Abstract
Background
To investigate the diagnostic efficacy of long noncoding RNA metastasis‐associated in lung adenocarcinoma transcript l (MALAT1) as a candidate serological biomarker for non‐small cell lung cancer (NSCLC).
Methods
Diagnostic studies relevant to circulation long noncoding RNA MALAT1 as a candidate serological biomarker for NSCLC were electronically systematically searched in PubMed, EMBASE, EBSCO and CNKI databases. Suitable studies were included in the meta‐analysis by pooling the diagnostic sensitivity, specificity, positive likelihood ratio (+LR), negative likelihood ratio (−LR), diagnostic odds ratio (DOR) and area under the symmetric ROC curve (AUC) through a random or fixed effects model. Deeks' funnel plot was applied for publication bias evaluation.
Results
Six studies with eight datasets were finally included in the meta‐analysis after a systematic search of the databases was performed. The pooled diagnostic sensitivity, specificity, +LR, −LR and DOR were 0.81 (95% CI:0.78–0.84), 0.67 (95% CI:0.63–0.71), 2.61 (95% CI:1.81–3.71), 0.28 (95% CI:0.19–0.43) and 13.73 (95% CI:6.19–30.44), respectively. The pooled area under the ROC curve (AUC) were 0.8663 and 0.8658, respectively by symmetric and asymmetric methods.
Conclusion
Based on the results of our study, serum long noncoding RNA MALAT1 is a promising biomarker for NSCLC screening. However, due to its low specificity, MALAT1 positive cases need further validation for NSCLC by other diagnostic methods such as radiology, cytology, etc.
Keywords: Long noncoding RNA, MALAT1, meta‐analysis, non‐small cell lung cancer: diagnosis, serological biomarker
Introduction
Carcinoma of the lung is the most diagnosed malignant tumor in men and second in women and the leading cause of cancer‐related deaths in both males and females.1, 2 According to the pathology type, lung cancer is generally divided into small cell lung cancer (SCLC) and non‐small cell lung cancer (NSCLC) which is further divided into squamous cell lung carcinoma and adenoma lung carcinoma. The general prognosis of NSCLC is poor due to the advanced stage at which it is first diagnosed in 80% of all NSCLC cases. Due to its advanced clinical stage, most patients do not have the option of surgery and chemoradiotherapy is the treatment of choice.3 Therefore, early diagnosis is important for the prognosis of patients diagnosed with NSCLC. However, early diagnosis or screening for lung cancer is difficult due to the lack of effective methods.4, 5, 6
Because of minimal invasion and easy sample collection, serological biomarkers for lung cancer diagnosis or screening are of great interest.7, 8, 9, 10 Several studies have recently evaluated long noncoding RNA MALAT1 expression in serum or plasma of NSCLC patients and discussed its diagnostic performance.11, 12 However, due to limited statistical power caused by a small sample size, the findings of the publications were inconclusive. Therefore, we performed a meta‐analysis by pooling the open published studies relevant long noncoding RNA MALAT1 as a candidate serological biomarker for NSCLC.
Methods
Publication search
Diagnostic clinical studies on long noncoding RNA MALAT1 as a candidate serological biomarker for the diagnosis of NSCLC were systematically searched in the electronic databases of PubMed, EMBASE, EBSCO and CNKI. The electronic search words used were as follows: “lung carcinoma/lung cancer/lung tumor/lung neoplasm/malignant neoplasm of lung/non‐small cell lung cancer/NSCLC” and “metastasis associated in lung adenocarcinoma transcript 1/MALAT1/MALAT‐1/lncRNA MALAT 1/lncRNA MALAT‐1.” The reference to include published data was also screened in order to find potential suitable studies.
Inclusion and exclusion criteria
The publication inclusion criteria was defined as: (i) The patients included in each original study were confirmed with non‐small cell lung cancer by cytology or pathology; (ii) the controls were healthy subjects or cases without malignant disease; (iii) MALAT1 in serum or plasma was examined by real‐time PCR assay; (iv) the number of true positive (tp), false positive (fp), false negative (fn) and true negative (tn) in NSCLC and controls could be drawn or calculated from the original included studies. Exclusion criteria included: (i) small cell lung cancer or other type of lung carcinoma; (ii) not enough data to draw the tp, fp, fn and tn and (iii) MALAT1 detected in tissue or bronchoalveolar lavage fluid other than serum or plasma.
Data collection
The data and main information from each included study was extracted by two reviewers (Jie P and Yuan B) independently and cross‐checked. The extracted information and data included the first and corresponding author, period of the work performed, area of the study performed, number of tp fp fn tn cases, control type and the cutoff value of serum MALAT1 level.
Statistical analysis
Statistical analysis was performed using MetaDiSc1.4 (http://www.biomedsearch.com) statistical software. χ2 and I2 test were applied in order to evaluate the statistical heterogeneity across the included publications. The diagnostic effect size (sensitivity, specificity, +LR, −LR, and DOR) were pooled through a random or fixed effects model according to the statistical heterogeneity evaluation. Deeks' funnel plot was applied for publication bias evaluation.
Results
General features of studies
Six studies11, 12, 13, 14, 15, 16 with eight datasets were finally included in the meta‐analyses after a systematic search of the databases was performed. The identification process of the studies is shown in Figure 1. Of the six diagnostic clinical studies included, two used non‐cancer subjects as the control and four used healthy subjects as the control. All NSCLC patients had been diagnosed by pathology or cytology. The main features of the included six studies are shown in Table 1.
Figure 1.
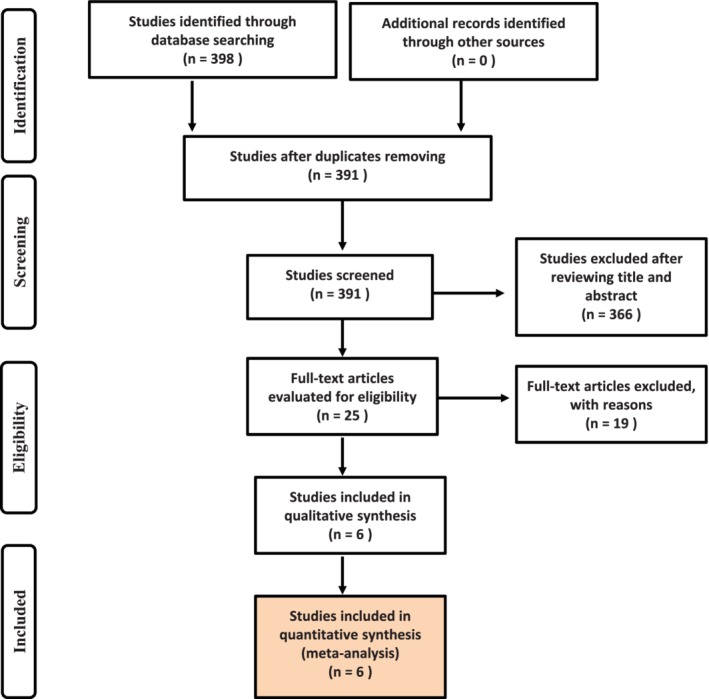
Flowchart of relevant studies from the electronic databases.
Table 1.
General characteristics of the 19 publications included in the study
| Sample size | Distribution | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| First author | Year | NSCLC | Control | TP | FP | FN | TN | Control type | Cutoff value |
| Shi | 2016 | 60 | 92 | 53 | 5 | 8 | 87 | Noncancer control | 0.62 |
| Guo | 2015 | 88 | 100 | 72 | 22 | 29 | 43 | Healthy control | 10.34 |
| Weber | 2013 | 45 | 25 | 21 | 0 | 24 | 25 | Noncancer control | Maximum Youden's Index:‐0.41 |
| Wen | 2016 | 60 | 60 | 50 | 21 | 10 | 99 | Healthy control | 0.03 |
| Peng | 2016 | 36 | 36 | 32 | 17 | 4 | 19 | Healthy control | 1.10 |
| Peng | 2016 | 120 | 71 | 119 | 46 | 1 | 25 | Healthy control | 2.08 |
| Guo | 2019 | 148 | 117 | 118 | 56 | 30 | 61 | Healthy control | Maximum Youden's Index:0.51 |
| Guo | 2019 | 120 | 103 | 95 | 41 | 25 | 62 | Healthy control | Maximum Youden's Index:0.51 |
NSCLC, non‐small cell lung cancer; TP, true positive; FP, false positive; FN, false negative; TN, true negative
Pooled results
Before pooling the diagnostic effect size of sensitivity, specificity, +LR and −LR, the statistical heterogeneity across the included studies was evaluated. The results indicated significant statistical heterogeneity existed for all the diagnostic effect size (P < 0.05). Therefore, the data was pooled using a random effects model. The pooled diagnostic sensitivity, specificity, +LR and −LR were 0.81(95% CI:0.78–0.84), 0.67(95% CI:0.63–0.71), 2.61(95% CI:1.81–3.71) and 0.28 (95% CI:0.19–0.43), respectively, Figure 2. The pooled DOR was 13.73 (95% CI:6.19–30.44) using a random effects model, Figure 3.
Figure 2.
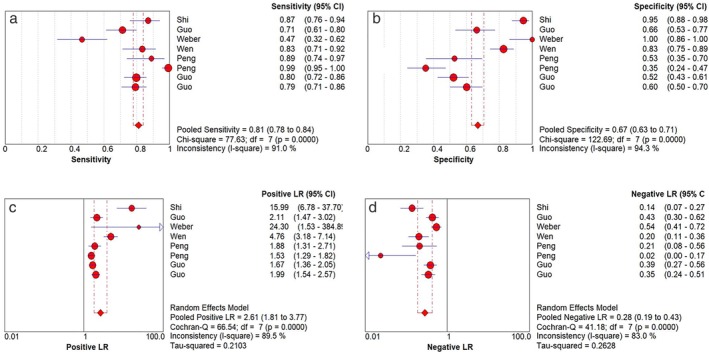
Forest plot of diagnostic effective size for MALAT1 as a candidate serological biomarker for non‐small cell lung cancer (NSCLC). (a) pooled diagnostic sensitivity; (b) pooled diagnostic specificity; (c) pooled +LR; (d) pooled −LR.
Figure 3.
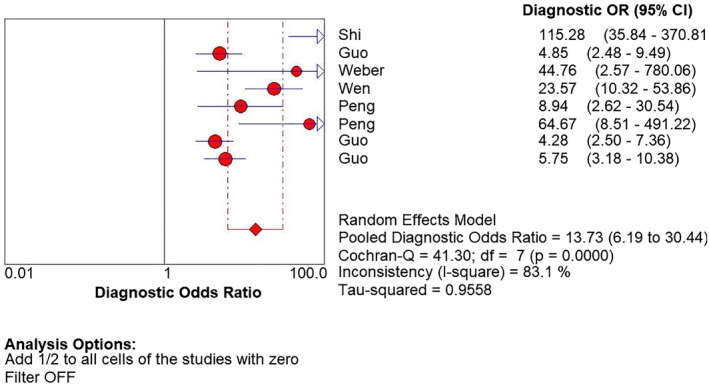
Forest plot of diagnostic odds ratio for MALAT1 as a candidate serological biomarker for non‐small cell lung cancer (NSCLC).
Pooled ROC
The area under the ROC curve (AUC) was pooled by symmetric or asymmetric methods with results of AUC = 0.8663 and 0.8658, respectively, Figure 4.
Figure 4.
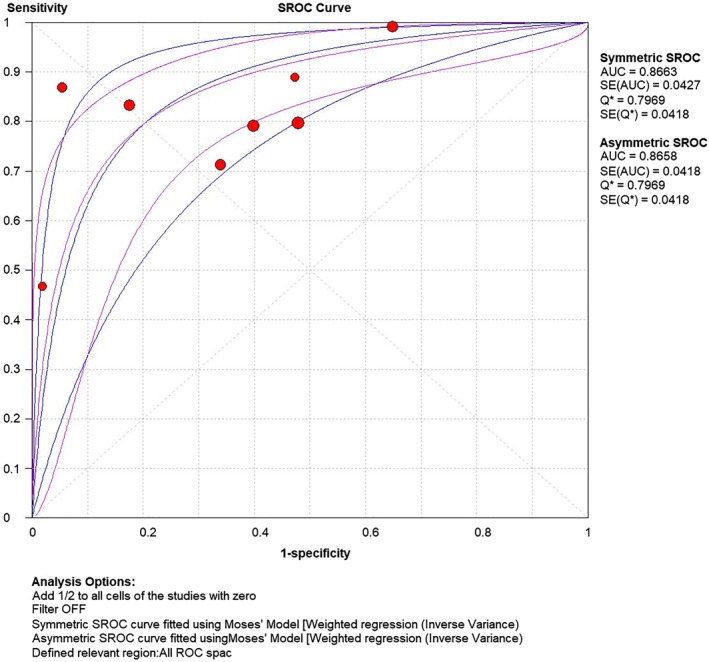
ROC curve of MALAT1 in serum as a biomarker for the diagnosis of non‐small cell lung cancer (NSCLC).
Subgroup analysis
According to the pathology type of NSCLC, subgroup analysis for squamous cell lung cancer and lung adenocarcinoma was made. The pooled sensitivity, specificity, +LR, −LR and DOR for squamous cell lung cancer and lung adenocarcinoma are shown in Table 2.
Table 2.
Subgroup analysis of MALAT1 in serum as a biomarker for non‐small cell lung cancer (NSCLC) diagnosis according to pathology type
| Diagnostic reference | Squamous cell carcinoma | Chi‐square/Cochran‐Q | P‐value | Adenocarcinoma | Chi‐square | P‐value |
|---|---|---|---|---|---|---|
| Sen | 0.74 (0.60–0.85) | 10.10 | 0.0015 | 0.62 (0.48–0.75) | 8.13 | 0.0043 |
| Sep | 0.87 (0.78–0.93) | 8.33 | 0.0039 | 0.88 (0.79–0.94) | 7.51 | 0.0061 |
| +LR | 8.01 (1.36–47.31) | 2.01 | 0.156 | 5.44 (2.01–14.70) | 1.19 | 0.2760 |
| −LR | 0.23 (0.03–1.84) | 9.10 | 0.0027 | 0.43 (0.17–1.10) | 7.10 | 0.0077 |
| DOR | 54.86 (13.65–220.43) | 0.00 | 0.9453 | 17.55 (6.57–46.89) | 0.20 | 0.6583 |
Publication bias evaluation
The publication bias was evaluated by Deeks' funnel plot and indicated no significant publication bias for MALAT1 as a candidate serological biomarker for NSCLC (P > 0.05), Figure 5.
Figure 5.
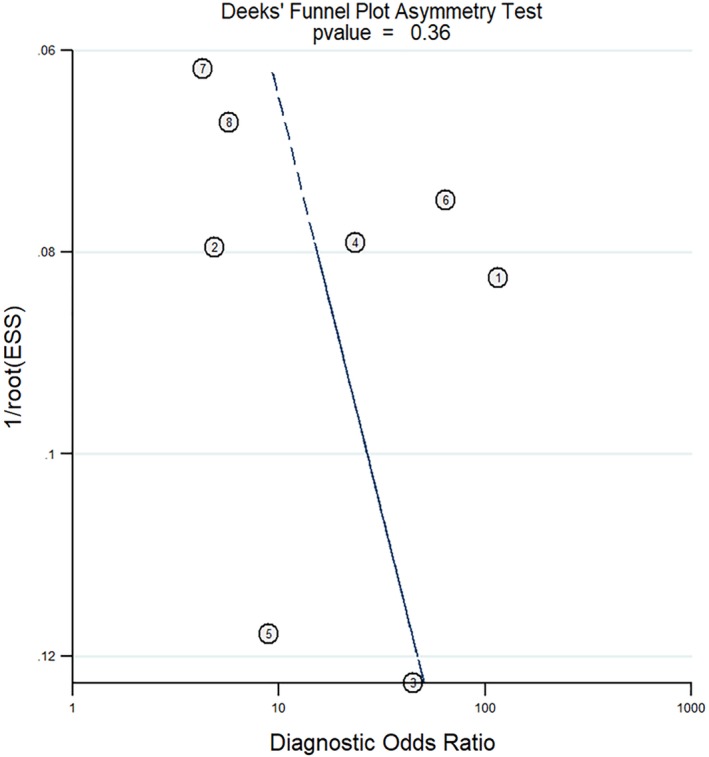
Deeks' funnel plot indicated no publication bias. ( ) Study and (
) Study and ( ) regression line.
) regression line.
Discussion
Long noncoding RNA belongs to one of the main types of noncoding RNA family. In recent years, many studies have found that abnormal expression of long noncoding RNA MALAT1 can be used as a potential biomarker of cancer diagnosis.12, 13 MALAT1 is one of the first identified long noncoding RNA related to post‐transcription modification. In 2003, Ji et al. found that the expression level of MALAT1 in metastatic tissue of NSCLC patients was significantly higher than that in nonmetastatic cancer tissue.17 The findings of the study by Zhao et al. indicated that MALAT1 may be an important molecule associated with lung cancer metastasis.18 At present, there are several meta‐analysis relevant to MALAT1 expression and patients’ prognosis with positive results.19, 20 However, there is no confirmed evidence of MALAT1 expression and cancer diagnosis. In the present meta‐analysis, the results showed that the sensitivity and specificity of serum MALAT1 in the diagnosis of lung cancer were 0.81 and 0.67, respectively, and the AUC was 0.6, indicating that the detection of MALAT1 in circulating blood was of high diagnostic value for lung cancer. Due to the high diagnostic sensitivity of serum MALAT1 as a reference for NSCLC, the misdiagnosis rate of lung cancer is low, which indicates that serum MALAT1 has better clinical application value in lung cancer screening. In addition, the diagnostic odds ratio (DOR) is also an important index to evaluate diagnostic efficiency. When the value is more than one, the larger the value is, the better the discrimination effect of the diagnostic test is.12 In this study, the overall combined DOR value was 13.73, suggesting that the detection of serological MALAT1 was more effective in the overall diagnosis of lung cancer. Furthermore, the combined positive likelihood ratio suggested that the probability of positive expression of MALAT1 in lung cancer patients was 2.6 times higher than that in the control group, which further reflected the potential value of circulating MALAT1 in the diagnosis of lung cancer.
The meta‐analysis in our study also had its limitations. First, only six studies with eight datasets were included in the present work. Second, the diagnostic sensitivity, specificity, +LR, −LR and DOR were pooled through a random effects model due to significant statistical heterogeneity. Third, only studies published in English or Chinese were screened and included in the meta‐analysis.
Disclosure
No authors report any conflict of interest.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 2. Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer 2019; 10: 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maconachie R, Mercer T, Navani N, McVeigh G, Guideline Committee. Lung cancer: diagnosis and management. summary of updated NICE guidance. BMJ 2019; 364: 1049. [DOI] [PubMed] [Google Scholar]
- 4. Duma N, Santana‐Davila R, Molina JR. Non‐small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc 2019; 94: 1623–40. [DOI] [PubMed] [Google Scholar]
- 5. Arenberg D. Update on screening for lung cancer. Transl Lung Cancer Res 2019; 8: S77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moffat J, Hiom S, Kumar HS, Baldwin DR. Lung cancer screening ‐ gaining consensus on next steps ‐ proceedings of a closed workshop in the UK. Lung Cancer 2018; 125: 121–7. [DOI] [PubMed] [Google Scholar]
- 7. Seijo LM, Peled N, Ajona D et al Biomarkers in lung cancer screening: Achievements, promises, and challenges. J Thorac Oncol 2019; 14: 343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu GCW, Lazare K, Sullivan F. Serum and blood based biomarkers for lung cancer screening: A systematic review. BMC Cancer 2018; 18: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tang ZM, Ling ZG, Wang CM, Wu YB, Kong JL. Serum tumor‐associated autoantibodies as diagnostic biomarkers for lung cancer: A systematic review and meta‐analysis. PLOS One 2017; 12: e0182117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakamura H, Nishimura T. History, molecular features, and clinical importance of conventional serum biomarkers in lung cancer. Surg Today 2017; 47: 1037–59. [DOI] [PubMed] [Google Scholar]
- 11. Guo F, Yu F, Wang J et al Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed Rep 2015; 3: 309–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weber DG, Johnen G, Casjens S et al Evaluation of long noncoding RNA MALAT1 as a candidate blood‐based biomarker for the diagnosis of non‐small cell lung cancer. BMC Res Notes 2013; 6: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi T, Gao G, Cao Y. Expression of MALAT1 in serum of the patients with non‐small cell lung cancer (NSCLC) and its clinical significance. Chin J Clin Lab Sci 2016; 34: 114–7. [Google Scholar]
- 14. Wen Z, Li X. Diagnostic value of long non‐coding RNA metastasis‐associated lung adenocarcinoma transcript 1 in serum for clinical diagnosis of lung cancer. J Clin Pulmon Med 2016; 21: 2154–8. [Google Scholar]
- 15. Guo X. The research for diagnostic significance of plasma lncRNAs for lung cancer. Guzhou Medical University 2019; 7: 17–21. [Google Scholar]
- 16. Peng H, Wang J, Li J et al A circulating non‐coding RNA panel as an early detection predictor of non‐small cell lung cancer. Life Sci 2016; 151: 235–42. [DOI] [PubMed] [Google Scholar]
- 17. Ji Q, Liu X, Fu X et al Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/β‐catenin signal pathway. PLOS One 2013; 8: e78700. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. MALAT1: A long non‐coding RNA highly associated with human cancers. Oncol Lett 2018; 16: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mei H, Liu Y, Zhou Q, Hu K, Liu Y. Long noncoding RNA MALAT1 acts as a potential biomarker in cancer diagnosis and detection: A meta‐analysis. Biomark Med 2019; 13: 45–54. [DOI] [PubMed] [Google Scholar]
- 20. Tang R, Jiang M, Liang L, Xiong D, Dang Y, Chen G. Long noncoding RNA MALAT‐1 can predict poor prognosis: A meta‐analysis. Med Sci Monit 2016; 22: 302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


