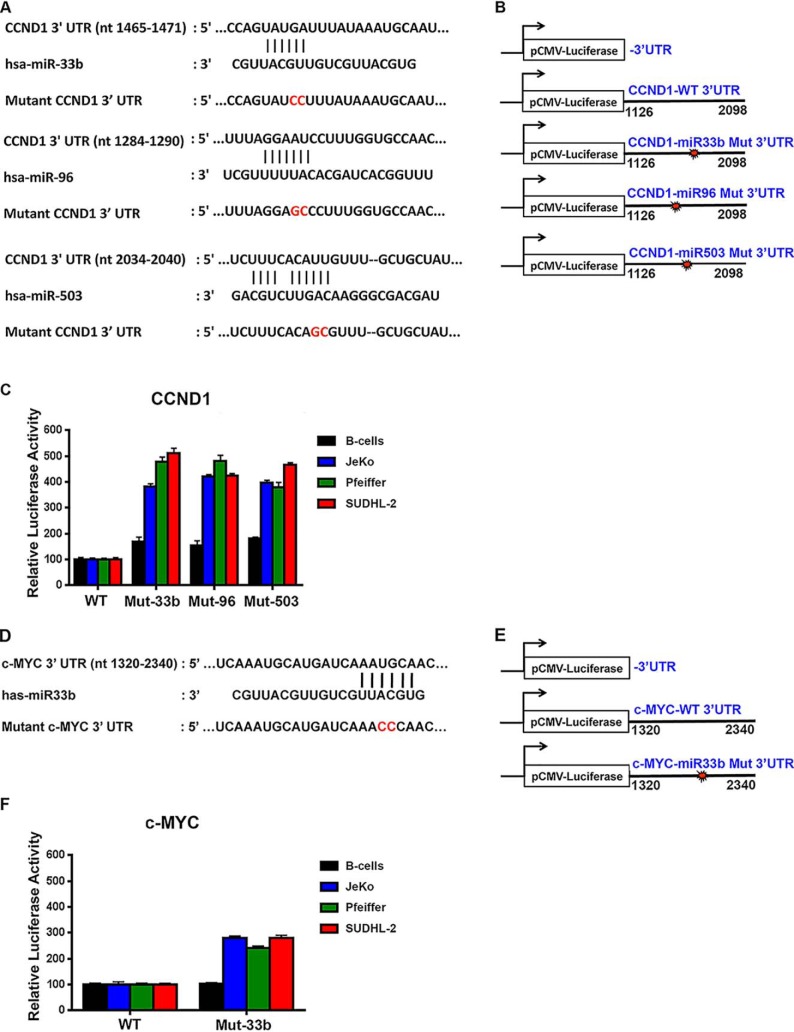
Functional analysis of miR33b-, miR96-, and miR503-binding sites in the 3′UTR of CYCLIN D1 and c-MYC mRNAs
In silico analysis showed that although miR33b binds to both c-MYC and CYCLIN D1 3′UTRs, miR96 and miR503 bind only to CYCLIN D1 3′UTR. To evaluate the contribution of each of these binding sites in translational regulation of both CYCLIN D1 and c-MYC, we performed luciferase assays with constructs containing either WT or mutant sequences at predicted bindings sites for each miR in the 3′UTRs. Sequence analysis of the 3′UTR of human CYCLIN D1 mRNA revealed the presence of conserved miR33b- (nucleotides 1465–1471), miR96-(nucleotides 1284–1290), and miR503 (nucleotides 2034–2040)-binding sites, whereas the 3′UTR of c-MYC mRNA harbored only one miR33b-binding site (nucleotides 1969–1974) (Fig. 4, A and D). To assess the effect of each miRNA on CYCLIN D1 and c-MYC expression, we cloned the WT 3′UTR region of either CYCLIN D1 mRNA (nucleotide 1126–2098) or c-MYC mRNA (nucleotides 1320–2340) downstream from the coding sequence for the firefly luciferase reporter gene. We also generated mutant constructs where each site was mutated to abolish miRNA binding (Fig. 4, B and E). Normal B cells and lymphoma cells (JeKo-1, Pfeiffer, and SUDHL-2) were transfected with either control pCMV-luciferase alone or pCMV-luciferase fused to WT or mutant CYCLIN D1, or WT or mutant c-MYC 3′UTR, and luciferase activity was measured 36 h later.
Figure 4.
miR96 and miR503 target CYCLIN D1, and miR33b targets both CYCLIN D1 and c-MYC. A, B, D, and E, schematic of putative miR33b, miR96, and miR503 target sites within the human CYCLIN D1 3′UTR (A) and c-MYC 3′UTR (D) as predicted by TargetScan algorithm. Mutations introduced into the human CYCLIN D1 and c-MYC 3′UTR are highlighted in red. C and F, normal B cells (25 × 106) or transformed JeKo-1, Pfeiffer, and SUDHL-2 (5 × 106) cells were electroporated with either pCMV-LUC, pCMV-LUC fused to WT or mutated CYCLIN D1 3′UTR, or c-MYC 3′UTR, and luciferase expression was measured using dual-luciferase reporter assay. Luciferase activity is represented relative to pCMV-LUC for each cell line and has been normalized using Renilla luciferase. Experiments were conducted twice in triplicate, and the data in each graph are represented as mean ± S.D.
When the construct containing WT CYCLIN D1 3′UTR was electroporated into normal B cells, which express miR33b, miR96, and miR503, a basal level of luciferase activity was observed (Fig. 4C). When the same construct was introduced into lymphoma cell lines (which express lower albeit detectable levels of miR33b, miR96, and miR503 compared with normal B cells), there was no significant increase in luciferase activity over that seen with normal B-cell control. When constructs containing mutant CYCLIN D1 3′UTR were used, there was a slight enhancement (1.5–1.8-fold) in luciferase activity in normal B cells compared with a 3.9–5-fold increase in luciferase activity in lymphoma cells, suggesting that mutation of miR33b-, miR96-, and miR503-binding sites prevented down-regulation of luciferase activity. Similar results were observed with luciferase constructs containing either WT or mutant c-MYC 3′UTR (Fig. 4F). These results indicate that the identified PRMT5 target miRNAs are involved in CYCLIN D1 and c-MYC translational regulation and that their suppression in lymphoma cells contributes to enhanced CYCLIN D1 and c-MYC protein expression.
Restored expression of miR33b, miR96, or miR503 reduces CYCLIN D1 and c-MYC expression in lymphoma cells
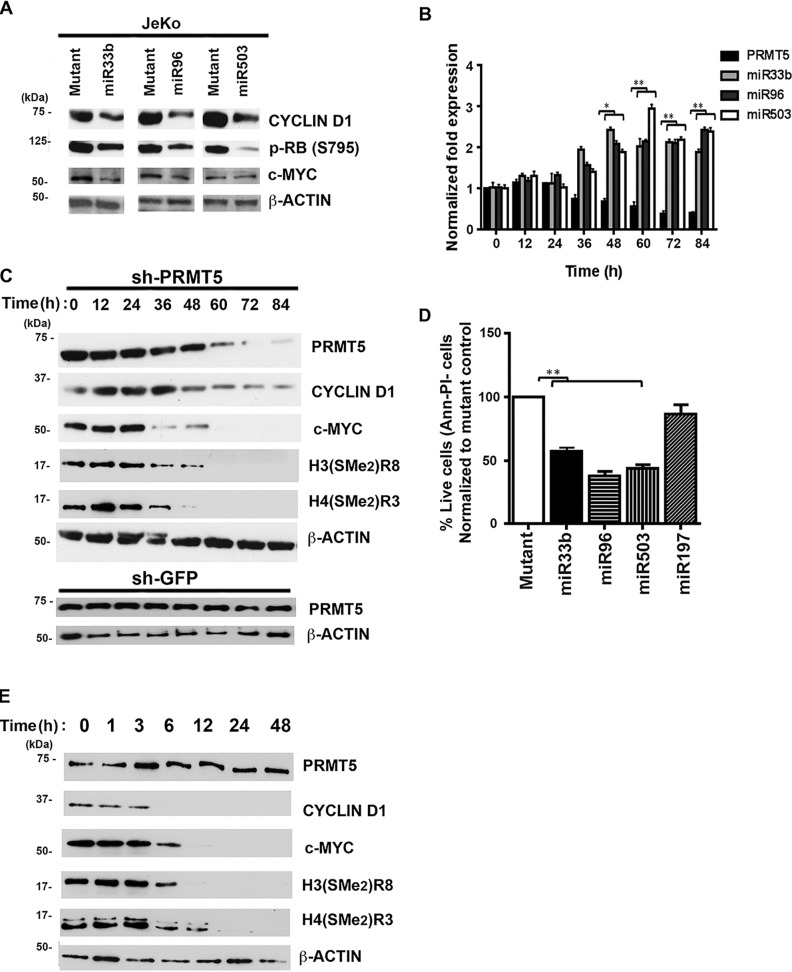
Having shown that specific miRNA-binding sites in the 3′UTR of either CYCLIN D1 or c-MYC mRNAs can regulate luciferase translation, we wanted to determine the consequences of re-introduction of either miR33b, miR96, or miR503 on CYCLIN D1 and c-MYC expression. Lymphoma cells were electroporated with either WT or mutant miRNAs, and whole-cell extracts were analyzed for CYCLIN D1 and c-MYC protein expression (Fig. 5A and Fig. S6, C and E). In accordance with the results of the luciferase assay, re-introduction of WT miR33b led to a decrease in CYCLIN D1 and c-MYC protein expression, whereas both miR96 and miR503 caused a significant drop in CYCLIN D1 protein levels. Because CYCLIN D1–CDK4/6 complexes are responsible for phosphorylation of the retinoblastoma protein (RB1), we checked whether the levels of phospho-RB (Ser-795) were altered in JeKo-1 cells upon reintroduction of each miRNA. As expected, re-introduction of miR33b, miR96, or miR503 triggered a reduction in phospho-RB (Ser-795) (Fig. 5A). We also analyzed CYCLIN D1 and c-MYC mRNA levels; however, we did not find any significant changes in the presence of each miRNA (Fig. S6, A, B, and D).
Figure 5.
PRMT5 target miRNAs regulates CYCLIN D1 and c-MYC expression. A, WT or mutated miR33b, miR96, and miR503 were individually electroporated as dsRNA (2.5 μg) in JeKo-1 cells, and 20 μg of RIPA extracts were analyzed by Western blotting using the indicated antibodies. B, JeKo-1 cells were infected with sh-PRMT5 lentivirus, and the mRNA levels of PRMT5 and its target miRNAs were measured by real-time RT-PCR at different time points. For comparison, the steady-state levels of the target miRNAs examined were determined before infection and are shown at time point 0 h. The experiment was performed twice in triplicate, and the data points in each graph are represented as means ± S.D. C and E, JeKo-1 cells were infected with sh-PRMT5 lentivirus (or sh-GFP control) (C) or treated with CMP5 (E), and RIPA extracts (20 μg) were analyzed for PRMT5 and its epigenetic marks CYCLIN D1 and c-MYC protein levels at different time points. For comparison, the protein levels of PRMT5 and its epigenetic marks CYCLIN D1 and c-MYC were determined before infection (and CMP5 treatment) and are shown at time point 0 h. D, re-expression of miR33b, miR96, and miR503 results in cellular apoptosis. Equal numbers (5 × 106) of JeKo-1 cells were electroporated with 2.5 μg of miR33b, miR96, and miR503, and cells were stained with FITC-annexin V antibody and propidium iodide before they were analyzed by flow cytometry. (*, p ≤ 0.005; **, p ≤ 0.0001.)
Because miR33b, miR96, and miR503 are direct PRMT5 targets, we sought to evaluate the effect of PRMT5 knockdown on CYCLIN D1 and c-MYC expression. JeKo-1 cells were infected with lentivirus designed to express a PRMT5-specific short hairpin RNA, and levels of PRMT5 mRNA as well as levels of all three miRNAs were monitored every 12 h over an 84-h time span (Fig. 5B). Similarly, kinetic analysis of PRMT5, its epigenetic marks, CYCLIN D1 and c-MYC protein expression, was also conducted following genetic knockdown of PRMT5 via shRNA (Fig. 5C) and pharmacological inhibition with CMP5 (Fig. 5E). Our findings showed that as the levels of PRMT5 mRNA began to decrease 36 h post-infection, the levels of miR33b, miR96, and miR503 increased gradually to reach maximal de-repression 48–60 h post-infection (Fig. 5B). Analysis of protein expression revealed that there was kinetic concordance between miRNA derepression and increased repression of CYCLIN D1 and c-MYC protein expression (Fig. 5C). These results indicate that PRMT5 knockdown leads to derepression of miR33b, miR96, and miR503, which in turn triggers inhibition of both CYCLIN D1 and c-MYC translation.
Given the importance of CYCLIN D1 and c-MYC in the control of cell growth and proliferation, we next determined whether re-introduction of individual miRNAs would induce apoptosis (Fig. 5D and Fig. S6, F and G). Electroporation of each individual miRNA into JeKo-1 cells, followed by fluorescence-activated cell sorting (FACS) analysis, demonstrated that re-introduction of any of the three miRNAs results in a 45–55% (p < 10−4 for each miRNA) lymphoma cell death. Similar findings were obtained when miR33b, miR96, or miR503 were electroporated into either Pfeiffer (Fig. S6F) or SUDHL-2 (Fig. S6G) cell lines. Collectively, these results show that through its ability to epigenetically silence expression of specific miRNAs, PRMT5 is able to promote lymphoma cell survival by up-regulating CYCLIN D1–CDK4/CDK6 proliferative signaling and c-MYC–driven oncogenic gene expression.
PRMT5 knockdown results in miR33b, miR96, and miR503 derepression through loss of repressive complex recruitment targeting miRNA promoters
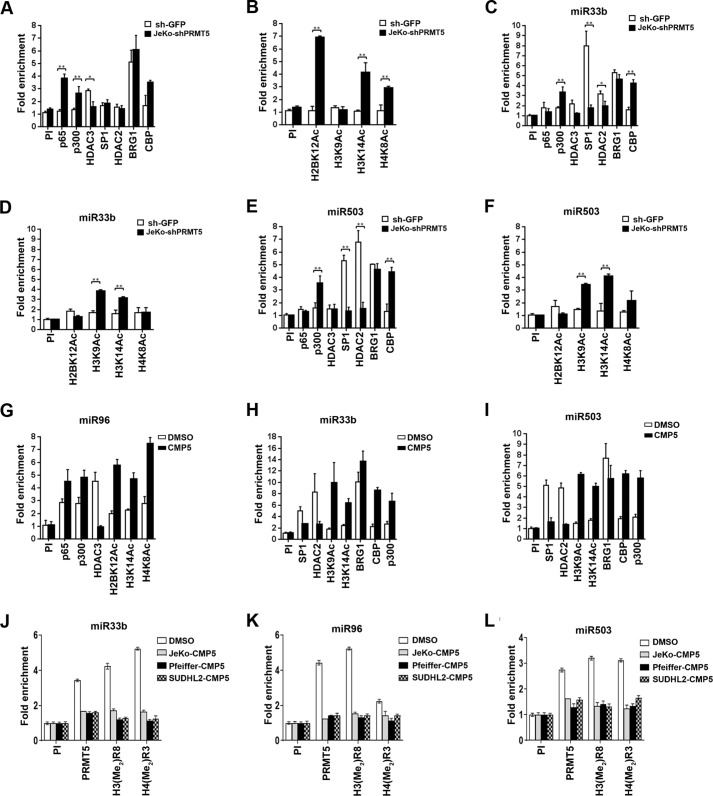
We have previously shown that PRMT5 interacts with different repressor complexes to regulate target gene expression (12). In light of these results, we monitored recruitment of transcription factors (NF-κB p65 and Sp1), co-activators (p300 and CBP), co-repressors (HDAC2 and HDAC3), and histone acetylation marks known to be regulated by these chromatin-modifying enzymes in the promoter region of miR33b, miR96, and miR503 (Fig. 6 and Fig. S7). Real-time quantitative ChIP-PCR analysis using cross-linked chromatin from JeKo-1 cells infected with lentivirus that expressed either control sh-GFP or sh-PRMT5 showed that PRMT5 knockdown led to a 4-fold (p = 2 × 10−4) increase in NF-κB p65 recruitment, loss of HDAC3 binding, a 2.5–3-fold (p = 5 × 10−4) enhanced binding of p300 and CBP, as well as a 3–7-fold increase in acetylation of histones H2BK12 (p = 3 × 10−4), H3K14 (p < 10−4), and H4K8 (p < 10−4) in the miR96 promoter region (Fig. 6, A and B). These molecular changes were consistent with restored miR96 transcription and were reproducible in Pfeiffer and SUDHL-2 lymphoma cell lines (Fig. S7, A and B). When we tested recruitment of NF-κB p65 and HDAC3 to the promoter region of miR33b and miR503, there were no significant changes (Fig. 6, C and E), suggesting that different complexes were involved in transcriptional control of PRMT5 target miRNAs.
Figure 6.
PRMT5 cooperates with distinct co-repressors to silence expression of miR33b, miR96, and miR503 target genes. A–F, cross-linked chromatin from either sh-GFP- or sh-PRMT5-infected JeKo-1 cells was immunoprecipitated using indicated antibodies, and miR96, miR33b, and miR503 promoter sequences were detected using specific primers and probes. ChIP assays were carried out twice in triplicate. Fold enrichment with each antibody was calculated relative to the PI sample. Data in each graph are represented as mean ± S.D. G–I, cross-linked chromatin from either DMSO- or CMP5-treated JeKo-1 cells was immunoprecipitated first using PI or anti-PRMT5 antibody. The immunoprecipitated complexes were then released in the presence of 20 mm DTT. Next, a second round of immunoprecipitation was carried out using the indicated antibodies, and miR96, miR33b, and miR503 promoter sequences were detected using specific primers and probes. ChIP assays were carried out twice in triplicate. J–L, cross-linked chromatin from either DMSO- or CMP5-treated lymphoma cell lines cells was immunoprecipitated using PI or the indicated antibodies, and miR96, miR33b, and miR503 promoter sequences were detected using specific primers and probes. ChIP assays were carried out twice in triplicate. Fold enrichment with each antibody was calculated relative to the PI sample. Data in each graph are represented as mean ± S.D.
Prior work has shown that SP1 and histone deacetylases can physically interact with PRMT5 and repress transcription of miR29b (33, 34). To assess whether SP1 was involved in transcriptional regulation of miR33b and miR503, we tested its recruitment to both miR33b and miR503 promoters in JeKo-1 (Fig. 6, C and E), Pfeiffer (Fig. S7, C and E), and SUDHL-2 (Fig. S7, D and F) cell lines. Although SP1 was not recruited to the miR96 promoter in JeKo-1 cells (Fig. 6A), its binding was enriched 8- and 7.5-fold (p < 10−4) on the miR33b and miR503 promoters, respectively (Fig. 6, C and E). Similarly, SP1 binding to these promoters was increased 5.5- and 4.5-fold (p < 10−4) in Pfeiffer and SUDHL-2 cells, respectively (Fig. S7).
Both HDAC2 and HDAC3 have been shown to interact with SP1 and are recruited to transcriptionally repressed SP1 target genes (34, 35). Because there was no significant recruitment of HDAC3 on both miR33b and miR503 promoters, we checked for the presence of HDAC2. We found that HDAC2 binding was enriched 3-fold (p < 3.4 × 10−2) on the miR33b promoter and 6.5-fold (p < 10−4) on the miR503 promoter in JeKo-1 cells (Fig. 6, C and E). Consistent with these results, HDAC2 recruitment was enhanced 3-fold (p < 10−4) in Pfeiffer and 4-fold (p < 10−4) in SUDHL-2 cells (Fig. S7, C–F) to miR33b and miR503 promoter regions. PRMT5 knockdown resulted in loss of HDAC2 recruitment on both miR33b and -503 promoters in all three lymphoma cell types, which in turn led to increased H3K9 (4-fold for miR33b and 3.5-fold for miR503, p < 10−4) and H3K14 (3-fold for miR33b and 4-fold for miR503, p < 10−4) acetylation in JeKo-1 cells (Fig. 6, D and F). ChIP-PCR analysis also showed that with PRMT5 knockdown there was an increase in both CBP (4-fold for miR33b and 4.3-fold for miR503, p < 10−4) and p300 (3.2-fold for miR33b, p < 10−3 and 3.5-fold for miR503, p < 10−4) binding to the promoter regions of miR33b and miR503 in JeKo-1 cells (Fig. 6, C and E). Similar results were observed in Pfeiffer and SUDHL-2 cell lines (Fig. S7, C–F). These dynamic changes in chromatin binding were very specific, because when we tested for recruitment of the hSWI/SNF ATPase, BRG1, there was no significant change in its binding despite its 4–5-fold enrichment in the promoter region of all three miRNAs. These results suggest that PRMT5 knockdown induces changes in the chromatin structure of all three target miRNA promoters to promote their transcription via distinct complexes. To test whether, PRMT5 and the co-activator complexes are co-recruited to promoters of each miRNA, we performed a ChIP-reChIP assay using chromatin from JeKo-1 cells treated with either DMSO or with CMP5 (Fig. 6, G–I) (12). DNA was first immunoprecipitated with anti-PRMT5 antibody and then re-immunoprecipitated using indicated antibodies in Fig. 6, G–I. Our results show that inhibition of PRMT5 causes gain of p65 and p300 and acetylated histone marks H2BK12, H3K14, and H4K8 as well as loss of HDAC3 on the promoter of miR96 (Fig. 6G). Inhibition of PRMT5 also resulted in gain of CBP, p300, and acetylated histones H3K9 and H3K14 and loss of SP1 and HDAC2 on promoters of miR33b and miR503 (Fig. 6, H and I). To confirm inhibition of PRMT5 activity via CMP5, Fig. 6, J–L, shows loss of PRMT5 and its epigenetic marks on the promoter of targeted miRs. In a separate control experiment, we showed that PRMT5 inhibition led to loss of the PRMT5 epigenetic histone mark H3(Me2)R8 and not total H3 recruitment (Fig. S8). Interestingly, PRMT5 inhibition did not result in any significant change to BRG1 recruitment on any of the promoters.
Discussion
Post-translational modification of conserved histone residues plays an essential role in controlling the transcription of genes that govern growth regulatory networks. Recent studies from our group and others have shown that aberrant methylation of histone arginine residues as well as nonhistone proteins by PRMT enzymes promotes lymphomagenesis (7, 36). We have previously shown that PRMT5-driven symmetric methylation of H3R8 is associated with transcriptional repression of tumor suppressor genes such as RBL2, ST7, and PTPROt (10–12, 21). Although PRMT5 has been mainly shown to repress gene transcription, recent work in acute myeloid leukemia (AML) has implicated PRMT5 in transcriptional activation of FLT3, suggesting that it plays a dual epigenetic role as a transcriptional activator and transcriptional repressor in cancer cells (10, 12, 33).
Genome-wide miRNA screens provide valuable information about small, noncoding RNAs that cells utilize to control expression of target genes. Differences in global miRNA profiles also help distinguish normal cells from transformed cells and help to further classify the different subtypes of cancer cells within each tumor. In this study, we have used a ChIP-Seq data set derived from normal B cells and two DLBCL cell lines to show that PRMT5 suppresses expression of miR33b, miR96, and miR503, which are involved in regulating CYCLIN D1 and c-MYC translation. CYCLIN D1 is a G1 to S cell cycle regulator that operates through phosphorylation and inactivation of RB1. c-MYC is an established proto-oncogene that functions as a transcriptional activator promoting cell survival and proliferation by controlling expression of key target genes needed for G1 to S transition. Our studies have identified three specific miRNAs, miR33b, miR96, and miR503, that can directly target the CYCLIN D1 3′UTR and regulate its expression in aggressive B-cell lymphomas. In MCL, CYCLIN D1 overexpression is driven by the t(11;14) translocation; however, our results show the importance of epigenetic silencing of CYCLIN D1-specific miRNAs to promote translation and overexpression. We have also found that miR33b can specifically target the c-MYC 3′UTR and that re-expression of each individual miRNA triggers down-regulation of CYCLIN D1 and c-MYC protein expression and leads to enhanced cell death. These findings were further substantiated using MCL primary tumor samples, which showed that miR33b, miR96, and miR503 levels are suppressed and that both CYCLIN D1 and c-MYC protein levels are overexpressed.
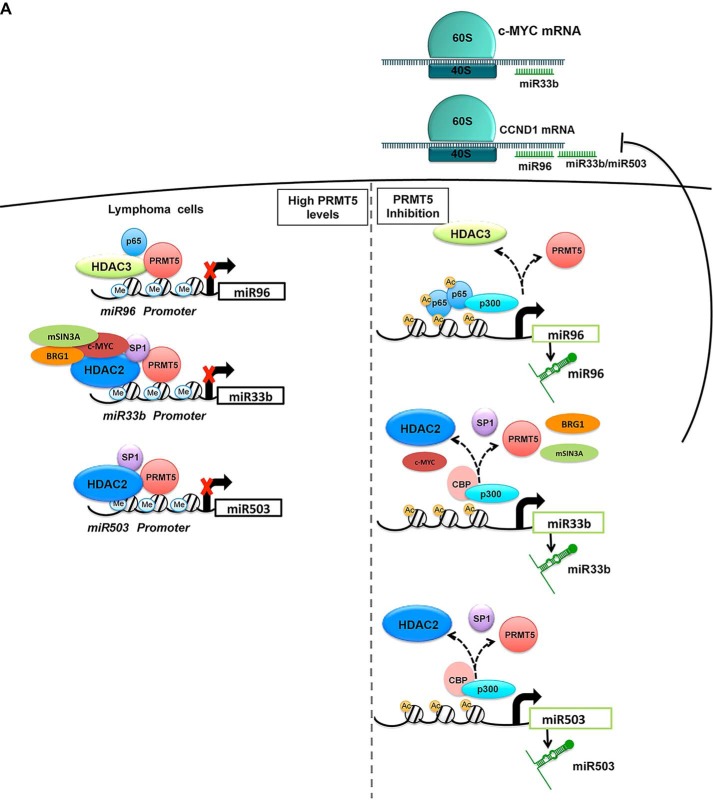
Mechanistic studies showed that PRMT5 is involved in transcriptional repression of target miRNAs through recruitment of two distinct repressor complexes (summarized in Fig. 7). At the miR96 promoter, PRMT5 promotes binding of HDAC3 to induce deacetylation of promoter histones H2BK12, H3K14, and H4K8, which is associated with PRMT5-driven enhanced symmetric methylation of H3R8 and H4R3 as evidenced by ChIP assays. PRMT5 knockdown led to miR96 derepression resulting from decreased HDAC3 recruitment and binding of the transcription activator NF-κB p65 and its co-activators CBP and p300. Interestingly, the mechanism by which PRMT5 suppresses transcription of miR33b and miR503 does not involve NF-κB p65, but instead it relies on removal of the selective protein SP1 and its associated co-repressor, HDAC2. PRMT5 has been shown to symmetrically methylate promoter histone H4R3 in the regulatory region of miR29b in AML patients (33). The mechanism by which PRMT5 silences miR-29b involves both SP1 and HDACs, and PRMT5 inhibition results in decreased H4(Me2)R3 in the miR29b regulatory region (33). Consistent with these results, we have found that PRMT5 is co-recruited with SP1 and HDAC2 to regulate expression of miR33b and miR503, suggesting that PRMT5 cooperates with different transcription factors and associated co-repressors to efficiently silence target gene expression. On both miR33b and miR503 promoters, PRMT5 co-localizes with SP1, BRG1, and HDAC2. PRMT5 knockdown leads to dissociation of the repressive complex and concomitant recruitment of the histone acetyltransferases CBP and p300, which hyperacetylate histones H3K9 and H3K14. Thus, it appears that when PRMT5 activity is inhibited, dynamic changes take place to promote transcriptional derepression through recruitment of transcriptional activators and co-activators.
Figure 7.
Schematic representation of PRMT5 associated co-repressors that regulate expression of miR33b, miR96, and miR503. PRMT5 engages in two different forms of repressor complexes to silence transcription of miR33b, miR96, and miR503.
A recent study has shown that miR33b is down-regulated in gastric cancer cells, and hypermethylation of CpG islands located upstream of the miR33b promoter is responsible for its decreased expression (37). Although DNA hyper-methylation is known for its ability to suppress gene expression in cancer cells, our results show that in three different types of aggressive lymphoma cells PRMT5-mediated histone modification contributes to target tumor suppressor miRNA gene repression. Interestingly, in another report, overexpression of miR33b was shown to have no impact on CYCLIN D1 mRNA or protein levels in multiple myeloma (MM) cells (38); however, ectopic expression of miR33b in MM cells induced their growth arrest and death.
The involvement of miR503 in the control of cancer cell growth and survival was recently highlighted in a study by Wang et al. (39), where overexpression of miR503 in a hepatocellular carcinoma cell line (HepG2/ADM), known for its resistance to the drug adriamycin, led to down-regulation of drug resistance-related proteins, including multidrug resistance 1, multidrug resistance–associated protein 1, DNA excision repair protein ERCC1, SURVIVIN, and BCL2. The net outcome of miR503 re-expression in HepG2/ADM cells was reversal of adriamycin resistance through drug efflux inhibition. Increased expression of miR503 was also associated with cell cycle blockade at G0/G1 and increased cell death (40). It is conceivable that PRMT5-mediated suppression of miR503 expression may render cancer cells more drug-resistant to therapeutic drugs and enhance survival by driving expression of growth-promoting genes such as CYCLIN D1 and c-MYC. Our results clearly show the direct and inverse relationship between PRMT5 and miR503 and demonstrate the effects of miR503 re-expression on promoting lymphoma cell death.
Although our findings show that PRMT5 modulates CYCLIN D1 and c-MYC through direct suppression of tumor suppressor miRs in aggressive B-cell lymphomas, we speculate that a similar mechanism could be operable in other types of cancer. For example, it is well-established that PRMT5 regulates cell-cycle–related proteins such as CYCLIN D1 in hepatocellular carcinoma and oropharyngeal squamous cell carcinoma in addition to modulation of c-Myc expression in breast cancer (41, 42). Our data support the notion that PRMT5 operates upstream of CYCLIN D1 and c-MYC and promotes overexpression through direct inhibition of regulatory miRNAs. Importantly, a study by Klier et al. (43) has shown that CYCLIN D1 knockdown leads to only moderate reduction in cell growth without induction of apoptosis in MCL cells, which was associated with weak induction of p27(Kip1), decreased RB1 (Ser-807/811) phosphorylation, and consistent up-regulation of CYCLIN D2 mRNA and protein expression. Interestingly, simultaneous knockdown of CYCLIN D1 and D2 did not intensify the effects observed with CYCLIN D1 knockdown alone, suggesting that compensatory cyclin-independent mechanisms governing proliferation are activated. These findings have important implications for MCL and perhaps DLBCL therapy, as strategies targeting only CYCLIN D1 function might be hampered by compensatory regulatory mechanisms, resulting in a low probability of treatment response. Because PRMT5 is overexpressed in MCL and DLBCL and directly controls expression of tumor suppressor miRNAs that target CYCLIN D1 and c-MYC, our results prompted us to focus on the development of anti-PRMT5 therapy (12). Recent findings indicate that PRMT5 regulates both histones and nonhistone proteins, including critical transcription factors such as NF-κB p65, E2F1, and p53 and chromatin remodelers such as PRC2, as well as cellular metabolism and cell migration (7). It is possible that each of these additional biological functions of PRMT5 contribute to tumorigenesis. Therefore, strategies aimed at targeting PRMT5, a pleiotropic enzyme regulating the activity of multiple cancer drivers, will likely be an attractive approach for aggressive B-cell lymphomas and potentially other cancers.
Experimental procedures
Cell culture and B-cell isolation
Jeko-1 is a cell line derived from the peripheral blood of a patient with blastoid MCL in leukemic phase. DLBCL is classified in two distinct molecular subtypes with different biological characteristics and clinical outcomes: germinal center B cell (GCB) and activated B cell (ABC), with the latter associated with a poorer prognosis. Pfeiffer is a GCB–DLBCL cell line, and SUDHL2 represents an ABC–DLBCL cell line. The PDX cell line was established from an ibrutinib-resistant MCL PDX mouse model developed in Dr. Alinari's laboratory. A20 is a murine DLBCL cell line. B-cell NHL cell lines (JeKo-1, Pfeiffer, SUDHL-2, PDX, and A20) were grown in RPMI 1640 medium supplemented with 10% FBS and 1 mm sodium pyruvate. All studies using patient lymphoma samples, which had no patient identifiers, abide by the declaration of Helsinki principles and were approved by the Ohio State University Comprehensive Cancer Center Institutional Review Board (IRB protocol no. 1997CO194) and conducted in agreement with the approved guidelines (IBC protocol no. 2006R0017-R1-AM6). Similarly, all animal studies were performed in compliance with guidelines approved by the Federal and the Ohio State University Institutional Animal Care and Use Committee (IACUC protocol no. 20009A0094-R3). To isolate normal B cells, tonsillar tissues were minced extensively in RPMI 1640 containing 10% FBS and strained through a collector sieve (Bellco Glass, Inc.) to remove tissue debris. Next, monocytes were removed by adhesion to tissue culture plates, and B cells were isolated by depletion of T lymphocytes. Cells were mixed with 8-fold excess of sheep red blood cells (Colorado Serum Co.) and incubated on ice for 1 h. To separate B cells from rosetted T lymphocytes, 10 ml of Ficoll-Paque (Amersham Biosciences) was added, and samples were spun at room temperature before collecting the layer containing B cells. B lymphocytes were washed with RPMI 1640 media, and the purity of the isolated B cells was determined by FACS analysis using anti-CD19–PE antibody.
Cell viability
To assess the effects of miRNA re-expression on lymphoma cell growth and death, 5 × 106 lymphoma cells (JeKo-1, Pfeiffer, and SUDHL-2) were electroporated with 2.5 μg of either miR33b, miR96, miR503, or miR197 and seeded into 6-cm-diameter plates before harvesting after 30 h. Next, cells were stained with 5 μl of FITC–annexin V antibody (BD Biosciences) and 5 μl of propidium iodide solution (BD Biosciences) for 15 min at room temperature before they were analyzed on a Beckman Coulter FC500 flow cytometer.
Plasmid construction, transfection of normal and transformed B lymphocytes, and luciferase assays
Plasmid pCMV-Luc/CCND1–3′UTR was generated by inserting the 3′UTR of CYCLIN D1 (nucleotide 1126–2098) as a blunt-end PCR fragment into SalI-linearized and Klenow-treated pCMV-Luc. To mutate the miR33b-, miR96-, and miR503-binding sites, pCMV-Luc/CYCLIN D1 3′UTR was used as a template in a QuikChange multisite–directed mutagenesis reaction as specified by the manufacturer (Stratagene, Inc.). To mutate the underlined miR33b, miR96, or miR503 seed sequence binding site, three distinct primers were used (5′-CCAGTATCCTTTATAAATGCAAT-3′), (5′-TTTAGGAGCCCTTTGGTGCCAAC-3′), and (5′-TCTTTCACAGCGTTTGCTGCTAT-3′), respectively (mutated nucleotides are shown in bold). Similarly, plasmid pCMV-Luc/cMYC-3′UTR was generated by inserting the 3′UTR of c-MYC (nucleotides 1320–2340) as an NdeI–XhoI PCR fragment into NdeI–XhoI-linearized pCMV-Luc. To mutate the underlined miR33b seed sequence–binding site, the following primer was used (5′-TCAAATGCATGATCAAACCCAAC-3′). All mutated sites were verified by Sanger DNA sequencing.
To study the effects of mutating the binding sites for miR33b, miR96, and miR533, ∼2.5 × 107 B cells or 5 × 106 MCL and DLBCL cells were transfected with 15 μg of total DNA (6 μg of pCMV-Luc construct, 2 μg of pRenilla-Luc control plasmid, and 7 μg of pBS(KS+)) with Lipofectamine 2000 (Invitrogen). After 30 h, cells were lysed in 100 μl of 1× passive lysis buffer provided in the kit (Promega, Inc.). Dual-luciferase reporter analysis was carried out using 40 μl of cell lysate as specified by the manufacturer (Promega, Inc.).
To evaluate the effects of miR33b, miR96, and miR503 on CYCLIN D1 or c-MYC protein expression, either WT or mutant miR33b-, miR96-, and miR503-specific dsRNAs were individually electroporated into each lymphoma cell lines using the Amaxa Biosystems electroporator. Approximately 5 × 106 cells were electroporated with 2.5 μg of dsRNA corresponding to each miR being studied in 100 μl of Amaxa Cell Line nucleofector kit V reagent (Lonza, Inc.). Cells were then plated in 3 ml of RPMI 1640 containing 10% FBS and lysed 48 h later in 100 μl of RIPA lysis buffer before proteins were analyzed by Western blotting. To generate WT and mutant miR33b, miR96, miR503, and miR197 dsRNAs, T7-driven single-strand DNA oligonucleotides (listed in Table S3) were transcribed individually using the Ambion silencer siRNA construction kit (catalogue number AM1620). Sense and antisense WT or mutant miRNA products were annealed for 14 h at 37 °C, and dsDNA templates along with RNA leader sequences were removed according to the manufacturer's protocol. Next, WT or mutant miR33b, miR96, miR503, and miR197 dsRNAs were purified through an Ambion filter cartridge before electroporation into JeKo-1, Pfeiffer, and SUDHL-2 cells. PRMT5 knockdown was conducted using a lentiviral construct that expresses PRMT5-specific shRNA, as described previously (21).
Reverse transcription, real time (RT)-PCR
Reverse transcription (RT) was performed on 1 μg of total RNA, isolated with TRIzol reagent (Life Technologies, Inc.), using a reverse transcription kit (Applied Biosystems, Inc.), as described previously (9). Briefly, a 20-μl reaction containing 1 μg of RNA, 1× TaqMan RT buffer, 5.5 μm MgCl2, 0.5 μm dNTPs, 2.5 m random hexamer, 0.4 units/μl RNase inhibitor, and 1.25 units/μl multiscribe reverse transcriptase was first incubated for 10 min at 25 °C followed by incubation for 1 h at 42 °C. After inactivation of the reaction for 5 min at 95 °C, real-time PCR was performed using the TaqMan system (Applied Biosystems, Inc.) in a 10-μl reaction containing 1 μl of RT reaction, 1× TaqMan universal master mix, 0.8 μm of each primer, and 0.2 μm of universal probe (Roche Applied Science). Gene-specific primers and probes were used in real-time RT-PCR to detect PRMT5 (forward, 5′-TATGTGGTACGGCTGCACA-3′, and reverse, 5′-TGGCTGAAGGTGAAACAGG-3′, probe 31), CYCLIN D1 (forward, 5′-GAAGATCGTCGCCACCTG-3′, and reverse, 5′-GACCTCCTCCTCGCACTTCT-3′, probe 67), and c-MYC (forward, 5′-GATCCAGACTCTGACCTTTTGC-3′, reverse, 5′-CACCAGCAGCGACTCTGA-3′, probe 34).
ChIP assay, ChIP-Seq library preparation, data processing, and analysis
Cross-linked chromatin was prepared from JeKo-1, Pfeiffer, or SUDHL-2 cell lines, as described previously (9); however, after sonication, cross-linked chromatin was digested with micrococcal nuclease (1 unit/ml) at 37 °C for 20 min before adding 200 μl of stop buffer (100 mm Tris-HCl (pH 8.6), 0.45% SDS, 2.5% Triton X-100, 5 mm EDTA (pH 8.0), protease inhibitors). Chromatin was analyzed by agarose gel electrophoresis to ensure that DNA fragment sizes did not exceed 500 bp, and ChIP assays were carried out essentially as described previously (10, 11). To amplify DNA sequences of PRMT5-regulated miRNA genes, pre-immune and immune antibodies raised against PRMT5, H3(Me2)R8, and H4(Me2)R3, were used to immunoprecipitate cross-linked chromatin from lymphoma cell lines that were infected with lentivirus-expressing shGFP or shPRMT5 RNAs (21). After extensive washing, nucleoprotein complexes were eluted in 200 μl of elution buffer (50 mm Tris-HCl (pH 8.0), 10 mm EDTA, 1% SDS), and cross-links were reversed by incubating samples at 65 °C for 12 h. Immunoprecipitated DNA was purified by phenol and chloroform extraction and resuspended in 40 μl of TE buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA). miRNA promoter sequences were amplified using the following specific primers and probes: miR33b (forward, 5′-GAAGGCCAGTGGGTACCTG-3′, and reverse, 5′-CCACCCCTCACCTTGTCA-3′, probe 20); miR96 (forward, 5′-ATCTGGTCTGGTTTGGGATG-3′, and reverse, 5′-GAAACAGAGCCCCAGGAAC-3′, probe 84); and miR503 (forward, 5′-TGGGGACAATGAATGAGACTT-3′, and reverse, 5′-GCCAGTTTCAAAGTCTTGGTG-3′, probe 4).
For ChIP-Seq assays, cross-linked chromatin was prepared from 5 × 106 normal B, Pfeiffer, and SUDHL2 cells, and immunoprecipitated using the anti-H3(Me2)R8 antibody as described previously (9, 10). For all cell types, we performed ChIP-Seq experiments using two biological replicates with two technical replicates each. Real-time ChIP assays were performed as described previously (11). After confirmation of target sequence enrichment in ChIP versus input samples by real-time PCR, libraries were created using Illumina Hi-Seq 2000 Prep Kit (Illumina, Inc). Gel size selection of the 300–400-bp fragments was conducted after the adapter ligation step, followed by 15 amplification cycles. Real-time PCR was performed again to confirm enrichment of target sequences and then the libraries were sequenced using an Illumina GAIIx for high-throughput sequencing.
ChIP data for normal B, Pfeiffer, and SUDHL2 cells were analyzed to understand genome-wide binding sites of PRMT5. First, 50-bp sequence reads were aligned to human reference genome (GRCh37) with Rsubread version 1.22.2 (44), and duplicates were marked with Picard tools version 1.94 (http://broadinstitute.github.io/picard). Genome-wide correlation and heatmap of ChIP signals were generated using bamCorrelate, computeMatrix, and plotHeatmap functions of deepTools version 3.0.2 (44) The PRMT5 load around TSS was generated using a series of in-house shell and R scripts utilizing GenomicRanges (45), Rsamtools (45) packages, and Ensembl GRCh37 annotation release 75. Next, the number of reads aligned to 2 kb upstream and downstream of the TSS regions was counted, and the PRMT5 load at each TSS was calculated as (number of reads around TSS × sequence length)/(total number of reads/106).
Target scan
To target prediction algorithms, TargetScan was used for the identification of potential miRNAs predicted to bind human CYCLIN D1 and c-Myc 3′UTR, as described previously (46). We blasted all the potential miRNAs that were predicted to bind CYCLIN D1 and c-Myc against our ChIP-Seq data and identified several miRNAs that are PRMT5 targets suggesting that PRMT5 may control translation of CYCLIN D1 and c-Myc by suppressing transcription of its miRNAs.
Statistical analysis
Results were expressed as the mean ± S.D., unless otherwise specified. All the experiments include at least three independent replicates unless otherwise specified. Two-sample t tests and analysis of variance were used to generate p values for comparisons between two groups and for comparisons among more than two groups, respectively, when data are independent. Otherwise, linear-mixed effects models were used for analysis to take account of correlation among observations when data are correlated; an example is the RT-PCR data in Fig. 5B that gene expression was repeatedly measured over the experiment time. SAS 9.4 (SAS Institute Inc.) software was used for analysis, and GraphPad Prism4 was used to generate graphs. p values < 0.05 were considered significant after Holm's procedure adjustment for multiple comparisons or outcomes (47).
Author contributions
V. K., H. G. O., J. C., and X. Z. data curation; V. K., H. G. O., and X. Z. software; V. K., H. G. O., J. C., X. Z., and S. S. formal analysis; V. K., H. G. O., J. C., X. Z., S. S., and R. A. B. validation; V. K., L. A., and S. S. investigation; V. K., L. A., J. C., S. S., and R. A. B. visualization; V. K., S. S., and R. A. B. methodology; V. K., L. A., S. S., and R. A. B. writing-original draft; V. K., L. A., S. S., and R. A. B. writing-review and editing; L. A., S. S., and R. A. B. supervision; L. A., S. S., and R. A. B. funding acquisition; L. A., S. S., and R. A. B. project administration; S. S. and R. A. B. conceptualization; R. A. B. resources.
Supplementary Material
Acknowledgment
We thank Selina Chen-Kiang from for providing cell lines and helpful discussions.
This work was supported by the American Association for Cancer Research Basic Cancer Research Fellowship (to L. A.), the Leukemia Lymphoma Society Mantle Cell Lymphoma Research Initiative (MCL7001-18) (to R. A. B.), National Institutes of Health NCI Grant P01 CA214274 (to R. A. B. and L. A.), and National Priorities Research Program (NPRP) Grant 8-617-3-13 (to S. S. and R. A. B.) from the National Research Fund (a member of Qatar Foundation). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S8 and Tables S1–S3.
- PRMT
- protein arginine methyltransferase
- TSS
- transcription start site
- TES
- transcription end site
- PI
- pre-immune
- FBS
- fetal bovine serum
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- HDAC
- histone deacetylase
- H3(Me2)R8
- histone 3 dimethylation at Arg-8
- H4(Me2)R8
- histone 4 dimethylation at Arg-8
- MCL
- Mantle cell lymphoma
- DLBCL
- diffuse large B-cell lymphoma
- NHL
- non-Hodgkin's lymphoma
- RB
- retinoblastoma protein
- AML
- acute myeloid leukemia
- miRNA
- microRNA
- miR
- microRNA
- ChIP-Seq
- ChIP-Sequence
- MM
- multiple myeloma
- GCB
- germinal center B cell
- ABC
- activated B cell
- RT
- reverse transcription.
References
- 1. Sif S. (2004) ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J. Cell. Biochem. 91, 1087–1098 10.1002/jcb.20005 [DOI] [PubMed] [Google Scholar]
- 2. Martin C., and Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 10.1038/nrm1761 [DOI] [PubMed] [Google Scholar]
- 3. Fischle W., Wang Y., and Allis C. D. (2003) Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15, 172–183 10.1016/S0955-0674(03)00013-9 [DOI] [PubMed] [Google Scholar]
- 4. Hake S. B., Xiao A., and Allis C. D. (2004) Linking the epigenetic language′ of covalent histone modifications to cancer. Br. J. Cancer 90, 761–769 10.1038/sj.bjc.6601575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pal S., and Sif S. (2007) Interplay between chromatin remodelers and protein arginine methyltransferases. J. Cell. Physiol. 213, 306–315 10.1002/jcp.21180 [DOI] [PubMed] [Google Scholar]
- 6. Bedford M. T., and Richard S. (2005) Arginine methylation an emerging regulator of protein function. Mol. Cell 18, 263–272 10.1016/j.molcel.2005.04.003 [DOI] [PubMed] [Google Scholar]
- 7. Karkhanis V., Hu Y. J., Baiocchi R. A., Imbalzano A. N., and Sif S. (2011) Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 36, 633–641 10.1016/j.tibs.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zurita-Lopez C. I., Sandberg T., Kelly R., and Clarke S. G. (2012) Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J. Biol. Chem. 287, 7859–7870 10.1074/jbc.M111.336271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., and Sif S. (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24, 9630–9645 10.1128/MCB.24.21.9630-9645.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pal S., Baiocchi R. A., Byrd J. C., Grever M. R., Jacob S. T., and Sif S. (2007) Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 26, 3558–3569 10.1038/sj.emboj.7601794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L., Pal S., and Sif S. (2008) Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 28, 6262–6277 10.1128/MCB.00923-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alinari L., Mahasenan K. V., Yan F., Karkhanis V., Chung J. H., Smith E. M., Quinion C., Smith P. L., Kim L., Patton J. T., Lapalombella R., Yu B., Wu Y., Roy S., De Leo A., et al. (2015) Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood 125, 2530–2543 10.1182/blood-2014-12-619783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dacwag C. S., Bedford M. T., Sif S., and Imbalzano A. N. (2009) Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol. Cell. Biol. 29, 1909–1921 10.1128/MCB.00742-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Campo E., and Rule S. (2015) Mantle cell lymphoma: evolving management strategies. Blood 125, 48–55 10.1182/blood-2014-05-521898 [DOI] [PubMed] [Google Scholar]
- 15. Non-Hodgkin's Lymphoma Classification Project. (1997) A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin's lymphoma. The Non-Hodgkin's Lymphoma Classification Project. Blood 89, 3909–3918 10.1182/blood.V89.11.3909 [DOI] [PubMed] [Google Scholar]
- 16. Tsujimoto Y., Yunis J., Onorato-Showe L., Erikson J., Nowell P. C., and Croce C. M. (1984) Molecular cloning of the chromosomal breakpoint of B-cell lymphomas and leukemias with the t(11;14) chromosome translocation. Science 224, 1403–1406 10.1126/science.6610211 [DOI] [PubMed] [Google Scholar]
- 17. Jares P., Colomer D., and Campo E. (2007) Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat. Rev. Cancer 7, 750–762 10.1038/nrc2230 [DOI] [PubMed] [Google Scholar]
- 18. Aguilera N. S., Bijwaard K. E., Duncan B., Krafft A. E., Chu W. S., Abbondanzo S. L., Lichy J. H., and Taubenberger J. K. (1998) Differential expression of cyclin D1 in mantle cell lymphoma and other non-Hodgkin's lymphomas. Am. J. Pathol. 153, 1969–1976 10.1016/S0002-9440(10)65710-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nasi S., Ciarapica R., Jucker R., Rosati J., and Soucek L. (2001) Making decisions through Myc. FEBS Lett. 490, 153–162 10.1016/S0014-5793(01)02118-4 [DOI] [PubMed] [Google Scholar]
- 20. O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., and Mendell J. T. (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- 21. Chung J., Karkhanis V., Tae S., Yan F., Smith P., Ayers L. W., Agostinelli C., Pileri S., Denis G. V., Baiocchi R. A., and Sif S. (2013) Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J. Biol. Chem. 288, 35534–35547 10.1074/jbc.M113.510669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen R. W., Bemis L. T., Amato C. M., Myint H., Tran H., Birks D. K., Eckhardt S. G., and Robinson W. A. (2008) Truncation in CCND1 mRNA alters miR-16–1 regulation in mantle cell lymphoma. Blood 112, 822–829 10.1182/blood-2008-03-142182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiestner A., Tehrani M., Chiorazzi M., Wright G., Gibellini F., Nakayama K., Liu H., Rosenwald A., Muller-Hermelink H. K., Ott G., Chan W. C., Greiner T. C., Weisenburger D. D., Vose J., Armitage J. O., et al. (2007) Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood 109, 4599–4606 10.1182/blood-2006-08-039859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ott G., Rosenwald A., and Campo E. (2013) Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood 122, 3884–3891 10.1182/blood-2013-05-498329 [DOI] [PubMed] [Google Scholar]
- 25. Meyer N., and Penn L. Z. (2008) Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- 26. Oberley M. J., Rajguru S. A., Zhang C., Kim K., Shaw G. R., Grindle K. M., Kahl B. S., Kanugh C., Laffin J., and Yang D. T. (2013) Immunohistochemical evaluation of MYC expression in mantle cell lymphoma. Histopathology 63, 499–508 10.1111/his.12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hernández L., Hernández S., Beà S., Pinyol M., Ferrer A., Bosch F., Nadal A., Fernández P. L., Palacín A., Montserrat E., and Campo E. (1999) c-myc mRNA expression and genomic alterations in mantle cell lymphomas and other nodal non-Hodgkin's lymphomas. Leukemia 13, 2087–2093 10.1038/sj.leu/2401599 [DOI] [PubMed] [Google Scholar]
- 28. Copie-Bergman C., Cuillière-Dartigues P., Baia M., Briere J., Delarue R., Canioni D., Salles G., Parrens M., Belhadj K., Fabiani B., Recher C., Petrella T., Ketterer N., Peyrade F., Haioun C., et al. (2015) MYC-IG rearrangements are negative predictors of survival in DLBCL patients treated with immunochemotherapy: a GELA/LYSA study. Blood 126, 2466–2474 10.1182/blood-2015-05-647602 [DOI] [PubMed] [Google Scholar]
- 29. Hu Z., Medeiros L. J., Chen Z., Chen W., Li S., Konoplev S. N., Lu X., Pham L. V., Young K. H., Wang W., and Hu S. (2017) Mantle cell lymphoma with MYC rearrangement: a report of 17 patients. Am. J. Surg. Pathol. 41, 216–224 10.1097/PAS.0000000000000758 [DOI] [PubMed] [Google Scholar]
- 30. Sesques P., and Johnson N. A. (2017) Approach to the diagnosis and treatment of high-grade B-cell lymphomas with MYC and BCL2 and/or BCL6 rearrangements. Blood 129, 280–288 10.1182/blood-2016-02-636316 [DOI] [PubMed] [Google Scholar]
- 31. Yi S., Zou D., Li C., Zhong S., Chen W., Li Z., Xiong W., Liu W., Liu E., Cui R., Ru K., Zhang P., Xu Y., An G., Lv R., et al. (2015) High incidence of MYC and BCL2 abnormalities in mantle cell lymphoma, although only MYC abnormality predicts poor survival. Oncotarget 6, 42362–42371 10.18632/oncotarget.5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koh C. M., Bezzi M., Low D. H., Ang W. X., Teo S. X., Gay F. P., Al-Haddawi M., Tan S. Y., Osato M., Sabò A., Amati B., Wee K. B., and Guccione E. (2015) MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523, 96–100 10.1038/nature14351 [DOI] [PubMed] [Google Scholar]
- 33. Tarighat S. S., Santhanam R., Frankhouser D., Radomska H. S., Lai H., Anghelina M., Wang H., Huang X., Alinari L., Walker A., Caligiuri M. A., Croce C. M., Li L., Garzon R., Li C., et al. (2016) The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia 30, 789–799 10.1038/leu.2015.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu S., Wu L. C., Pang J., Santhanam R., Schwind S., Wu Y. Z., Hickey C. J., Yu J., Becker H., Maharry K., Radmacher M. D., Li C., Whitman S. P., Mishra A., Stauffer N., et al. (2010) Sp1/NFκB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell 17, 333–347 10.1016/j.ccr.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Won J., Yim J., and Kim T. K. (2002) Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem. 277, 38230–38238 10.1074/jbc.M206064200 [DOI] [PubMed] [Google Scholar]
- 36. Yang Y., and Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 10.1038/nrc3409 [DOI] [PubMed] [Google Scholar]
- 37. Yin H., Song P., Su R., Yang G., Dong L., Luo M., Wang B., Gong B., Liu C., Song W., Wang F., Ma Y., Zhang J., Wang W., and Yu J. (2016) DNA methylation-mediated down-regulating of MicroRNA-33b and its role in gastric cancer. Sci. Rep. 6, 18824 10.1038/srep18824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tian Z., Zhao J. J., Tai Y. T., Amin S. B., Hu Y., Berger A. J., Richardson P., Chauhan D., and Anderson K. C. (2012) Investigational agent MLN9708/2238 targets tumor-suppressor miR33b in MM cells. Blood 120, 3958–3967 10.1182/blood-2012-01-401794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang D., Zhang N., Ye Y., Qian J., Zhu Y., and Wang C. (2014) Role and mechanisms of microRNA503 in drug resistance reversal in HepG2/ADM human hepatocellular carcinoma cells. Mol. Med. Rep. 10, 3268–3274 10.3892/mmr.2014.2591 [DOI] [PubMed] [Google Scholar]
- 40. Li Y., Li W., Ying Z., Tian H., Zhu X., Li J., and Li M. (2014) Metastatic heterogeneity of breast cancer cells is associated with expression of a heterogeneous TGFβ-activating miR424–503 gene cluster. Cancer Res. 74, 6107–6118 10.1158/0008-5472.CAN-14-0389 [DOI] [PubMed] [Google Scholar]
- 41. Jiang H., Zhu Y., Zhou Z., Xu J., Jin S., Xu K., Zhang H., Sun Q., Wang J., and Xu J. (2018) PRMT5 promotes cell proliferation by inhibiting BTG2 expression via the ERK signaling pathway in hepatocellular carcinoma. Cancer Med. 7, 869–882 10.1002/cam4.1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Z., Kong J., Wu Y., Zhang J., Wang T., Li N., Fan J., Wang H., Zhang J., and Ling R. (2018) PRMT5 determines the sensitivity to chemotherapy by governing stemness in breast cancer. Breast Cancer Res. Treat. 168, 531–542 10.1007/s10549-017-4597-6 [DOI] [PubMed] [Google Scholar]
- 43. Klier M., Anastasov N., Hermann A., Meindl T., Angermeier D., Raffeld M., Fend F., and Quintanilla-Martinez L. (2008) Specific lentiviral shRNA-mediated knockdown of cyclin D1 in mantle cell lymphoma has minimal effects on cell survival and reveals a regulatory circuit with cyclin D2. Leukemia 22, 2097–2105 10.1038/leu.2008.213 [DOI] [PubMed] [Google Scholar]
- 44. Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., and Manke T. (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M. T., and Carey V. J. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lewis B. P., Burge C. B., and Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 47. Holm S. (1979) A simple sequentially rejective multiple test procedure. Scand. J. Statist. 6, 65–70 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.