Abstract
Background
Non‐small‐cell lung cancer (NSCLC) is the most lethal type of cancer. Long non‐coding RNAs (lncRNAs) and microRNAs (miRNAs) have been identified as crucial regulators in the development of NSCLC. The aim of our study was to explore the molecular mechanism of SNHG1 to enable better treatment for NSCLC patients.
Methods
Quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed to detect the expression of Small nucleolar RNA host gene 1 (SNHG1), miR‐361‐3p and frequently rearranged in advanced T‐cell lymphomas 1 (FRAT1). The protein level of FRAT1 was measured by western blot assay. Cell proliferation was evaluated by methyl thiazolyl tetrazolium (MTT) assay. Cell apoptosis was assessed by flow cytometry assay. The number of migrated and invaded cells were counted by transwell assay. The relationship between miR‐361‐3p and SNHG1 or FRAT1 was confirmed by dual‐luciferase reporter assay.
Results
Our results indicated that SNHG1 and FRAT1 were highly expressed in NSCLC tissues and cells. SNHG1 silencing inhibited proliferation, induced apoptosis and blocked migration and invasion of NSCLC cells. Also, FRAT1 downregulation suppressed proliferation, promoted apoptosis and hindered migration and invasion of NSCLC cells. Further, FRAT1 could recover the effects of SNHG1 silencing on proliferation, apoptosis, migration and invasion of NSCLC cells. SNHG1 sponged miR‐361‐3p and negatively regulated miR‐361‐3p expression. Meanwhile, miR‐361‐3p targeted FRAT1 and inversely modulated FRAT1 expression. In addition, miR‐361‐3p inhibition abated the effect of SNHG1 knockdown on FRAT1 expression.
Conclusion
In conclusion, LncRNA SNHG1 promoted the proliferation, repressed apoptosis and enhanced migration and invasion of NSCLC cells by regulating FRAT1 expression via sponging miR‐361‐3p.
Keywords: FRAT1, miR‐361‐3p, NSCLC, SNHG1
Introduction
As the leading cause of cancer‐related deaths, lung cancer remains the greatest problem to global public health.1 Non‐small‐cell lung cancer (NSCLC) accounts for approximately 85% of all patients with lung cancer worldwide.2 Until recently, due to a lack of therapeutic targets for NSCLC, patients are usually diagnosed at an advanced stage of disease when they are illeligible to receive the standard treatment.3 Although there have been new therapeutic methods applied in clinical settings, the overall survival rate of NSCLC patients remains at 15%.4 Therefore, a better understanding of the underlying pathogenesis of NSCLC is urgently required in order to develop an effective therapeutic strategy to improve the survival rate of NSCLC patients.
Long noncoding RNAs (lncRNAs) play important roles in cancer and have been widely reported in the literature. They are transcripts longer than 200 nucleotides that are not translated into proteins. They act as important regulators in many human cancers, such as prostate cancer, bladder cancer and cervical cancer.5, 6, 7 Small nucleolar RNA host gene 1 (SNHG1) located at the region of chromosome 11 has been identified to be associated with the occurrence of multiple human cancers, including NSCLC.8, 9, 10 Zhang et al. discovered that SNHG1 might be an oncogene which promotes cell metastasis in lung squamous cell carcinoma.11 Also, Lu et al. proved the oncogenic role of SNHG1 in lung cancer, and found that SNHG1 sponged miR‐145‐5p to enhance cell progression in NSCLC.12 However, more research on the mechanism of SNHG1 is necessary to fully understand the role of SNHG1 in NSCLC.
Since the first microRNA (miRNA) was discovered in 1993, the research on the miRNA has attracted the attention of many scientists worldwide.13, 14 MiRNAs are small RNAs which exert their functions by targeting the 3′‐untranslated region (3′‐UTR) of mRNAs. MiRNAs can regulate a variety of cellular processes and signaling pathways.15, 16 Emerging studies have reported that the aberrant expression of miRNAs is related to the initiation and progression of NSCLC.17, 18 Furthermore, a previous study has indicated that miR‐361‐3p is involved in the regulation of several human cancers, including NSCLC.19 Chen et al. found that the oncogene circRNA 100 146 promoted cell progression of NSCLC by targeting miR‐361‐3p.20 However, the mechanism of SNHG1 regulating miR‐361‐3p in NSCLC remains unclear.
Frequently rearranged in advanced T‐cell lymphomas 1 (FRAT1) is transcribed from chromosome 10, and has been demonstrated to play a promotional role in the regulation of Wnt/β‐catenin signaling.21 several studies have determined that FRAT1 participates in the cell progression of diverse human cancers. For example, Fan et al. reported that FRAT1 knockdown hindered hypoxia‐induced cell migration and invasion through suppression of the Wnt/β‐catenin pathway in hepatocellular carcinoma.22 However, the role of FRAT1 and the regulatory network of SNHG1/miR‐361‐3p/FRAT1 axis need to be confirmed.
In this study, we explored the expression and role of SNHG1 in NSCLC and discovered the underlying mechanism of SNHG1/miR‐361‐3p/FRAT1 axis in the development of NSCLC. These results indicate that SNHG1 might serve as a potential therapeutic target for the treatment of NSCLC.
Methods
Patients and cell culture
All patients (n = 40) were recruited from the hospital of The First People's Hospital of Lianyungang. Prior to surgical resection, all patients had completed signed informed written consent for inclusion into the study. The procedure was conducted in our hospital and tissues immediately stored at −8°C following surgery. The sample tissue was located at least 5 cm away from the NSCLC site and were defined as normal. All experiments and protocols were approved by the Ethics Committee of The First People's Hospital of Lianyungang. The clinicopathological characteristics and SNHG1 expression in NSCLC patients are shown in Table 1.
Table 1.
The clinicopathological characteristics and lncRNA SNHG1 expression in NSCLC patients
| Parameters | Group | Total | LncRNA SNHG1 | P‐value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Gender | Male | 22 | 12 | 10 | 0.267 |
| Female | 18 | 8 | 10 | ||
| Age (years) | ≥55 | 24 | 11 | 13 | 0.512 |
| <55 | 16 | 9 | 7 | ||
| Tumor size (cm) | <3 cm | 14 | 9 | 5 | 0.051 |
| ≥3 cm | 26 | 11 | 15 | ||
| Histology | Adenoma | 17 | 9 | 8 | 0.241 |
| Squamous | 23 | 11 | 12 | ||
| Differentiation | Well | 13 | 8 | 5 | 0.082 |
| Moderate‐poor | 27 | 12 | 15 | ||
| TNM stage | I | 15 | 10 | 5 | 0.003* |
| II–III | 25 | 10 | 15 | ||
| Lymph node metastasis | Absence | 23 | 14 | 9 | 0.004* |
| Presence | 17 | 6 | 11 | ||
P < 0.05.
Human bronchial epithelial cell line (BEAS‐2B) and NSCLC cell line (H23) were provided by American Tissue Culture Collection (ATCC, Manassas, VA, USA). The H1299 cell line was purchased from the West China Hospital of Sichuan University (Chengdu, China). All cells were plated on Dulbecco modified Eagle medium (DMEM; Gibco, Carlsbad, CA, USA), which was added to 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA). The plates were placed in an incubator at 37°C
Cell transfection
Small interference RNA against SNHG1 and FRAT (si‐SNHG1 and si‐FRAT1), the negative control (si‐NC), miR‐361‐3p mimics (miR‐361‐3p), negative control mimics (miR‐NC), miR‐361‐3p inhibitor (anti‐miR‐361‐3p) and its negative control (anti‐miR‐NC) were synthesized by Genepharma Co., Ltd. (Shanghai, China). The empty vector (pcDNA) and FRAT1 overexpression vector (FRAT1) were obtained from RIBOBIO Co., Ltd. (Guangzhou, China). Lipofectamine 2000 transfection reagent, purchased from Invitrogen (Carlsbad, CA, USA), was then used to transfect all oligonucleotides and vectors into H23 and H1299 cells. The transfected cells were incubated at 37°C to enable analysis.
Quantitative real‐time polymerase chain reaction (qRT‐PCR)
NSCLC tissues and normal tissues were digested with trypsin and were then added to a TRIzol reagent (Invitrogen). NSCLC cells were harvested and mixed with TRIzol reagent based on the standard protocol. The quality of total RNA was detected by measuring the OD value (A260/A280) using Nanodrop 2000 (Thermo Scientific; Waltham, MA, USA). Subsequently, two reverse transcription kits, All‐in‐OneTM miRNA First stand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA) and M‐MLV reverse transcriptase kit (Invitrogen), were selected for expression detection. SYBR green (Applied Biosystems, Foster City, CA, USA) was used to perform the qRT‐PCR experiment. U6 was regarded as the internal control of miRNAs, and glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was used as the internal control of lncRNAs and mRNAs. All qRT‐PCR data was normalized using the 2‐ΔΔCt method. All primers were obtained from Sangon Biotech Co., Ltd. (Shanghai, China), and the primers sequences for SNHG1, miR‐361‐3p, FRAT1, GAPDH and U6 are listed as follows: SNHG1, 5′‐CCGCTCGAGATTTAGGTGACACTATAGAAGTTCTCATTTTTCTACTGCTCG‐3′ (Forward) and 5′‐ATAGTTTAGCGGCCGCTTTTTTTTTTTTTTTTATGTAATCAATCATTTTAT‐3′ (Reverse); miR‐361‐3p, 5′‐ACACTCCAGCTGGGTCCCCCAGGTGTGATTC‐3′ (Forward) and 5′‐CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAATCAGA‐3′ (Reverse); FRAT1, 5′‐GCCCTGTCTAAAGTGTATTTTCAG‐3′ (Forward) and 5′‐CGCTTGAGTAGGACTGCAGAG‐3′ (Reverse); GAPDH, 5′‐CGGAGTCAACGGATTTGGTCGT‐3′ (Forward) and 5′‐GGGAAGGATCTGTCTCTGACC‐3′ (Reverse); U6, 5′‐CTCGCTTCGGCAGCACA‐3′ (Forward), 5′‐AACGCTTCA CGAATTTGCGT‐3′ (Reverse).
Western blot assay
Total protein extraction was performed by using a radioimmunoprecipitation assay buffer (RIPA; Beyotime, Shanghai, China). The extracted proteins were quantified by BCA protein assay kit (Beyotime). Equal amount of proteins were then loaded into each lane of freshly prepared sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel. After two hour‐electrophoresis, proteins in the gel were transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Subsequently, the blocking experiment was carried out with 5% nonfat milk for one hour incubation at room temperature. Next, the antibody I against FRAT1 (ab137391; 1/500; Abcam, Cambridge, MA, USA) or GAPDH (ab181602; 1/10000; Abcam) was added into membranes for overnight incubation at 4°C. The following day, the antibody II (ab6721; 1/2000; Abcam) was added into membranes for another two hours. GAPDH was considered to be the internal control. Finally, the proteins on the membranes were examined by Image Laboratory software (Bio‐Rad, Hercules, CA, USA).
Methyl thiazolyl tetrazolium (MTT) assay
Transfected H23 and H1299 cells were placed into 96‐well plates, and put into an incubator at 37°C with 5% CO2. After 24 hour, 48 hour, and 72 hour cultivation, all cells were harvested and interacted with 20 μL MTT solution (Sigma, St. Louis, MO, USA). Following another 4 hour incubation, blue‐violet radial crystallizations (formazan) were observed and dissolved with 100 μL Dimethyl sulfoxide (DMSO; Sigma). A microplate reader (Bio‐Rad) at 490 nm was employed to measure the absorbance.
Flow cytometry assay
After 48‐hour cultivation in an incubator, transfected H23 and H1299 cells were collected and resuspended with 1 × binding buffer. An Annexin V‐fluorescein isothiocyanate/propidium iodide (FITC/PI) apoptosis detection kit (Solarbio, Beijing, China) was selected to evaluate the apoptosis of H23 and H1299 cells. The cell suspension was then added to Annexin V‐FITC and PI solution for 10‐minutes incubation. A flow cytometer (BD Biosciences, San Jose, CA, USA) was then used to analyze the apoptotic cells.
Transwell assay
For cell migration detection, transfected H23 and H1299 cells were placed into the upper chambers (Coring, New York, NY, USA). DMEM medium containing serum was added to the lower chambers. The chambers were then maintained in an incubator at 37°C with 5% CO2. Cells in the upper layers of upper chambers were gently wiped with a cotton bud, and cells through the membranes were interacted with 0.5% crystal violet (Sigma). Finally, a microscope (Olympus, Tokyo, Japan) was used to detect the migrated cells with at least three random views.
For cell invasion detection, upper chambers were firstly covered with Matrigel (Becton Dickinson, Franklin Lakes, NJ, USA), and then transfected H23 and H1299 cells were seeded into upper chambers. The subsequent protocols were the same as that of cell migration detection. Finally, the invaded cells were counted with a microscope (Olympus) with at least three random fields.
Dual‐luciferase reporter assay
StarBase online tool was used to predict the potential targets of SNHG1 and miR‐361‐3p. The partial sequences of SNHG1 and FRAT1 3′‐UTR containing the specific binding site with miR‐361‐3p were then amplified and cloned into the downstream of pGL3 luciferase vector (Promega, Madison, WI, USA), called WT‐SNHG1 and FRAT1 3′‐UTR‐WT. The mutant sequences of SNHG1 and FRAT1 3′‐UTR were also designed and cloned into pGL3 vector, called MUT‐SNHG1 and FRAT1 3′‐UTR‐MUT. The fusion vectors described above were then cotransfected into H23 and H1299 cells with miR‐361‐3p or miR‐NC. The vector (pRL‐TK) (Promega) expressing renilla luciferase was also transferred into H23 and H1299 cells as an internal reference to normalize firely luciferase activity. At 48 hours post‐transfection, the luciferase activities of H23 and H1299 cells were measured by luciferase reporter assay kit (Promega).
Statistical analysis
Statistical analysis was carried out using GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). All data is presented as mean ± standard deviation (SD) from three independent experiments. The differences between two groups were analyzed by student's t‐test, and one‐way analysis of variance (ANOVA) was performed to analyze the differences among more than two groups. A P‐value less than 0.05 was regarded as a statistically significant difference.
Results
Expression of SNHG1 and FRAT1 were increased in NSCLC tissues and cells
First, qRT‐PCR and western blot assays were performed to detect the expression of SNHG1 and FRAT1 in NSCLC tissues and cells. The results showed that SNHG1 expression was elevated in NSCLC tissues (n = 40) compared with normal tissues (n = 40) (Fig 1a). Also, SNHG1 expression was upregulated in H23 and H1299 cells relative to BEAS‐2B cells (Fig 1b). Similarly, mRNA and protein expression of FRAT1were greatly enhanced in OSCLC tissues (n = 40) in comparison with normal tissues (n = 40) (Fig 1c,d). In addition, a higher expression of FRAT1 was found in H23 and H1299 cells than in BEAS‐2B cells (Fig 1e,f). These results suggested that SNHG1 and FRAT1 were abnormally expressed in NSCLC tissues and cells, and they might be associated with the development of NSCLC.
Figure 1.
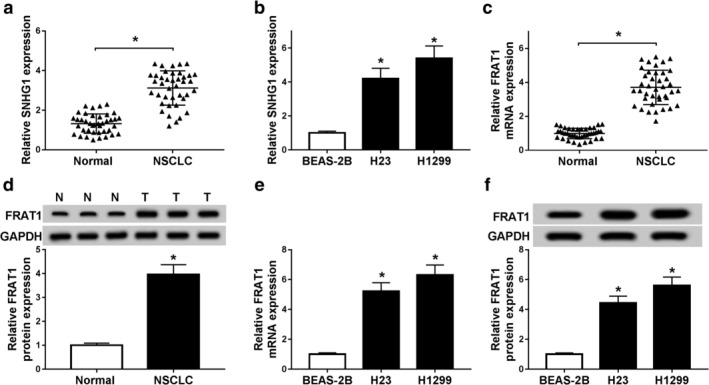
SNHG1 and FRAT1 expression were upregulated in NSCLC tissues and cells. (a) SNHG1 expression was detected by qRT‐PCR assay in NSCLC tissues (n = 40) and normal tissues (n = 40). (b) SNHG1 expression was measured by qRT‐PCR assay in BEAS‐2B, H23 and H1299 cells. (c) FRAT1 mRNA expression was examined by qRT‐PCR assay in NSCLC tissues and normal tissues. (d) FRAT1 protein level was detected by western blot assay in NSCLC tissues (n = 40) and normal tissues (n = 40). (e) FRAT1 expression was measured by qRT‐PCR assay in BEAS‐2B, H23 and H1299 cells. (f) FRAT1 protein expression was examined by western blot assay in BEAS‐2B, H23 and H1299 cells. *P < 0.05.
SNHG1 silencing repressed proliferation, induced apoptosis and suppressed migration and invasion of NSCLC cells
To explore the role of SNHG1 in NSCLC, qRT‐PCR assay was conducted to measure SNHG1 expression. The results determined that SNHG1 expression was sharply reduced in H23 and H1299 cells by SNHG1 knockdown (Fig 2a). MTT assay showed that the proliferation of H23 and H1299 cells was inhibited by SNHG1 silencing (Fig 2b,c). Further, flow cytometry assay indicated that silenced SNHG1 triggered the apoptosis of H23 and H1299 cells (Fig 2d). Also, both cell migration and invasion were blocked by transfection with si‐SNHG1 (Fig 2e,f). These data implied that SNHG1 knockdown hindered progression of NSCLC cells.
Figure 2.
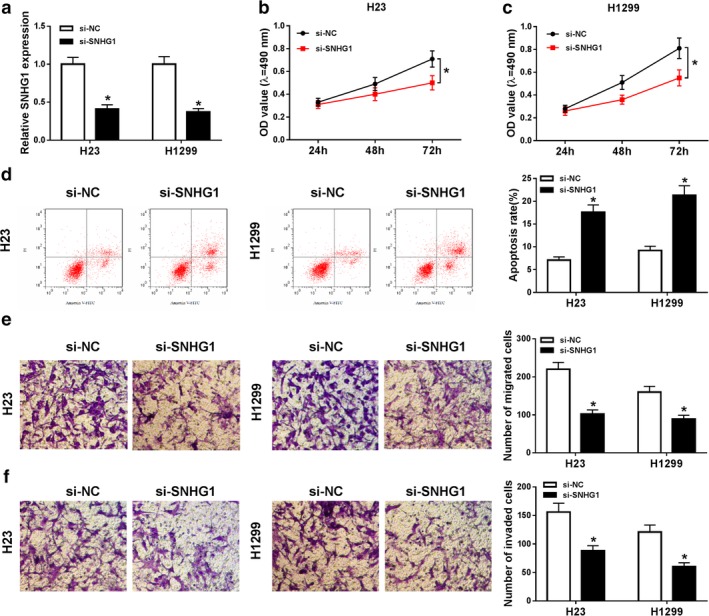
Silenced SNHG1 restrained the proliferation, induced apoptosis and hindered migration and invasion of NSCLC cells. (a) SNHG1 expression was measured by qRT‐PCR assay in H23 and H1299 cells transfected with si‐NC or si‐SNHG1. (b and c) The proliferation of transfected H23 and H1299 cells was detected by MTT assay. (d) The apoptosis of transfected H23 and H1299 cells was evaluated by flow cytometry assay. (e and f) The migration and invasion of transfected H23 and H1299 cells were assessed using transwell assay. *P < 0.05.
FRAT1 downregulation inhibited proliferation, enhanced apoptosis and impeded migration and invasion of NSCLC cells
To further explore the role of FRAT1 in NSCLC, H23 and H1299 cells were transfected with si‐FRAT1. QRT‐PCR and western blot assays confirmed that both mRNA and protein expression of FRAT1 were prominently decreased in H23 and H1299 cells by FRAT1 silencing (Fig 3a,b). MTT assay confirmed that the proliferation of H23 and H1299 cells was restrained by transfection with si‐FRAT1 (Fig 3c,d). The apoptosis of H23 and H1299 cells was also promoted by silenced FRAT1 (Fig 3e). In addition, the number of both migrated and invaded cells was reduced by FRAT1 knockdown (Fig 3f,g). All the above results demonstrated that FRAT1 knockdown suppressed progression of NSCLC cells.
Figure 3.
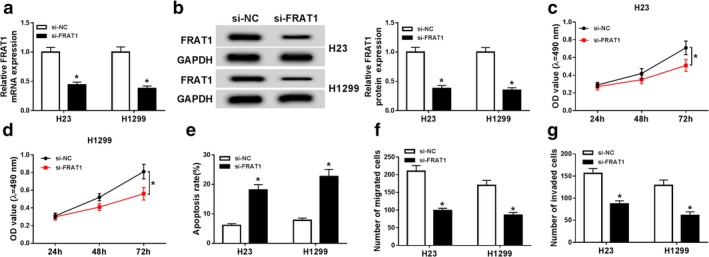
FRAT1 downregulation repressed proliferation, promoted apoptosis and suppressed migration and invasion of NSCLC cells. (a) FRAT1 mRNA expression was detected by qRT‐PCR assay in H23 and H1299 cells transfected with si‐NC or si‐FRAT1. (b) FRAT1 protein expression was measured by western blot assay in transfected H23 and H1299 cells. (c and d) The proliferation of transfected H23 and H1299 cells was evaluated by MTT assay. (e) The apoptosis of transfected H23 and H1299 cells was assessed by flow cytometry assay. (f and g) The migration and invasion of transfected H23 and H1299 cells were examined by transwell assay. *P < 0.05.
FRAT1 attenuated the effects of SNHG1 silencing in proliferation, apoptosis, migration and invasion of NSCLC cells
Based on the Pearson analysis, there was a positive correlation between SNHG1 expression and FRAT1 expression in NSCLC (Fig 4a). To illuminate the interaction between SNHG1 and FRAT1, we detected the expression of FRAT1 in si‐SNHG1 transfected NSCLC cells. QRT‐PCR and western blot assays revealed that both mRNA and protein levels of FRAT1 were largely reduced in H23 and H1299 cells transfected with si‐SNHG1 (Fig 4b,c). MTT assay then displayed that the proliferation of H23 and H1299 cells was repressed by SNHG1 silencing, and then FRAT1 overexpression promoted cell proliferation of H23 and H1299 cells transfected with si‐SNHG1 (Fig 4d,e). Moreover, the apoptosis of H23 and H1299 cells was enhanced by silenced SNHG1, which was abated by transfection with FRAT1 (Fig 4f,g). Transwell assay indicated that cell migration and invasion of H23 and H1299 cells were blocked by SNHG1 knockdown, while FRAT1 upregulation weakened the effects of SNHG1 knockdown (Fig 4h–k). In total, FRAT overexpression recovered SNHG1 silencing‐mediated antiproliferation, proapoptosis, antimigration and anti‐invasion effects of NSCLC cells.
Figure 4.
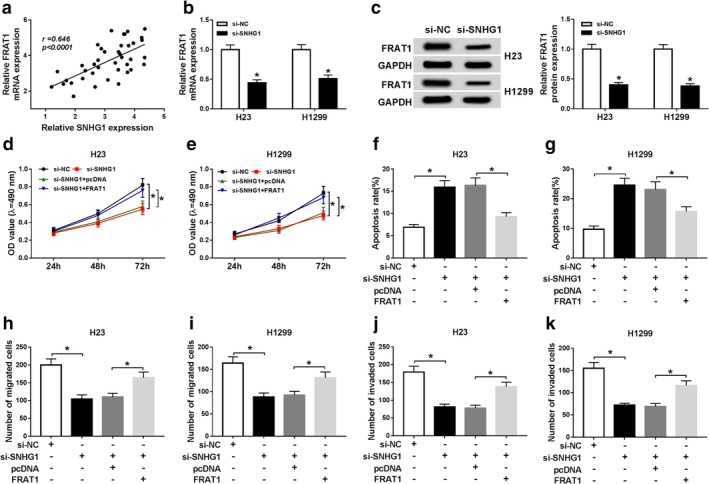
SNHG1 knockdown regulated proliferation, apoptosis, migration and invasion of NSCLC cells by modulating FRAT1 expression. (a) The correlation between SNHG1 expression and FRAT1 expression was determined by Pearson analysis. (b) FRAT1 mRNA expression was measured by qRT‐PCR assay in H23 and H1299 cells transfected with si‐NC or si‐SNHG1. (c) FRAT1 protein expression was detected by western blot assay in H23 and H1299 cells transfected with si‐NC or si‐SNHG1. (d and e) MTT assay was conducted to evaluate the proliferation of H23 and H1299 cells transfected with si‐NC, si‐SNHG1, si‐SNHG1 + pcDNA or si‐SNHG1 + FRAT1. (f and g) The apoptosis of transfected H23 and H1299 cells was assessed by flow cytometry assay. (h–j) The migration and invasion of transfected H23 and H1299 cells were examined by transwell assay. *P < 0.05.
SNHG1 sponged miR‐361‐3p and reversely regulated miR‐361‐3p expression
SNHG1 was predicted by the starBase online tool to search for its target. Interestingly, SNHG1 was able to bind to miR‐361‐3p (Fig 5a). Dual‐luciferase reporter assay was then performed to confirm the prediction. The results indicated that luciferase activities of H23 and H1299 cells were largely reduced by cotransfection with miR‐361‐3p and WT‐SNHG1, while there was no change of luciferase activities in MUT‐SNHG1 group (Fig 5b,c). Also, qRT‐PCR assay revealed that miR‐361‐3p expression was decreased in NSCLC tissues (n = 40) and cells (Fig 5d,e). Pearson analysis determined that SNHG1 expression was negatively correlated with miR‐361‐3p expression in NSCLC. Besides, miR‐361‐3p expression was elevated in H23 and H1299 cells transfected with si‐SNHG1 (Fig 5g). Totally, SNHG1 directly bound to miR‐361‐3p and negatively modulated miR‐361‐3p expression.
Figure 5.
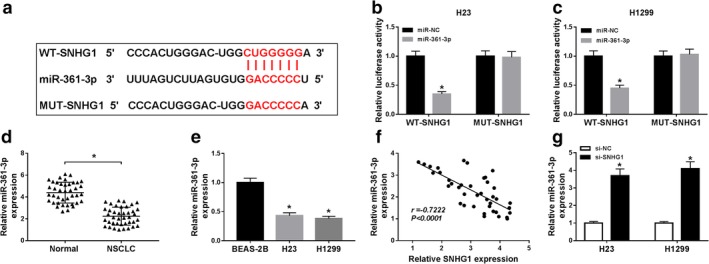
SNHG1 directly targeted miR‐361‐3p and reversely regulated miR‐361‐3p expression. (a) The binding sites between SNHG1 and miR‐361‐3p and the mutant sequences of SNHG1 were shown. (b and c) Dual‐luciferase reporter assay was conducted to detect the luciferase activities of H23 and H1299 cells transfected with miR‐NC or miR‐361‐3p and WT‐SNHG1 or MUT‐SNHG1. (d) The expression of miR‐361‐3p was measured by qRT‐PCR assay in NSCLC tissues (n = 40) and normal tissues (n = 40). (e) MiR‐361‐3p expression was examined by qRT‐PCR assay in BEAS‐2B, H23 and H1299 cells. (f) The correlation between SNHG1 expression and miR‐361‐3p expression was determined by Pearson analysis. (g) The expression of miR‐361‐3p was detected by qRT‐PCR assay in H23 and H1299 cells transfected with si‐NC or si‐SNHG1. *P < 0.05.
MiR‐361‐3p targeted FRAT1 and repressed FRAT1 expression
To determine the relationship between miR‐361‐3p and FRAT1, starBase online tool was utilized to predict the binding sites (Fig 6a). Dual‐luciferase reporter assay was carried out to verify their combination. The results demonstrated that miR‐361‐3p remarkably reduced luciferase activities of H23 and H1299 cells relative to miR‐NC in FRAT1 3′‐UTR‐WT group, while in FRAT1 3′‐UTR‐MUT group, luciferase activities remained unchanged when cells were transfected with miR‐NC or miR‐361‐3p (Fig 6b,c). Moreover, miR‐361‐3p expression was inversely correlated with FRAT1 expression (Fig 6d). QRT‐PCR assay showed that miR‐361‐3p mimics enhanced miR‐361‐3p expression, and miR‐361‐3p overexpression inhibited FRAT1 expression (Fig 6e,f). Western blot assay also indicated that FRAT1 protein expression was reduced by miR‐361‐3p overexpression (Fig 6g). In summary, miR‐361‐3p targetedly regulated FRAT1 expression.
Figure 6.
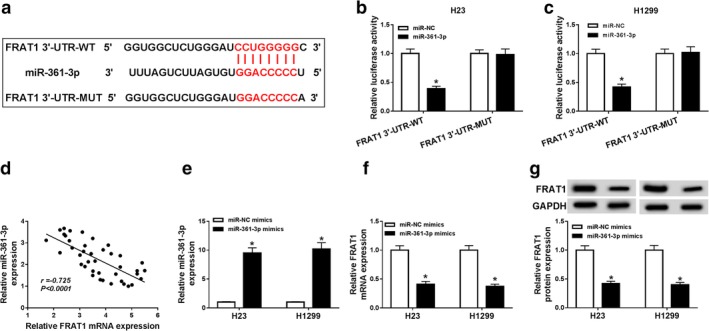
FRAT1 was a target of miR‐361‐3p. (a) The putative binding sequences between miR‐361‐3p and FRAT1 and the mutant sequences of FRAT1 were exhibited. (b and c) Dual‐luciferase reporter assay was conducted to detect the luciferase activities of H23 and H1299 cells transfected with miR‐NC or miR‐361‐3p and FRAT1 3′‐UTR‐WT or FRAT1 3′‐UTR‐MUT. (d) The correlation between miR‐361‐3p expression and FRAT1 expression was determined by Pearson analysis. (e and f) The expression levels of miR‐361‐3p and FRAT1 were measured by qRT‐PCR assay in H23 and H1299 cells transfected with miR‐NC mimics or miR‐361‐3p mimics. (g) FRAT1 protein expression was examined by western blot assay in transfected H23 and H1299 cells. *P < 0.05.
SNHG1 knockdown decreased FRAT1 expression by miR‐361‐3p
To elucidate the influence of SNHG1/miR‐361‐3p axis on FRAT1 expression, qRT‐PCR and western blot assays were performed to measure the expression of FRAT1. The results displayed that both mRNA and protein levels of FRAT1 were blocked by SNHG1 silencing in H23 and H1299 cells, which were reversed by transfection with anti‐miR‐361‐3p (Fig 7a–d). All the above data indicated that SNHG1 modulated FRAT1 expression by sponging miR‐361‐3p.
Figure 7.
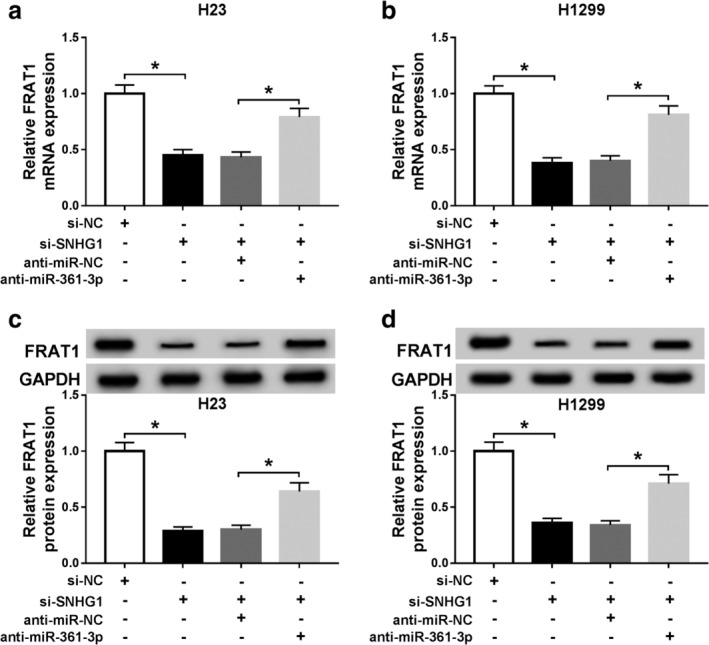
SNHG1 knockdown reduced FRAT1 expression by targeting miR‐361‐4p. (a and b) FRAT1 mRNA expression was detected by qRT‐PCR assay in H23 and H1299 cells transfected with si‐NC, si‐SNHG1, si‐SNHG1 + anti‐miR‐NC or si‐SNHG1 + anti‐miR‐361‐3p. (c and d) FRAT1 protein expression was measured by western blot assay in transfected H23 and H1299 cells. *P < 0.05.
Discussion
NSCLC is a common malignant tumor worldwide and a serious threat to human health. Patients with NSCLC have a very low survival rate because they usually have advanced disease at the time of diagnosis and there is a shortage of effective therapeutic targets. Many studies have shown that the dysregulation of lncRNAs is closely related to the occurrence and development of multiple human cancers, including NSCLC. Therefore, it is essential to investigate the molecular basis of lncRNAs to improve the survival rate of NSCLC patients.
Previous studies implicated that SNHG1 was upregulated in NSCLC tissues and cells, and SNHG1 knockdown inhibited NSCLC cell proliferation, migration and invasion.12, 23, 24 Consistent with these results, the expression of SNHG1 in NSCLC tissues was increased relative to that in normal tissues. Also, the higher expression of SNHG1 was found in NSCLC cells. These data implied that SNHG1 exerted vital roles in NSCLC development. Furthermore, in our study, we explored the functional effects of SNHG1 and found that SNHG1 silencing led to inhibition of cell proliferation, migration and invasion, and promotion of apoptosis in NSCLC cells. Therefore SNHG1 might play an oncogenic role in NSCLC development. It has been determined that lncRNAs functioned as competing endogenous RNAs (ceRNAs) to sponge miRNAs.25 To search for the downstream targets of SNHG1, we used the starBase website tool. Among these miRNAs, miR‐361‐3p attracted our attention because its expression and function had been mentioned in previous research. Moreover, dual‐luciferase reporter assay proved the relationship between SNHG1 and miR‐361‐3p. Interestingly, Chen et al. reported that miR‐361‐3p expression was decreased in NSCLC tissues and cells, and repressed NSCLC tumor cell proliferation and metastasis.26 In our study, the downregulation of miR‐361‐3p was found in NSCLC tissues and cells, which was in accordance with the previous study. In summary, SNHG1 sponged miR‐361‐3p to modulate the expression of miR‐361‐3p.
Further, the targets of miR‐361‐3p were predicted by starBase, and FRAT1 contained the binding sites of miR‐361‐3p. Meanwhile, this prediction was verified by dual‐luciferase reporter assay. Furthermore, the expression of FRAT1 was surprisingly negatively correlated with miR‐361‐3p expression. Therefore, FRAT1 was a target of miR‐361‐3p and used for further investigation. FRAT1 has been reported to be highly expressed in NSCLC, and FRAT1 downregulation restrained the invasion of NSCLC cells.27 In the present study, we also measured the expression of FRAT in NSCLC tissues and cells and discovered that FRAT1 expression was elevated in NSCLC tissues compared with that in nontumor tissues. Furthermore, silenced FRAT1 suppressed proliferation, facilitated apoptosis, and inhibited migration and invasion of NSCLC cells. The above data also supported the results of Zhang et al. and provided new evidence to elucidate the role of FRAT1 in NSCLC. SNHG1 and FRAT1 were highly expressed in NSCLC tissues and cells, while miR‐361‐3p was downregulated. SNHG1 downregulation inhibited proliferation, promoted apoptosis and repressed migration and invasion of NSCLC cells. Moreover, SNHG1 exerted its role in NSCLC cells by modulating FRAT1 expression via sponging miR‐361‐3p. However, all experiments in this study were conducted in vitro, and there is no in vivo data to support these results. Hence, further studies of SNHG1 in mice models need to be carried out. In summary, the results of our study highlights the existence of a novel regulatory mechanism of SNHG1 in NSCLC, which shows that SNHG1 might serve as a therapeutic target for the treatment of NSCLC.
Disclosure
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008; 359: 1367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Skjefstad K, Johannessen C, Grindstad T e a. A gender specific improved survival related to stromal miR‐143 and miR‐145 expression in non‐small cell lung cancer. Sci Rep 2018; 8: 8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoffman RM, Sanchez R. Lung cancer screening. Med Clin North Am 2017; 101: 769–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gu P, Chen X, Xie R e a. A novel AR translational regulator lncRNA LBCS inhibits castration resistance of prostate cancer. Mol Cancer 2019; 18: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma Q‐Y, Li S‐Y, Li X‐Z e a. Long non‐coding RNA DILC suppresses bladder cancer cells progression. Gene 2019; 710: 193–201. [DOI] [PubMed] [Google Scholar]
- 7. Song W, Wang J, Liu H e a. Effects of LncRNA Lnc‐LIF‐AS on cell proliferation, migration and invasion in a human cervical cancer cell line. Cytokine 2019; 120: 165–75. [DOI] [PubMed] [Google Scholar]
- 8. Zheng S, Li M, Miao K, Xu H. SNHG1 contributes to proliferation and invasion by regulating miR‐382 in breast cancer. Cancer Manag Res 2019; 11: 5589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang D, Wei Y, Zhu J, Wang F. Long non‐coding RNA SNHG1 functions as a competitive endogenous RNA to regulate PDCD4 expression by sponging miR‐195‐5p in hepatocellular carcinoma. Gene 2019; 714: 143994. [DOI] [PubMed] [Google Scholar]
- 10. You J, Fang N, Gu J e a. Noncoding RNA small nucleolar RNA host gene 1 promote cell proliferation in nonsmall cell lung cancer. Indian J Cancer 2014; 51 (Suppl. 3): e99–e102. [DOI] [PubMed] [Google Scholar]
- 11. Zhang H‐Y, Yang W, Zheng F‐S, Wang Y‐B, Lu J‐B. Long non‐coding RNA SNHG1 regulates zinc finger E‐box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in lung squamous cell carcinoma. Biomed Pharmacother 2017; 90: 650–8. [DOI] [PubMed] [Google Scholar]
- 12. Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non‐small cell lung cancer progression by up‐regulating MTDH via sponging miR‐145‐5p. FASEB J 2018; 32: 3957–67. [DOI] [PubMed] [Google Scholar]
- 13. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin‐14 by lin‐4 mediates temporal pattern formation in C. elegans . Cell 1993; 75: 855–62. [DOI] [PubMed] [Google Scholar]
- 14. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 1993; 75: 843–52. [DOI] [PubMed] [Google Scholar]
- 15. Bartel DP. Metazoan MicroRNAs. Cell 2018; 173: 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell 2012; 149: 515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z‐X, Bian H‐B, Wang J‐R, Cheng Z‐X, Wang K‐M, De W. Prognostic significance of serum miRNA‐21 expression in human non‐small cell lung cancer. J Surg Oncol 2011; 104: 748–51. [DOI] [PubMed] [Google Scholar]
- 18. Larzabal L, de Aberasturi AL, Redrado M e a. TMPRSS4 regulates levels of integrin α5 in NSCLC through miR‐205 activity to promote metastasis. Br J Cancer 2014; 110: 764–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roth C, Stückrath I, Pantel K, Izbicki JR, Tachezy M, Schwarzenbach H. Low levels of cell‐free circulating miR‐361‐3p and miR‐625* as blood‐based markers for discriminating malignant from benign lung tumors. PLOS One 2012; 7: e38248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L, Nan A, Zhang N e a. Circular RNA 100146 functions as an oncogene through direct binding to miR‐361‐3p and miR‐615‐5p in non‐small cell lung cancer. Mol Cancer 2019; 18: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jonkers J, Korswagen HC, Acton D, Breuer M, Berns A. Activation of a novel proto‐oncogene, Frat1, contributes to progression of mouse T‐cell lymphomas. EMBO J 1997; 16: 441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan W‐H, Du F‐J, Liu X‐J, Chen N. Knockdown of FRAT1 inhibits hypoxia‐induced epithelial‐to‐mesenchymal transition via suppression of the Wnt/β‐catenin pathway in hepatocellular carcinoma cells. Oncol Rep 2016; 36: 2999–3004. [DOI] [PubMed] [Google Scholar]
- 23. Li Z, Lu Q, Zhu D, Han Y, Zhou X, Ren T. Lnc‐SNHG1 may promote the progression of non‐small cell lung cancer by acting as a sponge of miR‐497. Biochem Biophys Res Commun 2018; 506: 632–40. [DOI] [PubMed] [Google Scholar]
- 24. Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non‐small cell lung cancer through inhibition of miR‐101‐3p and activation of Wnt/β‐catenin signaling pathway. Oncotarget 2017; 8: 17785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: The key to posttranscriptional regulation. ScientificWorldJournal 2014; 2014: 896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen W, Wang J, Liu S e a. MicroRNA‐361‐3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J Exp Clin Cancer Res 2016; 35: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, Yu JH, Lin XY e a. Overexpression of Frat1 correlates with malignant phenotype and advanced stage in human non‐small cell lung cancer. Virchows Arch 2011; 459: 255–63. [DOI] [PubMed] [Google Scholar]


