Abstract
Proteasome activity is required for diverse cellular processes, including transcriptional and epigenetic regulation. However, inhibiting proteasome activity can lead to an increase in transcriptional output that is correlated with enriched levels of trimethyl H3K4 and phosphorylated forms of RNA polymerase (Pol) II at the promoter and gene body. Here, we perform gene expression analysis and ChIP followed by sequencing (ChIP-seq) in MCF-7 breast cancer cells treated with the proteasome inhibitor MG132, and we further explore genome-wide effects of proteasome inhibition on the chromatin state and RNA Pol II transcription. Analysis of gene expression programs and chromatin architecture reveals that chemically inhibiting proteasome activity creates a distinct chromatin state, defined by spreading of the H3K4me3 mark into the gene bodies of differentially-expressed genes. The distinct H3K4me3 chromatin profile and hyperacetylated nucleosomes at transcription start sites establish a chromatin landscape that facilitates recruitment of Ser-5- and Ser-2–phosphorylated RNA Pol II. Subsequent transcriptional events result in diverse gene expression changes. Alterations of H3K36me3 levels in the gene body reflect productive RNA Pol II elongation of transcripts of genes that are induced, underscoring the requirement for proteasome activity at multiple phases of the transcriptional cycle. Finally, by integrating genomics data and pathway analysis, we find that the differential effects of proteasome inhibition on the chromatin state modulate genes that are fundamental for cancer cell survival. Together, our results uncover underappreciated downstream effects of proteasome inhibitors that may underlie targeting of distinct chromatin states and key steps of RNA Pol II–mediated transcription in cancer cells.
Keywords: histone modification, RNA polymerase II, proteasome, transcription, breast cancer
Introduction
The 26S proteasome is a large multiprotease component of the ubiquitin proteasome system (UPS)2 that recognizes and destroys ubiquitylated and misfolded proteins (1). Proteasome activity is required for multiple DNA transactions, and there is increasing evidence the 26S proteasome regulates transcription, chromatin organization, and ultimately the expression of genetic information that governs gene networks critical for cellular homeostasis. Dysfunction of the proteolytic activity of the proteasome disrupts many cellular processes that are important in health and disease (2).
A role for the 26S proteasome in transcription as a protein degradation machine follows from the impact of proteasomal degradation of activator or repressor proteins that influence gene transcription. Transcription is a highly-coordinated process involving multiple steps: transcription initiation, elongation, and termination, and the proteasome is involved in the control of each of these steps (3–8). Typically, the transcription cycle begins with the recognition of specific core promoter DNA sequences by DNA-binding transcription factors together with coregulatory factors that remodel chromatin to facilitate the recruitment of RNA Pol II and general transcription factors that form the preinitiation complex (PIC) (9, 10). Multiple studies have established the proteolytic activity of the proteasome to be an important component in the formation of the PIC and transcription initiation. For example, the widespread overlap between transcription activation domains and degradation signals of many transcription factors supports a direct involvement of proteasome degradation in the formation of the PIC and transcriptional activation (11–13). Furthermore, studies showing that critical core components of the 26S proteasome machinery were associated with the RNA Pol II holoenzyme and the mediator complex provided the first evidence supporting a role for the proteasome in PIC formation (14–16).
Transcription occurs in the context of chromatin. Organization of the genome into chromatin impedes transcription, and as a result, chromatin must undergo structural modifications for transcription to occur (17, 18). Posttranslational modifications (PTMs) of N-terminal histone tails and globular domains, such as acetylation, methylation, and ubiquitination of lysine residues, play a vital role in regulating the physical properties of chromatin and the accessibility of the underlying DNA (19–21). Specific histone modifications such as trimethylation of histone H3 lysine 4 (H3K4me3) and acetylation of H3 Lys-27 (H3K27ac), Lys-122 (H3K122ac), and Lys-9 and Lys-14 (H3K9/14ac) are commonly associated with active transcriptional response (22–24). The proteasome tightly regulates the turnover of the protein machinery that writes, reads, and erases these PTMs to modulate transcriptional response (25–29). Additionally, there are long-standing links between proteasome and chromatin-dependent modifications, through the ubiquitin proteasome system (30–32). Ubiquitylated histones have profound influence on chromatin structure and transcription (33). There are also several instances in biology where the UPS and chromatin-modifying machinery either cross-talk or are within the same biological complexes. For example, TBL1/TBLR1 E3 ligases are subunits of the N-COR/HDAC repressor complex (34, 35); the F-Box E3 ligases, KDM2A/B (FBXL 11/10), demethylate histone H3K36me2/3 and are in a complex with the E3 ligase NEDD4 that ubiquitinates RNA Pol II (36); and the SWI/SNF BAF subunit complex ARID1A (BAF250a) associates with Elongin C, a component of Skp1–Cul1–F box (SCF) E3 ligase that targets histone H2B (37).
Cancer cells exhibit unique chromatin states, and accumulating evidence indicates different cancer types exhibit characteristic histone modification patterns that drive specific gene expression programs. Global levels of many histone marks have been mapped in human cell lines and tissues, allowing correlation between individual patterns and gene expression. For example, genome-wide mapping of chromatin changes occurring during tumorigenesis revealed loss of histone H4K16 acetylation and H4K20 trimethylation as hallmarks of human cancers (38). Loss of H4K16ac in breast cancer may be an early sign of cancer, whereas low levels of H3K9ac and K14ac are prognostic of poor outcomes (39). The di- and trimethyl H3K27 marks are especially enriched in human breast cell lines (40). Thus, alterations in the chromatin landscape are an important hallmark of many types of cancers. Normal cells, as well as cancer cells, depend on the function of the proteasome to regulate many processes critical for cell survival. Whereas the proteasome has been implicated in the control of transcription and regulation of chromatin structure (30–32), the effects of proteasome inhibition on chromatin modifications and RNA Pol II transcription have not been extensively examined genome-wide in cancer cells. Exploring how histone modifications and proteasome activity cross-talk to regulate gene transcription may help to better understand how the proteasome pathway impacts cellular processes critical for cell survival.
In this study, we exposed MCF-7 breast cancer cells to the proteasome inhibitor MG132 to better understand how blocking protein turnover impacts chromatin state and subsequent gene-expression programs. We show that proteasome inhibition establishes a distinct hyperacetylated chromatin landscape characterized by the spreading of the H3K4me3 mark into the gene body. This chromatin environment facilitates the recruitment and processivity of RNA Pol II leading to the expression of genes whose functions are relevant to breast cancer pathology.
Results
Treatment with proteasome inhibitor MG132 causes widespread time-dependent gene expression changes in MCF-7 cells
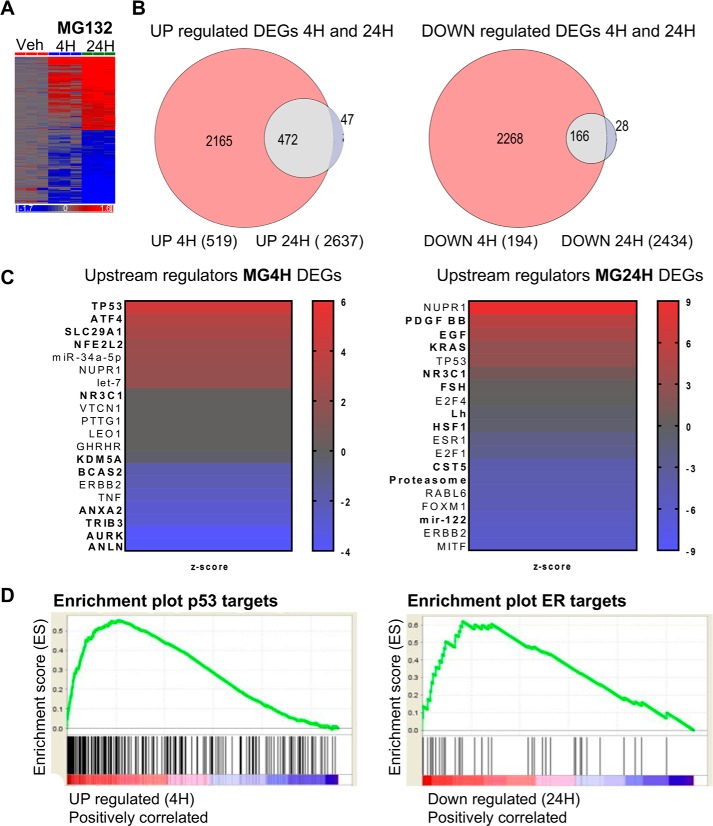
Proteasome inhibitors have emerged as powerful anti-cancer drugs, but downstream mechanisms of their antitumor effects are poorly understood. To begin to address mechanisms of proteasome inhibitors in tumor cells, we monitored the changes in gene expression and chromatin state in MCF-7 breast cancer cells exposed to MG132, a drug that effectively blocks the activity of the 26S proteasome complex. We performed microarray analysis of MCF-7 cells treated with vehicle (DMSO) or 1 μm MG132 for 4 h (MG4H) and 24 h (MG24H) and found profound time-dependent changes in gene expression, with ∼700 (519 up; 194 down) and ∼5000 (2637 up; 2434 down) genes being significantly changed (false discovery rate (FDR) <0.05 and fold change >|1.5|) at the 4- and 24-h time points, respectively (Fig. 1, A and B). A majority of genes (∼90%) changed at 4 h were also changed at 24 h, as indicated by the overlap on the Venn diagram (Fig. 1B). Genes shared between 4 and 24 h included induced genes PMAIP1 and GABARAPL1 and down-regulated genes CYP26A1 and METTL7A, which were also validated by quantitative PCR (Fig. S1A). To confirm the effectiveness of the proteasome inhibition, we first examined changes in the expression of proteasome subunits. As expected, genes encoding proteasomal subunits are up-regulated by MG132 treatment, demonstrating proteasome inhibition was effective in our experimental system (Fig. S1B). Conversely, treatment for 24 h elicits expression of an expanded set of genes, unique to the 24-h treatment (Fig. 1B). Examples of genes changed at 24 h include KLF6, IL6, ESR1, and E2F2 (Fig. S1A). Gene Ontology (GO) analysis of the differentially changed genes revealed significantly-enriched molecular terms that were time-dependent (Fig. S1C). As expected, GO terms representing the proteasome ubiquitin pathway and unfolded protein response were highly enriched in proteasome-inhibited cells (Table S1). Commonly-enriched (Z score >1) GO terms of genes up-regulated by MG132 at 4 and 24 h included NRF2-mediated oxidative stress response, hypoxia signaling in the cardiovascular system, death receptor signaling, and PI3K/AKT signaling (Fig. S1C). p53 signaling was an enriched term at 4 h, whereas downstream signaling pathways like IL-6, nerve growth factor (NGF), and NF-κB were enriched at 24 h (Fig. S1C). Down-regulated genes were predominantly enriched for terms representing cell cycle and DNA damage, including Wnt/β-catenin signaling, role of BRCA1 in DNA damage response, estrogen-mediated S-phase entry, and aryl hydrocarbon receptor signaling (Fig. S1C).
Figure 1.
Time-dependent gene expression changes in MCF-7 cells treated with MG132. A, hierarchical clustering heatmap of gene expression changes in MCF-7 cells treated with vehicle (Veh (DMSO)) or MG132 for 4 and 24 h. Red in the heatmap denotes up-regulation, and blue denotes down-regulation of genes. B, Venn diagrams showing numbers of DEGs in cells exposed to MG132 for 4 and 24 h. C, heatmaps showing upstream regulators of DEGs. MG132 DEGs encode biological information. The top 20 most-enriched upstream regulators of up-regulated (bold) and down-regulated (regular) DEGs in MCF-7 were cells treated with MG132 for 4 h (p value ≤9.88E−05) and 24 h (p value ≤5.12E−08). D, GSEA analysis of the TP53 and ESR1 pathways showing genes induced at 4 h are enriched with p53, and genes repressed at 24 h are enriched with ESR1 pathway signatures, respectively.
Ingenuity pathway analysis (IPA) upstream regulator analysis was done to predict relevant transcriptional regulators that could play a role in the observed gene expression changes. The analysis revealed several regulators of genes in the enriched GO pathways. Upstream regulators of genes differentially changed at the 4-h time point predominantly modulate cell cycle and oxidative stress (Fig. 1C, left panel). Upstream regulators of up-regulated genes include transcription factors, for example TP53, whose activity is most increased and nuclear factor erythroid 2 like 2 (NFE2L2, NRF2), a positive regulator of NRF2-mediated oxidative stress. The loss in activity of aurora kinase (AURK) and anillin actin-binding protein (ANLN) leads to up-regulation of genes that control cell cycle, cell proliferation, and migration (Fig. 1C, left panel, bold). The activation of the chromatin modifier, nuclear protein 1, transcriptional regulator (NUPR1), and miRNAs together with inhibition of TNF and ERBB2 activity in cells treated with MG132 for 4 h suggests gene repression may be mediated by these factors (Fig. 1C, left panel). Consistent with proteasome inhibition, the activity of the 26S proteasome is decreased (negative Z score) in cells treated for 24 h (Fig. 1C, right panel, bold), and this reduced activity results in up-regulation of a subset of genes, including those encoding a majority of proteasome subunits (Fig. S1B). Other upstream regulators of 24-h–induced DEGs are factors involved in cell proliferation and migration, including Erb-B2 receptor tyrosine kinase 2 (ERBB2, HER2/neu), Kirsten rat sarcoma (KRAS), and epidermal growth factor (EGF) (Fig. 1C, right panel, bold). Finally, gene repression at 24 h may be partly mediated by NUPR1 and transcription factors, melanocyte-inducing transcription factor, ERBB2, and FOXM1, whose activities primarily promote cell differentiation, proliferation, and survival, and hence their activity is decreased by MG132 treatment (Fig. 1C, right panel). Intriguingly, the latter transcription factors largely regulate the activity of the estrogen receptor α (ESR1), whose activity is also decreased by proteasome inhibition (Fig. 1C, right panel). Consistent with the finding that p53 and estrogen receptor (ERα) signaling pathways were activated and inhibited at 4 and 24 h, respectively (Fig. 1C and Fig. S1C), gene set enrichment analysis indicated that genes induced at 4 h were enriched in p53 targets, and genes repressed at 24 h were enriched in ER targets (Fig. 1D). Notably, in agreement with the gene expression data, p53 and ERα protein levels increased and decreased at 4 and 24 h, respectively (Fig. S1D). These data support the hypothesis that proteasome inhibition leads to significant changes in expression of multiple genes, including genes important for distinct pathways whose predicted upstream regulators are key drivers of biological processes relevant to breast cancer.
Proteasome inhibition increases H3K4me3 spreading downstream of TSS into gene bodies
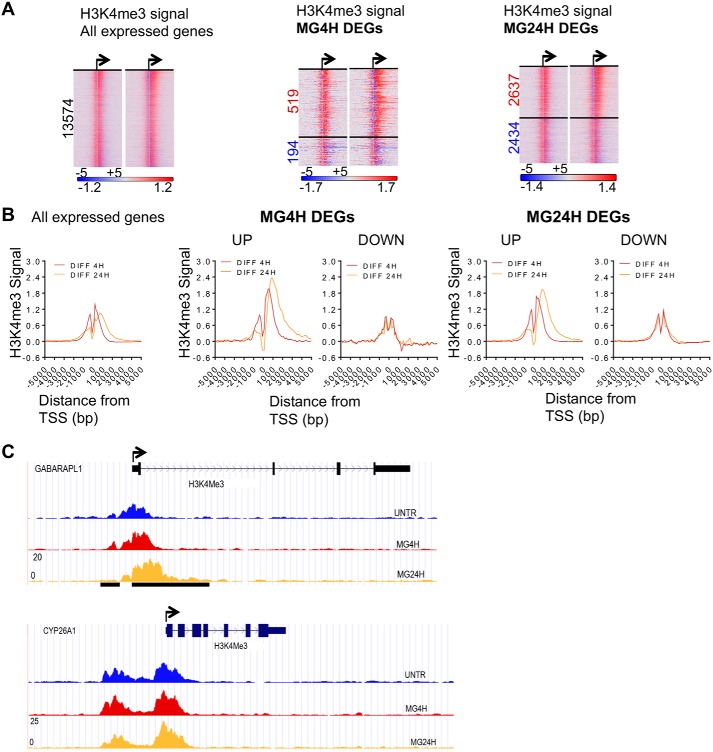
To investigate the functional relationship between chromatin landscape and gene expression changes that occur during proteasome inhibition, we performed at least two independent biological replicates of ChIP followed by sequencing (ChIP-seq) of histone marks generally associated with active transcription, such as H3K4me3 (41, 42). We first analyzed chromatin features of genes that were differentially expressed upon MG132 treatment, focusing on genomic regions surrounding the transcription start sites (TSS). H3K4me3 signal centered around the TSS (±2 kb) of all expressed (∼13,500) and differentially-expressed genes (up- and down-regulated) in MCF-7 cells were similar between vehicle (UNTR) and MG132-treated cells (Fig. S2A). However, in contrast to regions in close proximity to the TSS (∼+750 bp), a subset of genes up-regulated upon MG132 treatment showed a tendency of increased H3K4me3 signal downstream of TSS starting (∼+1 kb) and extending into the gene body, as is evident in the heatmaps showing the difference in H3K4me3 signal (±5 kb) of the up-regulated genes (Fig. 2A and Fig. S2B). We observed spreading of the H3K4me3 mark into the gene body of the up-regulated but not the down-regulated genes on the metagene plots representing the difference between H3K4me3 signal in control and cells treated for 4 and 24 h (Fig. 2B, DIFF 4H and DIFF 24H). Interestingly, close examination of the metagene plots reveal a loss in H3K4me3 (−1)-modified nucleosome signal and what seems to be a shift in the +1 H3K4me3-modified nucleosome into the gene body at 24 h (DIFF 24 h). The differences in the distribution of the H3K4me3 at induced compared with repressed genes can be observed on examples of UCSC genome browser tracks showing the spreading of H3K4me3 signal into the gene body of GABARAPL1, which is the induced and not the repressed CYP26A1 (Fig. 2C and Fig. S1A).
Figure 2.
Proteasome inhibition increases H3K4me3 spreading downstream of TSS into gene bodies. A, MG132 treatment causes an increase in H3K4me3 signal downstream of TSS of up-regulated genes. Heatmaps depicting differential normalized H3K4me3 ChIP-seq signal ranked by gene expression and centered around TSS ± 5 kb. All expressed genes are as follows: left panel: 13,574; MG4H DEGs, middle panel: UP, 519, and DOWN, 194; and MG24H DEGs: right panel: UP, 2637; DOWN, 2434. The two heatmaps for each expression group represent the difference in H3K4me3 signal at 4 and 24 h, respectively. Up-regulated genes are indicated in red, and down-regulated genes are indicated in blue. B, metagene plots of average H3K4me3 differential signal after 4 and 24 h of treatment. C, UCSC browser representation showing H3K4me3 spreading (black bar) at TSS of GABARAPL1 (shared up), but not at TSS of CYP26A1 (shared down). Black arrow represents putative TSS and direction of transcription.
Proteasome inhibition induces a distinct hyperacetylated chromatin landscape at TSS marked with extended H3K4me3 domain
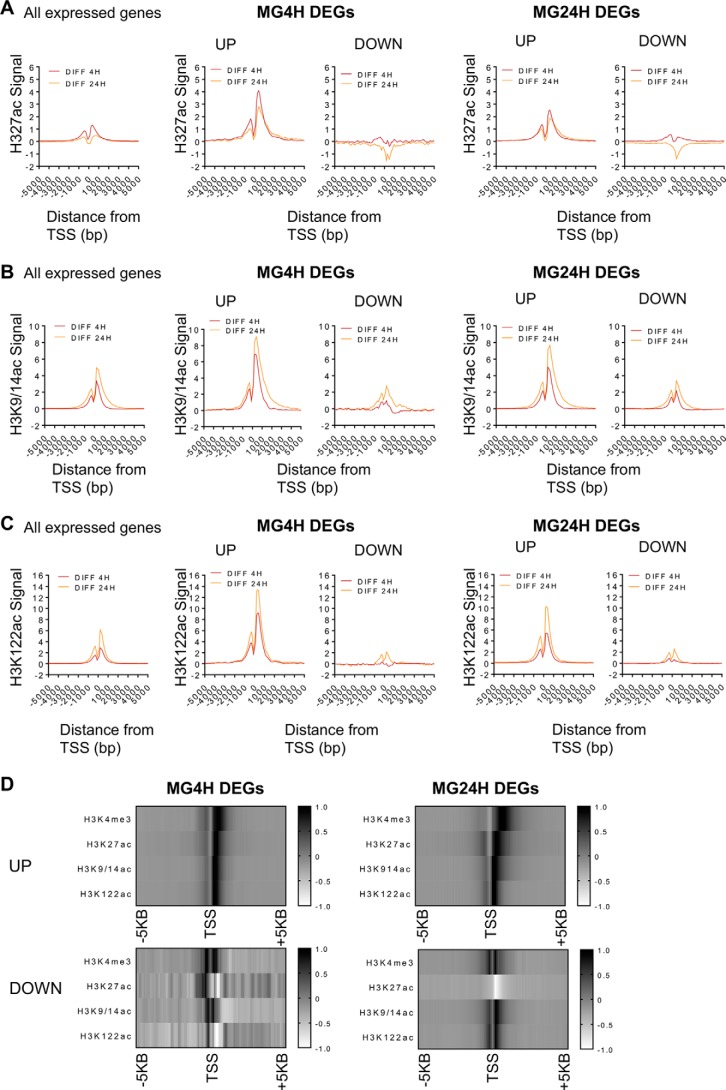
To further assess the chromatin state underlying proteasome inhibition-mediated spreading of H3K4me3 into gene bodies and gene expression changes, we examined the effect of MG132 treatment on the acetylation of H3K27ac, K9/14ac, and K122ac, marks associated with active transcription (22–24). We first asked whether the enriched ChIP-seq signal at TSSs (±2 kb) for each acetylated mark was different between control and MG132-treated cells. H3K27ac ChIP signal at TSSs of genes that were up-regulated increased within 4 h of treatment and remained enriched albeit at lower levels after 24 h (Fig. S3A, UP, compare 4- and 24-h signal). In contrast, the H3K27ac signals at the TSS of repressed genes were similar to those observed at all expressed genes and did not show substantial difference in H3K27ac signal compared with control (Fig. S3A, ALL, DOWN). Metagene analysis of the difference in H3K27ac signal at TSSs clearly showed enriched H3K27ac signal within 4 h in up-regulated genes (Fig. 3A, UP). In contrast, repressed genes show a loss of H3K27ac modified nucleosome (Fig. 3A, DOWN). Thus, the increase in H3K27ac signal upon MG132 treatment is specific to genes that are induced, because MG132 does not increase H3K27ac at all expressed TSSs (Fig. 3A and Fig. S3A, All genes).
Figure 3.
Proteasome inhibition establishes dynamic hyperacetylated chromatin landscape at TSS. A, metagene plots of H3K27ac average differential ChIP-seq signal centered around TSS ±5 kb. All expressed genes (left panel), MG4H DEGs (middle panels), and MG24H DEGs (right panels) are shown. Number of genes per each category are as indicated in Fig. 2. B, metagene plots of H3K9/14ac average differential signal centered around TSS ±5 kb. C, metagene plots of H3K122ac average differential signals centered around TSS ±5 kb. D, heatmap showing average density of H3K4me3 and acetylated H3 at TSS ±5 kb of genes differentially expressed at 4 and 24 h. The intensity of black and white represents the increase and decrease in signal centered around TSS ±5 kb, respectively.
H3K9/14 acetylation is another mark that has been associated with active transcription at many human promoters (23). We examined the effect of MG132 on H3K9/14ac at TSSs and found that nucleosomes at TSS of all genes expressed in MCF-7 cells were enriched with H3K9/14ac (Fig. 3B and Fig. S3B, left panel). However, TSSs of induced genes gained H3K9/14ac signal within 4 h, which progressively increased at 24 h (Fig. 3B and Fig. S3B, UP). The difference in H3K9/14ac signal between control and MG132-treated samples is evident in metagene plots (Fig. 3B, DIFF 4 H, DIFF 24 H). Additionally, although TSSs of repressed genes globally have less signal, they maintain H3K9/14ac nucleosomes particularly after 24 h of treatment, in contrast to what was observed for H3K27ac (Fig. 3, A and B, DOWN, DIFF 24 H). Notably, H3K9/14ac signal extends into the gene body of induced genes particularly in cells treated with MG132 for 24 h (Fig. 3B, UP, DIFF 24 H).
Acetylation of globular domain residue H3K122 correlates with active transcription (24, 43). We found that H3K122ac was enriched above basal levels in cells treated with MG132 compared with control (Fig. 3C and Fig. S3C, All expressed genes). At induced TSSs, H3K122ac signal increases within 4 h (DIFF4H) of treatment, progressively increasing at 24 h (DIFF24H) (Fig. 3C, UP, and Fig. S3C, UP). H3K122ac signal at repressed TSSs was much lower than the global average signal, although cells treated with MG132 for 24 h show some evidence of H3K122ac nucleosome (DIFF 24 h, Fig. 3C, DOWN). We note the kinetics of H3K122ac accumulation at induced genes are distinct from that of H3K27ac and H3K9/14ac in that there is a large increase of H3K122ac in MG132-treated samples compared with control (Fig. S3C, 4H and 24H signal). Furthermore, the H3K122ac footprint is discrete9 within TSS ±1 kb and does not extend into the gene body as observed for the H3K9/14ac (Fig. 3C and Fig. S3C).
Together, our data suggest that inhibiting proteasome activity causes the spreading of H3K4me3 signal into the gene body, and this chromatin landscape coincides with distinct acetylation of nucleosomes at gene TSS based on the level of gene expression. H3K27ac is specifically enriched only at TSS of induced genes and at an early time point (4 h), whereas the H3K9/14ac and H3K122ac signals at TSS progressively increase in a time-dependent manner. Notably H3K9/14ac spreading occurs within the H3K4me3 domain at induced genes. The combinatorial patterns of H3K4me3 and acetylated marks are observed on the heatmap showing the average density of the epigenetic marks at TSS (±5 kb) of up- and down-regulated genes (Fig. 3D) and browser tracks of GABARAPL1 and CYP26A1 (Fig. S3D).
Proteasome inhibition alters distribution of nonphosphorylated and Ser-5–phosphorylated Pol II CTD at expressed gene TSS
Recent studies show that chromatin landscape established by the extended H3K4me3 domains is a key step in the recruitment of RNA Pol II and transcription of genes that determine cell fate and cancer phenotypes (44–47). Based on these observations, we asked whether changes in the chromatin state observed upon MG132 treatment influenced genome-wide dynamics of RNA Pol II binding at TSSs of genes expressed in MCF-7 breast cancer cells. Analysis of nonphosphorylated Pol II within the ±5-kb TSS window revealed that treatment for 24 h caused global accumulation of nonphosphorylated Pol II at TSS of all genes expressed in MCF-7 cells (Fig. 4A and Fig. S4A). However, there was a significant time-dependent increase in the recruitment of nonphosphorylated RNA Pol II at TSS of up-regulated compared with down-regulated genes (Fig. 4, B and C, and Fig. S4, B and C). Genes up-regulated at both 4- and 24-h time points gain nonphosphorylated RNA Pol II at the TSS within 4 h of treatment, and Pol II occupancy is progressively maintained at the 24-h time point (Fig. 4, B and C). Interestingly, although basal levels of nonphosphorylated Pol II in the control cells were similar between the 4- and 24-h DEGs, genes induced during the 4-h treatment showed more RNA Pol II occupancy at 4 and 24 h (Fig. 4, B and C). Remarkably, in cells treated with MG132 for 24 h, nonphosphorylated RNA Pol II also accumulates at repressed genes (Fig. 4, B and C, DOWN). These data indicate that inhibiting proteasome activity causes accumulation of nonphosphorylated RNA Pol II at TSSs of all expressed genes.
Figure 4.
Proteasome inhibition affects the dynamics of RNA Pol II binding at TSS. Differential binding of nonphosphorylated and pSer-5 RNA Pol II at TSSs of up- and down-regulated genes. Metagene plots of average nonphosphorylated Pol II CTD metagene plots signal centered around TSS ±5 kb. A, all expressed genes; B, MG4H DEGs; C, MG24H DEGs. Metagene plots of average pSer-5 Pol II CTD signal centered around TSS ±5 kb. D, all expressed genes; E, MG4H DEGs; F, MG24H DEGS. Numbers of genes per each category are as indicated in Fig. 2.
To determine whether RNA Pol II occupancy resulted in productive transcription initiation, we performed ChIP-seq with an antibody against Ser-5–phosphorylated Pol II CTD (pSer-5), which is a marker of transcription initiation (48, 49). In contrast to nonphosphorylated RNA Pol II CTD, pSer-5 Pol II signal was not significantly enriched at the TSS of all genes (Fig. 4D, compare with Fig. 4A and Fig. S4D). Induced genes gained pSer-5 Pol II at their TSS compared with genes that are repressed (Fig. 4, E and F, and Fig. S4, E and F). Up-regulation of genes coincided with increased abundance of pSer-5 Pol II at the TSS for 4- and 24-h time points (Fig. 4, E and F). A close-up of the pSer-5 Pol II profiles revealed induced 4-h DEGs gained pSer-5 Pol II within 4 h with no further accumulation of pSer-5 Pol II signal during the 24-h treatment (Fig. 4E). We also note genes induced at 4 h have a substantial increase in pSer-5 Pol II enrichment compared with those induced at 24 h (Fig. 4, E and F, UP). Notably, unlike nonphosphorylated Pol II, pSer-5 Pol II is not enriched at TSSs of repressed genes (Fig. 4, E and F, DOWN). These observations suggest different dynamics of pSer-5 CTD recruitment in cells treated with MG132 for 4 h compared with 24 h. pSer-5 CTD recruitment may be an important bookmark for genes that undergo rapid transcription initiation and induction in proteasome-inhibited cells.
Inhibiting proteasome activity increases recruitment and processivity of RNA Pol II
Because proteasome-inhibited cells exhibited hyperphosphorylation of CTD on Ser-5, which suggested formation of a competent initiation complex, we asked whether proteasome inhibition results in the transition of RNA Pol II to productive elongation. We performed ChIP-seq for the pSer-2 phosphorylation, which is a marker of elongation (50, 51). Intriguingly, long-term MG132 treatment (24 h) causes a global accumulation of pSer-2 Pol II at TSS, gene bodies, and 3′ end of all genes as evident on the heatmap and in metagene plots of ChIP signal from over 13,000 genes expressed in MCF-7 cells (Fig. 5, A and B). There were significant differences in pSer-2 Pol II recruitment at genes up-regulated during 4 h (519) compared with 24 h (2637) of treatment (Fig. 5, C and D, E and F). Proteasome inhibition causes recruitment of pSer-2 Pol II at TSS within 4 h, and the signal remains high at TSS, gene body, and the 3′ end at the 24-h time point as is evident on the heat map and metagene analysis of pSer-2 signal at ∼500 up-regulated genes (Fig. 5, C and D, metaplot, MG4H DEGs, UP). In contrast, the ∼2600 genes induced at 24 h slowly gain pSer-2 Pol II within the transcription unit, with maximum signal at 24 h for both the 4- and 24-h DEGs (Fig. 5, E and F, metaplot, MG24H DEGs, UP).
Figure 5.
Proteasome inhibition increases recruitment and processivity of RNA Pol II. A, C, and E, genome-wide accumulation of pSer-2 Pol II around TSS and gene body. Heatmap analysis of pSer-2 Pol II signal in cells treated with vehicle (UNTR) or proteasome inhibitor for the region flanking ±5 kb relative to TSS (black arrow) and TTS (red triangle). Gene body regions were split into 100 total bins per gene. Rows represent genes, and Pol II signal is sorted according to fold gene expression for specific time of treatment. A, all expressed genes; C, MG4H DEGs; E, MG24H DEGs. B, D, and F, metagene plot pSer-2 Pol II occupancy indicating proteasome inhibition causes altered processivity of Pol II. B, all expressed genes; D, MG4H DEGs; F, MG24H DEGs. G, H, and I, expanded metagene plots of TTS region ±5 kb shows distinct differences in pSer-2 Pol II occupancy between up- and down-regulated genes.
Examination of repressed genes revealed distinct differences in pSer-2 Pol II occupancy compared with induced genes. First, we note a decrease in pSer-2 Pol II occupancy within 4 h compared with control cells as is evident from pSer-2 profiles at ∼200 and ∼2400 repressed genes (Fig. 5, D and F, metaplots, DOWN). Close examination of this group of genes revealed that at the 24-h time point, pSer-2 Pol II signal within the transcription unit is similar to control cells (Fig. 5, D and F, metagene plot, DOWN). In addition, the dynamics of pSer-2 Pol II binding at the 3′ end of the repressed and induced group of genes is distinct (Fig. 5, G–I, expanded 3′ end). We noted that in proteasome-inhibited cells, compared with repressed genes, pSer-2 Pol II binding at induced genes progressively increased upstream and downstream of the TTS (Fig. 5, H and I). However, this seems to be a global phenomenon, as the trend is observed at TTS of all expressed genes (Fig. 5G); metagene analysis of the pSer-2 Pol II signal at the 3′-end region (TTS ± 5 kb) shows a clear time-dependent increase in the upstream and downstream signal at induced compared with repressed genes (Fig. 5, H and I). Our results imply the following: 1) proteasome inhibition may cause increased or longer pSer-2 Pol II dwell times at TSS, and 2) pSer-2 Pol II in proteasome-inhibited cells can travel through the entire length of genes that are induced, suggesting proteasome inhibition also enhances pSer-2 Pol II processivity at a subset of differentially-expressed genes. Finally, we note that the differential enrichment of RNA Pol II at induced and repressed genes was not due to RNA Pol II abundance, as the protein levels of RNA Pol II forms in the nucleus remained relatively equal upon MG132 treatment (Fig. S5). However, cells treated with MG132 appear to have more nuclear polyubiquitinated Ser-5- and Ser-2–phosphorylated forms.
pSer-2 Pol II processivity during the elongation process has been coupled with the histone H3K36me3 chromatin modification mark (52–55). We next asked whether histone H3K36me3 deposition was affected in proteasome-inhibited cells. H3K36me3 ChIP-seq signal at all genes expressed in MCF-7 cells was similar between the control and proteasome-inhibited cells (Fig. 6, A and B). Analysis of the ChIP signal at differentially-expressed genes showed that proteasome inhibition enhanced H3K36me3 5′ to 3′ deposition at ∼500 and 2600 genes that were induced during the 4- and 24-h treatment, respectively (Fig. 6, C–F). This effect was evident in metagene plots of ∼500 and 2600 genes up-regulated after 4 and 24 h of treatment with proteasome inhibitor (Fig. 6, D and F). The H3K36me3 signal at ∼200 repressed genes during 4 h of treatment did not differ from those of control cells at 4 h, although the signal at the same genes decreased at 24 h (Fig. 6D, DOWN). Analysis of ∼2400 genes repressed during 24 h of treatment revealed that H3K36me3 5′ to 3′ signal decreased progressively in a time-dependent manner (Fig. 6F, metaplot, DOWN). We also noted differences in the H3K36me3 profiles at the 3′ end of genes (Fig. 6, G–I). The H3K36me3 signal remains high upstream and progressively decreases downstream of TTS at genes induced by proteasome inhibition (Fig. 6, H and I, UP), whereas an opposite effect is observed at repressed genes, where proteasome inhibition does not affect the progressive decrease in H3K36me3 downstream from the TTS (Fig. 6, H and I, DOWN). The differences in H3K36me3 deposition are specific for differentially-expressed genes because they are not observed at all genes expressed in MCF-7 cells (Fig. 6, B and G).
Figure 6.
H3K36me3 enrichment in gene bodies correlates with gene expression levels. A, C, and E, genome-wide accumulation of H3K36me3 around TSS and gene body. Heatmap analysis of H3K36me3 ChIP-seq signal in cells treated with vehicle (UNTR) or proteasome inhibitor for the region flanking ±5 kb relative to TSS (black arrow) and TTS (red triangle). A, all expressed genes; C, MG4H DEGs; E, MG24H DEGs. B, D, and F, metagene plot of H3K36me3 occupancy shows increased and decreased enrichment at up- and down-regulated genes, respectively. B, all expressed genes; D, MG4H DEGs; and F, MG24H DEGs. G, H, and I, expanded metagene plots TTS region ±5 show distinct differences in H3K36me3 occupancy between up- and down-regulated genes.
Intriguingly, the H3K36me3 5′ to 3′ profiles differ from those seen with pSer-2 Pol II in proteasome-inhibited cells. Proteasome inhibition causes global accumulation of the pSer-2 Pol II signal at all genes expressed in MCF-7 cells, particularly at 24 h (Fig. 5, A and B), whereas H3K36me3 deposition correlates with time-dependent changes in transcriptional output. Furthermore, as we observed for pSer-2 Pol II binding, the dynamics of H3K36me3 occupancy at the 3′ end of induced and repressed genes are distinct (Fig. 6, H and I). Inhibiting proteasome activity causes a distinct decoupling of pSer-2 Pol II and H3K36me3 upstream and downstream of TTS of induced (Fig. S6, B and C, UP) compared with repressed genes, and this effect is maximal in cells treated with inhibitor for 24 h (Fig. S6, D and E, DOWN). The different kinetics of pSer-2 Pol II and H3K36me3 at the 3′ end of induced and repressed genes suggests gene regulation mechanisms involving the 3′-end region mediate gene repression effects during proteasome inhibition. In addition, pSer-2 Poll II accumulation downstream of the TTS indicates proteasome activity is critical for RNA pol II transactions at the 3′ end of genes and Pol II accumulation or stalling at the transcription termination sites during proteasome inhibition could ultimately result in defects in transcriptional termination.
Discussion
The 26S proteasome system is essential for the integrity of the genome and proper gene expression, and dysregulation of proteasome function disrupts many basic cellular processes that are important in health and disease. We examined how proteasome inhibition affected chromatin modifications and RNA Pol II transcription by performing genome-wide gene expression and ChIP-seq studies in MCF-7 breast cancer cells treated with a chemical inhibitor of the 26S proteasome. We find that proteasome inhibition causes time-dependent and pathway-specific targeted changes in gene expression. The changes in gene expression are concomitant with a distinct reorganization of the chromatin architecture that involves spreading of the H3K4me3 into the gene bodies, together with dynamic recruitment and processivity of the RNA Pol II at differentially-expressed genes. Our analyses of chromatin state and RNA Pol II transcription in proteasome-inhibited breast cancer cells is a promising resource for future studies in the field.
Cancer cells depend on the activity of the 26S proteasome to maintain gene expression programs that preserve their tumor state (56). Indeed, gene expression analysis reveals that proteasome inhibition enhances antiproliferative while dampening cell-proliferative gene expression programs. In particular, MCF-7 breast cancer cells thrive by their estrogen receptor–dependent proliferative capacity (57). We show genes that are repressed encode pathways related to cell cycle, DNA damage, and estrogen-mediated S-phase entry. Conversely, genes encoding signaling pathways associated with cell death such as NF-κB, oxidative stress, and p53 signaling are up-regulated. Furthermore, we observe a general dysregulation of genes encoding proteasome subunits indicating treatment with MG132 effectively blocks proteasome activity (58). Our findings in breast cancer cells are in general agreement with other gene expression analysis in tumor cells treated with proteasome inhibitors. The majority of studies examining gene expression changes in tumor cells have been performed in multiple myeloma cells (59). Similar to our observations, proteasome-inhibited multiple myeloma cells predominantly express genes encoding cell cycle regulators, apoptosis/cell death, and NF-κB signaling (59).
Chromatin may act as a barrier to transcription, and the global changes in steady-state mRNA expression that we observe could be attributed to changes in chromatin architecture. We identify a distinct chromatin state, characterized by the spreading of the H3K4me3 mark into gene bodies and hyperacetylation of TSSs at genes altered by the proteasome inhibitor. H3K4me3 occupancy is normally restricted within a narrow genomic region around the active TSS (22, 42). However, recent studies show that the spreading of the H3K4me3 domain downstream of the TSS represents a distinct gene regulatory feature that is associated with diverse and cell-specific gene expression programs (44–46, 60–63). Here, we show that proteasome inhibition causes spreading of H3K4me3 marks into gene bodies of induced genes, several of which are targets of the tumor suppressor p53. Our finding is consistent with a recent study that examined genome-wide epigenetic signatures of cancer driver genes and found that broad H3K4me3 domains were associated with high expression of tumor suppressor genes (45).
In addition to the spreading of the H3K4me3 mark, proteasome inhibition also creates a hyperacetylated chromatin landscape at the TSS of altered genes. Combinatorial patterns of active marks like H3K27ac, H3K122ac, and H3K9/14ac and the spreading of H3K4me3 can create cross-talk between specific histone posttranslational modifications to influence gene expression changes (19, 64). Interestingly, although the marked H3K4me3 regions are hyperacetylated, the distribution of each acetylated mark is distinct. H3K27ac is strictly enriched at the TSS of induced but not repressed genes, whereas H3K9/14 and H3K122ac marks are globally enriched at all genes, with an increased association of the marks at induced TSSs suggestive of a more open chromatin architecture. The distinct distribution of each acetylated mark may indicate differential regulation of histone acetyltransferases by proteasome inhibition, a concept that warrants further investigation. Combinatorial occurrence of active marks, such as H3K27ac and spreading of H3K4me3, define open chromatin architecture at regulatory regions that are associated with higher transcriptional activity, transcriptional consistency, and cell type–specific gene expression networks (44, 45, 62, 63).
The extended open chromatin architecture, established during proteasome inhibition, may result in a loss of −1 and a shift of +1 H3K4me3-modified nucleosomes, particularly in cells treated with the inhibitor for 24 h. However, we have not confirmed alterations of nucleosome positioning under these conditions. Interestingly, a previous MNase-seq study showed that broad H3K4me3 domains exhibited fuzzier nucleosome positions when compared with sharp H3K4me3 peaks normally observed at TSSs (45). Furthermore, numerous elegant studies have shown that most transcriptionally-active gene promoters are characterized by the presence of a nucleosome-depleted region flanked by two well-positioned nucleosomes, −1 and +1 nucleosomes, followed by a nucleosomal array into the gene bodies (22, 65, 66). In general, −1 nucleosome is decorated with histone tail modifications, including acetylation and methylation, which are proposed to facilitate nucleosome eviction to allow formation of the pre-initiation complex and increased gene expression (17, 22, 67). Thus, we conclude that proteasome inhibition reorganizes chromatin at TSS of differentially-expressed genes primarily by weakening inter-nucleosomal interactions governed by spreading of H3K4me3 and key acetylated modifications downstream of TSS into the gene bodies of the affected genes.
One consequence of the open chromatin established by the spreading of the H3K4me3 mark is the recruitment of RNA Pol II, which coincides with increased levels of gene expression during proteasome inhibition. Broad H3K4me3 domains are associated with highly-transcribed genes with characteristically-frequent RNA Pol II initiation events and enriched binding of positive regulators of transcriptional elongation (44, 45, 63). RNA Pol II is a well-known target of the proteasome, and consequently, proteasome inhibition should have significant effects on RNA Pol II genomic distribution and underlying effects on gene expression (36, 68). We identify diverse responses of RNA Pol II in proteasome-inhibited cells. First, we show a global time-dependent accumulation of nonphosphorylated RNA Pol II at all TSSs independent of the level of gene expression. We predict these are stalled RNA Pol II complexes as proteasome activity is required to turn over RNA Pol II complexes (36, 68). By analyzing ChIP profiles at TSSs of expressed genes, we identify distinct RNA Pol II CTD phosphorylation patterns that may distinguish the stalled Pol II from transcriptionally productive RNA Pol II. The 5′ to 3′ profiles of Ser-5- and Ser-2–phosphorylated Pol II show a subset of genes in proteasome-inhibited cells undergo a full transcriptional cycle with production of mature mRNA, as induced and not repressed genes are enriched with Ser-5 and pSer-2 Pol II at TSSs and 5′ to 3′ of the full transcription unit. This correlation between the spreading H3K4me3 domain and increased recruitment of pSer-5 recruitment in proteasome-inhibited cells resonates well with recent findings showing TFIIH activity is associated with H3K4me3 spreading and the corresponding gene expression changes (47).
In general, eukaryotic transcription units are characterized by 5′ to 3′ profiles of specific Pol II CTD phospho-isoforms, pSer-5 at the 5′ end and pSer-2 at the 3′ end (4, 48, 69). The analysis of proteasome-inhibited cells reveals an uncharacteristic 5′ to 3′ profile of pSer-2 RNA Pol II. The pSer-2 signal in the gene body and 3′ end of genes reflected gene expression levels in proteasome-inhibited cells as expected. In addition, we observed two uncharacteristic profiles different from the conserved canonical pSer-2 5′ low to 3′ high profile: 1) a large accumulation of pSer-2 at TSS of all expressed genes particularly at 24 h, and 2) an extended accumulation of pSer-2 at 3′ end and downstream of TTSs. Recent elegant studies that combine RNA Pol II ChIP and other assays to monitor RNA Pol II transcription have detected the presence of noncanonical pSer-2 at the 5′ end of human genes (52, 70). pSer-2 at TSSs is associated with longer dwell times at start sites, which is plausible in proteasome-inhibited cells, because turnover of RNA Pol II complexes is inhibited (52). In contrast, pSer-2 at TSSs was also associated with reduced transcriptional polarity and antisense transcription at promoters of human cells (52). Reduced transcriptional polarity at promoters of proteasome-inhibited cells seems unlikely because there is a concomitant enhancement of pSer-2 in the entire transcriptional unit of expressed genes, suggesting these genes undergo competent transcriptional initiation and elongation. The current studies are not sufficient to determine whether pSer-2 at TSS denotes divergent transcription; however, analysis of metagene ChIP signal profiles do not detect the characteristic bimodal distribution of nonphosphorylated Pol II normally associated with divergent transcription (71, 72). Alternatively, the uncharacteristic 5′ to 3′ pSer-2 profiles may result from an additional modification to the CTD, such as ubiquitination, established during proteasome inhibition that then alters the dynamics of pSer-2 binding in the genome.
The appearance of the pSer-2 signal downstream of TTSs in proteasome-inhibited cells suggests proteasome inhibition may increase readthrough transcription that is reminiscent of the recruitment of proteasome activity in resolving RNA complexes at termination sites (73, 74). Moreover, analysis of pSer-2 signal at 3′ end also uncovers uncoupling of pSer-2 and H3K36me3. pSer-2 Pol II and H3K36me3 increase in the gene bodies of up-regulated genes, although the ChIP profiles are distinct. pSer-2 and H3K36me3 downstream of TTSs are decoupled supporting the notion that proteasome inhibition causes defects in both transcriptional termination and 3′ end mRNA processing (73, 75). Indeed, recent studies have discovered a noncanonical function of H3K36me3 in transcriptional termination and 3′ end mRNA processing (76–78).
The observations that pSer-2 and H3K36me3 downstream of TTSs are decoupled suggest proteasome-inhibited cells may use alternative mechanisms to modulate chromatin state and gene expression. A switch from H3K36me3-active chromatin state to a repressive mark at down-regulated genes is an attractive mechanism for gene repression. In this regard, binding of the heterochromatic mark H3K9me3 at gene bodies and transcription termination sites correlates with the gene repression (79, 80). Interestingly, the deposition of H3K9me2 at transcription termination sites and the 3′ end also results in RNAi-dependent gene repression of mammalian protein-coding genes (81). Furthermore, the analyses uncovering miRNAs as upstream regulators of a subset of genes affected by MG132 treatment supports an RNAi-dependent mechanism for regulating gene repression in proteasome-inhibited cells. A mechanism whereby H9Kme3 deposition modulates the chromatin state at the 3′ end to regulate mRNA processing and gene expression in proteasome-inhibited cells remains to be tested.
These findings may have important implications for the mechanism of action by proteasome inhibitors as anti-cancer therapy. The proteasome is clearly a valuable drug target for many diseases (82). Our results suggest that proteasome inhibitors may represent a class of potential drugs that specifically target key steps of RNA pol II transcriptional processes in various cancers. This would be consistent with recent studies where small molecule inhibitors of the transcriptional machinery have shown promising antiproliferative effects in cancer cells (83–85). Although this study does not identify the specific epigenetic and transcriptional regulators that are targeted by the proteasome, our findings demonstrate that downstream effects of proteasome inhibitors involve epigenetic mechanisms that regulate specific gene networks. Overall, these findings support an intriguing mechanism, where dysregulation of the proteasome function causes alterations in chromatin modifications that modulate RNA Pol II transcription to regulate gene expression patterns relevant to breast cancer.
Experimental procedures
For essential reagents and resources used in this publication refer to the key resource table (Table S2).
Cell culture
MCF-7 (ATCC® HTB-22TM) breast cancer cells were grown in a humidified incubator at 37 °C with 5% CO2 in modified Eagle's medium (MEM, GIBCO) supplemented with 10% fetal bovine serum (Atlanta Biologicals), 100 μg/ml penicillin/streptomycin, 2 mm glutamine, and 10 mm HEPES (GIBCO). For MG132 treatment, cells were seeded overnight in phenol red-free MEM supplemented with 5% charcoal-stripped calf serum (Atlanta Biologicals) and 2 mm glutamate. The next day, cells were treated with vehicle (DMSO, Sigma) or 1 μm MG132 (Calbiochem) for 4 or 24 h.
Gene expression profiling and analysis
Total RNA samples were prepared from three biological replicates of MCF-7 cells treated with vehicle (DMSO) or 1 μm MG132 for 4 or 24 h using total RNA, mirVana RNA isolation kit (AM1560, Applied Biosystems). Gene expression analysis was conducted using Agilent whole-human genome 4 × 44 multiplex format oligonucleotide arrays (Agilent Technologies) following the Agilent one-color microarray-based gene expression analysis according to the manufacturer's protocol. Data were obtained using the Agilent Feature Extraction software (version 9.5), using the one-color defaults for all parameters. The Agilent Feature Extraction Software performed error modeling, adjusting for additive and multiplicative noise. The resulting data were processed using the Rosetta Resolver® system (version 7.2) (Rosetta Biosoftware, Kirkland, WA).
Preprocessing of the data
Pixel intensity data were log2-transformed and quantile-normalized. To remove noisy genes at the low end of the intensity distribution, genes above the 20th percentile in 13 of 15 samples were retained. Missing data were imputed with the value at the 20th percentile = 3.17501.
Statistical data analysis
The following two-way analysis of variance (Equation 1) was used to model the log2 quantile-normalized data from the 29,503 genes passing the low intensity filter:
| (Eq. 1) |
where μ is grand mean of the experiment; Yijk represents the kth gene expression observation on the ith cell line (C) and jth time (T), and ϵijk is the random error assumed to be normally and independently distributed with mean 0 and standard deviation δ for all measurements. Fisher's least significant difference t test was performed for each gene to compare the mean of the treated samples to the mean of the control (0 h) matched by cell line type. DEGs were detected at a Benjamini and Hochberg false discovery rate (FDR) <0.05 and absolute fold change >1.5 (86).
Gene set enrichment analysis (GSEA)
GSEA version 3.0 was run using default parameters (87). Input data were the microarray probe expression values for all expressed genes, normalized to log2 fold change from the average at time 0. Probe values were collapsed to gene values internally within GSEA using the default of maximum probe. The gene sets tested were those in collection c2.cp from the Molecular Signatures Database (MSigDB). Enrichment plots were obtained to demonstrate the degree of positive or negative enrichment of the gene sets.
Quantitative RT-PCR
Total RNA was extracted from MCF-7 cells treated with vehicle (DMSO) or 1 μm MG132 for 4 or 24 h using the Qiagen kit (Qiagen, Valencia, CA). For reverse transcriptase–PCR (RT-PCR) analysis, cDNA was synthesized according to standard protocols after DNase I treatment (Invitrogen). Following reverse transcription, cDNA was used for real-time PCR employing SYBR Green detection. Real-time PCR was performed with CFX96 real-time PCR Detection System (Bio-Rad). Ribosomal protein L13A (RPL13A) and TATA-binding protein were used to normalize the differences in the amounts of mRNA in each reaction mixture. PCR primer pairs used in this study are available upon request.
Western blot analysis
For whole-cell extracts, cells were lysed as described previously (88). Cytosolic and nuclear extracts from cells treated with vehicle (control) or 1 μm MG132 for 4 and 24 h were prepared using NE-PERTM kit (Thermo Fisher Scientific). Twenty micrograms of protein were fractionated by SDS-PAGE and transferred to polyvinylidene difluoride membrane according to standard methods (88). Proteins were immunoblotted using the following primary antibodies: from ABCAM, pSer-5 (ab5131) and pSer-2 (ab5095); from Santa Cruz Biotechnology, nonphosphorylated RNA Pol II CTD (8WG16, sc-56767), GR (sc-393232), Lamin A/C (sc-20681), p53 (sc-126), and ERα (Sc-8004); and from Cell Signaling Technology, GR (CST catalog no. 3660). After washing, membranes were incubated with IRDye secondary antibodies following the manufacturer's recommendations (LI-COR, Biosciences). Membranes were washed, and signal was detected using LI-COR odyssey CLX imaging system (LICOR, Biosciences).
ChIP-seq
MCF-7 cells were treated with vehicle (DMSO) or proteasome inhibitor (MG132) as specified above. ChIP for histone modifications was done as follows. MCF-7 cells were briefly cross-linked in 1% formaldehyde for 5 min in phosphate-buffered saline (PBS) and quenched with glycine (125 mm) for 5 min. The quenched cross-linker was quickly discarded in a waste beaker. Cells were rinsed twice with PBS supplemented with protease inhibitors and harvested in 5 ml of PBS and an additional 5 ml of PBS rinse to collect the majority of the cells. The cells were then pelleted at 4 °C and followed by nuclei isolation, as described previously (88, 92). To fragment chromatin, nuclei were resuspended in MNase (Worthington) digestion buffer (10 mm Tris-HCl, pH 7.4, 15 mm NaCl, 60 mm KCl, 1 mm CaCl2, 0.15 mm spermidine, 0.5 mm spermidine) on ice. Nuclei were digested with 50 units of MNase nuclease on a temperature-controlled block at 25 °C for 20 min with gentle mixing at 400 rpm. Following digestion, samples were quickly placed on ice, and the reaction was stopped by adding EDTA/EGTA stop buffer (100 mm, EDTA, 10 mm EGTA, pH 7.5), gently mixing by pipetting, followed by addition of SDS lysis buffer (4% SDS, 40 mm EDTA, 200 mm Tris, pH 8.0), final concentration 1% SDS supplemented with HaltTM Protease Inhibitor Mixture (Thermo Fisher Scientific). To further release chromatin, nuclei were briefly disrupted using a mini homogenizer for 5 s and further incubated on the Hulamixer sample mixer (Invitrogen) for 10 min at 4 °C. Chromatin was recovered by centrifugation at 14,000 rpm for 10 min.
Following centrifugation, an aliquot of MNase-fragmented chromatin was diluted 10× with immune precipitation buffer (20 mm Tris, 8.0, 150 mm NaCl, 0.5% Triton X-100, 2 mm EDTA, 10% glycerol) and supplemented with protease inhibitors (Roche Applied Science). Antibodies against histone modification of interest were added, and chromatin was incubated overnight at 4 °C on a slow-rotating nutator. Next day, 20 μl of protein A and G magnetic Dynabeads (Invitrogen) was added, and samples were incubated for 2 h at 4 °C on a nutator to capture DNA/protein immunocomplexes. Following incubation, protein/DNA immunocomplexes were recovered by subsequent washes and eluted as described previously (8). Eluted immunoprecipitated complexes were digested with RNase A (Qiagen) followed by proteinase K digestion and reverse cross-linking as described previously (8). DNA was recovered using the Qiagen PCR kit purification system (Qiagen)and quantified using Quant-iTTM dsDNA HS assay kit with QubitTM fluorometer (Invitrogen).
For RNA Pol II ChIP, cells were cross-linked, pelleted, and harvested as described above. Pelleted cells were resuspended in sonication buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 0.5% Triton X-100, 2 mm EDTA, 10% glycerol) supplemented with protease inhibitors and incubated on ice for 10 min. Chromatin was fragmented in 15-ml tubes using the bioruptor (Diagenode) for 12 cycles (30 s on/30 s off), total sonication time 6 min. After sonication, the fragmented chromatin was spun briefly in a cooled centrifuge, transferred to 1.5-ml Eppendorf tubes, and centrifuged at 14,000 rpm for 10 min. Following centrifugation, an aliquot of the chromatin diluted with immunoprecipitation buffer was incubated with various RNA Pol II antibodies, and DNA/protein immunocomplexes were recovered as described above. Following DNA recovery, all libraries were prepared with Ilumina-compatible NEXTflex Rapid DNA-seq kit and sequenced on a NextSeq2000 (Illumina). At least two independent biological replicates were performed for all histone modifications and RNA Pol II ChIP assays.
ChIP-Seq processing (histone modifications and RNA Pol II)
The FASTQ files for each biological replicate were concatenated for each sample. Raw reads were quality-filtered to only include those with a mean Phred quality score of 20 or greater. Adapter was trimmed using Cutadapt version 1.12 (89). The preprocessed reads were aligned to the hg19 assembly using Bowtie version 1.2 and parameters -v 2 -m -best -strata (90). Aligned reads were deduplicated by only keeping one read pair when multiple pairs had both mates aligned to the same position. Bound locations were obtained from the aligned reads by extracting the entire length of the aligned fragment. Coverage tracks were generated from these bound locations using the genomecov tool from the bedtools suite version 2.17.0 (91). The coverage tracks were normalized to depth per 10 million mapped reads.
Author contributions
H. K. K. and T. K. A. conceptualization; H. K. K., B. D. B., and P. R. B. formal analysis; H. K. K. validation; H. K. K., B. D. B., and P. R. B. visualization; H. K. K. methodology; H. K. K. writing-original draft and final manuscript; H. K. K., B. D. B., and P. R. B. review and editing manuscript; T. K. A. writing-review and editing final manuscript; H. K. K. and B. D. B. data curation; T. K. A. resources; T. K. A. supervision; T. K. A. funding acquisition; T. K. A. project administration.
Supplementary Material
Acknowledgments
We are grateful to Drs. Paul Wade, Joseph Rodriguez, Laura Bisogno, Jackson Hoffman, and Justin Kosak for critical review of the manuscript. We thank members of the Archer group and the NIEHS Integrative Bioinformatics Group for their ongoing support, advice, and constructive criticism. We thank Greg Solomon, Nikki Reeves, and Jason Malphurs of the NIEHS Epigenomics Core Laboratory for their next-generation sequencing expertise, and Kevin Gerrish and the Molecular Genomics Core Laboratory for performing microarray analysis.
This work was supported by the Intramural Research Program NIEHS Grant Z01 ES071006-19 from the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6, Tables S1–S2.
- UPS
- ubiquitin proteasome system
- DEG
- differentially expressed gene
- GSEA
- Gene Set Enrichment Analysis
- TSS
- transcription start site
- Pol
- polymerase
- CTD
- C-terminal domain
- TTS
- transcription termination site
- PIC
- preinitiation complex
- PTM
- posttranslational modification
- FDR
- false discovery rate
- GO
- Gene Ontology
- ER
- estrogen receptor
- DIFF
- difference
- ChIP-seq
- ChIP sequencing
- MNase
- micrococcal nuclease
- MEM
- modified Eagle's medium
- UNTR
- untreated.
References
- 1. Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 2. Schmidt M., and Finley D. (2014) Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 10.1016/j.bbamcr.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Archer C. T., Burdine L., Liu B., Ferdous A., Johnston S. A., and Kodadek T. (2008) Physical and functional interactions of monoubiquitylated transactivators with the proteasome. J. Biol. Chem. 283, 21789–21798 10.1074/jbc.M803075200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buratowski S. (2009) Progression through the RNA polymerase II CTD cycle. Mol. Cell 36, 541–546 10.1016/j.molcel.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheung A. C., and Cramer P. (2012) A movie of RNA polymerase II transcription. Cell 149, 1431–1437 10.1016/j.cell.2012.06.006 [DOI] [PubMed] [Google Scholar]
- 6. Durairaj G., and Kaiser P. (2014) The 26S proteasome and initiation of gene transcription. Biomolecules 4, 827–847 10.3390/biom4030827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferdous A., Gonzalez F., Sun L., Kodadek T., and Johnston S. A. (2001) The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7, 981–991 10.1016/S1097-2765(01)00250-7 [DOI] [PubMed] [Google Scholar]
- 8. Kinyamu H. K., and Archer T. K. (2007) Proteasome activity modulates chromatin modifications and RNA polymerase II phosphorylation to enhance glucocorticoid receptor-mediated transcription. Mol. Cell. Biol. 27, 4891–4904 10.1128/MCB.02162-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwon H., Imbalzano A. N., Khavari P. A., Kingston R. E., and Green M. R. (1994) Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370, 477–481 10.1038/370477a0 [DOI] [PubMed] [Google Scholar]
- 10. Roeder R. G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 10.1016/S0968-0004(96)10050-5 [DOI] [PubMed] [Google Scholar]
- 11. Rogers S., Wells R., and Rechsteiner M. (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234, 364–368 10.1126/science.2876518 [DOI] [PubMed] [Google Scholar]
- 12. Salghetti S. E., Muratani M., Wijnen H., Futcher B., and Tansey W. P. (2000) Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. U.S.A. 97, 3118–3123 10.1073/pnas.97.7.3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wallace A. D., Cao Y., Chandramouleeswaran S., and Cidlowski J. A. (2010) Lysine 419 targets human glucocorticoid receptor for proteasomal degradation. Steroids 75, 1016–1023 10.1016/j.steroids.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J. W., Ryan F., Swaffield J. C., Johnston S. A., and Moore D. D. (1995) Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature 374, 91–94 10.1038/374091a0 [DOI] [PubMed] [Google Scholar]
- 15. Rubin D. M., Coux O., Wefes I., Hengartner C., Young R. A., Goldberg A. L., and Finley D. (1996) Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature 379, 655–657 10.1038/379655a0 [DOI] [PubMed] [Google Scholar]
- 16. Swaffield J. C., Melcher K., and Johnston S. A. (1995) A highly conserved ATPase protein as a mediator between acidic activation domains and the TATA-binding protein. Nature 374, 88–91 10.1038/374088a0 [DOI] [PubMed] [Google Scholar]
- 17. Li B., Carey M., and Workman J. L. (2007) The role of chromatin during transcription. Cell 128, 707–719 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 18. Wolffe A. P., and Hayes J. J. (1999) Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720 10.1093/nar/27.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gardner K. E., Allis C. D., and Strahl B. D. (2011) Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409, 36–46 10.1016/j.jmb.2011.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rothbart S. B., and Strahl B. D. (2014) Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 1839, 627–643 10.1016/j.bbagrm.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., and Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 23. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., and Young R. A. (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 10.1016/j.cell.2007.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tropberger P., Pott S., Keller C., Kamieniarz-Gdula K., Caron M., Richter F., Li G., Mittler G., Liu E. T., Bühler M., Margueron R., and Schneider R. (2013) Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 152, 859–872 10.1016/j.cell.2013.01.032 [DOI] [PubMed] [Google Scholar]
- 25. Fuchs S. M., Kizer K. O., Braberg H., Krogan N. J., and Strahl B. D. (2012) RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. J. Biol. Chem. 287, 3249–3256 10.1074/jbc.M111.273953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Y., Mayo M. W., Nagji A. S., Hall E. H., Shock L. S., Xiao A., Stelow E. B., and Jones D. R. (2013) BRMS1 suppresses lung cancer metastases through an E3 ligase function on histone acetyltransferase p300. Cancer Res. 73, 1308–1317 10.1158/0008-5472.CAN-12-2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poizat C., Sartorelli V., Chung G., Kloner R. A., and Kedes L. (2000) Proteasome-mediated degradation of the coactivator p300 impairs cardiac transcription. Mol. Cell. Biol. 20, 8643–8654 10.1128/MCB.20.23.8643-8654.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J., Qiu Z., and Wu Y. (2018) Ubiquitin regulation: the histone modifying enzyme's story. Cells 7, E118 10.3390/cells7090118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu K., Lei P. J., Ju L. G., Wang X., Huang K., Yang B., Shao C., Zhu Y., Wei G., Fu X. D., Li L., and Wu M. (2017) SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing. Nucleic Acids Res. 45, 92–105 10.1093/nar/gkw814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geng F., Wenzel S., and Tansey W. P. (2012) Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 81, 177–201 10.1146/annurev-biochem-052110-120012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keppler B. R., Archer T. K., and Kinyamu H. K. (2011) Emerging roles of the 26S proteasome in nuclear hormone receptor-regulated transcription. Biochim. Biophys. Acta 1809, 109–118 10.1016/j.bbagrm.2010.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCann T. S., and Tansey W. P. (2014) Functions of the proteasome on chromatin. Biomolecules 4, 1026–1044 10.3390/biom4041026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weake V. M., and Workman J. L. (2008) Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663 10.1016/j.molcel.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 34. Guenther M. G., Lane W. S., Fischle W., Verdin E., Lazar M. A., and Shiekhattar R. (2000) A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14, 1048–1057 [PMC free article] [PubMed] [Google Scholar]
- 35. Perissi V., Aggarwal A., Glass C. K., Rose D. W., and Rosenfeld M. G. (2004) A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116, 511–526 10.1016/S0092-8674(04)00133-3 [DOI] [PubMed] [Google Scholar]
- 36. Tan Y., Jin C., Ma W., Hu Y., Tanasa B., Oh S., Gamliel A., Ma Q., Yao L., Zhang J., Ohgi K., Liu W., Aggarwal A. K., and Rosenfeld M. G. (2018) Dismissal of RNA polymerase II underlies a large ligand-induced enhancer decommissioning program. Mol. Cell 71, 526–539.e8 10.1016/j.molcel.2018.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li X. S., Trojer P., Matsumura T., Treisman J. E., and Tanese N. (2010) Mammalian SWI/SNF–a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol. Cell. Biol. 30, 1673–1688 10.1128/MCB.00540-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fraga M. F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Iyer N. G., Pérez-Rosado A., Calvo E., Lopez J. A., Cano A., et al. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37, 391–400 10.1038/ng1531 [DOI] [PubMed] [Google Scholar]
- 39. Elsheikh S. E., Green A. R., Rakha E. A., Powe D. G., Ahmed R. A., Collins H. M., Soria D., Garibaldi J. M., Paish C. E., Ammar A. A., Grainge M. J., Ball G. R., Abdelghany M. K., Martinez-Pomares L., Heery D. M., and Ellis I. O. (2009) Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 69, 3802–3809 10.1158/0008-5472.CAN-08-3907 [DOI] [PubMed] [Google Scholar]
- 40. Leroy G., Dimaggio P. A., Chan E. Y., Zee B. M., Blanco M. A., Bryant B., Flaniken I. Z., Liu S., Kang Y., Trojer P., and Garcia B. A. (2013) A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin 6, 20 10.1186/1756-8935-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bernstein B. E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D. K., Huebert D. J., McMahon S., Karlsson E. K., Kulbokas E. J. 3rd., Gingeras T. R., Schreiber S. L., and Lander E. S. (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 10.1016/j.cell.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 42. ENCODE Project Consortium. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pradeepa M. M., Grimes G. R., Kumar Y., Olley G., Taylor G. C., Schneider R., and Bickmore W. A. (2016) Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet. 48, 681–686 10.1038/ng.3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benayoun B. A., Pollina E. A., Ucar D., Mahmoudi S., Karra K., Wong E. D., Devarajan K., Daugherty A. C., Kundaje A. B., Mancini E., Hitz B. C., Gupta R., Rando T. A., Baker J. C., Snyder M. P., et al. (2014) H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158, 673–688 10.1016/j.cell.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen K., Chen Z., Wu D., Zhang L., Lin X., Su J., Rodriguez B., Xi Y., Xia Z., Chen X., Shi X., Wang Q., and Li W. (2015) Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat. Genet. 47, 1149–1157 10.1038/ng.3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dhar S. S., Zhao D., Lin T., Gu B., Pal K., Wu S. J., Alam H., Lv J., Yun K., Gopalakrishnan V., Flores E. R., Northcott P. A., Rajaram V., Li W., Shilatifard A., et al. (2018) MLL4 is required to maintain broad H3K4me3 peaks and super-enhancers at tumor suppressor genes. Mol. Cell 70, 825–841.e6 10.1016/j.molcel.2018.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ebmeier C. C., Erickson B., Allen B. L., Allen M. A., Kim H., Fong N., Jacobsen J. R., Liang K., Shilatifard A., Dowell R. D., Old W. M., Bentley D. L., and Taatjes D. J. (2017) Human TFIIH kinase CDK7 regulates transcription-associated chromatin modifications. Cell Rep. 20, 1173–1186 10.1016/j.celrep.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heidemann M., Hintermair C., Voss K., and Eick D. (2013) Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta 1829, 55–62 10.1016/j.bbagrm.2012.08.013 [DOI] [PubMed] [Google Scholar]
- 49. Zhou Q., Li T., and Price D. H. (2012) RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fuda N. J., Ardehali M. B., and Lis J. T. (2009) Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461, 186–192 10.1038/nature08449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peterlin B. M., and Price D. H. (2006) Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 10.1016/j.molcel.2006.06.014 [DOI] [PubMed] [Google Scholar]
- 52. Fong N., Saldi T., Sheridan R. M., Cortazar M. A., and Bentley D. L. (2017) RNA Pol II dynamics modulate co-transcriptional chromatin modification, CTD phosphorylation, and transcriptional direction. Mol. Cell 66, 546–557.e3 10.1016/j.molcel.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li B., Howe L., Anderson S., Yates J. R. 3rd., and Workman J. L. (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278, 8897–8903 10.1074/jbc.M212134200 [DOI] [PubMed] [Google Scholar]
- 54. Wagner E. J., and Carpenter P. B. (2012) Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiao T., Hall H., Kizer K. O., Shibata Y., Hall M. C., Borchers C. H., and Strahl B. D. (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17, 654–663 10.1101/gad.1055503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Balch W. E., Morimoto R. I., Dillin A., and Kelly J. W. (2008) Adapting proteostasis for disease intervention. Science 319, 916–919 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- 57. Levenson A. S., and Jordan V. C. (1997) MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res. 57, 3071–3078 [PubMed] [Google Scholar]
- 58. Motosugi R., and Murata S. (2019) Dynamic regulation of proteasome expression. Front. Mol. Biosci. 6, 30 10.3389/fmolb.2019.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gandolfi S., Laubach J. P., Hideshima T., Chauhan D., Anderson K. C., and Richardson P. G. (2017) The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev. 36, 561–584 10.1007/s10555-017-9707-8 [DOI] [PubMed] [Google Scholar]
- 60. Dahl J. A., Jung I., Aanes H., Greggains G. D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A. Y., Preissl S., Jermstad I., Haugen M. H., Suganthan R., Bjørås M., et al. (2016) Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dai Z., Mentch S. J., Gao X., Nichenametla S. N., and Locasale J. W. (2018) Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat. Commun. 9, 1955 10.1038/s41467-018-04426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H., Zhang Y., Gao Y., and Gao S. (2016) Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 10.1038/nature19362 [DOI] [PubMed] [Google Scholar]
- 63. Zhao Y. T., Kwon D. Y., Johnson B. S., Fasolino M., Lamonica J. M., Kim Y. J., Zhao B. S., He C., Vahedi G., Kim T. H., and Zhou Z. (2018) Long genes linked to autism spectrum disorders harbor broad enhancer-like chromatin domains. Genome Res. 28, 933–942 10.1101/gr.233775.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee J. S., Smith E., and Shilatifard A. (2010) The language of histone crosstalk. Cell 142, 682–685 10.1016/j.cell.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lai W. K. M., and Pugh B. F. (2017) Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18, 548–562 10.1038/nrm.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schones D. E., Cui K., Cuddapah S., Roh T. Y., Barski A., Wang Z., Wei G., and Zhao K. (2008) Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 10.1016/j.cell.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 68. Wilson M. D., Harreman M., and Svejstrup J. Q. (2013) Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim. Biophys. Acta 1829, 151–157 10.1016/j.bbagrm.2012.08.002 [DOI] [PubMed] [Google Scholar]
- 69. Bentley D. L. (2014) Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schwartz J. C., Ebmeier C. C., Podell E. R., Heimiller J., Taatjes D. J., and Cech T. R. (2012) FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 26, 2690–2695 10.1101/gad.204602.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Erickson B., Sheridan R. M., Cortazar M., and Bentley D. L. (2018) Dynamic turnover of paused Pol II complexes at human promoters. Genes Dev. 32, 1215–1225 10.1101/gad.316810.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Seila A. C., Calabrese J. M., Levine S. S., Yeo G. W., Rahl P. B., Flynn R. A., Young R. A., and Sharp P. A. (2008) Divergent transcription from active promoters. Science 322, 1849–1851 10.1126/science.1162253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gillette T. G., Gonzalez F., Delahodde A., Johnston S. A., and Kodadek T. (2004) Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. U.S.A. 101, 5904–5909 10.1073/pnas.0305411101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Proudfoot N. J. (2016) Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352, aad9926 10.1126/science.aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Laroia G., Cuesta R., Brewer G., and Schneider R. J. (1999) Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284, 499–502 10.1126/science.284.5413.499 [DOI] [PubMed] [Google Scholar]
- 76. Grosso A. R., Leite A. P., Carvalho S., Matos M. R., Martins F. B., Vítor A. C., Desterro J. M., Carmo-Fonseca M., and de Almeida S. F. (2015) Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife 4, e09214 10.7554/eLife.09214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Meers M. P., Henriques T., Lavender C. A., McKay D. J., Strahl B. D., Duronio R. J., Adelman K., and Matera A. G. (2017) Histone gene replacement reveals a post-transcriptional role for H3K36 in maintaining metazoan transcriptome fidelity. Elife 6, e23249 10.7554/eLife.23249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Simon J. M., Hacker K. E., Singh D., Brannon A. R., Parker J. S., Weiser M., Ho T. H., Kuan P. F., Jonasch E., Furey T. S., Prins J. F., Lieb J. D., Rathmell W. K., and Davis I. J. (2014) Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 24, 241–250 10.1101/gr.158253.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Becker J. S., Nicetto D., and Zaret K. S. (2016) H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 32, 29–41 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Taneja N., Zofall M., Balachandran V., Thillainadesan G., Sugiyama T., Wheeler D., Zhou M., and Grewal S. I. (2017) SNF2 family protein Fft3 suppresses nucleosome turnover to promote epigenetic inheritance and proper replication. Mol. Cell 66, 50–62.e6 10.1016/j.molcel.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Skourti-Stathaki K., Kamieniarz-Gdula K., and Proudfoot N. J. (2014) R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516, 436–439 10.1038/nature13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cromm P. M., and Crews C. M. (2017) The proteasome in modern drug discovery: second life of a highly valuable drug target. ACS Cent. Sci. 3, 830–838 10.1021/acscentsci.7b00252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liang K., Smith E. R., Aoi Y., Stoltz K. L., Katagi H., Woodfin A. R., Rendleman E. J., Marshall S. A., Murray D. C., Wang L., Ozark P. A., Mishra R. K., Hashizume R., Schiltz G. E., and Shilatifard A. (2018) Targeting processive transcription elongation via SEC disruption for MYC-induced cancer therapy. Cell 175, 766–779.e17 10.1016/j.cell.2018.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sampathi S., Acharya P., Zhao Y., Wang J., Stengel K. R., Liu Q., Savona M. R., and Hiebert S. W. (2019) The CDK7 inhibitor THZ1 alters RNA polymerase dynamics at the 5′ and 3′ ends of genes. Nucleic Acids Res. 47, 3921–3936 10.1093/nar/gkz127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang Y., Zhang T., Kwiatkowski N., Abraham B. J., Lee T. I., Xie S., Yuzugullu H., Von T., Li H., Lin Z., Stover D. G., Lim E., Wang Z. C., Iglehart J. D., Young R. A., et al. (2015) CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 10.1016/j.cell.2015.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate- a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 10.1007/978-1-4419-9863-7_1215 [DOI] [Google Scholar]
- 87. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kinyamu H. K., and Archer T. K. (2003) Estrogen receptor-dependent proteasomal degradation of the glucocorticoid receptor is coupled to an increase in mdm2 protein expression. Mol. Cell. Biol. 23, 5867–5881 10.1128/MCB.23.16.5867-5881.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17, 10–12 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 90. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Quinlan A. R., and Hall I. M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Trotter K. W., and Archer T. K. (2012) Assaying chromatin structure and remodeling by restriction enzyme accessibility. Methods Mol. Biol. 833, 89–102 10.1007/978-1-61779-477-3_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.