Abstract
Background
TP73 antisense RNA 1 (TP73‐AS1) is a long noncoding RNA which has been shown to be involved in the progression of multiple malignant tumors. Previous studies have demonstrated the oncogenic role of TP73‐AS1 in breast cancer. However, its molecular mechanism remains largely unknown in breast tumorigenesis.
Methods
Expression of TP63‐AS1, miRNA‐125a‐3p (miR‐125a) and metadherin (MTDH) was detected by real‐time quantitative PCR and western blotting. The malignancy was evaluated by cell counting kit 8 (CCK‐8), transwell assays, flow cytometry and western blotting. The target binding was confirmed by dual luciferase reporter assay. Xenograft tumor model was performed to detect tumor growth in vivo.
Results
Expression of TP73‐AS1 was higher in breast cancer tissues and cell lines. Biologically, its knockdown could promote cell apoptosis rate, and inhibit proliferative capacity, migration and invasion ability in HCC‐70 and MB231 cells, accompanied with higher cleaved caspase 3 level and lower Ki67, N‐cadherin and Vimentin level. Moreover, TP73‐AS1 downregulation restrained the tumor growth of HCC‐70 cells in vivo. Mechanically, TP73‐AS1 functioned as a molecular “sponge” for miR‐125a to modulate MTDH, a downstream target of miR‐125a. Intriguingly, both miR‐125a overexpression and MTDH silencing exerted a tumor‐suppressive effect in the malignant progression of HCC‐70 and MB231 cells, which was counteracted by TP73‐AS1 upregulation and miR‐125a downregulation, respectively.
Conclusion
Knockdown of TP73‐AS1 inhibited cell proliferation, migration and invasion, but facilitated apoptosis in breast cancer cells in vitro through targeting miR‐125a and upregulating MTDH, suggesting a novel TP73‐AS1/miR‐125a/MTDH pathway in the malignant progression of breast cancer.
Keywords: Breast cancer, malignant progression, miR‐125a, MTDH, TP73‐AS1
Introduction
Breast cancer is a highly heterogeneous disease and the most overwhelmingly occurring malignancy in women globally. Different breast cancers may originate from different cells within the breast tissue.1 In malignant breast cancers, it is pathologically classified according to their expression status of key proteins estrogen receptor α (ERα), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2).2 The breast tumors lacking these three proteins are referred to as triple negative breast cancer (TNBC), which generally affects young women.3 In terms of strategies towards breast cancer treatment, therapies including chemotherapy, radiotherapy, surgical mastectomy, hormone based therapies and antiangiogenic therapies are well‐recognized approaches4; nevertheless, undesired effects such as acquired resistance, metastasis and recurrence always accompany these antitumor strategies. Moreover, lack of early diagnosis methods and biomarkers are also attributed to the high mortality of patients with breast cancer.
Recently, competing endogenous RNA (ceRNA) network has become the newest, most popular molecular mechanism in the pathogenesis of human cancers including breast cancer.5, 6 It is well characterized with regulatory dialogues between different types of noncoding RNAs (ncRNAs) such as long and short ncRNAs (lncRNAs and microRNAs). LncRNAs are a class of endogenous transcripts with more than 200 nucleotides, whereas microRNAs (miRNAs) are transcripts which consist of approximately 22 nucleotides. In breast cancer, both lncRNAs and miRNAs participate in the initiation and development of malignant breast tumors.7 Many ncRNAs have been identified and histopathologically linked to breast cancer; however, only a few have been implicated in key cellular function.8 Therefore, it is imperative and essential to uncover the role of ncRNAs in regulating cell behaviors of breast cancer cells, as well as the novel lncRNAs‐associated ceRNA network.
P73 antisense RNA 1T (TP73‐AS1, also known as PDAM or KIAA0495), is a newly discovered lncRNA on human chromosomal band 1p36.9 Considerable evidence has identified TP73‐AS1 as an oncogenic gene in several human malignant tumors such as hepatocellular carcinoma, glioma and non‐small cell lung cancer.10, 11, 12 In breast cancer, the link of TP73‐AS1 to cell proliferation, migration and invasion in vitro, as well as vasculogenic mimicry formation, has been previously reported,13–15 while TP73‐AS1 functioning as molecular “sponge” for miRNAs remains largely unclear in breast cancer. miRNA‐125a‐3p (miR‐125a) is a new member of the miR‐125a family which plays a role in different cancers.16 Expression and the role of miR‐125a are poorly stated compared to miRNA‐125a‐5p. Current evidence reports the tumor‐suppressive role of miR‐125a in multiple solid tumors including breast cancer.17, 18, 19 Metadherin (MTDH, also known as AEG‐1 or Lyric) is located at chromosome 8q22.20 MTDH is usually absent in human breast tissues, whereas abundantly expressed in breast tumors and cell lines. Functionally, MTDH as a cell surface protein in breast tumors has been found to mediate its metastasis and chemoresistance,21, 22 thus serving as a prognostic biomarker during breast carcinogenesis and survival.23 However, whether there is an interaction between TP73‐AS1, miR‐125a and MTDH remains to be verified.
In this study, we examined the expression of TP73‐AS1, miR‐125a and MTDH in human breast cancer tissues and cell lines. Their biological role was determined in the malignant progression of breast cancer cells in vitro and in vivo including cell proliferation, apoptosis, migration, invasion and tumor growth. Importantly, the regulatory effect between TP73‐AS1, miR‐125a and MTDH was further confirmed.
Methods
Clinical sample collection
A total of 45 patients (average age: 55 years) with breast cancer were recruited into the study. Pathological diagnosis was made according to the histology of tumor specimens or biopsy and examined by experienced pathologists. The patients in the study had not received any radiotherapy and chemotherapy prior to surgery. The breast cancer and adjacent normal tissues (≥5 cm away from breast cancer tissues) were collected during the operation in Changji Huizu People's Hospital of Xinjiang, and immediately preserved in liquid nitrogen. The ethical approval of this study protocol was authorized by the Ethics Committee of Changji Huizu People's Hospital of Xinjiang, and detailed written consent was obtained from each subject enrolled.
Cell culture
The normal human breast cell line MCF‐10A (CRL‐10137) and human breast cancer cell lines HCC‐70 (CRL‐2315) and MB231 (HTB‐26) were purchased from American Type Culture Collection (Manassas, VA, USA). All the cells were cultivated in Roswell Park Memorial Institute 1640 medium (HyClone, South Logan, UT, USA) and supplemented with 10% fetal bovine serum (FBS; Hyclone), 100 U/mL penicillin and 0.1 mg/mL streptomycin in 5% CO2 at 37°C. The cells were passaged every three days.
RNA extraction and real‐time quantitative PCR (RT‐qPCR)
Total RNA from tissue sample and cultured cells was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) and an RNeasy kit (QIAGEN, Hilden, Germany) with DNase I digestion. The reverse transcription was performed using HiFiScript cDNA Synthesis Kit (CWBIO, Beijing, China), and then SYBR Green Master Mix (CWBIO) was utilized to amplify cDNA with special primers synthesized by GENEWIZ (Suzhou, China). The amplification condition was followed the default program of ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). 18S and U6 small nuclear RNA (U6) acted as the endogenous control. The primers were listed as follows: TP73‐AS1, 5′‐CCGGTTTTCCAGTTCT TGCAC‐3′ (sense) and 5′‐GCCTCACAGGGAAACTTCATGC‐3′ (antisense)24; miR‐125a, 5′‐GGCGACAGGTGAGGTTCTT‐3′ (sense) and 5′‐GCAGGGTCCGAGGTATTC‐3′ (antisense); MTDH, 5′‐ACTGGGGGCTTTCGGGCTCTGCGT‐3′ (sense) and 5′‐AGTGCGTGTCGTGGAGTC‐3′ (antisense); 18S, 5′‐ TAACCCGTTGAACCCCATT‐3′ (sense) and 5′‐CCATCCAATCGGTAGTAGC‐3′ (antisense); U6, 5′‐GCTTCGGCAGCACATATACTAAAAT‐3′ (sense) and 5′‐CGCTTCACGAATTTGCGTGTCAT‐3′ (antisense). With 2−ΔΔCT method, the relative expression of TP73‐AS1, miR‐125a and MTDH was presented as fold changes.
Cell transfection
Small interfering RNA (siRNA) against TP73‐AS1 (si‐TP73‐AS1) and against MTDH (si‐MTDH), miR‐125a mimic and inhibitor were constituted by GENEWIZ, as well as negative controls. The overexpressing vectors TP73‐AS1 was constructed depending on pIRES2‐EGFP plasmid (Invitrogen). HCC‐70 and MB231 cells were preloaded in a six‐well plate (Corning, NY, USA) for 24 hours. All the nucleotides were separately transfected into the cells in logarithmic phase using Lipofectamine 2000 (Invitrogen) according to the instruction manual. In rescue experiments and dual luciferase reporter assay, half nucleotides were cotransfected in cells. Generally, transfected cells were harvested for further analysis after 24 hours. The sequence of siRNAs was as follows: si‐TP73‐AS1, 5′‐ GATCGCGTTCTGTGTGGAACTTACTGGATCAAGAGTCCAGTAAGTTCCACACAGAATTTTTTCCAAA‐3′ (sense), 5′‐AGCTTTTGGAAAAAATTCTGTGTGGAACTTACTGGACTCTTGATCCAGTAAGTTC CACACAGAACGC‐3′ (antisense)24; si‐MTDH, 5′‐ATGAACCAGAATCAGTCAGC‐3′.
Cell counting kit 8 (CCK‐8)
Transfected HCC‐70 and MB231 cells were reseeded in a 96‐well plate (Corning) with 5000 cells per well. After transfection for 0 hours, 24 hours, 48 hours and 72 hours, 10 μL of CCK‐8 solution (Dojindo, Kumamoto, Japan) was added to each well. After incubation for two hours, the optical density (OD) values was measured at 490 nm using a microplate reader and recorded in cell growth curve. The reactions were performed in sextuplicate for each group.
Flow cytometry
Transfected HCC‐70 and MB231 cells were harvested and resuspended in 500 μL of binding buffer (BD Biosciences, Detroit, MI, USA). The apoptotic cells were double‐labeled with 5 μL of FITC‐Annexin V (BD Biosciences) and 5 μL of propidium iodide (BD Biosciences) for 30 minutes in the dark. The cells were then analyzed on BD FACSCalibur flow cytometer (BD Biosciences) within one hour using the matched software. Apoptosis rate was recorded as percentage of apoptotic cells in Annexin V+/PI− and Annexin V+/PI+ quadrants. Each sample was repeated three times.
Transwell assays
The ability of invasion was measured using a Transwell chamber (8 μm pore size, Corning) coated with matrigel (Becton Dickinson, Franklin Lakes, USA). Transfected HCC‐70 and MB231 cells (4000 cells) in 200 μL medium without FBS were implanted in the upper chamber and 500 μL of medium with 10% FBS in the lower chamber. For cell migration analysis, the chamber was not coated with matrigel and other procedures were the same as the invasion analysis. All transwell assays were incubated for 37°C for 24 hours. The migrated and invaded cells on the lower surface were stained with crystal violet and photographed under a microscope in five predetermined fields (×200). Three independent experiments were carried out.
Western blotting
Total protein from cultivated and transfected HCC‐70 and MB231 cells was isolated in ice‐cold radioimmunoprecipitation lysis buffer (Pierce, Rockford, Alabama, USA) containing phenylmethanesulfonylfluoride. A total of 20 μg protein samples 5× loading buffer (Pierce) was separated by 8%–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) for the standard procedures, followed by transfer onto PVDF membranes (Millipore, Billerica, Massachusetts, USA). After incubation with primary and secondary antibodies, the protein bands were visualized by enhanced chemiluminescence (Millipore). The primary antibodies cleaved caspase 3 (#9664), N‐cadherin (#4061), Vimentin (#3932) and β‐actin (#4967) were provided by Cell Signaling Technology (Danvers, Massachusetts, USA) and anti‐Ki67 (#2586) and anti‐MTDH (#227981) were acquired from Abcam (Cambridge, UK). All the antibodies were diluted at 1:1000, and β‐actin was the loading control.
Dual luciferase reporter assay
The wild‐type (WT‐) and mutant (MUT‐) of TP73‐AS1 and MTDH 3′ untranslated region (3′ UTR) containing the complementary binding site of miR‐125a “seed sequence” were fused to pmirGLO (Promega, Madison, WI, USA) dual luciferase vector. HCC‐70 and MB231 cells were cotransfected with miR‐125a mimic or miR‐control with either WT‐TP73‐AS1/MTDH or MUT‐TP73‐AS1/MTDH. At 24 hours after transfection, the Firefly luciferase activity was performed using GloMax LUMINOMETER (Promega). Renilla luciferase activity was constitutively expressed in pmirGLO vectors and served as the internal control. All data were the average of at least three independent transfections.
Xenograft tumor model
HCC‐70 cells were infected by lentiviral particles encoding shRNA against TP73‐AS1 (sh‐TP73‐AS1; Neuron Biotech, Shanghai, China) or the negative control (sh‐NC) using Polybrene reagent (Sigma, St Louis, MO, USA). The six‐week‐old BALB/c nude mice were purchased from HFK Bio‐Technology (Beijing, China), and randomly divided into two groups (n = 6). Stably transfected HCC‐70 cells (2.5 × 106 cells) were then subcutaneously injected into the right flanks of mice. After inoculation, tumor volume was monitored every week with the formula: 0.5 × length × width.2 On the fifth week, the mice were euthanatized and the tumors surgically separated. The tumor weight was subsequently recorded and the tumors snap‐frozen in liquid nitrogen for further RNA or protein detection. All animal procedures were carried out in accordance with the Animal Care Committee of Changji Huizu People's Hospital of Xinjiang.
Statistical analysis
Data were from three independent experiments and expressed as mean ± standard error of the mean. Statistical analysis was performed using two‐tailed Student's t‐test or one‐way analysis of variance using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). Spearman's rank correlation analysis was used to estimate the correlation between the expression of TP73‐AS1, miR‐125a and MTDH in breast cancer tissue samples. Chi‐square test was used to analyze the relationship between TP73‐AS1 expression and clinical features. P < 0.05 was considered as statistically significant.
Results
TP73‐AS1 expression upregulated in breast cancer tissues and cell lines
Initially, the expression of TP73‐AS1 in breast cancer samples was detected by RT‐qPCR. As shown in Figure 1a, TP73‐AS1 level was distinctively higher in 45 tumor tissues than the paired adjacent normal tissues from clinical patients with breast cancer. According to the mean of TP73‐AS1 level, this cohort of patients was divided into high TP73‐AS1 group (n = 25) and low TP73‐AS1 group (n = 25). We noted that TP73‐AS1 high expression was associated with distant metastasis, instead of ER, PR or HER2 status (Table 1). RT‐qPCR analysis also estimated that TP73‐AS1 was highly expressed in human breast cancer cell lines HCC‐70 and MB231 compared to that in normal breast cell line MCF‐10A (Fig 1b). These results showed the dysregulation of TP73‐AS1 in breast cancer tissues and cells, suggesting a potential oncogenic role of TP73‐AS1 in breast cancer cells.
Figure 1.
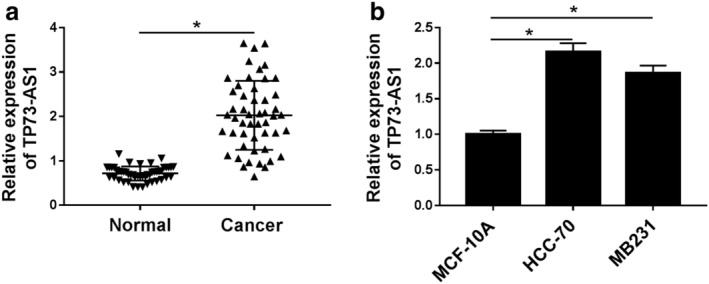
Expression of lncRNA TP73 antisense RNA 1 (TP73‐AS1) in breast cancer tissues and cell lines. (a) RT‐qPCR detected TP73‐AS1 expression level in paired tumor tissues (Cancer) and adjacent normal tissues (Normal) from breast cancer patients (n = 45). Fold change was analyzed using the formula 2−ΔΔCT. (b) RT‐qPCR estimated TP73‐AS1 level in human breast cancer cell lines (HCC‐70 and MB231) and the normal breast cell line MCF‐10A. Data represent mean ± standard error of the mean (SEM) and *P < 0.05.
Table 1.
Relationship between TP73‐AS1 expression and clinicopathological features of breast cancer patients. The patients were divided into TP73‐AS1 high expression group (n = 25) and low expression group (n = 20) according to mean. High TP73‐AS1 expression was associated with distant metastasis
| Clinicopathological features | n | TP73‐AS1 expression | P‐value | |
|---|---|---|---|---|
| High (n = 25) | Low (n = 20) | |||
| ER status | ‐ | ‐ | ‐ | ‐ |
| Negative | 24 | 16 | 9 | 0.109 |
| Positive | 21 | 9 | 11 | ‐ |
| PR status | ‐ | ‐ | ‐ | ‐ |
| Negative | 23 | 10 | 13 | 0.095 |
| Positive | 22 | 15 | 7 | ‐ |
| HER2 status | ‐ | ‐ | ‐ | ‐ |
| Negative | 25 | 14 | 11 | 0.947 |
| Positive | 20 | 11 | 9 | ‐ |
| Distant metastasis | ‐ | ‐ | ‐ | ‐ |
| Yes | 27 | 20 | 7 | 0.002* |
| No | 18 | 5 | 13 | ‐ |
Means the significant difference.
ER, estrogen receptor α; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Knockdown of TP73‐AS1 suppressed malignant progression of breast cancer cells in vitro
Given that TP73‐AS1 is upregulated in human breast cancer, we further investigated the detailed biological role of TP73‐AS1 in the malignant progression of breast cancer cells in vitro. Firstly, we forcedly knocked‐down its expression in HCC‐70 and MB231 cells by transfection with si‐TP73‐AS1. Luckily, we achieved a high transfection efficiency as verified by RT‐qPCR with TP73‐AS1 level lowered below 0.5 fold (Fig 2a). Due to TP73‐AS1 silencing, the proliferative capacity of HCC‐70 and MB231 cells was significantly impaired after transfection for 72 hours according to CCK‐8 assay (Fig 2b). Meanwhile, apoptosis rate was prominently elevated when si‐TP73‐AS1 transfected for 24 hours (Fig 2c) with little cell proliferation inhibition. The effect of TP73‐AS1 dysregulation on metastasis ability of breast cancer cells was also measured. As depicted by transwell assays, numbers of migrated cells and invaded cells were both attenuated after TP73‐AS1 was knocked‐down in HCC‐70 and MB231 cells at 24 hours (Fig 2d,e) with little cell proliferation inhibition. In addition, expression of biomarkers in cell processes were examined. In si‐TP73‐AS1‐transfected HCC‐70 and MB231 cells, expression of Ki67, N‐cadherin and Vimentin was observably lower, but cleaved caspase 3 expression was higher at 24 hours than that in si‐control‐transfected cells (Fig 2f). These results demonstrated that knockdown of TP73‐AS1 could inhibit cell proliferation, migration and invasion, but promote apoptosis in HCC‐70 and MB231 cells, suggesting a tumor‐suppressive role of TP73‐AS1 knockdown in the malignant progression of breast cancer cells in vitro.
Figure 2.
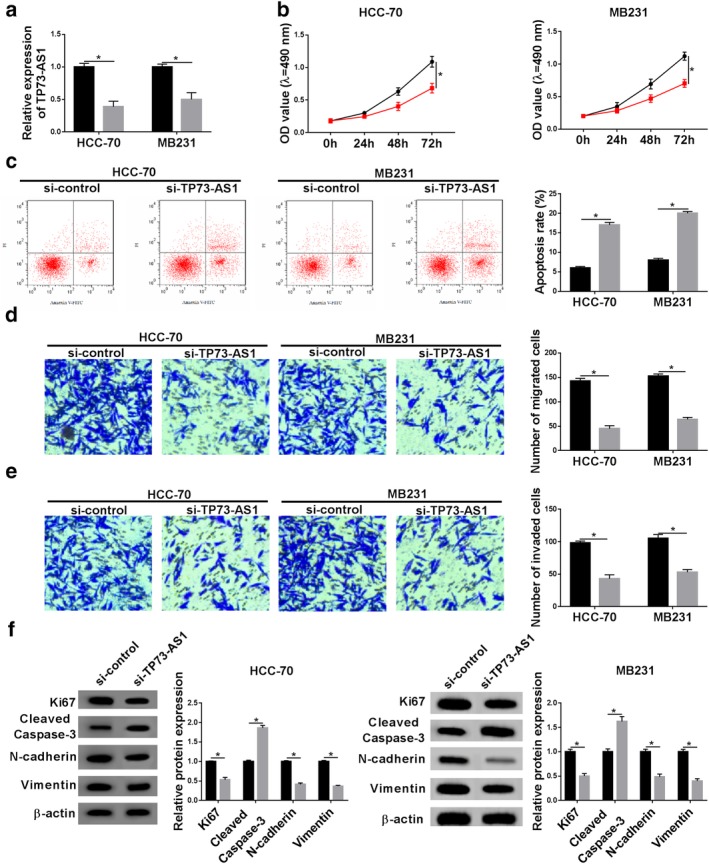
The effect of TP73‐AS1 knockdown on the malignant progression of breast cancer cells in vitro. HCC‐70 and MB231 cells were transfected with siRNA against TP73‐AS1 (si‐TP73‐AS1) or its control (si‐control). (a) RT‐qPCR was used to analyze TP73‐AS1 level after transfection for 24 hours. (b, c) Cell counting kit 8 (CCK‐8) was utilized to determine cell proliferative capacity after transfection at 0 hour, 24 hours, 48 hours and 72 hours. (C) Flow cytometry was conducted to examine apoptosis rate after transfection at 24 hours. The percentage of apoptotic cells in quadrants of Annexin V+/PI− and Annexin V+/PI+ was statistically recorded. (d, e) Transwell assays were performed to evaluate cell migration and invasion abilities at 24 hours. The number of migrated cells and invaded cells was statistically recorded. (f) Western blotting was implemented to test protein expression of Ki67, cleaved caspase‐3, N‐cadherin and Vimentin after transfection at 24 hours. Protein bands of western blotting were quantified by densitometry and presented as fold changes with normalization to β‐actin. Data represent mean ± SEM and *P < 0.05. (a) ( ) si‐control and (
) si‐control and ( ) si‐TP73‐AS1. (b) HCC (
) si‐TP73‐AS1. (b) HCC ( ) si‐control and (
) si‐control and ( ) si‐TP73‐AS1. MB231 (
) si‐TP73‐AS1. MB231 ( ) si‐control and (
) si‐control and ( ) si‐TP73‐AS1. (c–e) (
) si‐TP73‐AS1. (c–e) ( ) si‐control and (
) si‐control and ( ) si‐TP73‐AS1. (f) HCC‐70 (
) si‐TP73‐AS1. (f) HCC‐70 ( ) si‐control and (
) si‐control and ( ) si‐TP73‐AS1. MB231 (
) si‐TP73‐AS1. MB231 ( ) si‐control and (
) si‐control and ( ) si‐TP73‐AS1.
) si‐TP73‐AS1.
TP73‐AS1 regulated miR‐125a expression by targeting
We hypothesized that TP73‐AS1 upregulation contributes to the tumorigenesis and development of breast cancer. The lncRNA‐related ceRNA network was subsequently explored. As predicted by bioinformatics starbase Tools (version 2.0; http://starbase.sysu.edu.cn/starbase2/mirLncRNA.php), TP73‐AS1 contained a site which was complementary bound to the “seed sequence” of miR‐125a, as shown in Figure 3a. To testify the interaction between TP73‐AS1 and miR‐125a, the wild‐type of TP73‐AS1 (WT‐TP73‐AS1) containing UCACCUG was site‐directed mutated and dual luciferase reporter assay was launched in HCC‐70 and MB231 cells. The relative luciferase activity of vector expressing WT‐TP73‐AS1 was abundantly decreased only with cotransfection with miR‐125a mimic (Fig 3b); whereas, vector expressing its mutant (MUT‐TP73‐AS1) showed no significant difference between cotransfection of miR‐125a mimic or miR‐control. This finding confirmed the potential binding relationship between TP73‐AS1 and miR‐125a. Similarly, expression of miR‐125a in breast cancer was evaluated utilizing RT‐qPCR. In these patients, miR‐125a expression was downregulated in tumor tissues comparing to the adjacent normal tissues, and its expression was inversely correlated with TP73‐AS1 (R = −0.8145, P < 0.05) (Fig 3c,d). Besides, lower expression of miR‐125a was also observed in human breast cancer cell lines HCC‐70 and MB231 versus the normal breast cell line MCF‐10A (Fig 3e). RT‐qPCR analysis showed miR‐125a expression was extremely lower with pIRES2‐EGFP‐TP73‐AS1 transfection in HCC‐70 and MB231 cells (Fig 3f,g), and higher with si‐TP73‐AS1 transfection (Fig 2a and 3g). These outcomes indicated miR‐125a expression was negatively regulated by TP73‐AS1 via binding.
Figure 3.
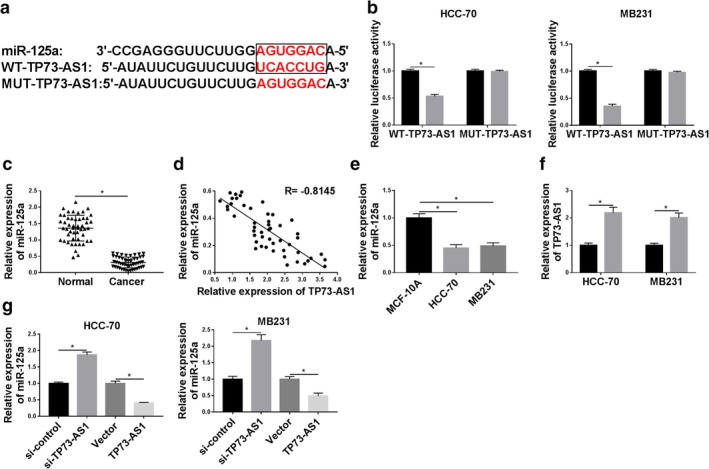
TP73‐AS1 regulated miRNA‐125a‐3p (miR‐125a) expression by targeting. (a) Prediction of binding sites between TP73‐AS1 and miR‐125a (in the box) were shown according to starbase Tools. The corresponding mutant of TP73‐AS1 (MUT‐TP73‐AS1) was presented as well. (b) Luciferase activity of wild‐type of TP73‐AS1 (WT‐TP73‐AS1) and MUT‐TP73‐AS1 was confirmed by dual luciferase reporter assay in HCC‐70 and MB231 cells when cotransfected with miR‐125a mimic (miR‐125a) or its control (miR‐control). HCC ( ) miR‐control and (
) miR‐control and ( ) miR‐125a. MB231 (
) miR‐125a. MB231 ( ) miR‐control and (
) miR‐control and ( ) miR‐125a. (c) RT‐qPCR measured level of miR‐125a in breast cancer tissues (n = 45) compared with the paired normal tissues. (d) Spearman's rank correlation analysis clarified the association between miR‐125a and TP73‐AS1 expression in breast cancer tissues (n = 45). (e) RT‐qPCR measured miR‐125a level in breast cancer cell lines (HCC‐70 and MB231) comparing to the normal cell line MCF‐10A. (f) RT‐qPCR determined the transfection efficiency of pIRES2‐EGFP empty vector (vector) and recombinant vector containing TP73‐AS1 (TP73‐AS1) in HCC‐70 and MB231 cells. (
) miR‐125a. (c) RT‐qPCR measured level of miR‐125a in breast cancer tissues (n = 45) compared with the paired normal tissues. (d) Spearman's rank correlation analysis clarified the association between miR‐125a and TP73‐AS1 expression in breast cancer tissues (n = 45). (e) RT‐qPCR measured miR‐125a level in breast cancer cell lines (HCC‐70 and MB231) comparing to the normal cell line MCF‐10A. (f) RT‐qPCR determined the transfection efficiency of pIRES2‐EGFP empty vector (vector) and recombinant vector containing TP73‐AS1 (TP73‐AS1) in HCC‐70 and MB231 cells. ( ) Vector and (
) Vector and ( ) TP73‐AS1. (g) RT‐qPCR detected miR‐125a expression level in HCC‐70 and MB231 cells when transfected with si‐TP73‐AS1, si‐control, TP73‐AS1 and vector. Data represent mean ± SEM and *P < 0.05.
) TP73‐AS1. (g) RT‐qPCR detected miR‐125a expression level in HCC‐70 and MB231 cells when transfected with si‐TP73‐AS1, si‐control, TP73‐AS1 and vector. Data represent mean ± SEM and *P < 0.05.
TP73‐AS1 upregulation could counteract the tumor‐suppressive role of miR‐125a in breast cancer cells progression in vitro
Rescue experiments were carried out to identify whether TP73‐AS1 influences the biological role of miR‐125a in breast cancer cells. On one hand, HCC‐70 and MB231 cells were transfected with miR‐125a mimic or miR‐control, followed by cell behaviors detection. The upregulation of miR‐125a was mediated by miR‐125a mimic transfection in HCC‐70 and MB231 cells (Fig 4a), and this upregulation was partially reversed by TP73‐AS1 overexpression vectors transfection (Fig S1a). CCK‐8 revealed that miR‐125a mimic transfection decreased the cell proliferative capacity of HCC‐70 and MB231 cells, accompanied by a lower Ki67 level (Fig 4b,f,g). On the contrary, flow cytometry revealed that the apoptosis rate was facilitated by miR‐125a upregulation along with higher cleaved caspase 3 level (Fig 4c,f,g). Transwell assays consistently illuminated that the ability of cell migration and invasion was reduced after miR‐125a overexpressed in HCC‐70 and MB231 cells (Fig 4d,e) in line with lower expression of N‐cadherin and Vimentin (Fig 4f,g). Therefore, we discovered a suppressive effect of miR‐125a on malignant progression of breast cancer cells in vitro, which was similar to the TP73‐AS1 knockdown role (Fig 2). On the other hand, HCC‐70 and MB231 cells were transfected with miR‐125a mimic, together with either pIRES2‐EGFP‐TP73‐AS1 or vector to determine the changes of cell progressions, while cotransfection of pIRES2‐EGFP‐TP73‐AS1 led to a partial but obvious recovery of the malignancy of HCC‐70 and MB231 cells in miR‐125a‐overexpressed cells compared to cotransfection of empty vector (Fig 4b–g). These results showed TP73‐AS1 upregulation could counteract the tumor‐suppressive role of miR‐125a in breast cancer cells progression in vitro.
Figure 4.
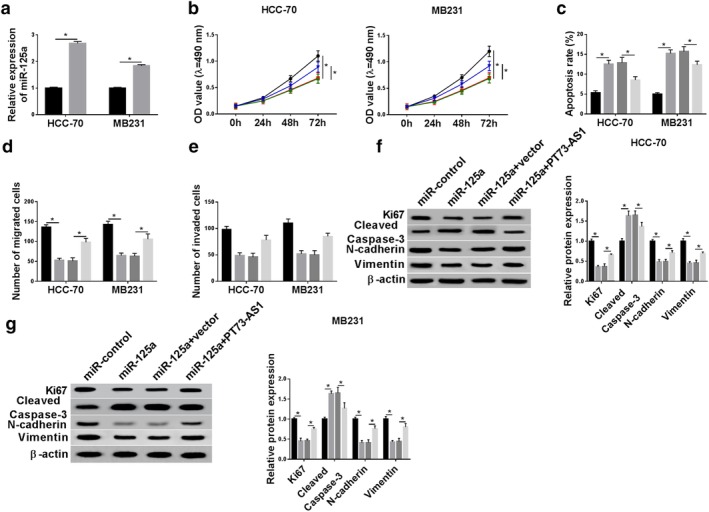
The influence of TP73‐AS1 upregulation on the role of miR‐125a in breast cancer cell malignancy in vitro. (a) RT‐qPCR determined the transfection efficiency of miR‐125a and miR‐control in HCC‐70 and MB231 cells. ( ) miR‐control and (
) miR‐control and ( ) miR‐125a. (b–f) HCC‐70 and MB231 cells were transfected with miR‐125a or miR‐control, and cotransfected with miR‐125a and either TP73‐AS1 or vector. (b) CCK‐8 determined cell proliferative capacity after transfection at 0 hour, 24 hours, 48 hours and 72 hours. HCC‐70 (
) miR‐125a. (b–f) HCC‐70 and MB231 cells were transfected with miR‐125a or miR‐control, and cotransfected with miR‐125a and either TP73‐AS1 or vector. (b) CCK‐8 determined cell proliferative capacity after transfection at 0 hour, 24 hours, 48 hours and 72 hours. HCC‐70 ( ) miR‐control, (
) miR‐control, ( ) miR‐125a, (
) miR‐125a, ( ) miR‐125a+vector and (
) miR‐125a+vector and ( ) miR‐125a+TP73‐AS1. MB231 (
) miR‐125a+TP73‐AS1. MB231 ( ) miR‐control, (
) miR‐control, ( ) miR‐125a, (
) miR‐125a, ( ) miR‐125a+vector and (
) miR‐125a+vector and ( ) miR‐125a+TP73‐AS1. (c) Flow cytometry examined apoptosis rate after transfection at 24 hours. (
) miR‐125a+TP73‐AS1. (c) Flow cytometry examined apoptosis rate after transfection at 24 hours. ( ) miR‐control, (
) miR‐control, ( ) miR‐125a, (
) miR‐125a, ( ) miR‐125a+vector and (
) miR‐125a+vector and ( ) miR‐125a+TP73‐AS1. (d, e) Transwell assays were performed to evaluate cell migration and invasion abilities at 24 hours. (
) miR‐125a+TP73‐AS1. (d, e) Transwell assays were performed to evaluate cell migration and invasion abilities at 24 hours. ( ) miR‐control, (
) miR‐control, ( ) miR‐125a, (
) miR‐125a, ( ) miR‐125a+vector and (
) miR‐125a+vector and ( ) miR‐125a+TP73‐AS1. (f, g) Western blotting tested protein expression of Ki67, cleaved caspase‐3, N‐cadherin and Vimentin after transfection at 24 hours. (f) HCC‐70 (
) miR‐125a+TP73‐AS1. (f, g) Western blotting tested protein expression of Ki67, cleaved caspase‐3, N‐cadherin and Vimentin after transfection at 24 hours. (f) HCC‐70 ( ) miR‐control, (
) miR‐control, ( ) miR‐125a, (
) miR‐125a, ( ) miR‐125a+vector and (
) miR‐125a+vector and ( ) miR‐125a+TP73‐AS1. (g) MB231 (
) miR‐125a+TP73‐AS1. (g) MB231 ( ) miR‐control, (
) miR‐control, ( ) miR‐125a, (
) miR‐125a, ( ) miR‐125a+vector and (
) miR‐125a+vector and ( ) miR‐125a+TP73‐AS1. Data represent mean ± SEM and *P < 0.05.
) miR‐125a+TP73‐AS1. Data represent mean ± SEM and *P < 0.05.
MTDH a downstream target gene of miR‐125a
Furthermore, the downstream target genes of miR‐125a were sought and identified. Online bioinformatics TargetScan tools (release 7.1; http://www.targetscan.org/vert_71) revealed a potential site of miR‐125a in the 3′ UTR of MTDH at position 2608‐2614. As shown in Figure 5a, the sequence UCACCUG in the wild‐type of MTDH 3′ UTR (WT‐MTDH) was mutated to AGUGGAC. Dual luciferase reporter assay demonstrated only luciferase activity of vector containing the WT‐MTDH was significantly decreased when miR‐125a was overexpressed by transfection (Fig 5b). This finding confirmed the potential binding between MTDH and miR‐125a. Expression of MTDH in breast cancer tissues was evaluated utilizing RT‐qPCR. In these patients, MTDH mRNA expression was upregulated in tumor tissues compared to the adjacent normal tissues, and its expression was negatively correlated with miR‐125a (R = −0.7560, P < 0.05) (Fig 5c,d). RT‐qPCR analysis and western blotting also showed a higher MTDH expression in human breast cancer cell lines HCC‐70 and MB231 than that in MCF‐10A cells (Fig 5e,f). Besides, MTDH mRNA expression and protein expression was upregulated with anti‐miR‐125a transfection (Fig 5g–i) in HCC‐70 and MB231 cells, and downregulated with miR‐125a mimic transfection (Figs 4a and 5h,i). These outcomes indicated MTDH served as a downstream target gene of miR‐125a and was negatively regulated by miR‐125a via direct binding.
Figure 5.
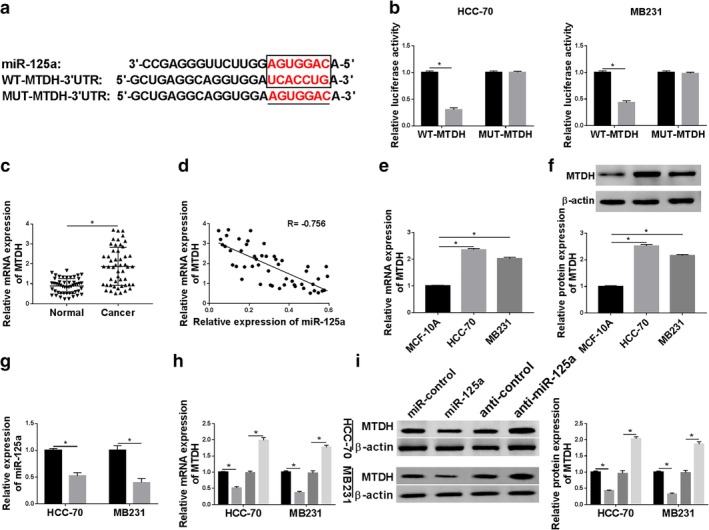
Metadherin (MTDH) was a downstream target gene of miR‐125a. (a) Prediction of binding sites between miR‐125a and 3′ UTR of MTDH (in the box) were shown according to TargetScan tools. The corresponding mutant of MTDH 3′ UTR (MUT‐MTDH) was also presented. (b) Luciferase activity of wild‐type of MTDH (WT‐MTDH) and MUT‐MTDH was confirmed by dual luciferase reporter assay in HCC‐70 and MB231 cells when cotransfected with miR‐125a or miR‐control. HCC‐70 ( ) miR‐control and (
) miR‐control and ( ) miR‐125a. MB231 (
) miR‐125a. MB231 ( ) miR‐control and (
) miR‐control and ( ) miR‐125a. (c) RT‐qPCR measured MTDH mRNA level in breast cancer tissues (n = 45) compared with the paired normal tissues. (d) Spearman's rank correlation analysis clarified the association between miR‐125a and MTDH expression in breast cancer tissues (n = 45). (e, f) RT‐qPCR and western blotting measured MTDH levels in breast cancer cell lines (HCC‐70 and MB231) comparing to MCF‐10A. (g) RT‐qPCR determined the transfection efficiency of miR‐125a inhibitor (anti‐miR‐125a) and its control (anticontrol) in HCC‐70 and MB231 cells. (
) miR‐125a. (c) RT‐qPCR measured MTDH mRNA level in breast cancer tissues (n = 45) compared with the paired normal tissues. (d) Spearman's rank correlation analysis clarified the association between miR‐125a and MTDH expression in breast cancer tissues (n = 45). (e, f) RT‐qPCR and western blotting measured MTDH levels in breast cancer cell lines (HCC‐70 and MB231) comparing to MCF‐10A. (g) RT‐qPCR determined the transfection efficiency of miR‐125a inhibitor (anti‐miR‐125a) and its control (anticontrol) in HCC‐70 and MB231 cells. ( ) anticontrol and (
) anticontrol and ( ) anti‐miR‐125a. (h, i) RT‐qPCR and western blotting detected MTDH expression levels in HCC‐70 and MB231 cells when transfected with miR‐125a, miR‐control, anti‐miR‐125a and anticontrol. (
) anti‐miR‐125a. (h, i) RT‐qPCR and western blotting detected MTDH expression levels in HCC‐70 and MB231 cells when transfected with miR‐125a, miR‐control, anti‐miR‐125a and anticontrol. ( ) miR‐control, (
) miR‐control, ( ) miR‐125a, (
) miR‐125a, ( ) anticontrol and (
) anticontrol and ( ) anti‐miR‐125a. Data represent mean ± SEM and *P < 0.05.
) anti‐miR‐125a. Data represent mean ± SEM and *P < 0.05.
Silencing of miR‐125a could abrogate the antitumor role of MTDH downregulation in breast cancer cells progression in vitro
Rescue experiments were carried out to identify whether miR‐125a influenced the biological role of MTDH in breast cancer cells. On one hand, HCC‐70 and MB231 cells were transfected with si‐MTDH or si‐control, followed by cell behaviors detection. The downregulation of MTDH mRNA and protein expression was mediated by si‐MTDH transfection (Fig 6a,b). This downregulation was rescued in the presence of anti‐miR‐125a (Fig S1b). CCK‐8 revealed that si‐MTDH transfection decreased cell proliferative capacity of HCC‐70 and MB231 cells, accompanied with lower Ki67 level (Fig 6c,g). Flow cytometry declared that the apoptosis rate was facilitated by MTDH downregulation along with a higher cleaved caspase 3 level (Fig 6d,g). In terms of cell metastasis ability, the numbers of migrated cells and invaded cells were reduced after MTDH was deleted as illuminated by transwell assays (Fig 6e,f); in addition, expression of N‐cadherin and Vimentin was inhibited in si‐MTDH‐transfected HCC‐70 and MB231 cells (Fig 6g). These results demonstrated a suppressive effect of MTDH downregulation on malignant progression of breast cancer cells in vitro which was similar to the TP73‐AS1 knockdown role (Fig 2). On the other hand, HCC‐70 and MB231 cells were transfected with si‐MTDH combined with anti‐miR‐125a and this cotransfection partially reversed the suppressive effect of MTDH knockdown on the malignancy of HCC‐70 and MB231 cells (Fig 6c–g). These results showed miR‐125a silencing could abrogate the antitumor role of MTDH downregulation in breast cancer cells progression in vitro.
Figure 6.
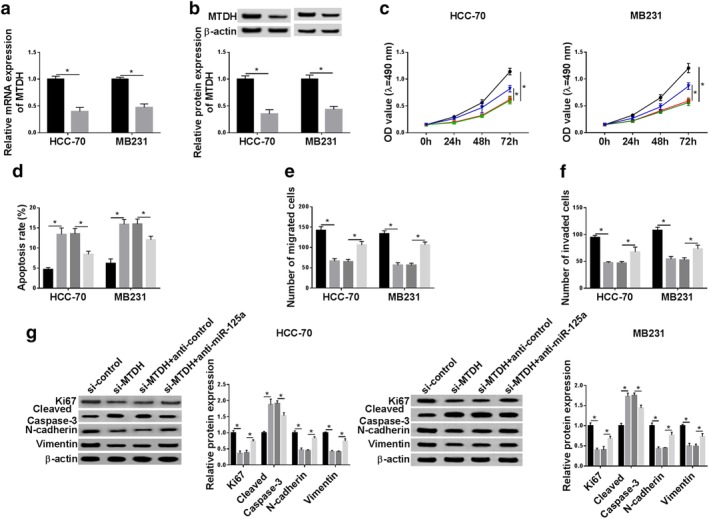
The impact of miR‐125a silencing on the role of MTDH downregulation in breast cancer cell malignancy in vitro. (a, b) RT‐qPCR and western blotting determined the transfection efficiency of siRNA against MTDH (si‐MTDH) and si‐control in HCC‐70 and MB231 cells. ( ) si‐control and (
) si‐control and ( ) si‐MTDH. (c–g) HCC‐70 and MB231 cells were transfected with si‐control or si‐MTDH, and cotransfected with si‐MTDH and either anticontrol or anti‐miR‐125a. (c) CCK‐8 determined cell proliferative capacity after transfection at 0 hour, 24 hours, 48 hours and 72 hours. HCC‐70 (
) si‐MTDH. (c–g) HCC‐70 and MB231 cells were transfected with si‐control or si‐MTDH, and cotransfected with si‐MTDH and either anticontrol or anti‐miR‐125a. (c) CCK‐8 determined cell proliferative capacity after transfection at 0 hour, 24 hours, 48 hours and 72 hours. HCC‐70 ( ) si‐control, (
) si‐control, ( ) si‐MTDH, (
) si‐MTDH, ( ) si‐MTDH+anticontrol and (
) si‐MTDH+anticontrol and ( ) si‐MTDH+anti‐miR‐125a. MB231 (
) si‐MTDH+anti‐miR‐125a. MB231 ( ) si‐control, (
) si‐control, ( ) si‐MTDH, (
) si‐MTDH, ( ) si‐MTDH+anticontrol and (
) si‐MTDH+anticontrol and ( ) si‐MTDH+anti‐miR‐125a. (d) Flow cytometry examined apoptosis rate after transfection at 24 hours. (
) si‐MTDH+anti‐miR‐125a. (d) Flow cytometry examined apoptosis rate after transfection at 24 hours. ( ) si‐control, (
) si‐control, ( ) si‐MTDH, (
) si‐MTDH, ( ) si‐MTDH+anticontrol and (
) si‐MTDH+anticontrol and ( ) si‐MTDH+anti‐miR‐125a. (e, f) Transwell assays were performed to evaluate cell migration and invasion abilities at 24 hours. (
) si‐MTDH+anti‐miR‐125a. (e, f) Transwell assays were performed to evaluate cell migration and invasion abilities at 24 hours. ( ) si‐control, (
) si‐control, ( ) si‐MTDH, (
) si‐MTDH, ( ) si‐MTDH+anticontrol and (
) si‐MTDH+anticontrol and ( ) si‐MTDH+anti‐miR‐125a. (g) Western blotting tested protein expression of Ki67, cleaved caspase‐3, N‐cadherin and Vimentin after transfection at 24 hours. HCC‐70 (
) si‐MTDH+anti‐miR‐125a. (g) Western blotting tested protein expression of Ki67, cleaved caspase‐3, N‐cadherin and Vimentin after transfection at 24 hours. HCC‐70 ( ) si‐control, (
) si‐control, ( ) si‐MTDH, (
) si‐MTDH, ( ) si‐MTDH+anticontrol and (
) si‐MTDH+anticontrol and ( ) si‐MTDH+anti‐miR‐125a. MB231 (
) si‐MTDH+anti‐miR‐125a. MB231 ( ) si‐control, (
) si‐control, ( ) si‐MTDH, (
) si‐MTDH, ( ) si‐MTDH+anticontrol and (
) si‐MTDH+anticontrol and ( ) si‐MTDH+anti‐miR‐125a. Data represent mean ± SEM and *P < 0.05.
) si‐MTDH+anti‐miR‐125a. Data represent mean ± SEM and *P < 0.05.
MTDH expression positively regulated by TP73‐AS1 through sponging miR‐125a
We confirmed TP73‐AS1/miR‐125a axis and miR‐125a/MTDH axis adjusted the malignancy of breast cancer cells, but whether TP73‐AS1 modulates MTDH expression remains to be confirmed. With transfection of si‐TP73‐AS1, we found that expression of MTDH mRNA and protein was consistently lowered when TP73‐AS1 was knocked‐down in HCC‐70 (Fig 7a,b) and MB231 cells (Fig 7c,d). Notably, HCC‐70 and MB231 cells exhibited a significantly improved MTDH expression when cotransfected with si‐TP73‐AS1 and anti‐miR‐125a (Fig 7a–d). In addition, MTDH expression was elevated in HCC‐70 and MB231 cells transfected with TP73‐AS1, which was further suppressed in the presence of miR‐125a mimic (Fig 7e–h). These findings showed MTDH expression was downregulated by TP73‐AS1 knockdown, and upregulated by TP73‐AS1 overexpression through miR‐125a, indicating a TP73‐AS1/miR‐125a/MTDH pathway in the malignant progression of breast cancer.
Figure 7.
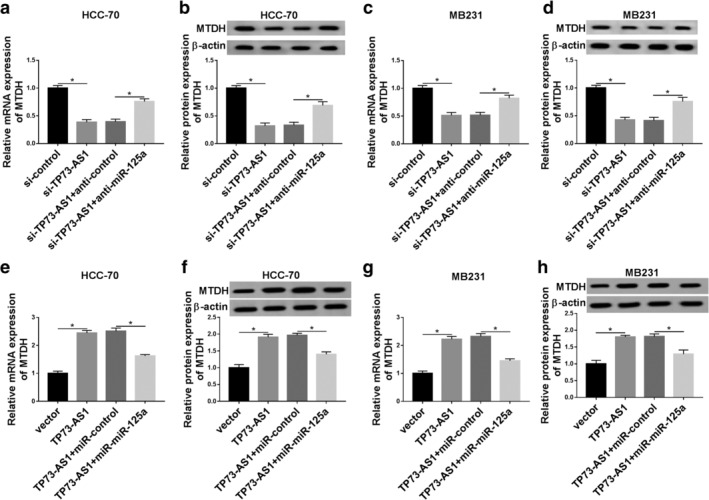
MTDH expression was positively regulated by TP73‐AS1 through sponging miR‐125a. RT‐qPCR and western blotting determined MTDH expression levels in (a, b) HCC‐70 cells and (c, d) MB231 cells transfected with si‐TP73‐AS1 alone or together with anti‐miR‐125a or anticontrol. MTDH expression levels in (e, f) HCC‐70 cells and (g, h) MB231 cells transfected with TP73‐AS1 alone or combined with miR‐125a or miR‐control were also measured. Data represent mean ± SEM and *P < 0.05.
Knockdown of TP73‐AS1 restrained tumor growth of breast cancer cells in vivo
To verify the functions of TP73‐AS1/miR‐125a/MTDH axis in breast cancer cells in vivo, we established a xenograft mice model stimulated by HCC‐70 cells stably expressing sh‐TP73‐AS1 or sh‐NC. After inoculation for weeks, tumors were detected in each mice in both groups. However, the tumor growth was significantly inhibited in sh‐TP73‐AS1 group, as evidenced by smaller tumor volume and lighter tumor weight (Fig 8a,b). On the fifth week, the tumors were harvested and gene expression status monitored. As seen in Fig 8c–e, TP73‐AS1 and MTDH mRNA showed lower expression, and miR‐125a higher expression in mice breast cancer tumors (n = 6). Moreover, the protein expression of MTDH was examined in one randomly selected xenograft tumor, and the data showed that MTDH was downregulated in xenograft tumor tissue (Fig 8f). These results indicated an antitumor role of TP73‐AS1 knockdown in vivo presumably through upregulating miR‐125a and downregulating MTDH.
Figure 8.
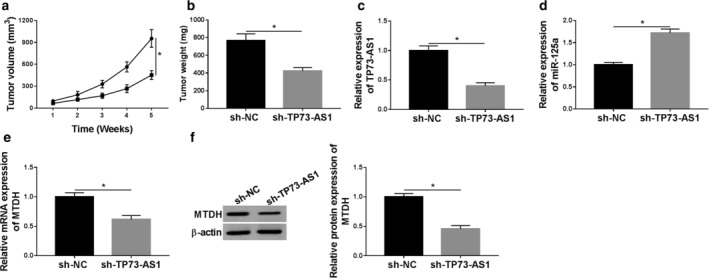
Knockdown of TP73‐AS1 restrained tumor growth of breast cancer cells in vivo. HCC‐70 cells were stably expressed shRNA against TP73‐AS1 (sh‐H TP73‐AS1) or its negative control (sh‐NC), and were then subcutaneously injected into the right flanks of BALB/c nude mice (n = 6). (a) Tumor volume was measured every week after inoculation, and tumor growth curve was drawn. ( ) sh‐NC and (
) sh‐NC and ( ) sh‐TP73‐AS1. (b) Tumor weight was recorded on the last week. (c–e) RT‐qPCR analysis testified the relative expression of TP73‐AS1, miR‐125a and MTDH in xenograft tumors. (f) Western blotting determined MTDH protein expression in randomly selected one xenograft tumor. Data represent mean ± SEM and *P < 0.05.
) sh‐TP73‐AS1. (b) Tumor weight was recorded on the last week. (c–e) RT‐qPCR analysis testified the relative expression of TP73‐AS1, miR‐125a and MTDH in xenograft tumors. (f) Western blotting determined MTDH protein expression in randomly selected one xenograft tumor. Data represent mean ± SEM and *P < 0.05.
Discussion
Breast tumorigenesis is a consequence of a combination of a number of molecular mechanisms including ceRNA network. Among these, lncRNAs‐associated ceRNA network play a crucial role in oncogenic pathways by sponging miRNAs. Several functional genes in breast cancer have been reviewed by Ergun et al. such as FOXO1, PTEN and CD44.25 The oncogene MTDH could modulate the apoptotic pathway, angiogenesis, growth and metastasis in breast cancer cells presumably through being targeted by miRNAs.26, 27, 28 Here, we attempted to determine a novel lncRNA and miRNA that would target MTDH and then regulate the malignant progression of breast cancer cells.
TP73‐AS1 is always upregulated in patients with varying cancer types and different cancer cell lines, and further participates in multiple cellular events (eg, cell proliferation, apoptosis, metastasis and drug resistance) to accelerate tumor progression through multiple mechanisms including serving as miRNA sponges.29 In this study, we noticed a higher expression of TP73‐AS1 in breast cancer tissues and cell lines (HCC‐70 and MB231) than that in paired adjacent normal tissues and normal breast cell line MCF‐10A. Functionally, knockdown of TP73‐AS1 was depicted to inhibit cell proliferative capacity, migration and invasive ability, but enhance cell apoptosis rate in HCC‐70 and MB231 cells through segregating miR‐125a, accompanied with lower level of Ki67, N‐cadherin and Vimentin, and higher cleaved caspase 3 level. Molecularly, miR‐125a was found to be negatively regulated by TP73‐AS1 via target binding. Combined with an inhibition of TP73‐AS1 knockdown on tumor growth of HCC‐70 cells in vivo, we propose a promising, novel TP73‐AS1/miR‐125a axis in breast cancer cells (Fig S2), suggesting that TP73‐AS1 might serve as an oncogene to modulate breast cancer cells behavior. Moreover, this finding had been reported in previous studies. For example, Yao et al.14 appointed TP73‐AS1 knockdown suppressed human DNA synthesis capability and cell viability of MDA‐MB‐231 and MCF‐7 cells in vitro by sponging miRNA‐200a via reducing TFAM expression. Meanwhile, these researchers also evaluated the effect of TP73‐AS1/miRNA‐200a axis on breast cancer migration and invasion.13 As a result, transwell and cell scratch assays determined a suppressive activity on cell migration and invasion when downregulating TP73‐AS1 by transfection of siRNA; ZEB1, a primary metastasis‐associated transcription factor, was targeted and regulated by miRNA‐200a, whose expression was sponged by TP73‐AS1. Similarly, miRNA‐490‐3p was another miRNA annotated to be targeted by TP73‐AS1.15 Furthermore, knockdown of TP73‐AS1/miRNA‐490‐3p axis led to vasculogenic mimicry formation inhibition in MDA‐MB‐231 cells through downregulating Twist1, which indicated a TP73‐AS1/miRNA‐490‐3p/Twist1 pathway in TNBC management. Taken together, TP73‐AS1 mediated miRNAs/mRNAs network contributes to breast tumorigenesis and development. According to data from Yao et al.,14 high expression of TP73‐AS1 was related to advanced TNM stage instead of ER status, PR status, lymph node micrometastases and tumor size; TP73‐AS1 high expression could predict a shorter overall survival. In addition, we observed a significant association between TP73‐AS1 expression and distant metastasis in clinic.
The concentration of miRNAs is an important factor for ceRNA activity. LncRNAs MALAT1 and NORAD were declared to compete miR‐125a and then took part in cancer cell proliferation and metastasis.30, 31 Here, we confirmed an interaction between TP73‐AS1 and miR‐125a (Fig S2). Functionally, overexpression of miR‐125a by transfection could decrease cell proliferation, migration and invasion, whereas increase cell apoptosis in HCC‐70 and MB231 cells. Mechanically, MTDH downregulation underlies the antitumor role of miR‐125a in breast cancer cells in vitro. Except for antiproliferation and antimetastasis, miR‐125a has been proven to function in other breast cancer progressions. For example, Zheng et al.32 stated that increasing the 3′ UTR of the pseudogene CYP4Z2P could introduce angiogenesis‐promoting properties in MCF‐7 and MDA‐MB‐231 cells through acting as a ceRNA for miRNAs including miR‐125a in regulating functional gene CYP4Z1. The drug‐resistance had also been demonstrated to be altered by miR‐125a dysregulation in breast cancer. For instance, Xu et al.33 discovered expression of miR‐125a was downregulated in docetaxel‐resistant clinical samples and MDA‐MB‐468 and MCF‐7 cells, and manipulation of miR‐125a inversely related to docetaxel resistance. Zheng et al.34 also revealed a lower expression level of miR‐125a in chemoresistant breast cells, and upregulation of miR‐125a sensitized the acquired resistant MCF‐7 cells to tamoxifen both in vitro and in vivo. Thereby, miR‐125a deregulation is widely spread in cell behaviors, angiogenesis and drug‐resistance in breast cancer, suggesting miR‐125a as a potential and promising target for breast cancer treatment. Furthermore, the essential role of miR‐125a in chemoresistance was probably mediated by lncRNAs sponging; however, this hypothesis should be further testified.
At present, the identification of circulating lncRNAs and miRNAs in breast cancer patients in order to diagnose and monitor therapeutic response is of great interest. However, this study did not investigate the expression of TP73‐AS1 and miR‐125a in serum and could be the focus of further research.
In conclusion, we showed TP73‐AS1 and MTDH expression were upregulated, and miR‐125a expression was downregulated in breast cancer tissues and cell lines. All three alterations with knockdown of TP73‐AS1, overexpression of miR‐125a and silencing of MTDH could suppress breast cancer cell proliferation, migration and invasion, but facilitate cell apoptosis in HCC‐70 and MB231 cells. Mechanically, TP73‐AS1 negatively regulated MTDH expression by functioning as a molecular “sponge” for miR‐125a. Therefore, we recommended a novel TP73‐AS1/miR‐125a/MTDH pathway in the malignant progression of breast cancer cells in vitro. Our results could provide a promising approach to progress the development of human breast cancer.
Disclosure
The authors declare that there are no competing interests associated with the manuscript.
Supporting information
Figure S1 The effect of TP73‐AS1 on miR‐125a expression, and miR‐125a on MTDH expression. (a) RT‐qPCR detected miR‐125a expression in HCC‐70 and MB231 cells cotransfected with miR‐125a mimic and either TP73‐AS1 or empty vectors. (b) Western blotting detected MTDH expression in HCC‐70 and MB231 cells cotransfected with miR‐125a mimic and either si‐MTDH and either anti‐miR‐125a or anticontrol. Data represent mean ± SEM and *P < 0.05.
Figure S2 The regulatory mechanism of TP73‐AS1 in breast cancer cells. TP73‐AS1 functioned as an oncogene in breast cancer through modulating TP73‐AS1/miR‐125a/MTDH pathway in cell proliferation, apoptosis, migration, invasion, and tumor growth.
Acknowledgment
None.
Contributor Information
Yuxiong Liu, Email: hnliuyuxiong@126.com.
Yanyan Han, Email: danlinghuatilpr@163.com.
References
- 1. Boyd NF, Martin LJ, Bronskill M, Yaffe MJ, Duric N, Minkin S. Breast tissue composition and susceptibility to breast cancer. J Natl Cancer Inst 2010; 102 (16): 1224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hammond ME, Hayes DF, Dowsett M et al American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 2010; 28 (16): 2784–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rakha EA, Ellis IO. Triple‐negative/basal‐like breast cancer: Review. Pathology 2009; 41 (1): 40–7. [DOI] [PubMed] [Google Scholar]
- 4. Samadi P, Saki S, Dermani FK, Pourjafar M, Saidijam M. Emerging ways to treat breast cancer: Will promises be met? Cell Oncol (Dordr) 2018; 41 (6): 605–21. [DOI] [PubMed] [Google Scholar]
- 5. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly‐Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol 2019; 234 (7): 10080–100. [DOI] [PubMed] [Google Scholar]
- 6. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci 2018; 19 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lo PK, Wolfson B, Zhou X, Duru N, Gernapudi R, Zhou Q. Noncoding RNAs in breast cancer. Brief Funct Genomics 2016; 15 (3): 200–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodriguez Bautista R, Ortega Gomez A, Hidalgo Miranda A et al Long non‐coding RNAs: Implications in targeted diagnoses, prognosis, and improved therapeutic strategies in human non‐ and triple‐negative breast cancer. Clin Epigenetics 2018; 10: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wong KY, Li Z, Zhang X, Leung GKK, Chan GCF, Chim CS. Epigenetic silencing of a long non‐coding RNA KIAA0495 in multiple myeloma. Mol Cancer 2015; 14: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mazor G, Levin L, Picard D et al The lncRNA TP73‐AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis 2019; 10 (3): 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L, Fang F, He X. Long noncoding RNA TP73‐AS1 promotes non‐small cell lung cancer progression by competitively sponging miR‐449a/EZH2. Biomed Pharmacother 2018; 104: 705–11. [DOI] [PubMed] [Google Scholar]
- 12. Li S, Huang Y, Huang Y et al The long non‐coding RNA TP73‐AS1 modulates HCC cell proliferation through miR‐200a‐dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res 2017; 36 (1): 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zou Q, Zhou E, Xu F, Zhang D, Yi W, Yao J. A TP73‐AS1/miR‐200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J Cell Biochem 2018; 119 (2): 2189–99. [DOI] [PubMed] [Google Scholar]
- 14. Yao J, Xu F, Zhang D et al TP73‐AS1 promotes breast cancer cell proliferation through miR‐200a‐mediated TFAM inhibition. J Cell Biochem 2018; 119 (1): 680–90. [DOI] [PubMed] [Google Scholar]
- 15. Tao W, Sun W, Zhu H, Zhang J. Knockdown of long non‐coding RNA TP73‐AS1 suppresses triple negative breast cancer cell vasculogenic mimicry by targeting miR‐490‐3p/TWIST1 axis. Biochem Biophys Res Commun 2018; 504 (4): 629–34. [DOI] [PubMed] [Google Scholar]
- 16. Jiang L, Huang Q, Zhang S et al Hsa‐miR‐125a‐3p and hsa‐miR‐125a‐5p are downregulated in non‐small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer 2010; 10: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hashiguchi Y, Nishida N, Mimori K et al Down‐regulation of miR‐125a‐3p in human gastric cancer and its clinicopathological significance. Int J Oncol 2012; 40 (5): 1477–82. [DOI] [PubMed] [Google Scholar]
- 18. Huang B, Luo W, Sun L et al MiRNA‐125a‐3p is a negative regulator of the RhoA‐actomyosin pathway in A549 cells. Int J Oncol 2013; 42 (5): 1734–42. [DOI] [PubMed] [Google Scholar]
- 19. Nakano M, Fukami T, Gotoh S, Nakajima M. A‐to‐I RNA editing up‐regulates human dihydrofolate reductase in breast cancer. J Biol Chem 2017; 292 (12): 4873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dhiman G, Srivastava N, Goyal M et al Metadherin: A therapeutic target in multiple cancers. Front Oncol 2019; 9: 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell 2004; 5 (4): 365–74. [DOI] [PubMed] [Google Scholar]
- 22. Hu G, Chong RA, Yang Q et al MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor‐prognosis breast cancer. Cancer Cell 2009; 15 (1): 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li J, Zhang N, Song LB et al Astrocyte elevated gene‐1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res 2008; 14 (11): 3319–26. [DOI] [PubMed] [Google Scholar]
- 24. Zang W, Wang T, Wang Y et al Knockdown of long non‐coding RNA TP73‐AS1 inhibits cell proliferation and induces apoptosis in esophageal squamous cell carcinoma. Oncotarget 2016; 7 (15): 19960–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ergun S, Oztuzcu S. Oncocers: ceRNA‐mediated cross‐talk by sponging miRNAs in oncogenic pathways. Tumour Biol 2015; 36 (5): 3129–36. [DOI] [PubMed] [Google Scholar]
- 26. Liu P, Tang H, Chen B et al miR‐26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett 2015; 357 (1): 384–92. [DOI] [PubMed] [Google Scholar]
- 27. Liu Y, Kong X, Li X, Li B, Yang Q. Knockdown of metadherin inhibits angiogenesis in breast cancer. Int J Oncol 2015; 46 (6): 2459–66. [DOI] [PubMed] [Google Scholar]
- 28. Zhang N, Wang X, Huo Q et al The oncogene metadherin modulates the apoptotic pathway based on the tumor necrosis factor superfamily member TRAIL (Tumor Necrosis Factor‐related Apoptosis‐inducing Ligand) in breast cancer. J Biol Chem 2013; 288 (13): 9396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chu F, Xue L, Miao H. Long noncoding rna tp73‐as1 in human cancers. Clin Chim Acta 2019; Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 30. Li H, Wang X, Wen C et al Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia‐induced epithelial‐mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer 2017; 16 (1): 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan L, Zhang J, Guo D, Ma J, Shui SF, Han XW. IL‐21R functions as an oncogenic factor and is regulated by the lncRNA MALAT1/miR‐125a‐3p axis in gastric cancer. Int J Oncol 2019; 54 (1): 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng L, Li X, Gu Y, Lv X, Xi T. The 3'UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res Treat 2015; 150 (1): 105–18. [DOI] [PubMed] [Google Scholar]
- 33. Xu X, Lv YG, Yan CY, Yi J, Ling R. Enforced expression of hsa‐miR‐125a‐3p in breast cancer cells potentiates docetaxel sensitivity via modulation of BRCA1 signaling. Biochem Biophys Res Commun 2016; 479 (4): 893–900. [DOI] [PubMed] [Google Scholar]
- 34. Zheng L, Meng X, Li X et al miR‐125a‐3p inhibits ERalpha transactivation and overrides tamoxifen resistance by targeting CDK3 in estrogen receptor‐positive breast cancer. FASEB J 2018; 32 (2): 588–600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The effect of TP73‐AS1 on miR‐125a expression, and miR‐125a on MTDH expression. (a) RT‐qPCR detected miR‐125a expression in HCC‐70 and MB231 cells cotransfected with miR‐125a mimic and either TP73‐AS1 or empty vectors. (b) Western blotting detected MTDH expression in HCC‐70 and MB231 cells cotransfected with miR‐125a mimic and either si‐MTDH and either anti‐miR‐125a or anticontrol. Data represent mean ± SEM and *P < 0.05.
Figure S2 The regulatory mechanism of TP73‐AS1 in breast cancer cells. TP73‐AS1 functioned as an oncogene in breast cancer through modulating TP73‐AS1/miR‐125a/MTDH pathway in cell proliferation, apoptosis, migration, invasion, and tumor growth.


