Abstract
Claudin (CLDN)-4 expression has been associated with malignancy in various cancers. When CLDN4 expression was examined in oral squamous cell carcinoma (OSCC), 22 out of 57 (39%) cases showed immunoreactivity in the nucleus. Nuclear CLDN4-positive cases showed a stronger correlation with cancer progression than the negative cases. Intratumoral anaerobic bacterial DNA examination revealed nuclear CLDN4 expression in 81% of Clostridium perfringens-positive cases. Treatment of human oral squamous cell carcinoma cell lines HSC3 and HSC4 with Clostridium perfringens enterotoxin (CPE), induced CLDN4 nuclear translocation to enhance epithelial-mesenchymal transition (EMT), stemness, cell proliferation and invasive ability. In addition, CPE treatment suppressed phosphorylation of yes-associated protein-1 (YAP1) and promoted YAP1 nuclear translocation, resulting in increased expression of YAP1 target genes; cyclin D1 and connective tissue growth factor. Moreover, it was revealed that the complex of YAP1, CLDN4 and zona occludens-2 (ZO-2) was formed by CPE treatment, further suppressing YAP1 phosphorylation by LATS1 and activating it. Thus YAP activation in OSCC was regarded important in promoting malignant phenotypes. Our research suggested that the control of oral anaerobic bacteria may suppress YAP activation and in turn tumor progression.
Keywords: clostridium perfringens enterotoxin, claudin-4, YAP, Hippo signal, oral squamous cell carcinoma, Pathology
INTRODUCTION
More than 300 species of bacteria are found in the oral cavity, 10% of which are anaerobic in nature [1]. The number of bacteria in the oral cavity is determined by scraping the gingiva. Mostly the normal count of bacteria is about 1011 to 1012 cfu/ml and of these 0.1% are anaerobic [1]. Anaerobic bacteria in oral bacteria are regarded as the cause of periodontal diseases [2]. The number of anaerobes like Peptostreptococcus, Prevotella, and Fusobacterium are increased in the periodontal pocket of smokers and a strong correlation with periodontitis has been noted in such cases [3].
The anaerobic bacteria Clostridium is found in the plaque-associated bacterial flora in the oral cavity and is involved in the formation of dental caries [4]. It is also involved in the establishment of bacterial flora in the stomach and small intestine [1]. However, it is also known to damage the intestinal mucosa and cause enteritis by binding tightly to claudin (CLDN)-3 and CLDN4 and inhibiting intestinal tight junction barriers [5, 6].
According to many studies CLDN4 is overexpressed in many epithelial malignancies and it has been correlated with cancer progression [7–14]. CLDN4 expression is considered as a marker of epithelial differentiation [15–17], and its reduced expression is associated with epithelial-mesenchymal transition (EMT) [18, 19]. It forms tight junctions, maintains the cancer microenvironment, accumulates growth factors, and inhibits the penetration of anticancer drugs in the cells [12–14, 20]. In contrast, CLDN4 proteins which do not form tight junctions, act as ligand for integrin signaling and promote survival signals and stemness as found in undifferentiated gastric cancer cells [14].
Expression of CLDN4 is also affected by inflammatory cytokines. For instance, TNF-α suppresses the expression of CLDN4 in colorectal cancer, and the level of TNF-α is increased by Clostridium perfringens type A enterotoxin (CPE) [13]. In gastric cancer, the expression of CLDN4 is modulated by Helicobacter pylori [14, 21]. Thus, expression of CLDN4 is thought to be affected by the presence of bacteria within the tumor environment.
The oral microbiome population might promote carcinogenesis by increasing oxidative stress in the oral cavity of people with adverse lifestyle habits such as smoking, alcohol drinking and betel chewing [22]. But it is still unclear what role the oral bacteria play exactly in the development of oral cancer.
CLDN along with occludin are important components of tight junctions. Tight junctions activate Hippo signaling through cell adhesion and suppress proliferation of cells [23, 24]. In contrast, zonula occludens (ZO)-1 and ZO-2, which are lining proteins in tight junctions, activate yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) by inhibiting the Hippo signaling pathway [25, 26], promoting cancer cell growth and epithelial-mesenchymal transition (EMT) to enhance cancer metastasis [27–29].
In this study, we examined the role of the bacteria in the oral cavity, especially anaerobic bacteria, in the development of oral cancer through the action on CLDN protein. We showed that anaerobic bacteria impair tight junctions and promote cancer progression through YAP1 activation in oral squamous cell carcinomas (OSCCs).
RESULTS
Expression of CLDN4 in OSCCs
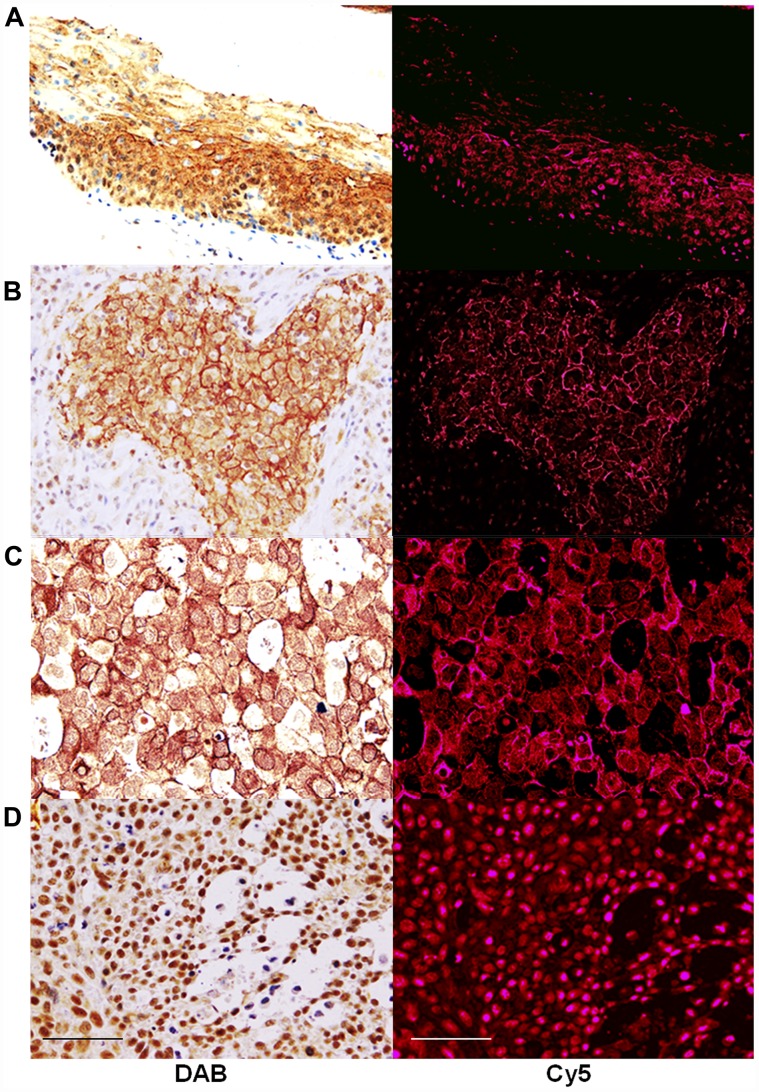
Immunostaining was performed to examine the expression of CLDN4 in the samples collected from 57 cases of OSCCs (Figure 1). In the squamous epithelium of the non-cancerous oral mucosa, the staining of the nucleus was observed in the basal region and the staining of the cell membrane was observed in the surface layer (Figure 1A). On the other hand, OSCCs showed CLDN4 immunoreactivity in the cytoplasmic membrane (Figure 1B), the cytoplasm (Figure 1C), and the nuclei (Figure 1D). In cases where clear expression was observed in the cell membrane, nuclear CLDN4 expression was not observed.
Figure 1. Expression of CLDN4 in oral squamous cell carcinomas.
Immunohistochemical evaluation was carried out to identify CLDN4 using anti-CLDN4 antibody, 4D3. CLDN4 was visualized by peroxidase-diaminobenzidine (DAB) method (left column) or Cy5-labeled secondary antibody (right column). Cy5 images were semi-quantified for evaluation of CLDN4 protein levels. (A) non-cancerous tongue epithelium (B) OSCC, G2, pT2pN0pM0, (C) OSCC, G2, pT3pN1pM0, (D) OSCC, G3, pT4PN2pM0. Bar, 100 μm.
CLDN4 protein expression was semi-quantitatively determined by fluorescence intensity (Figure 1 and Table 1). No correlation was found between the differentiation grade, pathological stage, tumor progression and CLDN4 expression. In contrast, there was a correlation between lymph node metastasis and CLDN4 expression in the nodal metastasis foci, which was higher than in the primary lesion.
Table 1. Expression of CLDN4 in oral squamous cell carcinomas.
| Tumor stages/Histological grades | n | CLDN4 intensity | P |
|---|---|---|---|
| Primary | 57 | 72 ± 10 | 0.006 |
| Lymph node | 8 | 161 ± 39 | |
| G1 | 10 | 54 ± 23 | NS |
| G2 | 32 | 84 ± 16 | |
| G3 | 15 | 60 ± 14 | |
| pStage 1–2 | 34 | 50 ± 12 | NS |
| pStage 3–4 | 23 | 89 ± 25 | |
| pT1-2 | 25 | 53 ± 12 | NS |
| pT3-4 | 32 | 87 ± 16 | |
| pN0 | 40 | 68 ± 13 | 0.0098 |
| pN1-2 | 17 | 81 ± 19 |
Tumor stages and histological grades were determined according to the guidelines of Union for International Cancer Control TNM classification system [60].
Abbreviations: NS, not significant.
Nuclear CLDN4 expression in OSCC cells
As observed in Figure 1, CLDN4 protein was frequently observed in the nuclei of OSCC cases. Cancer progression was compared between the cases in which nuclear CLDN4 was observed (including cases with positive expression in the nucleus and cytoplasm) and the cases in which CLDN4 expression was found in the cell membrane (Table 2). We observed that the progression, including tumor invasion and nodal metastasis, was more pronounced in nuclear CLDN4-positive cases than in cases where CLDN4 were observed in the cell membranes. Thus, CLDN4 expression in the nucleus was found to be associated with the progression of OSCC.
Table 2. Relationship between nuclear and membrane CLDN4 expression and clinicopathological parameters.
| Tumor stage/Histopathological grade | n | CLDN4 expression | P | |
|---|---|---|---|---|
| Nuclear | Membrane | |||
| n | 22 | 35 | ||
| G1 | 6 | 6 | 4 | NS |
| G2 | 9 | 9 | 23 | |
| G3 | 7 | 7 | 8 | |
| pStage 1–2 | 14 | 7 | 22 | 0.0307 |
| pStage 3–4 | 8 | 15 | 13 | |
| pT1-2 | 19 | 3 | 21 | 0.0008 |
| pT3-4 | 3 | 19 | 14 | |
| pN0 | 13 | 9 | 25 | 0.029 |
| pN1-2 | 9 | 13 | 10 | |
Tumor stages and histological grades were determined according to the guidelines of Union for International Cancer Control TNM classification system [60].
Abbreviations: NS, not significant.
Production of nuclear CLDN4 by CPE
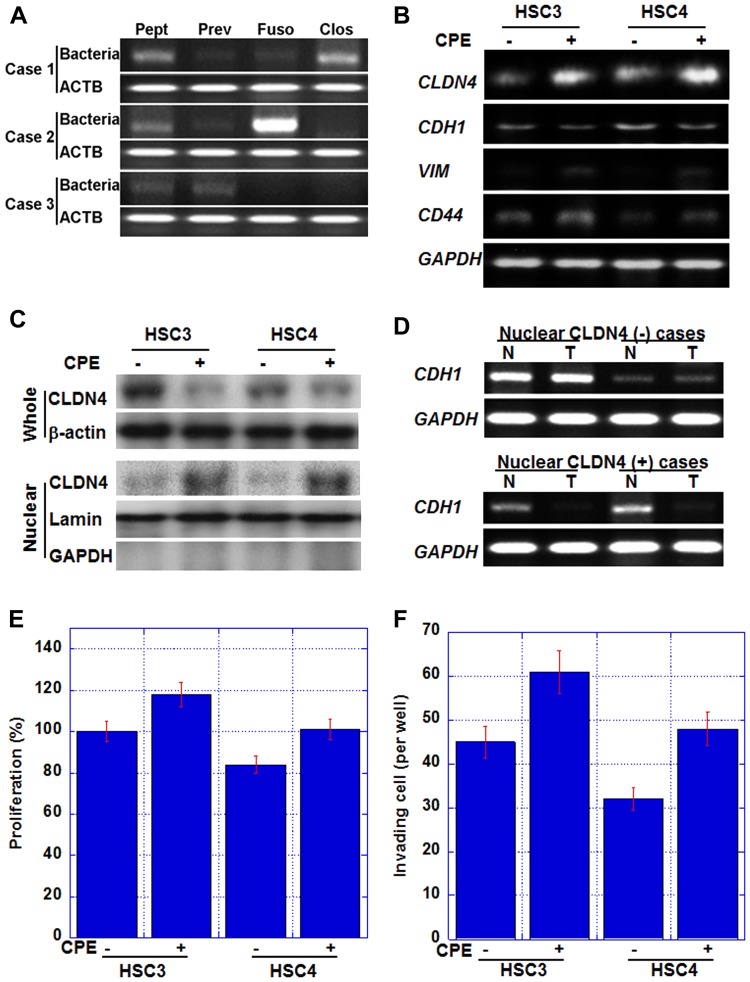
In nuclear CLDN4-positive cases, CLDN4 was not observed in the cell membrane (Figure 1), suggesting the possibility of some damage to the tight junction CLDN4. Since Clostridium perfringens enterotoxin (CPE) is a well-known factor that impairs CLDN4 [38], bacterial genome-specific sequences were amplified by PCR and examined for the presence of anaerobic bacteria in tumor tissues (Figure 2A). The presence of Peptostreptococcus, Prevotella, Fusobacterium, and Clostridium was examined in the tumor cells, and these anaerobic bacteria were found to thrive in the tumor cells in varying numbers. We examined the association of nuclear CLDN4 with Clostridium-negative and Clostridium-positive tumors. In 22 Clostridium-positive tumors, 18 (82%) showed nuclear CLDN4, whereas Clostridium-negative tumors showed no nuclear CLDN4 (P < 0.0001).
Figure 2. Effect of Clostridium perfringens enterotoxin (CPE) in human OSCC cell lines.
(A) Intratumoral bacterial DNAs were detected by PCR. Peptostreptcoccus (Pept); Prevotella (Prev); Fusobacterium (Fuso); C. perfringens (Clos); ACTB, β-actin. (B) Gene expression in HSC3 and HSC4 human OSCC cells treated with CPE (10 μg/ml). CDH1, E-cadherin; VIM, vimentin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. GAPDH, loading control. (C) Nuclear translocation of CLDN4 in CPE-treated OSCC cells. Whole, whole cell lysate; Nuclear, nuclear fraction. β-actin, loading control for whole cell lysate; lamin, loading control for nuclear protein; GAPDH, a marker for cytoplasmic protein. (D) CDH1 expression in nuclear CLDN4-positive and negative OSCCs. T, tumor tissue; N, non-neoplastic epithelium; CDH1, E-cadherin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. GAPDH, loading control. (E, F) Effect of CPE on cell proliferation (E) and in vitro invasion (F). Bar, S. D. from 3 independent trials.
Next, we examined whether CPE was involved in the production of nuclear CLDN4 using human OSCC cells (Figure 2B and 2C). Our analysis revealed that CLDN4 mRNA levels and protein levels were increased in the nuclear fraction by CPE treatment in both the cell lines (Figure 2C); however, CLDN4 levels in whole cell lysate were decreased. These findings suggested that nuclear CLDN4 was produced by CPE.
The effects of nuclear CLDN4 on cancer cells by CPE treatment were also examined (Figure 2B). CPE treatment decreased the expression of E-cadherin in both the cell lines, while the mRNA expression of vimentin and CD44 increased suggesting EMT. We also examined E-cadherin mRNA expression in 2 nuclear CLDN4-positive and 2 CLDN4-negative OSCCs in Figure 2D. Nuclear CLDN4-positive tumors showed decreased E-cadherin mRNA expression in comparison with the non-neoplastic epithelium, whereas Nuclear CLDN4-negative tumors showed retained E-cadherin expression. Moreover, cell proliferation and invasive ability were enhanced by CPE (Figure 2E and 2F).
Inhibition of Hippo suppression system by CPE
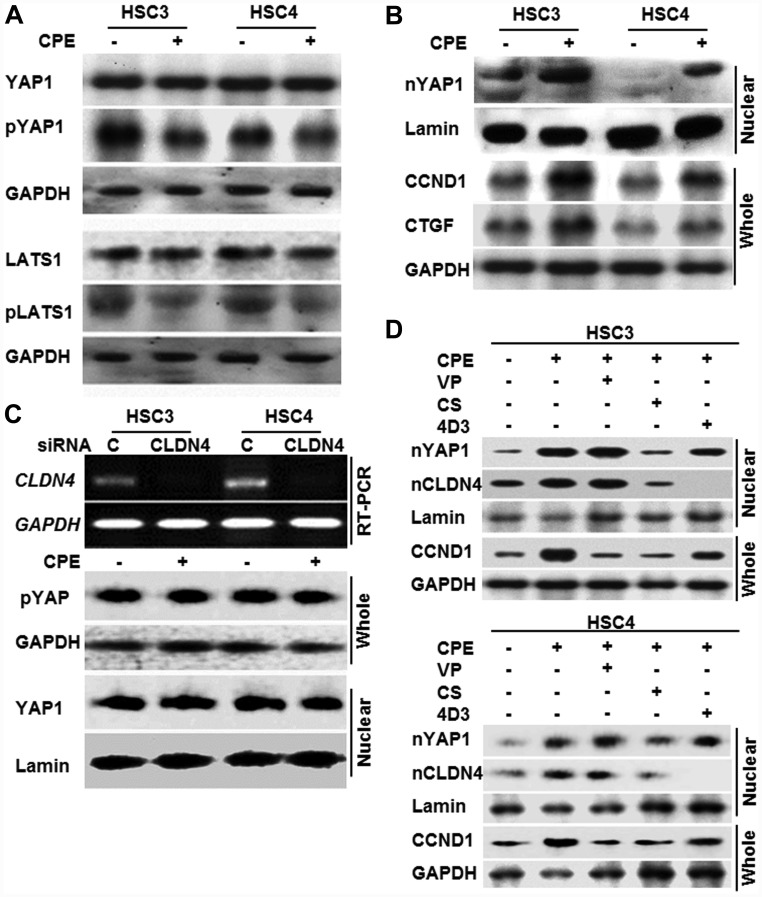
In contrary of CLDN4 mRNA, CPE decreased the expression of CLDN4 protein and E-cadherin mRNA, suggesting decrease of cell adhesion. Since Hippo signaling pathway is activated by cell adhesion and suppresses YAP, the effect of CPE on the Hippo-YAP system was examined (Figure 3).
Figure 3. Effect of CPE on YAP1 activation in OSCC cells.
(A) Phosphorylation of YAP1 and LATS1 in whole cell lysates of CPE (10 μg/ml)-treated OSCC cells. (B) Effect of CPE on expression of nuclear YAP1 (nYAP) and cyclin D1 (CCND1) and connecting tissue growth factor (CTGF). (C) Effect of CLDN4 knockdown on YAP1 activation. pYAP, phosphorylated YAP1. (D) Effect of YAP1 inhibitors and anti-CLDN4 antibody (4D3). VP, verteporfin; CS, cytostatin. Whole, whole cell lysate; Nuclear, nuclear fraction; lamin, loading control for nuclear protein; GAPDH, loading control for whole cell lysate.
In both HSC3 and HSC4 cell lines, the levels of phosphorylated YAP1 and phosphorylated LATS were reduced by CPE treatment (Figure 3A). Along with this the nuclear YAP1 increased, in turn inducing its target genes, cyclin D1 (CCND1) and connective tissue growth factor (CTGF) (Figure 3B). In contrast, when CLDN4 was knocked down, its mRNA almost disappeared, and CPE-induced alterations in phosphorylated YAP1 and nuclear YAP1 were also abrogated (Figure 3C).
Next, the effect of the YAP inhibitors and anti-CLDN4 antibody (4D3) on the activation of YAP1 by CPE was examined (Figure 3D). CPE increased the nuclear YAP1 and CLDN4 and induced the YAP1 target gene CCND1. When treated with verteporfin, which inhibits YAP1-TEAD interaction [39], nuclear translocation of YAP1 and CLDN4 was maintained; however, CCND1 expression was suppressed. In contrast, treatment with cytostatin, a dephosphorylation inhibitor of phosphorylated YAP1 [40], suppressed both nuclear translocation of YAP1 and CLDN4 and decreased CCND1 expression. In contrast, when treated with the 4D3 antibody, CLDN4 nuclear translocation disappeared, and YAP1 nuclear translocation and CCND1 expression decreased.
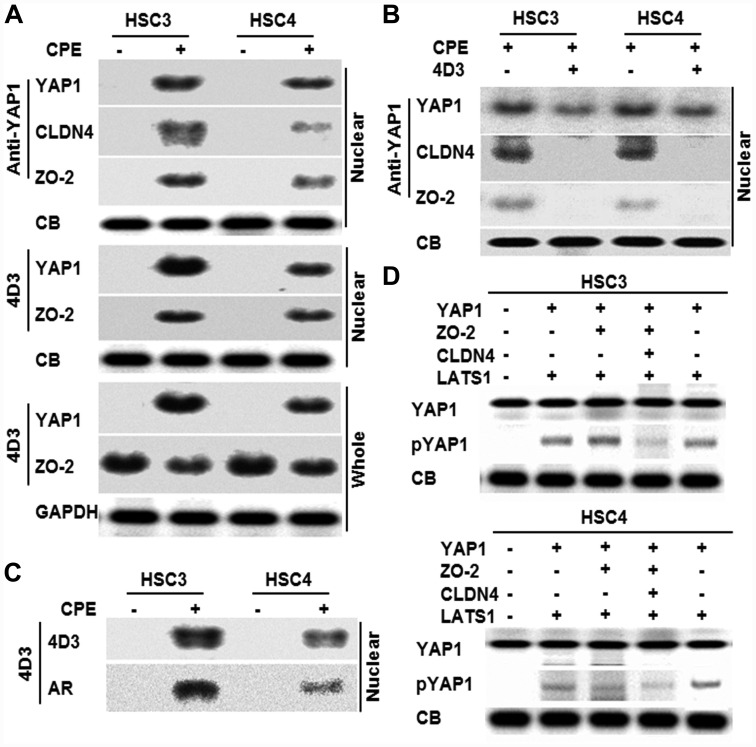
Interaction between YAP1 and CLDN4
Since interaction between YAP1 and CLDN4 was suggested above, immunoprecipitation was performed for these proteins and ZO-2, which had been reported to interact with YAP1 [25] (Figure 4A). When immunoprecipitation was performed with anti-YAP1 antibody in the nuclear fraction to detect YAP1, CLDN4, and ZO-2, their levels were found to be increased following CPE treatment. When the same nuclear fraction was immunoprecipitated with 4D3 antibody to detect YAP1 and ZO-2, the protein levels were found to be increased following CPE treatment. Furthermore, when the whole cell lysates were immunoprecipitated with 4D3 antibody to detect YAP1 and ZO-2, ZO-2 was also detected in CPE-untreated and CPE-treated cells; however, YAP1 was detected in only CPE treated cells. When nuclear fractions of cells treated with 4D3 antibody were precipitated with anti-YAP1 antibody to detect YAP1, CLDN4, and ZO-2 in CPE-treated OSCC cells, CLDN4 and ZO-2 were not detected. YAP1 was reduced by 4D3 treatment (Figure 4B). In order to examine whether CLDN4 on the cell membrane surface translocate to the nuclei, 125I was added to the culture medium simultaneously with the CPE treatment to label the protein on the cell membrane surface (Figure 4C). When the nuclear fraction was precipitated with the 4D3 antibody, an autoradiographic signal was detected in consistent with the signal detected with the 4D3 antibody. In order to examine the role of CLDN4 on YAP1 activation, in vitro phosphorylation of YAP1 was examined with or without CLDN4 (Figure 4D). YAP1 was phosphorylated by LATS1 with or without ZO-2, whereas mixture of YAP1, ZO-2 and CLDN4 showed reduction of YAP1 phosphorylation. In contrast, mixture of YAP1 and CLDN4 showed no reduction of YAP1 phosphorylation.
Figure 4. Effect of CPE on interaction of CLDN4 and YAP1 in OSCC cells.
(A) Effect of CPE (10 μg/ml) on protein interaction among CLDN4, YAP1 and ZO-2 was examined by immunoprecipitation. (B) Nuclear translocation of CLDN4, YAP1 and ZO-2 in CPE-treated OSCC cells with or without 4D3 treatment. (C) CLDN4 nuclear translocation in the 131I-surface labeled OSCC cells. Nuclear fraction was immunoprecipitated with 4D3 for detection with 4D3 and autoradiography (AR). (D) In vitro phosphorylation assay using recombinant human YAP1, ZO-2, CLDN4 and LATS1. Reaction solution was subjected to immunoblotting. Whole, whole cell lysate; Nuclear, nuclear fraction; CB, Coomassie blue; lamin, loading control for nuclear protein; GAPDH, loading control for whole cell lysate.
DISCUSSION
In this study, OSCCs showing nuclear CLDN4 expression were found more strongly correlated with cancer progression than those showing cell membrane CLDN4 expression. Previous studies have shown that CLDN4 forms tight junctions that preserve the cancer microenvironment to accumulate growth factors and maintain extracellular pH in an acidic range [12–14, 20]. Tight junctions also inhibit the penetration of anticancer drugs into this microenvironment and develop resistance against anticancer drugs [12–14, 20]. Among the 57 cases of oral cancer examined in this study, intraoral CLDN4 was found in 22 (39%) of them. It was also observed that expression of cell membrane CLDN4 was never detected in nuclear CLDN4-positive cases. We assumed that this might be because cytoplasmic membrane CLDN4 damage was associated with CLDN4 nuclear translocation.
CPE is known as a factor that impedes tight junctions. Clostridia that produce CPE are part of the intestinal microflora and their numbers increase with age. However, Clostridium has also been reported to be associated with dental caries in the oral cavity [4]. Likewise, the presence of anaerobic bacteria such as Peptostreptococcus, Prevotella, and Fusobacterium in oral flora exacerbate periodontal disease [2, 3, 41].
Since the detection of Clostridium by culture methods is insufficient [42], we employed PCR-amplification of bacterial DNA to determine the frequency of Clostridium in oral flora. In general, Clostridium spp. are detected in the bronchial lavage of pneumonia patients by 16S ribosomal DNA-based bacterial flora analysis [43–46]. From these literature [43–46], we decided to detect four types of bacteria including Clostridium as anaerobic bacteria that are commonly found in the oral cavity using a similar approach. In the present study, tumor tissue was examined because the patient’s saliva could not be available. In the future, it is desired to clarify the relationship between bacterial flora and tumor progression through studies using saliva, which is easy to obtain without any pain for patients.
CLDN4 is bound to ZO-1 and ZO-2 in tight junctions [47]. Since CPE implicates homotypic binding of CLDN4 [38], it is thought to internalize in the cytoplasm through binding to ZO-1 or ZO-2. In addition, ZO-2 has binding properties with YAP1 through PDZ domain, and promotes nuclear translocation of YAP1. Our results suggested that the CLDN4-ZO-2 complex, which was internalized in the cytoplasm by CPE, further promoted YAP1 nuclear translocation by binding to YAP1. It was shown that YAP1 is less susceptible to phosphorylation by LATS1 when present in YAP1-ZO-2-CLDN4 or YAP1-ZO-2 complex. It is possible that the addition of CLDN4 to the YAP1-ZO-2 complex masks the phosphorylation site of YAP1.
Our data showed that YAP1-ZO-2-CLDN4 complex might activate YAP1. Tight junction-Hippo system contains molecular redundancy; CLDN3 instead of CLDN4, TAZ instead of YAP1, and ZO-1 instead of ZO-2 might form a complex protein reacting to CPE. For this reason, TAZ, ZO-1, and CLDN3 were examined in the nuclear fraction of OSCC cells treated with CPE as in Figure 3A, but were not detected (Supplementary Figure 1A). In addition, TAZ, ZO-1 and CLDN3 were examined by immunoprecipitation with the same anti-YAP1 antibody as in Figure 4A, but were not detected (Supplementary Figure 1B). In addition, TAZ was not detected in the precipitate from 4D3 antibody (Supplementary Figure 1B). These results suggested that a complex of YAP1-ZO-2-CLDN4 was formed preferentially for CPE. This specificity needs further study.
It has been reported that while the combination of YAP1-ZO-2 promotes cell death [46], the combination of YAP2-ZO-2 enhances tumor promoting activity [47] In our immunoprecipitation study, contrary to previous findings, ZO-2 and YAP1 formed a tumor-promoting complex and no ZO-2-YAP2 complex was observed (DNS). Regarding this difference, our results suggested that the formation of a complex of YAP1-ZO-2 and CLDN4 had some pro-tumoral effect. Tumor-promoting role of YAP is attributed to the induction of tumor-promoting gene expression via TEADS [40]. According to our findings, the expression of cyclin D1 and CTGF has a definite association with the formation of YAP1-ZO-2-CLDN4 complex.
Our results showed that Cl. perfringens might be a potent accelerator of malignant phenotype in OSCC. In a number of previous studies, Clostridium sp. have shown a strong relationship with cancer. For instance, Cl. septicum is known to be associated with colorectal cancer. It functions as an immunosuppressant and is frequently detected in the blood of colorectal cancer patients [48, 49]. The association between Clostridium in the blood and Cl. perfringens, and colorectal cancer risk has been reported as P = 0.13 and P = 0.17, respectively [48]. Inhibition of Cl. leptum subgroup by antibiotics has been reported to inhibit tumorigenesis in azoxymethane-dextran sulfate colon carcinogenesis model through reduction of aberrant DNA methylation [50]. In addition, colon carcinogenesis caused by a high-fat diet is associated with an increase in the intestinal bacterium Clostridium subcluster XIVa [51]. Another anaerobic bacteria, Bacteroides fragilis, promotes colon carcinogenesis by its enterotoxin through activation of Wnt signal by β-catenin nuclear translocation with E-cadherin cleavage and secretion of inflammatory cytokines [52]. Thus, anaerobic bacteria have shown a good association with colorectal cancer. However, there is no report of an association between anaerobic bacteria and cancer in tissues other than the colorectum. However, in our study, the activation of YAP1 by CPE promoted cell proliferation, invasion ability, stemness, EMT, and consequent cancer malignancy. Further it is needed to examine the association of Cl. perfringens with oral carcinogemesis.
CPE is regarded as a therapeutic tool for epithelial malignant tumors by utilizing the damage of CLDN4 and CLDN3 [11, 53, 54]. The 50%-inhibitory concentration of CPE for colon cancer cell lines was between 50-100 ng/ml [53]. The concentration of CPE used in this study was 10 ng/ml and no cytotoxicity was observed (DNS) at this concentration. Our data suggested that such low concentration of CPE was not cytotoxic to cancer cells and instead promoted malignant phenotypes.
YAP has been reported to be involved in the development and progression of lung cancer, liver cancer, and various other cancers [55–58]. However, recently, its involvement in squamous cell carcinoma and head and neck cancer has attracted attention [55]. Some studies have also shown an association between YAP and radiation resistance and poor prognosis [29, 59]. In head and neck cancer, YAP correlated with poor prognosis and resistance to treatment, suggesting a potential molecular target [29]. Our study inferred that Clostridium induced YAP1 activation, which in turn suggested that the suppression of Clostridium by antibiotics and/or oral hygiene might contribute to the suppression of carcinogenesis and cancer progression. Extensive clinical studies are required to prove this hypothesis in future.
MATERIALS AND METHODS
Surgical specimens
We reviewed the pathological diagnosis and clinical data of 57 patients diagnosed with OSCCs from 2004 to 2016, in the Department of Molecular Pathology, Nara Medical University, Japan. As written informed consents were not obtained from the patients, any identifying information was removed from the samples prior to analysis, in order to ensure strict privacy protection (unlinkable anonymization). All procedures were performed in accordance with the Ethical Guidelines for Human Genome/Gene Research issued by the Japanese Government and were approved by the Ethics Committee of Nara Medical University (approval number 937).
Human OSCC cell lines
HSC-3 and HSC-4 human OSCC cell lines were purchased from Dainihon Pharmaceutical Co. (Tokyo, Japan). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37° C in 5% CO2. Cell growth was assessed using tetrazolium (MTT) dye assay, as previously described [30].
Antibody and reagents
The anti-human CLDN4 extracellular domain antibody, 4D3, was developed by immunizing rats with a plasmid vector encoding human CLDN4 [12]. CPE was purchased from Sigma, USA (Sigma, St. Louis, MO, USA).
Immunohistochemistry
Consecutive sections of 4 μm of OSCC were immunohistochemically stained using 0.2 μg/ml of 4D3 antibody by a previously described immunoperoxidase technique [31]. Secondary antibodies (peroxidase-conjugated and cy5-conjugated anti-mouse IgG rabbit antibodies, Medical and Biological Laboratories, Nagoya, Japan) were used at a concentration of 0.2 μg/ml. Tissue sections were color-developed with diamine benzidine hydrochloride (DAKO, Glastrup, Denmark) and counterstained with Meyer’s hematoxylin (Sigma, USA). Primary antibody was also detected by fluorescence microscopy (All-in-one fluorescence microscopy, BZ-X800, Keyence Japan, Osaka, Japan). Fluorescence intensity was measured according to the manufacturer’s instructions. The fluorescence intensity in the non-cancerous squamous cell epithelium was standardized and set to 100. For a negative control, non-immunized rat IgG (Santa-Cruz Biotechnology, Santa-Cruz, CA, USA) was used as the primary antibody.
Immunoblot analysis
Whole-cell lysates of OSCC cells were prepared as previously described elsewhere [32]. Lysates (20 μg) were subjected to immunoblot analysis using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, 12.5%), followed by electrotransfer onto nitrocellulose filters. The filters were incubated with primary antibodies, followed by peroxidase-conjugated IgG antibodies (Medical and Biological Laboratories). Anti-tubulin antibody was used to assess the levels of protein loaded per lane (Oncogene Research Products, Cambridge, MA, USA). The immune complex was visualized using an Enhanced Chemiluminescence Western-blot detection system (Amersham, Aylesbury, UK). Antibodies for CLDN4 (4D3), YAP1, phosphorylated YAP1 (pS127), zona occludens-2 (ZO-2), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), cyclin D1 (CCND1), connective tissue growth factor (CTGF) (Abcam, Cambridge, UK), large tumor suppressor kinase 1 (LATS1), phosphorylated LATS (pThr1079) (Cell Signaling Technology, Beverly, MA, USA), β-actin, lamin (Zymed Laboratories Inc., South San Francisco, CA, USA), (Proteintech Group Inc., Rosemont, IL, USA) were used as primary antibodies.
Bacterial DNA amplification
Bacterial DNA was extracted from OSCC specimen (10 thin-sliced paraffin-embedded tumor specimen, depraffinized, and hydrated) using the QIAamp DNA mini kit (Qiagen, GmbH, Hilden, Germany) according to the instructions of the manufacturer. The extracted DNA samples were stored at −20° C. PCR was carried out for 35 cycles and each cycle consisted of the following steps; denaturation (94° C for 5 min), annealing (50° C for 1 min) and primer extension (72° C for 1.5 min). Amplified PCR products were analyzed by 1.5% agar gel electrophoresis in Tris–Borate-EDTA buffer. The gel was stained with 0.5 μg/ml ethidium bromide. The primer sets used for the amplification of bacterial DNA for the following bacteria were as follows; Peptostreptococcus productus, forward, 5′-AAC TCC GGT GGT ATC AGA TG-3′ and reverse, 5′-GGG GCT TCT GAG TCA GGT A-3′ [33]; Fusobacterium prausnitzii, forward, 5′-AGA TGG CCT CGC GTC CGA-3′ and reverse, 5′-CCG AAG ACC TTC TTC CTC C-3′ [33]; Prevotella nigrescens, forward, 5′-GTG TTT CAT TGA CGG CAT CCG ATA TGA AAC-3′ and reverse, 5′-CCA CGT CTC TGT GGG CTG CGA-3′ [34]; Clostridium perfringens (CPE gene), forward, 5′-TCC CCT TTC TAG ATA ACG ATT AAC AC-3′ and reverse, 5′-GTT AGC ATG CTG TTT TCT AAG TTA AAA CC-3′ [35]. Primers were synthesized by Sigma Genosys (Ishikari, Japan).
Reverse transcription-polymerase chain reaction (RT-PCR)
To assess human CLDN4 mRNA expression, RT-PCR was performed with 0.5 μg total RNA extracted from HSC3 and HSC4 cells using an RNeasy kit (Qiagen, Germantown, MD, USA). The primer sets used were as follows; CLDN4, forward, 5′-CTC CAT GGG GCT ACA GGT AA-3′ and reverse, 5′-AGC AGC GAG TCG TAC ACC TT-3′ (NCBI reference sequence: NM_001305.4); CDH1, forward, 5′-TGC CCA GAA AAT GAA AAA GG-3′ and reverse, 5′-GTG TAT GTG GCA ATG CGT TC-3′ (Z13009.1); vimentin (VIM), forward, 5′-GAG AAC TTT GCC GTT GAA GC-3′ and reverse, 5′-TCC AGC AGC TTC CTG TAG GT-3′ (NM_003380.3); CD44, forward, 5′-CAT TCA AAT CCG GAA GTG CT-3′ and reverse, 5′-GTT GCC AAA CCA CTG TTC CT-3′ (FJ216964.1). Primers were synthesized by Sigma Genosys. The GAPDH mRNA was also amplified for use as an internal control (GenBank Accession No. NM_001101).
Protein extraction
For preparing whole cell lysate, HSC3 and HSC4 cells were washed twice with cold PBS, harvested and lysed with 0.1% SDS-added RIPA-buffer (Thermo Fisher Scientific, Tokyo, Japan) [32]. Cell fractions were extracted by using a Cell Fractionation Kit (Abcam, Cambridge, MA, USA), according to the manufacturer’s instructions [36]. Protein assay was performed using a Protein Assay Rapid Kit (Wako Pure Chemical Corporation, Osaka, Japan).
Immunoprecipitation
Immunoprecipitation was performed according to the method described previously [37]. Briefly, whole cell lysates were pre-cleaned in lysis buffer with protein A/G agarose (Santa Cruz) for 1 h at 4° C and subsequently centrifuged. The supernatants were incubated with antibodies against YAP1 (Abcam) or CLDN4 (4D3) or ZO-2 and protein A/G agarose for 3 h at 4° C. Precipitates were collected via centrifugation, washed five times with lysis buffer, solubilized with sample buffer (Sigma, 40 μg), and subjected to an immunoblot analysis with antibodies against CLDN4 (4D3), YAP1 (Abcam) or ZO-2 (Abcam).
Cell surface labeling
Cell surface proteins were iodized with Na131I (Amersham, Aylesbury, UK) with iodination reagent (Pierce, Rockford, IL, USA) in HSC3 and HSC4 cells, which were added into the culture media and incubated with cells for 1 h. Then cells were washed with cold PBS for 3 times. Cells were subjected to protein extraction mentioned above.
In vitro phosphorylation assay of recombinant proteins
Recombinant human YAP1 protein (Abcam, 1 μg) was mixed with recombinant human ZO-2 protein (Abcam, 1 μg) and/or recombinant human CLDN4 protein (Abcam, 1 μg) in a kinase buffer (20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 5 mM MnCl2, 1×phosphatase inhibitor, 1×protease inhibitor mixture) and incubated at 30° C for 30 min. Then recombinant human LATS1 protein (Abcam, 1 μg) and 20 μM ATP were added into the above given recombinant protein mixture and incubated at 30° C for 30 min. The reaction was stopped by adding 7 μl of 5× SDS sample dye, boiled at 100° C for 5 min, and subjected to SDS-PAGE.
Statistical analysis
Statistical significance was calculated using chi-square, Fisher’s square test, and Kruskal-Wallis test with InStat software (GraphPad, Los Angeles, CA, USA). Statistical significance was defined as a two-sided p-value of < 0.05.
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS AND FUNDING
This work was supported by MEXT KAKENHI Grant Numbers 16H05164 (HK), 17K19923 (HK), 18K09539 (KY) and 19K19332 (CN), Natural Science Foundation of Jiangsu Education Department Project (17KJB320010)(YL), and National Natural Science Foundation of China (81702723)(YL). The authors thank Ms. Tomomi Masutani for expert assistance with the preparation of this manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
REFERENCES
- 1. Evaldson G, Heimdahl A, Kager L, Nord CE. The normal human anaerobic microflora. Scand J Infect Dis Suppl. 1982; 35:9–15. [PubMed] [Google Scholar]
- 2. Stefanopoulos PK, Kolokotronis AE. The clinical significance of anaerobic bacteria in acute orofacial odontogenic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004; 98:398–408. 10.1016/j.tripleo.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 3. van Winkelhoff AJ, Bosch-Tijhof CJ, Winkel EG, van der Reijden WA. Smoking affects the subgingival microflora in periodontitis. J Periodontol. 2001; 72:666–671. 10.1902/jop.2001.72.5.666. [DOI] [PubMed] [Google Scholar]
- 4. Li Y, Zou CG, Fu Y, Li Y, Zhou Q, Liu B, Zhang Z, Liu J. Oral microbial community typing of caries and pigment in primary dentition. BMC Genomics. 2016; 17:558. 10.1186/s12864-016-2891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eichner M, Augustin C, Fromm A, Piontek A, Walther W, Bucker R, Fromm M, Krause G, Schulzke JD, Gunzel D, Piontek J. In colon epithelia, clostridium perfringens enterotoxin causes focal leaks by targeting claudins which are apically accessible due to tight junction derangement. J Infect Dis. 2017; 217:147–157. 10.1093/infdis/jix485. [DOI] [PubMed] [Google Scholar]
- 6. Shinoda T, Shinya N, Ito K, Ohsawa N, Terada T, Hirata K, Kawano Y, Yamamoto M, Kimura-Someya T, Yokoyama S, Shirouzu M. Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin. Sci Rep. 2016; 6:33632. 10.1038/srep33632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000; 60:6281–6287. [PubMed] [Google Scholar]
- 8. Rangel LB, Agarwal R, D’Souza T, Pizer ES, Alò PL, Lancaster WD, Gregoire L, Schwartz DR, Cho KR, Morin PJ. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003; 9:2567–2575. [PubMed] [Google Scholar]
- 9. Michl P, Buchholz M, Rolke M, Kunsch S, Lohr M, McClane B, Tsukita S, Leder G, Adler G, Gress TM. Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology. 2001; 121:678–684. 10.1053/gast.2001.27124. [DOI] [PubMed] [Google Scholar]
- 10. Sato N, Fukushima N, Maitra A, Iacobuzio-Donahue CA, van Heek NT, Cameron JL, Yeo CJ, Hruban RH, Goggins M. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am J Pathol. 2004; 164:903–914. 10.1016/S0002-9440(10)63178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kojima T, Kyuno D, Sawada N. Targeting claudin-4 in human pancreatic cancer. Expert Opin Ther Targets. 2012; 16:881–887. 10.1517/14728222.2012.708340. [DOI] [PubMed] [Google Scholar]
- 12. Kuwada M, Chihara Y, Luo Y, Li X, Nishiguchi Y, Fujiwara R, Sasaki T, Fujii K, Ohmori H, Fujimoto K, Kondoh M, Kuniyasu H. Pro-chemotherapeutic effects of antibody against extracellular domain of claudin-4 in bladder cancer. Cancer Lett. 2015; 369:212–221. 10.1016/j.canlet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 13. Fujiwara-Tani R, Sasaki T, Luo Y, Goto K, Kawahara I, Nishiguchi Y, Kishi S, Mori S, Ohmori H, Kondoh M, Kuniyasu H. Anti-claudin-4 extracellular domain antibody enhances the antitumoral effects of chemotherapeutic and antibody drugs in colorectal cancer. Oncotarget. 2018; 9:37367–37378. 10.18632/oncotarget.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nishiguchi Y, Fujiwara-Tani R, Sasaki T, Luo Y, Ohmori H, Kishi S, Mori S, Goto K, Yasui W, Sho M, Kuniyasu H. Targeting claudin-4 enhances CDDP-chemosensitivity in gastric cancer. Oncotarget. 2019; 10:2189–2202. 10.18632/oncotarget.26758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001; 2:285–293. 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 16. Turksen K, Troy TC. Junctions gone bad: claudins and loss of the barrier in cancer. Biochim Biophys Acta. 2011; 1816:73–79. 10.1016/j.bbcan.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 17. Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006; 6:186. 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Z, Zhang X, Gang H, Li X, Li Z, Wang T, Han J, Luo T, Wen F, Wu X. Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem Biophys Res Commun. 2007; 358:925–930. 10.1016/j.bbrc.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 19. Lin X, Shang X, Manorek G, Howell SB. Regulation of the epithelial-mesenchymal transition by claudin-3 and claudin-4. PLoS One. 2013; 8:e67496. 10.1371/journal.pone.0067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sasaki T, Fujiwara-Tani R, Kishi S, Mori S, Luo Y, Ohmori H, Goto K, Nishiguchi Y, Mori T, Sho M, Kondo M, Kuniyasu H. Targeting claudin-4 enhances chemosensitivity of pancreatic ductal carcinomas. Cancer Med. 2019; 8:6700–6708. 10.1002/cam4.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chavarría-Velázquez CO, Torres-Martínez AC, Montaño LF, Rendón-Huerta EP. TLR2 activation induced by H. pylori LPS promotes the differential expression of claudin-4, -6, -7 and -9 via either STAT3 and ERK1/2 in AGS cells. Immunobiology. 2018; 223:38–48. 10.1016/j.imbio.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 22. Islam S, Muthumala M, Matsuoka H, Uehara O, Kuramitsu Y, Chiba I, Abiko Y. How each component of betel quid is involved in oral carcinogenesis: mutual interactions and synergistic effects with other carcinogens-a review article. Curr Oncol Rep. 2019; 21:53. 10.1007/s11912-019-0800-8. [DOI] [PubMed] [Google Scholar]
- 23. Karaman R, Halder G. Cell junctions in hippo signaling. Cold Spring Harb Perspect Biol. 2018; 10:a028753. 10.1101/cshperspect.a028753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gumbiner BM, Kim NG. The Hippo-YAP signaling pathway and contact inhibition of growth. J Cell Sci. 2014; 127:709–717. 10.1242/jcs.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. González-Mariscal L, Gallego-Gutiérrez H, González-González L, Hernández-Guzmán C. ZO-2 is a master regulator of gene expression, cell proliferation, cytoarchitecture, and cell size. Int J Mol Sci. 2019; 20:4128. 10.3390/ijms20174128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haymond A, Dey D, Carter R, Dailing A, Nara V, Nara P, Venkatayogi S, Paige M, Liotta L, Luchini A. Protein painting, an optimized MS-based technique, reveals functionally relevant interfaces of the PD-1/PD-L1 complex and the YAP2/ZO-1 complex. J Biol Chem. 2019; 294:11180–11198. 10.1074/jbc.RA118.007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janse van Rensburg HJ, Yang X. The roles of the Hippo pathway in cancer metastasis. Cell Signal. 2016; 28:1761–1772. 10.1016/j.cellsig.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 28. Park JH, Shin JE, Park HW. The role of hippo pathway in cancer stem cell biology. Mol Cells. 2018; 41:83–92. 10.14348/molcells.2018.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Segrelles C, Paramio JM, Lorz C. The transcriptional co-activator YAP: A new player in head and neck cancer. Oral Oncol. 2018; 86:25–32. 10.1016/j.oraloncology.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 30. Kuniyasu H, Yano S, Sasaki T, Sasahira T, Sone S, Ohmori H. Colon cancer cell-derived high mobility group 1/amphoterin induces growth inhibition and apoptosis in macrophages. Am J Pathol. 2005; 166:751–760. 10.1016/S0002-9440(10)62296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuniyasu H, Yasui W, Shinohara H, Yano S, Ellis LM, Wilson MR, Bucana CD, Rikita T, Tahara E, Fidler IJ. Induction of angiogenesis by hyperplastic colonic mucosa adjacent to colon cancer. Am J Pathol. 2000; 157:1523–1535. 10.1016/S0002-9440(10)64790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuniyasu H, Oue N, Wakikawa A, Shigeishi H, Matsutani N, Kuraoka K, Ito R, Yokozaki H, Yasui W. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J Pathol. 2002; 196:163–170. 10.1002/path.1031. [DOI] [PubMed] [Google Scholar]
- 33. Wang RF, Cao WW, Cerniglia CE. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996; 62:1242–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okamoto M, Maeda N, Kondo K, Leung KP. Hemolytic and hemagglutinating activities of Prevotella intermedia and Prevotella nigrescens. FEMS Microbiol Lett. 1999; 178:299–304. 10.1111/j.1574-6968.1999.tb08691.x. [DOI] [PubMed] [Google Scholar]
- 35. Kim S, Labbe RG, Ryu S. Inhibitory effects of collagen on the PCR for detection of Clostridium perfringens. Appl Environ Microbiol. 2000; 66:1213–1215. 10.1128/AEM.66.3.1213-1215.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matsushima-Otsuka S, Fujiwara-Tani R, Sasaki T, Ohmori H, Nakashima C, Kishi S, Nishiguchi Y, Fujii K, Luo Y, Kuniyasu H. Significance of intranuclear angiotensin-II type 2 receptor in oral squamous cell carcinoma. Oncotarget. 2018; 9:36561–36574. 10.18632/oncotarget.26337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuniyasu H, Yasui W, Pettaway CA, Yano S, Oue N, Tahara E, Fidler IJ. Interferon-alpha prevents selection of doxorubicin-resistant undifferentiated-androgen-insensitive metastatic human prostate cancer cells. Prostate. 2001; 49:19–29. 10.1002/pros.1114. [DOI] [PubMed] [Google Scholar]
- 38. Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J Cell Biol. 1999; 147:195–204. 10.1083/jcb.147.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu C, Cheng X, Chen J, Wang Y, Wu X, Tian R, Liu B, Ding X, Sun Q, Gong W. Suppression of YAP/TAZ-Notch1-NICD axis by bromodomain and extraterminal protein inhibition impairs liver regeneration. Theranostics. 2019; 9:3840–3852. 10.7150/thno.33370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ni L, Luo X. Structural and biochemical analyses of the core components of the hippo pathway. Methods Mol Biol. 2019; 1893:239–256. 10.1007/978-1-4939-8910-2_18. [DOI] [PubMed] [Google Scholar]
- 41. Sutter VL. Anaerobes as normal oral flora. Rev Infect Dis. 1984; 6:S62–S66. 10.1093/clinids/6.Supplement_1.S62. [DOI] [PubMed] [Google Scholar]
- 42. MacDougall LK, Broukhanski G, Simor A, Johnstone J, Mubareka S, McGeer A, Daneman N, Garber G, Brown KA. Comparison of qPCR versus culture for the detection and quantification of Clostridium difficile environmental contamination. PLoS One. 2018; 13:e0201569. 10.1371/journal.pone.0201569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kawanami T, Fukuda K, Yatera K, Kido M, Mukae H, Taniguchi H. A higher significance of anaerobes: the clone library analysis of bacterial pleurisy. Chest. 2011; 139:600–608. 10.1378/chest.10-0460. [DOI] [PubMed] [Google Scholar]
- 44. Yamasaki K, Kawanami T, Yatera K, Fukuda K, Noguchi S, Nagata S, Nishida C, Kido T, Ishimoto H, Taniguchi H, Mukae H. Significance of anaerobes and oral bacteria in community-acquired pneumonia. PLoS One. 2013; 8:e63103. 10.1371/journal.pone.0063103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heiskala M, Peterson PA, Yang Y. The roles of claudin superfamily proteins in paracellular transport. Traffic. 2001; 2:93–98. 10.1034/j.1600-0854.2001.020203.x. [DOI] [PubMed] [Google Scholar]
- 46. Oka T, Schmitt AP, Sudol M. Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene. 2012; 31:128–134. 10.1038/onc.2011.216. [DOI] [PubMed] [Google Scholar]
- 47. Oka T, Remue E, Meerschaert K, Vanloo B, Boucherie C, Gfeller D, Bader GD, Sidhu SS, Vandekerckhove J, Gettemans J, Sudol M. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem J. 2010; 432:461–472. 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- 48. Kwong TNY, Wang X, Nakatsu G, Chow TC, Tipoe T, Dai RZW, Tsoi KKK, Wong MCS, Tse G, Chan MTV, Chan FKL, Ng SC, Wu JCY, et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology. 2018; 155:383–90.e8. 10.1053/j.gastro.2018.04.028. [DOI] [PubMed] [Google Scholar]
- 49. Sidhu JS, Mandal A, Virk J, Gayam V. Early detection of colon cancer following incidental finding of clostridium septicum bacteremia. J Investig Med High Impact Case Rep. 2019; 7:2324709619832050. 10.1177/2324709619832050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hattori N, Niwa T, Ishida T, Kobayashi K, Imai T, Mori A, Kimura K, Mori T, Asami Y, Ushijima T. Antibiotics suppress colon tumorigenesis through inhibition of aberrant DNA methylation in an azoxymethane and dextran sulfate sodium colitis model. Cancer Sci. 2019; 110:147–156. 10.1111/cas.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Higashimura Y, Naito Y, Takagi T, Uchiyama K, Mizushima K, Ushiroda C, Ohnogi H, Kudo Y, Yasui M, Inui S, Hisada T, Honda A, Matsuzaki Y, et al. Protective effect of agaro-oligosaccharides on gut dysbiosis and colon tumorigenesis in high-fat diet-fed mice. Am J Physiol Gastrointest Liver Physiol. 2016; 310:G367–G375. 10.1152/ajpgi.00324.2015. [DOI] [PubMed] [Google Scholar]
- 52. Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest. 2014; 124:4166–4172. 10.1172/JCI72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pahle J, Menzel L, Niesler N, Kobelt D, Aumann J, Rivera M, Walther W. Rapid eradication of colon carcinoma by Clostridium perfringens Enterotoxin suicidal gene therapy. BMC Cancer. 2017; 17:129. 10.1186/s12885-017-3123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matsuhisa K, Kondoh M, Suzuki H, Yagi K. Comparison of mucosal absorption-enhancing activity between a claudin-3/-4 binder and a broadly specific claudin binder. Biochem Biophys Res Commun. 2012; 423:229–233. 10.1016/j.bbrc.2012.05.060. [DOI] [PubMed] [Google Scholar]
- 55. Dong X, Meng L, Liu P, Ji R, Su X, Xin Y, Jiang X. YAP/TAZ: a promising target for squamous cell carcinoma treatment. Cancer Manag Res. 2019; 11:6245–6252. 10.2147/CMAR.S197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hsu PC, Jablons DM, Yang CT, You L. Epidermal growth factor receptor (EGFR) pathway, yes-associated protein (YAP) and the regulation of programmed death-ligand 1 (PD-L1) in non-small cell lung cancer (NSCLC). Int J Mol Sci. 2019; 20:3821. 10.3390/ijms20153821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li N, Lu N, Xie C. The Hippo and Wnt signalling pathways: crosstalk during neoplastic progression in gastrointestinal tissue. FEBS J. 2019; 286:3745–3756. 10.1111/febs.15017. [DOI] [PubMed] [Google Scholar]
- 58. Zhang S, Zhou D. Role of the transcriptional coactivators YAP/TAZ in liver cancer. Curr Opin Cell Biol. 2019; 61:64–71. 10.1016/j.ceb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 59. Akervall J, Nandalur S, Zhang J, Qian CN, Goldstein N, Gyllerup P, Gardinger Y, Alm J, Lorenc K, Nilsson K, Resau J, Wilson G, Teh B. A novel panel of biomarkers predicts radioresistance in patients with squamous cell carcinoma of the head and neck. Eur J Cancer. 2014; 50:570–581. 10.1016/j.ejca.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 60. UICC TNM Classification 7th edition, eds: Sobin LH, Gospodarowicz MK, Wittekind C. Wily-Blackwell, 2009; page 100–105. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.