Abstract
Background
Non‐small cell lung cancer (NSCLC) is an intractable malignant lung cancer with high rates of metastasis and mortality. Currently, long noncoding RNA nuclear RNA host gene 7 (SNHG7) is recognized as a biomarker of multiple cancers. However, the role of SNHG7 in NSCLC requires further understanding.
Methods
The expression of SNHG7, miR‐449a and TGIF2 in NSCLC tumors and cells was examined by quantitative real time polymerase chain reaction (qRT‐PCR). Cell viability was measured by MTT assay. Cell migration and invasion was conducted using transwell assay. Protein expression of TGIF2, vimentin, N‐cadherin and E‐cadherin was detected by western blot. The interaction between miR‐449a and SNHG7 or TGIF2 was determined by luciferase reporter system, RIP and RNA pull‐down assay, respectively. Xenograft mice models were established by subcutaneously injecting A549 cells transfected with sh‐SNHG7 and sh‐control.
Results
SNHG7 expression was upregulated in NSCLC tumors and cells compared with normal tissues and cells. SNHG7 silencing repressed cell proliferation, migration, invasion and epithelial to mesenchymal transition (EMT) in NSCLC. Consistently, SNHG7 knockdown hindered tumor growth in vivo. The subsequent luciferase reporter system, RIP and RNA pull‐down assay validated the interaction between miR‐449a and SNHG7 or TGIF2. The rescue experiments displayed that miR‐449a inhibitor counteracted SNHG7 silencing induced inhibition on proliferation, migration, invasion and EMT. Similarly, restoration of TGIF2 reversed miR‐449a mediated inhibition on cell progression. In addition, the results indicated that SNHG7 could regulate cell progression by targeting miR‐449a/TGIF2 axis.
Conclusion
SNHG7 contributed to cell proliferation, migration, invasion and EMT in NSCLC by upregulating TGIF2 via sponging miR‐449a, representing a novel targeted therapy method for NSCLC.
Keywords: miR‐449a, NSCLC, progression, SNHG7, TGIF2
Background
Non‐small cell lung cancer (NSCLC) is an intractable malignant lung cancer and is ranked as the leading cause of cancer related deaths globally.1, 2 NSCLC is characterized by multistage progression, distant metastasis and high mortality.3 Therapeutic outcomes vary along with the clinical stages of NSCLC: five‐year survival rate for patients at stage I + II is higher than 75% whereas lower than 15% for stage III + IV.4, 5 A delayed diagnosis or distant metastasis in NSCLC make it inadequate for surgical resection.6 Therefore, investigation of the molecular mechanism of NSCLC tumorigenesis and progression is urgently required.
Long noncoding RNAs (lncRNAs) are fundamental regulatory molecules in multiple diseases which participate in genomic imprinting, chromatin dynamics regulation, cell cycle and apoptosis.7, 8, 9 Nuclear RNA host gene 7 (SNHG7) with 984 base pair (bp) in length is a novel oncogene mapped in chromosome 9q34.3 and exerts a promotive function in a variety of cancers, such as bladder, gastric cancer and NSCLC.10, 11, 12 For example, SNHG7 was reported to contribute to cell proliferation and induced cell cycle arrest by upregulating STAT2 though sponging miR‐653‐5p in neuroblastoma.13 Consistently, SNHG7 acted as a sponger of miR‐34a to facilitate tumor progression of breast cancer by inducing epithelial mesenchymal transition (EMT) via Notch‐1 pathway.14 Overexpression of SNHG7 also contributed to cell growth in osteosarcoma by interacting with p53/DNMT1.15 By contrast, loss of SNHG7 attenuated cell viability, EMT, migration and invasion in prostate cancer by binding to miR‐324‐3p and altering WNT2B expression.16 However, the underlying molecular mechanism of SNHG7 in NSCLC cell progression requires in‐depth exploration.
MicroRNAs (miRNAs), critical regulators in many diseases, are involved in cell metabolism, survival, differentiation, autophagy and apoptosis by targeting the corresponding messenger RNA (mRNA) to initiate gene alteration, mRNA degradation and protein translation blockage.17, 18, 19 Ectopic expression of miR‐449a is considered an important factor in many different diseases, such as arthritis and cancer.20, 21 As a tumor suppressor, miR‐449a has been reported to repress EMT, migration and invasion of NSCLC by interacting with ADAM10.22 In addition, in the study by Hu et al. miR‐449a was found to be closely associated with cancer cell chemosensitivity; for example, overexpression of miR‐449a improved cisplatin mediated cytotoxicity in gastric adenocarcinoma cancer by regulating cyclin D1.23 Likewise, miR‐449a alleviated drug resistance of tamoxifen against breast cancer via regulation of ADAM22.24 Nevertheless, how miR‐449a affects NSCLC development requires further understanding.
Transforming growth factor‐beta‐induced 2 (TGIF2) has been recognized as an effective regulator in many cancer types. For example, in the study by Flum et al. TGIF2 was a target of miR‐129‐5p to reduce glioma cell cycle, growth, migration and invasion.25 Consistently, miR‐34 has been reported to restrain cell growth and stimulate cell apoptosis by interacting with the target TGIF2 in osteosarcoma.26 Therefore, we assume that TGIF2 acts as a target of miR‐449a to regulate NSCLC cell progression.
In our study, we attempted to illuminate the molecular mechanism of gene expression during NSCLC tumorigenesis and progression. The interaction among SNHG7, miR‐449a and TGIF2 were evaluated by luciferase reporter system, RIP and RNA pull‐down assay. The regulatory effects of SNHG7, miR‐449a and TGIF2 on NSCLC cell proliferation, migration, invasion and EMT were investigated by MTT, transwell and western blot assay.
Methods
Tissue samples
A total of 42 NSCLC patients were recruited from Yantai Yuhuangding Hospital. The participants gave their signed informed consent and claimed no previous family history. NSCLC tumors and normal tissues were obtained by surgery from the patients in our study. Our protocols were approved by Ethics Committee of Yantai Yuhuangding Hospital.
Cell transfection
A549, H1299 cells and human bronchial epithelial cells BEAS‐2B were purchased from ATCC (Manassas, VA, USA). The cells were cultured in DMEM medium supplemented with 10% FBS and 0.05% penicillin/streptomycin. Small interfering RNA (siRNA) targeting SNHG7 (si‐SNHG7), siRNA control (si‐control), SNHG7 overexpression vector (SNHG7), TGIF2 overexpression vector (TGIF2) were synthesized by Genepharma (Shanghai, China). MiR‐449a mimics, miR‐449a inhibitor (anti‐miR‐449a), miRNA control (miR‐control) and miRNA control inhibitor (anti‐miR‐control) were purchased from RIBOBIO (Guangzhou, China). The vectors were transfected in A549 and H1299 cells using Lipofectamine 2000 (11668019, Invitrogen, Carlsbad, CA, USA).
Quantitative real‐time polymerase chain reaction (qRT‐PCR)
The tissues and cells were incubated with TRIzol reagent (15596018, Invitrogen) to obtain total RNA. The cDNA for SNHG7 and TGIF2 was synthesized by All‐in‐One First‐Strand cDNA Synthesis Kit (AORT‐0050, FulenGen, Guangzhou, China). qRT‐PCR was then performed using SYBR green (A25742, Applied Biosystems, Foster City, CA, USA). GAPDH and U6 were exploited as internal reference. The primers for SNHG7, miR‐449a and TGIF2 were listed: SNHG7, (Forward, 5′‐GCCCTGCAGCCTCGC‐3′; Reverse, 5′‐CAGCGGCGCCTCCTC‐3′); miR‐449a, (Forward, 5′‐TGGCAGTGTATTGTTA‐3′; Reverse, 5′‐ATCCAGTGCAGGGTCCGAGG‐3′); TGIF2 (Forward, 5′‐TCTCTGTGTTGCCTCCCTCT‐3′; Reverse, 5′‐CCACCTCAGCCCAATACACT‐3′); GAPDH, (Forward, 5′‐AGGTCGGTGTGAACGGATTTG‐3′; Reverse, 5′‐GGGGTCGTTGATGGCAACA‐3′); U6, (Forward, 5′‐ACCCTGAGAAATACCCTCACAT‐3′; Reverse, 5′‐GACGACTGAGCCCCTGATG‐3′).
MTT assay
Transfected A549 and H1299 cells were placed on 96‐well plates for 24, 48 and 72 hours. After that, 10 μL MTT (C0009, Beyotime, Shanghai, China) was added to each well for four hours to react with the cells. After rinsing them with PBS, 100 μL DMSO was added to each well for two hours, and the optical density (OD) value (450 nm) was then measured with a spectrophotometer.
Transwell assay
For cell migration detection, transfected A549 and H1299 cells were placed in the upper chamber of transwell for 24 hours. Subsequently, migrated cells in the lower chamber were stained with 0.1% crystal violet (C0775, Sigma, St. Louis, MO, USA). With regard to the invasion assay, the upper chamber of transwell was precoated with Matrigel for four hours. The following experiments was carried out in accordance with migration assay. Finally, the migration and invasion rate were counted using a microscope.
Western blot
Western blot was carried out as previously reported.27 The primary antibodies against TGIF2 (ab155948), vimentin (ab8069), N‐cadherin (ab18203) and E‐cadherin (ab194982) were purchased from Abcam (Cambridge, MA, USA) and HRP‐conjugated secondary antibody was obtained from Sangon (Shanghai, China).
Luciferase reporter assay
Wild‐type (SNHG7 WT, TGIF2 3′UTR WT) and mutant type (SNHG7 MUT, TGIF2 3′UTR MUT) luciferase vectors were constructed. Those vectors were cotransfected with miR‐449a or miR‐control in A549 and H1299 cells. Luciferase activities were determined by dual‐luciferase assay. The primers for SNHG7 WT and TGIF2 3′UTR WT were listed: SNHG7 WT, (Forward, 5′‐CCCAAGCTTCTCTGCGTGCGCCGGAGGCT‐3′; Reverse, 5′‐CCCGGGAACGTTTTCCATTCAAGACCAATTTA‐3′); TGIF2 3′UTR WT (Forward, 5′‐CCCAAGCTTGGGTGGCGAATTCGGCACGA‐3′; Reverse, 5′‐CATGCCATGGTGGTAAACCAAACCAATGATAAA‐3′);
RNA immunoprecipitation (RIP) assay
EZ‐Magna RIP RNA‐binding protein immunoprecipitation kit (17–701, Millipore, Billerica, MA, USA) was exploited for Ago2 RIP assay. In brief, A549 and H1299 cells were lysed by RIP buffer and centrifuged at 10 000 g for five minutes. The cell lysis was then incubated with magnetic beads coated with anti‐Ago2 or IgG antibody.28 The enrichment of SNHG7 was analyzed by qRT‐PCR.
RNA pull‐down assay
Biotinylated miR‐449a (Bio‐miR‐449a), Input‐miR‐449a, Input negative control (Input‐NC) and biotinylated negative control (Bio‐NC) (Santa Cruz Biotechnology, Dallas, Texas, USA) were transfected into A549 and H1299 cells. The cells were then incubated with Dynabeads M‐280 Streptavidin (60 210, Invitrogen) for 10 minutes. Finally, SNHG7 level was measured by qRT‐PCR.
Murine xenograft assay
Female nude mice (n = 6) age five weeks were purchased from Shanghai Laboratory Animals Center (Shanghai, China). Xenograft mice models were established by subcutaneously injecting A549 cells transfected with sh‐SNHG7 and sh‐control. After 28 days measurement of tumor volume, tumor tissues were collected from the mice. All the animal experiment protocols were approved by the Animal Ethics Committee of Yantai Yuhuangding Hospital.
Statistical analysis
Data are presented as means ± standard deviation (SD). Statistical analysis was performed by SPSS software and GraphPad Prism 7. The correlation between miR‐449a and SNHG7 or TGIF2 was analyzed by Pearson's correlation coefficient. A P‐value less than 0.05 (P < 0.05) was considered statistically significant.
Results
Overexpression of SNHG7 in NSCLC
The functional role of SNHG7 in NSCLC was initially evaluated by qRT‐PCR. The correlations between SNHG7 expression and clinical characteristics in NSCLC patients are shown in Table 1. We found that SNHG7 was overexpressed in NSCLC patients with these features: large tumor size, advanced stage and lymph node metastasis. As illustrated in Fig 1a, SNHG7 expression was dramatically upregulated in NSCLC tumors compared with the corresponding normal tissues. Synchronously, SNHG7 expression was much higher in NSCLC cell lines (A549, H1299) than that of human bronchial epithelial cells BEAS‐2B (Fig 1b). Hence, these results suggest that SNHG7 might act as an oncogene in NSCLC.
Table 1.
Correlations between SNHG7 expression and clinical characteristics in patients with NSCLC
| SNHG7 expression | ||||
|---|---|---|---|---|
| Parameter | Case | Low (n = 20) | High (n = 22) | P‐value† |
| Age (years) | 0.303 | |||
| ≤60 | 26 | 14 | 12 | |
| >60 | 16 | 6 | 10 | |
| Sex | 0.204 | |||
| Female | 23 | 13 | 10 | |
| Male | 19 | 7 | 12 | |
| Smoking | 0.108 | |||
| No | 24 | 14 | 10 | |
| Yes | 18 | 6 | 12 | |
| Tumor size | 0.011* | |||
| ≤3 cm | 30 | 18 | 12 | |
| >3 cm | 12 | 2 | 10 | |
| TNM stages | 0.0007* | |||
| I–II | 20 | 15 | 5 | |
| III–IV | 22 | 5 | 17 | |
| Lymph node metastasis | 0.0009* | |||
| Negative | 27 | 18 | 9 | |
| Positive | 15 | 2 | 13 | |
P < 0.05.
Chi‐square test.
TNM, tumor‐node‐metastasis.
Figure 1.
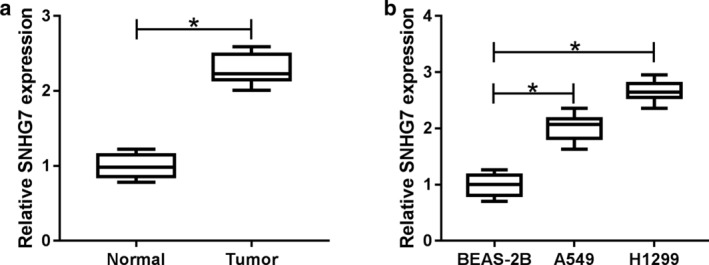
SNHG7 was upregulated in NSCLC tumors and cells. a SNHG7 expression in NSCLC tumors compared with normal tissues. b SNHG7 expression in NSCLC cell lines (A549, H1299) compared with human bronchial epithelial cells BEAS‐2B. *P < 0.05.
SNHG7 depletion inhibited proliferation, migration, invasion and EMT in NSCLC
Loss‐of‐function experiments were conducted by silencing SNHG7 to further investigate the function of SNHG7 in NSCLC. A great decline of SNHG7 expression was noticed in A549 and H1299 cells transfected with si‐SNHG7, indicating the transfection efficiency was relatively high (Fig 2a). The subsequent MTT results revealed that SNHG7 knockdown distinctly repressed NSCLC cell growth (Fig 2b,c). Consistently, cell migration and invasion were restrained after SNHG7 silencing compared with control groups (Fig 2d,e). The influences of SNHG7 on NSCLC cell EMT was examined by analyzing EMT associated protein (vimentin, N‐cadherin and E‐cadherin) expression using western blot. The results showed that the expression of vimentin and N‐cadherin was reduced whereas E‐cadherin was enhanced by SNHG7 silencing (Fig 2f,g). Altogether, SNHG7 knockdown inhibited proliferation, migration, invasion and EMT in NSCLC.
Figure 2.
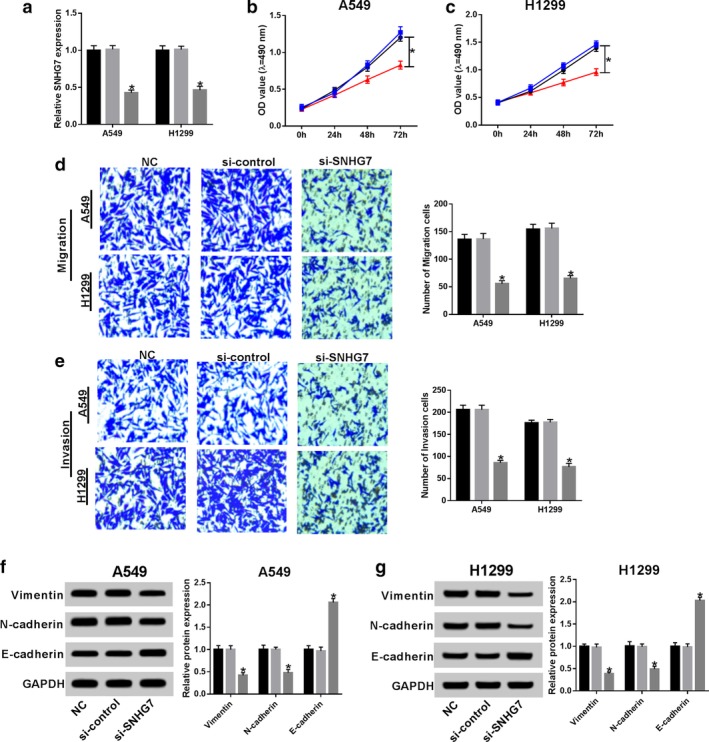
SNHG7 knockdown repressed proliferation, migration, invasion and EMT in NSCLC. A549 and H1299 cells were transfected with si‐SNHG7 and si‐control. (a) SNHG7 expression in transfected A549 and H1299 cells ( ) NC, (
) NC, ( ) si‐control, and (
) si‐control, and ( ) si‐SNHG7. Cell viability of (b) transfected A549 and (c) H1299 cells (
) si‐SNHG7. Cell viability of (b) transfected A549 and (c) H1299 cells ( ) NC, (
) NC, ( ) si‐control, and (
) si‐control, and ( ) si‐SNHG7 measured by MTT assay (
) si‐SNHG7 measured by MTT assay ( ) NC, (
) NC, ( ) si‐control, and (
) si‐control, and ( ) si‐SNHG7. (d) Cell migration and (e) invasion(
) si‐SNHG7. (d) Cell migration and (e) invasion( ) NC, (
) NC, ( ) si‐control, and (
) si‐control, and ( ) si‐SNHG7 (e) of transfected A549 and H1299 cells were examined by transwell assay(
) si‐SNHG7 (e) of transfected A549 and H1299 cells were examined by transwell assay( ) NC, (
) NC, ( ) si‐control, and (
) si‐control, and ( ) si‐SNHG7. Protein expression of vimentin, N‐cadherin and E‐cadherin in (f) transfected A549 and (g) H1299 cells (
) si‐SNHG7. Protein expression of vimentin, N‐cadherin and E‐cadherin in (f) transfected A549 and (g) H1299 cells ( ) NC, (
) NC, ( ) si‐control, and (
) si‐control, and ( ) si‐SNHG7 (evaluated by western blot (
) si‐SNHG7 (evaluated by western blot ( ) NC, (
) NC, ( ) si‐control, and (
) si‐control, and ( ) si‐SNHG7. GAPDH was used as internal reference. *P < 0.05.
) si‐SNHG7. GAPDH was used as internal reference. *P < 0.05.
SNHG7 was a sponger of miR‐449a
By searching the online prediction tool starBase, we discovered that miR‐449a contained the binding sites of SNHG7 (Fig 3a). Luciferase activity reduction in A549 and H1299 cells cotransfected with SNHG7 WT and miR‐449a certified the interaction between SNHG7 and miR‐449a (Fig 3b,c). As expected, SNHG7 and miR‐449a enrichment were markedly higher in A549 (Fig 3d,e) and H1299 (Fig 3g,h) cells which were bound with Ago2 antibody than that of IgG. Similarly, a considerable amount of SNHG7 was detected in Input‐miR‐449a group by RNA pull‐down assay compared with the control groups (Fig 3f,i). Furthermore, miR‐449a expression was boosted by SNHG7 silencing while inhibited by overexpression of SNHG7 (Fig 3j). Meanwhile, miR‐449a expression in cells was elevated by miR‐449a transfection and blocked by miR‐449a inhibitor (Fig 3k). Pearson's correlation coefficient analysis revealed that SNHG7 was inversely correlated with miR‐449a (R2 = 0.891, P < 0.0001) (Fig 3l). The findings demonstrated that SNHG7 acted as a sponge‐ of miR‐449a in NSCLC.
Figure 3.
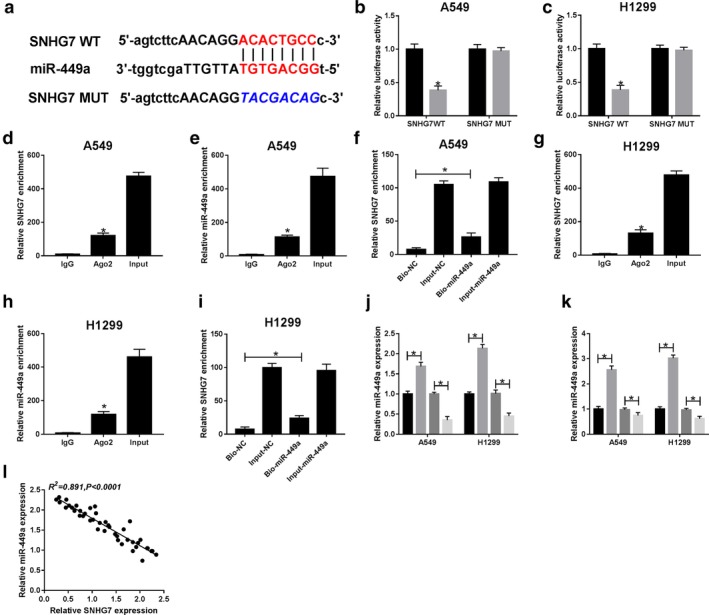
SNHG7 directly interacted with miR‐449a. (a) The putative binding sites between SNHG7 and miR‐449a. Luciferase activity of (b) A549 and (c) H1299 cells ( ) miR‐contol, and (
) miR‐contol, and ( ) miR‐449a cotransfected with SNHG7 WT or SNHG7 MUT and miR‐449a or miR‐control (
) miR‐449a cotransfected with SNHG7 WT or SNHG7 MUT and miR‐449a or miR‐control ( ) miR‐contol, and (
) miR‐contol, and ( ) miR‐449a. (d) SNHG7 and (e) miR‐449a enrichment in A549 cells were analyzed by RIP assay. SNHG7 enrichment in (f) A549 and (i) H1299 cells transfected with Bio‐NC, Input‐NC, Bio‐miR‐449a and Input‐miR‐449a was detected using RNA pull‐down assay. (g) SNHG7 and (h) miR‐449a enrichment in H1299 cells were analyzed by RIP assay. (j) The expression of miR‐449a in A549 and H1299 cells transfected with si‐control, si‐SNHG7, vector and SNHG7 (
) miR‐449a. (d) SNHG7 and (e) miR‐449a enrichment in A549 cells were analyzed by RIP assay. SNHG7 enrichment in (f) A549 and (i) H1299 cells transfected with Bio‐NC, Input‐NC, Bio‐miR‐449a and Input‐miR‐449a was detected using RNA pull‐down assay. (g) SNHG7 and (h) miR‐449a enrichment in H1299 cells were analyzed by RIP assay. (j) The expression of miR‐449a in A549 and H1299 cells transfected with si‐control, si‐SNHG7, vector and SNHG7 ( ) si‐control, (
) si‐control, ( ) si‐SNHG7, (
) si‐SNHG7, ( ) vector, and (
) vector, and ( ) SNHG7. (k) The expression of miR‐449a in A549 and H1299 cells transfected with miR‐NC, miR‐449a, anti‐miR‐NC and anti‐miR‐449a (
) SNHG7. (k) The expression of miR‐449a in A549 and H1299 cells transfected with miR‐NC, miR‐449a, anti‐miR‐NC and anti‐miR‐449a ( ) miR‐NC, (
) miR‐NC, ( ) miR‐449a, (
) miR‐449a, ( ) anti‐miR‐NC, and (
) anti‐miR‐NC, and ( ) anti‐miR‐449a. (l) The correlation between SNHG7 and miR‐449a (R2 = 0.891, P < 0.0001). *P < 0.05.
) anti‐miR‐449a. (l) The correlation between SNHG7 and miR‐449a (R2 = 0.891, P < 0.0001). *P < 0.05.
SNHG7 regulated cell proliferation, migration, invasion and EMT by targeting miR‐449a in NSCLC
Rescue experiments were conducted by transfecting si‐control, si‐SNHG7, si‐SNHG7 + anti‐miR‐control and si‐SNHG7 + anti‐miR‐449a in A549 and H1299 cells to disclose the association between SNHG7 and miR‐449a in NSCLC cell progression. As exhibited in Fig 4a,b, miR‐449a expression was increased after SNHG7 silencing and miR‐449a inhibitor reversed the trend. Cell viability results displayed that miR‐449a inhibitor attenuated SNHG7 silencing induced inhibition on NSCLC cell proliferation (Fig 4c,d). Consistently, cell migration and invasion were reduced in cells transfected with si‐SNHG7 and enhanced in si‐SNHG7 + anti‐miR‐449a transfection cells (Fig 4e,f). At the same time, miR‐449a inhibitor rescued SNHG7 silencing mediated suppression on vimentin, N‐cadherin expression and acceleration on E‐cadherin expression (Fig 4g,h). Collectively, SNHG7 contributed to cell proliferation, migration, invasion and EMT by regulating miR‐449a in NSCLC.
Figure 4.
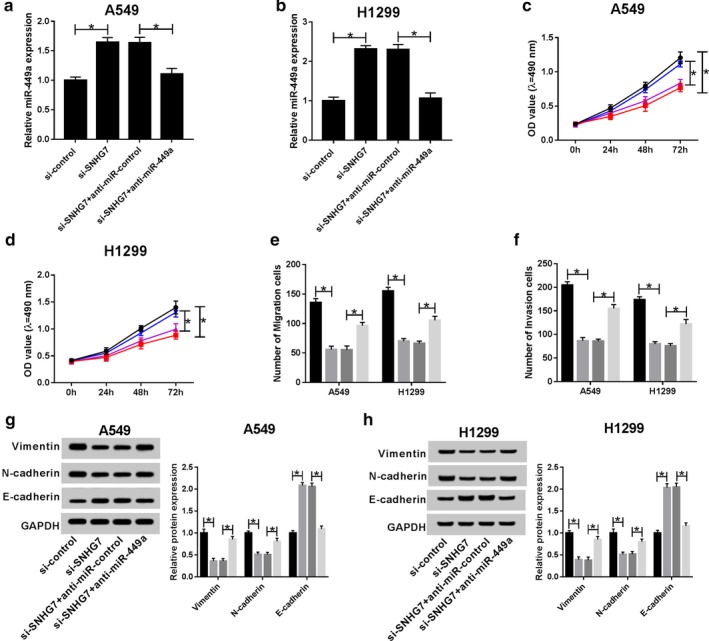
MiR‐449a inhibitor abrogated SNHG7 silencing induced inhibition on proliferation, migration, invasion and EMT in NSCLC. A549 and H1299 cells were transfected with si‐control, si‐SNHG7, si‐SNHG7 + anti‐miR‐control and si‐SNHG7 + anti‐miR‐449a. The expression of miR‐449a in (a) transfected A549 and (b) H1299 cells. Cell viability of (c) transfected A549 ( ) si‐control, (
) si‐control, ( ) si‐SNHG7, (
) si‐SNHG7, ( ) si‐SNHG7+anti‐miR‐control, and (
) si‐SNHG7+anti‐miR‐control, and ( ) si‐SNHG7+anti‐miR‐449a and (d) H1299 cells (
) si‐SNHG7+anti‐miR‐449a and (d) H1299 cells ( ) si‐control, (
) si‐control, ( ) si‐SNHG7, (
) si‐SNHG7, ( ) si‐SNHG7+anti‐miR‐control, and (
) si‐SNHG7+anti‐miR‐control, and ( ) si‐SNHG7+anti‐miR‐449a. (e) Cell migration (
) si‐SNHG7+anti‐miR‐449a. (e) Cell migration ( ) si‐control, (
) si‐control, ( ) si‐SNHG7, (
) si‐SNHG7, ( ) si‐SNHG7+anti‐miR‐control, and (
) si‐SNHG7+anti‐miR‐control, and ( ) si‐SNHG7+anti‐miR‐449a and (f) invasion of transfected A549 and H1299 cells (
) si‐SNHG7+anti‐miR‐449a and (f) invasion of transfected A549 and H1299 cells ( ) si‐control, (
) si‐control, ( ) si‐SNHG7, (
) si‐SNHG7, ( ) si‐SNHG7+anti‐miR‐control, and (
) si‐SNHG7+anti‐miR‐control, and ( ) si‐SNHG7+anti‐miR‐449a. Protein expression of vimentin, N‐cadherin and E‐cadherin in (g) transfected A549 (
) si‐SNHG7+anti‐miR‐449a. Protein expression of vimentin, N‐cadherin and E‐cadherin in (g) transfected A549 ( ) si‐control, (
) si‐control, ( ) si‐SNHG7, (
) si‐SNHG7, ( ) si‐SNHG7+anti‐miR‐control, and (
) si‐SNHG7+anti‐miR‐control, and ( ) si‐SNHG7+anti‐miR‐449a and (h) H1299 cells (
) si‐SNHG7+anti‐miR‐449a and (h) H1299 cells ( ) si‐control, (
) si‐control, ( ) si‐SNHG7, (
) si‐SNHG7, ( ) si‐SNHG7+anti‐miR‐control, and (
) si‐SNHG7+anti‐miR‐control, and ( ) si‐SNHG7+anti‐miR‐449a. GAPDH was used as internal reference. *P < 0.05.
) si‐SNHG7+anti‐miR‐449a. GAPDH was used as internal reference. *P < 0.05.
TGIF2 was a target of miR‐449a
Targetscan tool showed that miR‐449a could bind to 3’ untranslated regions (3′UTR) of TGIF2 (Fig 5a). Dual‐luciferase reporter assay was exploited to confirm the relationship between TGIF2 and miR‐449a and resulted in reduced luciferase activity in cells cotransfected with TGIF2 3′UTR WT and miR‐449a. However, luciferase activity in cells transfected with TGIF2 3′UTR MUT remained unchanged (Fig 5b,c). In addition, the expression of TGIF2 mRNA was repressed by miR‐449a and promoted by miR‐449a inhibitor (Fig 5d). As expected, the expression of TGIF2 protein showed the same trend (Fig 5e,f). We also discovered that TGIF2 was negatively correlated with miR‐449a (R2 = 0.739, P < 0.0001) (Fig 5g). All the data implicated that TGIF2 was a target of miR‐449a.
Figure 5.
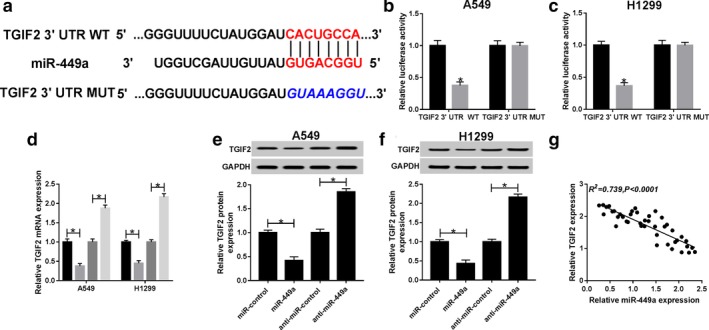
TGIF2 is a target of miR‐449a. (a) The putative binding sites between TGIF2 and miR‐449a predicted by Targetscan. Luciferase activity of (b) A549 ( ) miR‐control, and (
) miR‐control, and ( ) miR‐449a and (c) H1299 cells cotransfected with TGIF2 3′UTR WT or TGIF2 3′UTR MUT and miR‐449a or miR‐control (
) miR‐449a and (c) H1299 cells cotransfected with TGIF2 3′UTR WT or TGIF2 3′UTR MUT and miR‐449a or miR‐control ( ) miR‐control, and (
) miR‐control, and ( ) miR‐449a. (d) TGIF2 mRNA expression in A549 and H1299 cells transfected with miR‐control, miR‐449a, anti‐miR‐control and anti‐miR‐449a (
) miR‐449a. (d) TGIF2 mRNA expression in A549 and H1299 cells transfected with miR‐control, miR‐449a, anti‐miR‐control and anti‐miR‐449a ( ) miR‐control, (
) miR‐control, ( ) miR‐449a, (
) miR‐449a, ( ) anti‐miR‐control, and (
) anti‐miR‐control, and ( ) anti‐miR‐449a. TGIF2 protein expression (e) in A549 and (f) H1299 cells transfected with miR‐control, miR‐449a, anti‐miR‐control and anti‐miR‐449a. (g) The correlation between TGIF2 and miR‐449a (R2 = 0.739, P < 0.0001). *P < 0.05.
) anti‐miR‐449a. TGIF2 protein expression (e) in A549 and (f) H1299 cells transfected with miR‐control, miR‐449a, anti‐miR‐control and anti‐miR‐449a. (g) The correlation between TGIF2 and miR‐449a (R2 = 0.739, P < 0.0001). *P < 0.05.
Restoration of TGIF2 counteracted the suppression of miR‐449a on cell proliferation, migration, invasion and EMT in NSCLC
Given that TGIF2 was a target of miR‐449a, we hypothesized that miR‐449a modulates NSCLC cell progression by interacting with TGIF2. To certify the hypothesis, A549 and H1299 cells were transfected with miR‐control, miR‐449a, miR‐449a + vector and miR‐449a + TGIF2. Declined expression of TGIF2 mRNA was observed in miR‐449a transfection cells. However, expression of TGIF2 mRNA was enhanced by TGIF2 (Fig 6a,b). The expression of TGIF2 protein in A549 and H1299 cells exhibited the same trend (Fig 6c,d). Moreover, cell proliferation was retarded by miR‐449a and expedited by TGIF2 (Fig 6e,f). Similarly, restoration of TGIF2 attenuated the suppressive effects induced by miR‐449a on NSCLC cell migration and invasion (Fig 6g,h). Besides, deficiency of TGIF2 inhibited vimentin, N‐cadherin expression and boosted E‐cadherin expression. However, abundance of TGIF2 exerted the opposite effects (Fig 6i,j). Collectively, miR‐449a modulated NSCLC cell proliferation, migration, invasion and EMT by interacting with TGIF2.
Figure 6.
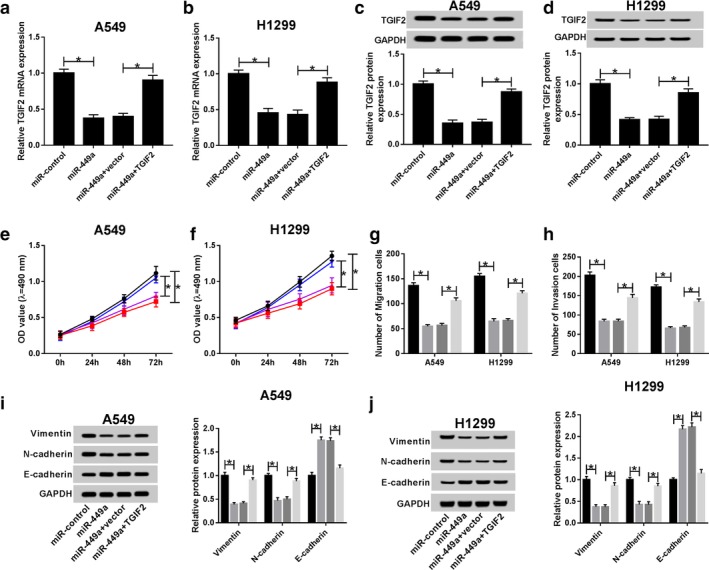
TGIF2 attenuated the suppression of miR‐449a on proliferation, migration, invasion and EMT in NSCLC. A549 and H1299 cells were transfected with miR‐control, miR‐449a, miR‐449a + vector and miR‐449a + TGIF2. TGIF2 mRNA expression in (a) transfected A549 and (b) H1299 cells. TGIF2 protein expression in (c) transfected A549 and (d) H1299 cells. GAPDH was used as internal reference. Cell viability of (e) transfected A549 ( ) miR‐control, (
) miR‐control, ( ) miR‐449a, (
) miR‐449a, ( ) miR‐449a+vector, and (
) miR‐449a+vector, and ( ) miR‐449a+TGIF2 and (f) H1299 cells (
) miR‐449a+TGIF2 and (f) H1299 cells ( ) miR‐control, (
) miR‐control, ( ) miR‐449a, (
) miR‐449a, ( ) miR‐449a+vector, and (
) miR‐449a+vector, and ( ) miR‐449a+TGIF2. (g) Cell migration (
) miR‐449a+TGIF2. (g) Cell migration ( ) miR‐control, (
) miR‐control, ( ) miR‐449a, (
) miR‐449a, ( ) miR‐449a+vector, and (
) miR‐449a+vector, and ( ) miR‐449a+TGIF2 and (h) invasion of transfected A549 and H1299 cells (
) miR‐449a+TGIF2 and (h) invasion of transfected A549 and H1299 cells ( ) miR‐control, (
) miR‐control, ( ) miR‐449a, (
) miR‐449a, ( ) miR‐449a+vector, and (
) miR‐449a+vector, and ( ) miR‐449a+TGIF2. Protein expression of vimentin, N‐cadherin and E‐cadherin in (i) transfected A549 (
) miR‐449a+TGIF2. Protein expression of vimentin, N‐cadherin and E‐cadherin in (i) transfected A549 ( ) miR‐control, (
) miR‐control, ( ) miR‐449a, (
) miR‐449a, ( ) miR‐449a+vector, and (
) miR‐449a+vector, and ( ) miR‐449a+TGIF2 and (j) H1299 cells (
) miR‐449a+TGIF2 and (j) H1299 cells ( ) miR‐control, (
) miR‐control, ( ) miR‐449a, (
) miR‐449a, ( ) miR‐449a+vector, and (
) miR‐449a+vector, and ( ) miR‐449a+TGIF2. GAPDH was used as internal reference. *P < 0.05.
) miR‐449a+TGIF2. GAPDH was used as internal reference. *P < 0.05.
Association of SNHG7, miR‐449a and TGIF2 in NSCLC
To illuminate the relationship of SNHG7, miR‐449a and TGIF2 during NSCLC cell progression, A549 and H1299 cells were transfected with si‐control, si‐SNHG7, si‐SNHG7 + anti‐miR‐control and si‐SNHG7 + anti‐miR‐449a. First, we analyzed the correlation between SNHG7 and TGIF2. The results indicated that there was a positive linear relationship between SNHG7 and TGIF2 (R2 = 0.908, P < 0.0001) (Fig 7a). In addition, TGIF2 mRNA expression was declined by SNHG7 silencing and elevated by miR‐449a inhibitor (Fig 7b,c). Meanwhile, miR‐449a inhibitor neutralized SNHG7 silencing mediated inhibition on TGIF2 protein production (Fig 7d,e). Therefore, we considered that SNHG7 could regulate TGIF2 expression by sponging miR‐449a in NSCLC.
Figure 7.
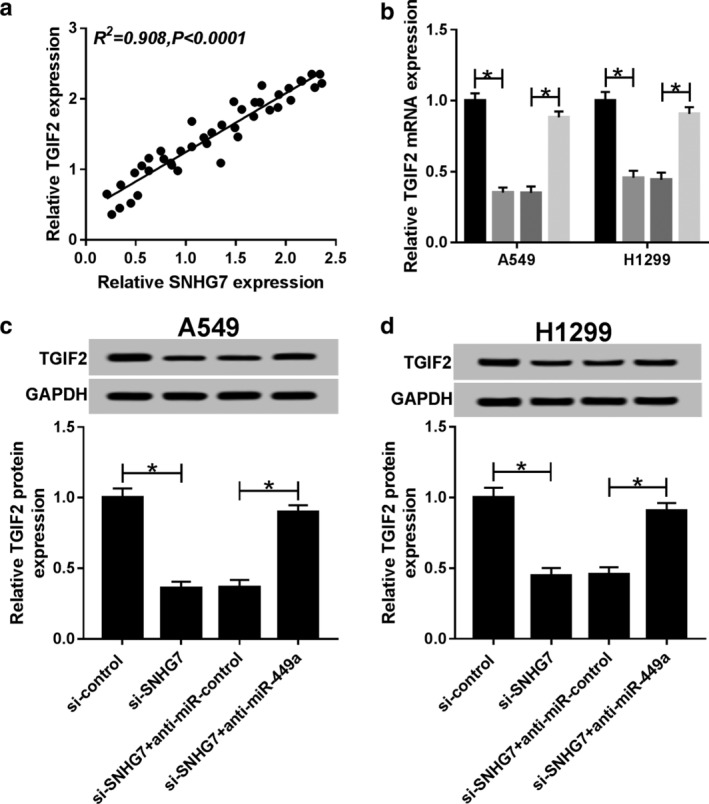
SNHG7 regulated TGIF2 expression by sponging miR‐449a in NSCLC. A549 and H1299 cells were transfected with si‐control, si‐SNHG7, si‐SNHG7 + anti‐miR‐control and si‐SNHG7 + anti‐miR‐449a. (a) The correlation between TGIF2 and SNHG7 (R2 = 0.908, P < 0.0001). (b) TGIF2 mRNA expression in transfected A549 and H1299 cells ( ) si‐control, (
) si‐control, ( ) si‐SNHG7, (
) si‐SNHG7, ( ) si‐SNHG7+anti‐miR‐control, and (
) si‐SNHG7+anti‐miR‐control, and ( ) si‐SNHG7+anti‐miR‐449a. TGIF2 protein expression in (c) transfected A549 and (d) H1299 cells. GAPDH was used as internal reference. *P < 0.05.
) si‐SNHG7+anti‐miR‐449a. TGIF2 protein expression in (c) transfected A549 and (d) H1299 cells. GAPDH was used as internal reference. *P < 0.05.
Interference of SNHG7 hindered tumor growth in vivo
The regulatory effects of SNHG7 on cell growth in vivo were determined by measuring tumor growth of SNHG7 low expression xenograft mice. As illustrated in Fig 8a, tumor growth was retarded significantly in sh‐SNHG7 transfection mice in comparison with sh‐control group. After 28 days measurement, tumor tissues were harvested when the mice were sacrificed. Tumor weight was dramatically lower in sh‐SNHG7 xenograft mice than that of sh‐control mice (Fig 8b). More importantly, SNHG7 expression was reduced whereas miR‐449a expression was enhanced in sh‐SNHG7 xenograft mice tumors (Fig 8c,d). Meanwhile, TGIF2 mRNA and protein expression was downregulated after SNHG7 knockdown in mice (Fig 8e,f). Thus, our findings implied that SNHG7 regulated NSCLC tumor growth in vivo by targeting miR‐449a/TGIF2 axis.
Figure 8.
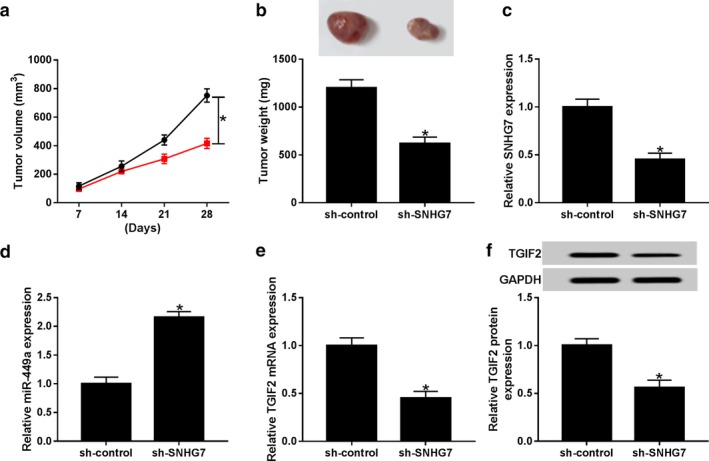
SNHG7 knockdown hindered tumor growth in vivo. (a) Tumor volume was measured every seven days ( ) sh‐control, and (
) sh‐control, and ( ) sh‐SNHG7. (b) Tumor weight was measured when the xenograft mice were sacrificed at day 28. (c) SNHG7 and (d) miR‐449a expression in tumors collected from the xenograft mice. The expression of (e) TGIF2 mRNA and (f) protein in tumors. *P < 0.05.
) sh‐SNHG7. (b) Tumor weight was measured when the xenograft mice were sacrificed at day 28. (c) SNHG7 and (d) miR‐449a expression in tumors collected from the xenograft mice. The expression of (e) TGIF2 mRNA and (f) protein in tumors. *P < 0.05.
Discussion
Growing evidence has identified that SNHG7 is implicated in tumorigenesis of multiple cancers and dysregulation of SNHG7 is the cause of various diseases.29 Until now, SNHG7 has been recognized as an essential biomarker in different cancers. For instance, abundance of SNHG7 expression has been reported to expedite cell cycle and proliferation in prostate cancer via absorbing miR‐503 to enhance cyclin D1.30 In addition, in the study by Sun et al. SNHG7 was upregulated in hepatocellular carcinoma and promoted cell metastasis process through blocking RBM5 expression. 31 SNHG7‐miR‐342‐3p‐ID4 feedback loop has also been reported to accelerate tumor growth in vitro and in vivo in pancreatic cancer.32 Similarly, in the report by Li et al. it was found that upregulation of SNHG7 could accelerate cell cycle, proliferation as well as restrain apoptosis in colorectal cancer via targeting miR‐34a/GALNT7 axis through activation of PI3K/Akt/mTOR pathway.33 On the contrary, deficiency of SNHG7 led to decreased cell proliferation and increased apoptosis in thyroid cancer by blocking BDNF.34 However, the exact role of SNHG7 during NSCLC progression is unclear.
Previous studies have clarified that miR‐449a was a significant biomarker and promising therapeutic target for cancer.35 For instance, miR‐449a expression was reduced in breast cancer as well as NSCLC and miR‐449a retarded cell viability, migration and invasion in those cancer types by regulating the PLAGL2 and NF‐kB pathway, respectively.36, 37 Interestingly, a reduced expression of miR‐449a contributed to growth and metastasis of prostate cancer cells by targeting PrLZ.38 Moreover, antitumor compounds ailanthone and neferine were capable of cascading the production of miR‐449a, thereby suppressing cell growth in acute myeloid leukemia and gastrointestinal stromal cancer.39, 40 In addition, Zhang et al. demonstrated that upregulation of miR‐449a strengthened prostate cancer cell radiosensitivity in relation to ionizing radiation and enhanced apoptosis by targeting the pRb/E2F1 pathway.41 Therefore, we suggest that SNHG7 could interact with miR‐449a to modulate NSCLC progression.
We initially discovered that the expression of SNHG7 was considerably higher in NSCLC tumors and cells than that of normal tissues and cells. Silencing of SNHG7 showed negative effects on cell viability, migration, invasion and EMT. Consistently, animal experiments exhibited that SNHG7 knockdown restricted tumor growth significantly in vivo. Luciferase reporter system, RIP and RNA pull‐down assay confirmed the interaction between miR‐449a and SNHG7 or TGIF2. The rescue experiments clarified that miR‐449a inhibitor abrogated SNHG7 silencing induced inhibition on proliferation, migration, invasion and EMT in NSCLC. Similarly, restoration of TGIF2 inversed miR‐449a mediated inhibition on cell proliferation, migration, invasion and EMT. Moreover, we observed that SNHG7 could regulate TGIF2 expression by sponging miR‐449a.
In summary, our study demonstrated that SNHG7 was able to regulate cell progression in NSCLC by targeting miR‐449a/TGIF2 axis. In contrast, depletion of SNHG7 inhibited tumor growth in vitro and in vivo. Therefore, our study discloses the underlying mechanism of NSCLC progression and provides novel biomarkers for NSCLC diagnosis.
Disclosure
The authors have no conflict of interest to declare.
Supporting information
Appendix S1. Supporting information.
Acknowledgments
Not applicable.
[Correction added on 13 December 2019, after first online publication: The first author's name has been corrected from ‘Pang Lingling’ to ‘Lingling Pang’.]
References
- 1. Keskin S, Kutluk AC, Tas F. Prognostic and predictive role of angiogenic markers in non‐ small cell lung cancer. Asian Pac J Cancer Prev 2019; 20 (3): 733–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gantenbein N, Bernhart E, Anders I et al Influence of eukaryotic translation initiation factor 6 on non‐small cell lung cancer development and progression. Eur J Cancer 2018; 101: 165–80. [DOI] [PubMed] [Google Scholar]
- 3. Li W, Li N, Kang X, Shi K. Circulating long non‐coding RNA AFAP1‐AS1 is a potential diagnostic biomarker for non‐small cell lung cancer. Clin Chim Acta 2017; 475: 152–6. [DOI] [PubMed] [Google Scholar]
- 4. Tian S. Identification of monotonically differentially expressed genes for non‐small cell lung cancer. BMC Bioinformatics 2019; 20 (1): 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shin JY, Yoon JK, Marwaha G. Progress in the treatment and outcomes for early‐stage non‐small cell lung cancer. Lung 2018; 196 (3): 351–8. [DOI] [PubMed] [Google Scholar]
- 6. da Silva GT, Bergmann A, Thuler LCS. Incidence and risk factors for bone metastasis in non‐small cell lung cancer. Asian Pac J Cancer Prev 2019; 20 (1): 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu F, Dong P, Mao Y, Zhao B, Huang Z, Zheng J. Loss of lncRNA‐SNHG7 promotes the suppression of hepatic stellate cell activation via miR‐378a‐3p and DVL2. Mol Ther Nucleic Acids 2019; 17: 235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun X, Xin Y, Wang M et al Overexpression of long non‐coding RNA KCNQ1OT1 is related to good prognosis via inhibiting cell proliferation in non‐small cell lung cancer. Thorac Cancer 2018; 9 (5): 523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shu T, He L, Wang X et al Long noncoding RNA UCA1 promotes chondrogenic differentiation of human bone marrow mesenchymal stem cells via miRNA‐145‐5p/SMAD5 and miRNA‐124‐3p/SMAD4 axis. Biochem Biophys Res Commun 2019; 514 (1): 316–22. [DOI] [PubMed] [Google Scholar]
- 10. Wang MW, Liu J, Liu Q et al LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci 2017; 21 (20): 4613–22. [PubMed] [Google Scholar]
- 11. She K, Yan H, Huang J, Zhou H, He J. miR‐193b availability is antagonized by LncRNA‐SNHG7 for FAIM2‐induced tumour progression in non‐small cell lung cancer. Cell Prolif 2018; 51 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhong X, Long Z, Wu S, Xiao M, Hu W. LncRNA‐SNHG7 regulates proliferation, apoptosis and invasion of bladder cancer cells assurance guidelines. J BUON 2018; 23 (3): 776–81. [PubMed] [Google Scholar]
- 13. Chi R, Chen X, Liu M et al Role of SNHG7‐miR‐653‐5p‐STAT2 feedback loop in regulating neuroblastoma progression. J Cell Physiol 2019; 234 (8): 13403–12. [DOI] [PubMed] [Google Scholar]
- 14. Sun X, Huang T, Liu Z, Sun M, Luo S. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR‐34a through EMT initiation and the Notch‐1 pathway. Eur J Pharmacol 2019; 856: 172407. [DOI] [PubMed] [Google Scholar]
- 15. Zhang GD, Gai PZ, Liao GY, Li Y. LncRNA SNHG7 participates in osteosarcoma progression by down‐regulating p53 via binding to DNMT1. Eur Rev Med Pharmacol Sci 2019; 23: 3602–10. [DOI] [PubMed] [Google Scholar]
- 16. Han Y, Hu H, Zhou J. Knockdown of LncRNA SNHG7 inhibited epithelial‐mesenchymal transition in prostate cancer though miR‐324‐3p/WNT2B axis in vitro. Pathol Res Pract 2019; 215: 152537. [DOI] [PubMed] [Google Scholar]
- 17. Baek D, Lee KM, Park KW e a. Inhibition of miR‐449a promotes cartilage regeneration and prevents progression of osteoarthritis in in vivo rat models. Mol Ther Nucleic Acids 2018; 13: 322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhong Z, Dong Z, Yang L, Chen X, Gong Z. MicroRNA‐31‐5p modulates cell cycle by targeting human mutL homolog 1 in human cancer cells. Tumour Biol 2013; 34 (3): 1959–65. [DOI] [PubMed] [Google Scholar]
- 19. Yang X, Qu X, Meng X e a. MiR‐490‐3p inhibits osteogenic differentiation in thoracic ligamentum flavum cells by targeting FOXO1. Int J Biol Sci 2018; 14 (11): 1457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cai Y, Jiang C, Zhu J et al miR‐449a inhibits cell proliferation, migration, and inflammation by regulating high‐mobility group box protein 1 and forms a mutual inhibition loop with yin Yang 1 in rheumatoid arthritis fibroblast‐like synoviocytes. Arthritis Res Ther 2019; 21 (1): 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L, Liu H, Du L et al miR‐449a suppresses LDHA‐mediated glycolysis to enhance the sensitivity of non‐small cell lung cancer cells to ionizing radiation. Oncol Res 2018; 26 (4): 547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meng H, Huang Q, Zhang X, Huang J, Shen R, Zhang B. MiR‐449a regulates the cell migration and invasion of human non‐small cell lung carcinoma by targeting ADAM10. Onco Targets Ther 2019; 12: 3829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu J, Fang Y, Cao Y, Qin R, Chen Q. miR‐449a regulates proliferation and chemosensitivity to cisplatin by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig Dis Sci 2014; 59 (2): 336–45. [DOI] [PubMed] [Google Scholar]
- 24. Li J, Lu M, Jin J, Lu X, Xu T, Jin S. miR‐449a suppresses Tamoxifen resistance in human breast cancer cells by targeting ADAM22. Cell Physiol Biochem 2018; 50 (1): 136–49. [DOI] [PubMed] [Google Scholar]
- 25. Flum M, Kleemann M, Schneider H et al miR‐217‐5p induces apoptosis by directly targeting PRKCI, BAG3, ITGAV and MAPK1 in colorectal cancer cells. J Cell Commun Signal 2018; 12 (2): 451–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang W, Tian Y, Dong S et al The long non‐coding RNA SNHG3 functions as a competing endogenous RNA to promote malignant development of colorectal cancer. Oncol Rep 2017; 38 (3): 1402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diao Y, Jin B, Huang L, Zhou W. MiR‐129‐5p inhibits glioma cell progression in vitro and in vivo by targeting TGIF2. J Cell Mol Med 2018; 22 (4): 2357–67. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Xi L, Zhang Y, Kong S, Liang W. miR‐34 inhibits growth and promotes apoptosis of osteosarcoma in nude mice through targetly regulating TGIF2 expression. Biosci Rep 2018; 38 (3): BSR20180078. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Chen Y, Peng Y, Xu Z et al Knockdown of lncRNA SNHG7 inhibited cell proliferation and migration in bladder cancer through activating Wnt/beta‐catenin pathway. Pathol Res Pract 2019; 215 (2): 302–7. [DOI] [PubMed] [Google Scholar]
- 30. Qi H, Wen B, Wu Q et al Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR‐503. Biomed Pharmacother 2018; 102: 326–32. [DOI] [PubMed] [Google Scholar]
- 31. Sun BZ, Ji DG, Feng ZX, Wang Y. Long noncoding RNA SNHG7 represses the expression of RBM5 to strengthen metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 2019; 23: 5699–704. [DOI] [PubMed] [Google Scholar]
- 32. Cheng D, Fan J, Ma Y et al LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR‐342‐3p. Cell Biosci 2019; 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Li Y, Zeng C, Hu J et al Long non‐coding RNA‐SNHG7 acts as a target of miR‐34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol 2018; 11 (1): 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang YH, Huo BL, Li C, Ma G, Cao W. Knockdown of long noncoding RNA SNHG7 inhibits the proliferation and promotes apoptosis of thyroid cancer cells by downregulating BDNF. Eur Rev Med Pharmacol Sci 2019; 23: 4815–21. [DOI] [PubMed] [Google Scholar]
- 35. Yong‐Ming H, Ai‐Jun J, Xiao‐Yue X, Jian‐Wei L, Chen Y, Ye C. miR‐449a: A potential therapeutic agent for cancer. Anticancer Drugs 2017; 28 (10): 1067–78. [DOI] [PubMed] [Google Scholar]
- 36. Wu D, Liu J, Chen J, He H, Ma H, Lv X. miR‐449a suppresses tumor growth, migration, and invasion in non‐small cell lung cancer by targeting a HMGB1‐mediated NF‐kappaB signaling pathway. Oncol Res 2019; 27 (2): 227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu B, Zhang X, Wang S, Shi B. MiR‐449a suppresses cell migration and invasion by targeting PLAGL2 in breast cancer. Pathol Res Pract 2018; 214 (5): 790–5. [DOI] [PubMed] [Google Scholar]
- 38. Chen W, Liu Y, Chen H, Ning H, Ding K. Loss of miR‐449a‐caused PrLZ overexpression promotes prostate cancer metastasis. Int J Oncol 2017; 51 (2): 435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, Zhang C, Min D. Ailanthone up‐regulates miR‐449a to restrain acute myeloid leukemia cells growth, migration and invasion. Exp Mol Pathol 2019; 108: 114–20. [DOI] [PubMed] [Google Scholar]
- 40. Xue F, Liu Z, Xu J, Xu X, Chen X, Tian F. Neferine inhibits growth and migration of gastrointestinal stromal tumor cell line GIST‐T1 by up‐regulation of miR‐449a. Biomed Pharmacother 2019; 109: 1951–9. [DOI] [PubMed] [Google Scholar]
- 41. Mao A, Liu Y, Wang Y et al miR‐449a enhances radiosensitivity through modulating pRb/E2F1 in prostate cancer cells. Tumour Biol 2016; 37 (4): 4831–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting information.


