Introduction
The nematode worm Caenorhabditis elegans is an ideal tool for functional genomics. It has been characterised in great detail in terms of its development, morphology and physiology at the cellular level and its genome has been completely sequenced (1). A powerful genetic toolset for C. elegans is available and a large collection of mutants are available. In addition, C. elegans is one of the few animals for which high-throughput in vivo RNA interference (RNAi) screening has been established (2). Many of the genes involved in human disease, including those involved in diabetes, obesity, skeletomuscular disorders, neurodegeneration and the cell cycle, have homologues in C. elegans. Study of these in C. elegans has made a large contribution to our current understanding of human disease gene pathways.
In functional genomic studies of yeast, Raamsdonk and colleagues (3) suggested a functional genomics approach based on metabolomics to reveal the phenotypes of silent mutations (3). This has been further demonstrated in the excreted metabolites produced by yeast (‘metabolic footprinting’;(4)). In a similar manner, metabolomics has been shown to be highly discriminatory for a variety of mutants in different organisms including Arabidopsis (5) and mice (6).
In this study we have examined the metabolome of C. elegans using a combination of 1H Nuclear Magnetic Resonance (NMR) spectroscopy to examine aqueous soluble metabolites and Gas Chromatography Mass Spectrometry (GC-MS) to examine the total fatty acid composition. These tools have been used with multivariate statistics to examine NHR-49. While this nuclear hormone receptor has significant homology with hepatocyte nuclear factor 4 (HNF4) in mammals, Van Gilst and colleagues have proposed the function is closer to the mammalian peroxisome proliferator-activated receptors (PPARs) (7, 8). The metabolic phenotype of C. elegans nhr-49 mutants has then been compared with the metabolic changes in the PPAR-α null mouse to demonstrate the translational potential of metabolomics-based functional genomics.
Methods
Nematode methods
C. elegans was grown using standard conditions at 20 °C on plates seeded with Escherichia coli strain OP50 (9). The wild-type strain was var. Bristol N2 (10). The nhr-49 mutant strain used was VC870 (nhr-49(gk405)), which was generated by the C. elegans knockout consortium (11) and distributed by the Caenorhabditis Genetics Center, University of Minnesota, Twin Cities, MN. For each experiment approximately 2000 mixed-stage animals were harvested and nine repeats were made. The animals were washed twice in dd H2O, centrifuged (2000 rpm, 20 minutes) and the worm pellets were stored at -80 °C until extraction.
Mouse husbandry
Tissues from SVEV/129 mice, and PPAR-α null mice (n=5; 5 month age) were obtained from stable colonies at the University of Oxford. The UK Home Office approved all procedures. Mice were fed standard laboratory chow ad libitum prior to death (Special Diet Services, Essex, UK). Animals were killed by terminal anaesthesia and liver collected rapidly. Livers were stored at -80 °C until extraction.
Metabolite extraction
Metabolites from mouse liver tissue or whole nematodes were extracted using a methanol-chloroform extraction procedure. Frozen tissue (~100 mg) was pulverised with dry ice, 600 μl methanol-chloroform (2:1) was added and samples were sonicated for 15 minutes. 200 μl of chloroform and water were added, and the samples centrifuged. The aqueous layer was dried overnight in an evacuated centrifuge and the lipid fraction was dried overnight under nitrogen.
High resolution 1H NMR spectroscopy of aqueous fraction
The dried extracts were rehydrated in 600 μl D2O, buffered in 0.24 M sodium phosphate (pH 7.0) containing 1 mM (sodium-3-(tri-methylsilyl)-2, 2, 3, 3-tetradeuteriopropionate (TSP) (Cambridge Isotope Laboratories MA, USA) as an internal standard. The samples were analysed using an AVANCE II+ NMR spectrometer operating at 500.13 MHz for the 1H frequency (Bruker, Germany) using a 5 mm TXI probe. Spectra were collected using a solvent suppression pulse sequence based on a 1-dimensional NOESY pulse sequence to saturate the residual 1H water signal (relaxation delay = 2 s, t1= 3 μs, mixing time= 150 ms, solvent presaturation applied during the relaxation time and the mixing time). 128 transients were collected into 16 K data points over a spectral width of 12 p.p.m at 27 °C. In addition representative samples of nematodes were also examined by two dimensional spectroscopy, including COSY and HSQC spectroscopy (12), in conjunction with reference to previous literature and on-line databases ((13, 14); Madison Metabolomics Consortium Database, http://mmcd.nmrfam.wisc.edu/index.html and the database with the Chenomx software (Chenomx, Alberta, Canada)).
GC-MS of lipid fraction
Lipids were dissolved in 0.25 ml of chloroform/methanol (1:1 v/v). 100 μl BF3/methanol (Sigma-Aldrich) was added and the vials were incubated at 80 °C for 90 minutes. 0.3 ml H2O and 0.6 ml hexane was added and each vial vortex mixed. The organic layer was evaporated to dryness before reconstitution in 200 μl hexane for analysis.
The derivatised organic metabolites were injected onto a ZB-WAX column (30m x 0.25 mm ID x 0.25 μm df; 100% polyethylene glycol). The initial column temperature was 60 °C and was held for 2 minutes. This was increased by 15 °C min-1 to 150 °C then increased at a rate of 4 °C min-1 to 230 °C. The column eluent was introduced into a Trace DSQ quadrupole mass spectrometer (Thermo Electron) (transfer line temperature= 240 °C, ion source temperature= 250 °C, electron beam= 70eV). The detector was turned on after 240 s and data was collected in full scan mode using 3 scans s-1 across a mass range of 50-650 m/z.
Multivariate statistical analysis
NMR spectra were processed using ACD 1D NMR processor (vers. 8, ACD, Toronto, Canada). Free induction decays were Fourier transformed following multiplication by a line broadening of 1 Hz, and referenced to TSP at 0.0 p.p.m. Spectra were phased and baseline corrected manually. Each spectrum was integrated using 0.04 p.p.m. integral regions between 0.5-4.5, and 5.1-10.0 p.p.m. Each spectral region was normalised to a total integral value of 10000. In addition the Chenomx software was used to peak fit individual metabolites, and a dataset was produced consisting of these fitted resonances.
GC-MS Chromatograms were analysed using Xcalibur, version 2.0 (Thermo Fisher), integrating each peak individually. Each integrated peak was normalised so that the total sum of peaks was set to 10000. Deconvolution of overlapping peaks was achieved by traces of single ions. Mass spectra were assigned using the NIST database of mass spectra and standard compound analysis.
Datasets were imported into SIMCA-P 11.0 (Umetrics, Umeå, Sweden) for processing using Partial Least Squares-Discriminate analysis (PLS-DA; a regression extension of Principal Components Analysis used for supervised classification). GC-MS data was scaled to unit variance by dividing each variable by 1/(Sk) where Sk is the standard deviation of the variable. 1H NMR spectroscopy data was Pareto scaled, in which each variable was centred and multiplied by 1/(Sk)1/2. Identification of major metabolic perturbations within the pattern recognition models was achieved by analysis of corresponding loadings plots, with metabolites selected which are considered to have a significant contribution to that PC (p<0.05 following a jack knifing routine). Additionally, R2 and Q2 were used as measures for the robustness of a pattern recognition model. R2 is the fraction of variance explained by a component, and cross validation of R2 gives Q2 which reveals the fraction of the total variation predicted by a component. In addition a leave-one-out analysis was performed on each C.elegans dataset to examine how well class membership was predicted.
Results
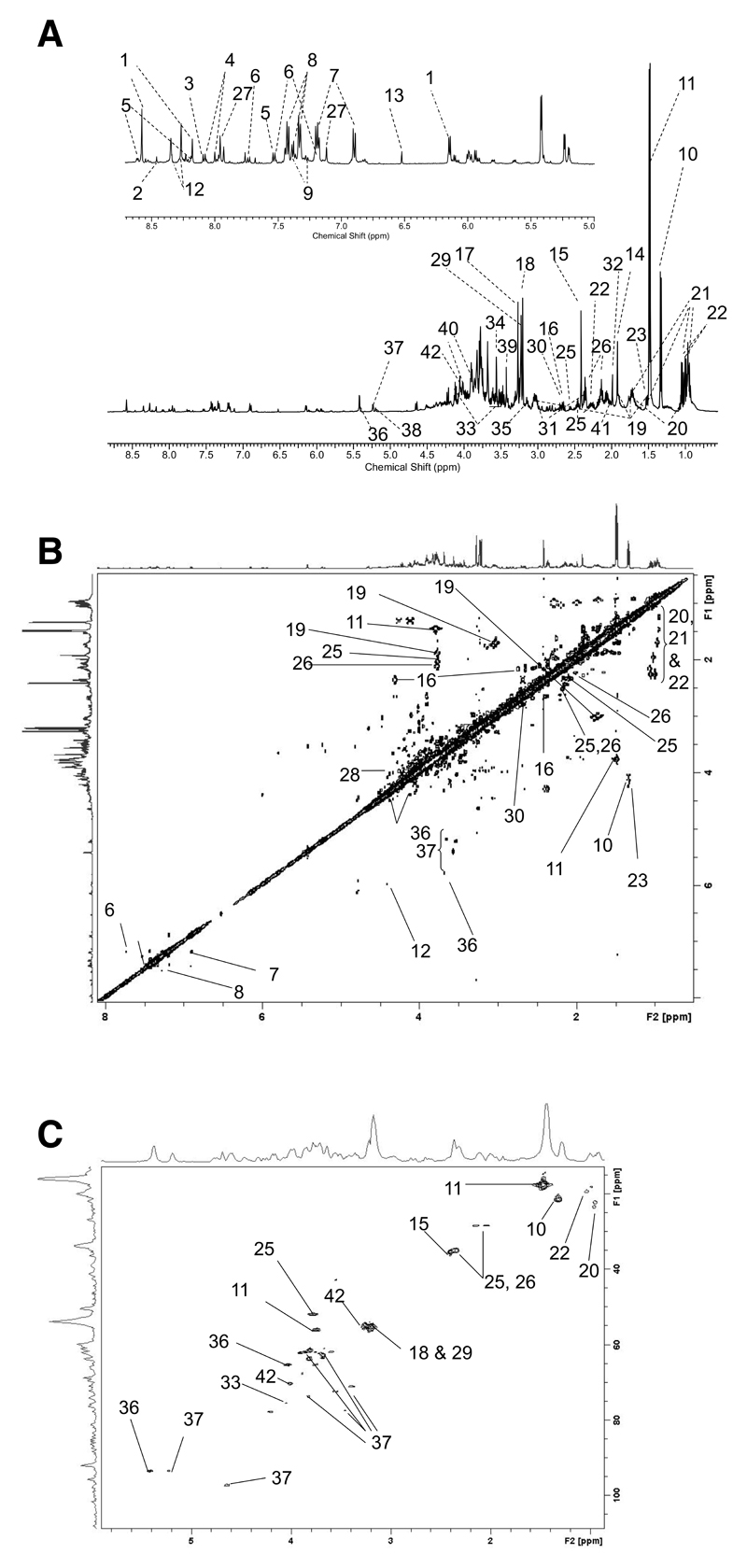
Figure 1a shows a high resolution 1H NMR spectrum acquired in ~14 minutes of the aqueous metabolites extracted from C. elegans. Using the library and peak fitting routine within the Chenomx software package a total of 55 metabolites were identified and quantified, and confirmed by the use of 2-D NMR spectroscopy and GC-MS [Supplementary Table 1 and Figure 1].
Figure 1.
(a). Annotated 1H NMR spectrum showing aqueous soluble metabolites extracted from C. elegans. Assignments were confirmed through the use of COSY (b) and HSQC (c) spectra. Key: 1. ANP, 2. formate, 3. GTP, 4. hypoxanthine; 5. nicotinamide; 6. Indoxyl sulphate; 7. 4-Hydroxyphenylacetic acid. 8. phenylalanine; 9. phenylacetylglycine; 10. lactate; 11 alanine, 12. inosine; 13. fumarate. 14 acetate; 15. succinate; 16 malate; 17. TMAO, 18. choline, 19 lysine, 20. leucine; 21. iso-leucine, 22. valine, 23 threonine; 24. beta-alanine; 25. glutamine; 26. glutamate; 27. histidine; 28 tyrosine; 29. phosphocholine, 30. aspartate, 31 alpha-ketoglutarate, 32. methionine; 33. myo-inositol; 34. glycine, 35 melonate. 36. glucose-6-phosphate; 37. glucose, 38. fructose, 39. methanol; 40. serine, 41. proline; 42. betaine. In addition there are numerous sugar and amino acid resonances between ~3.3-4.0 ppm.
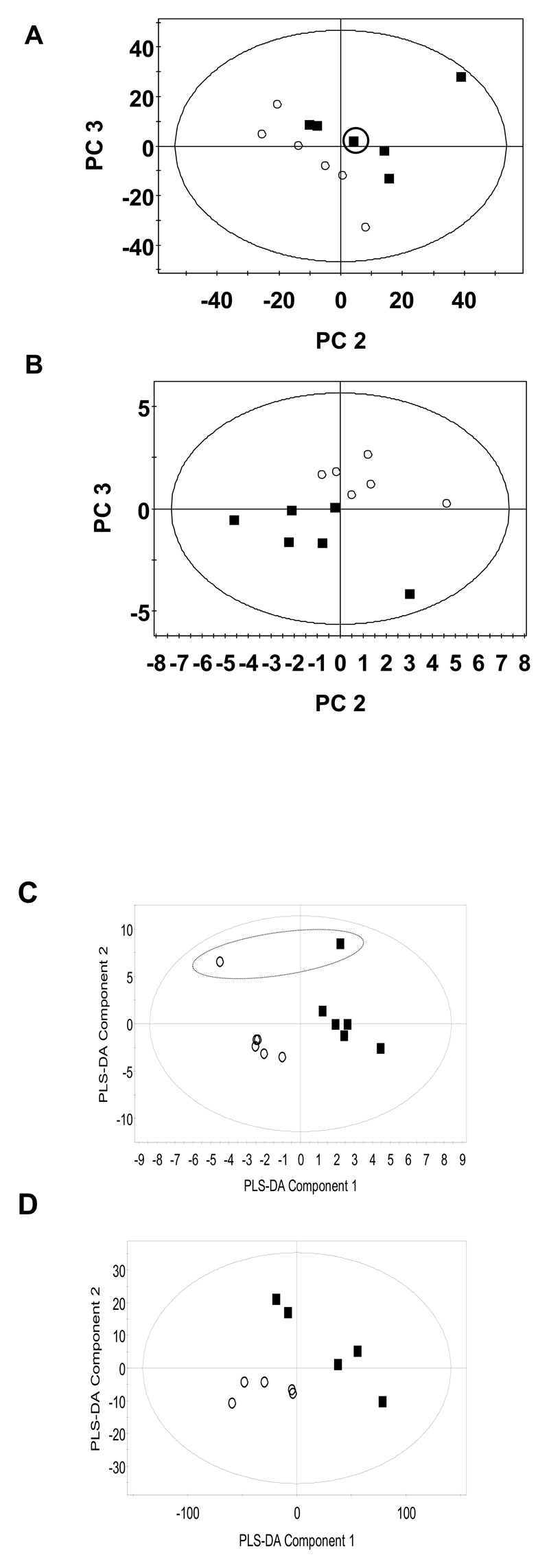
Using a bucketing routine to integrate the NMR spectra and applying PLS-DA to the resultant dataset, classification into wild type and nhr-49 mutants was achieved (Figure 2a). Using a leave one out analysis of the data set all samples except one was correctly predicted to belong to the correct group. On inspection this sample belonged to a spectrum with low signal to noise. Examining the loadings plots the metabolites which were responsible for this classification were increases in lactate (δ 1.30-1.34 signifying the bucket represented in the loadings plot), alanine (δ 1.46-1.50), leucine (δ 1.62-1.66) and decreases in lysine (δ 2.98-3.06), glucose (δ 3.26-3.34; δ 3.50-3.62), malonate (δ 3.10-3.14) and aspartate (δ 2.70-2.74) in the mutant worms.
Figure 2.
Score plots showing the differentiation of nhr-49 (○) and N2 control C. elegans nematodes (■) following (a) PLS-DA of the aqueous soluble metabolites by 1H NMR spectroscopy (R2(X)=; R2(Y)=; Q2=) and (b) PLS-DA analysis of the total lipid compliment by GC-MS following methyl esterification (R2(X) = 54%, R2(Y)=92%; Q2=76%). (c) PLS-DA score plots showing the differentiation of PPAR-α null tissue (○) and control tissue (■) following analysis of the aqueous soluble metabolites by 1H NMR spectroscopy, and (d) following PLS-DA analysis of the total lipid compliment by GC-MS following methyl esterification.
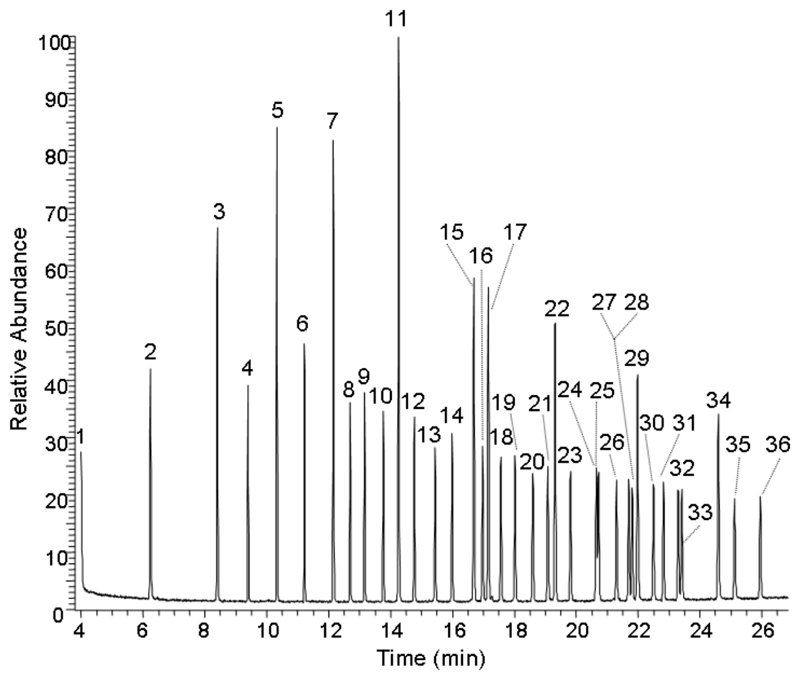
Examining the total fatty acid complement of C. elegans a total of 42 peaks were detected in the chromatogram with 31 of these being identified through a combination of the NIST library and retention time matching of known standards [Supplementary Table 2 and supplementary Figure 1]. Applying PLS-DA to the integrated peaks of the chromatograms the two groups were separated across 2 PLS-DA components, [Figure 2b]. Using a leave-one-out analysis the class membership of all samples were correctly predicted, although two samples were identified as being outliers to the model according to the distance to model criteria. These samples contained high concentrations of palmitoleidic acid which is found in high concentrations within E. coli. Examining the loadings plots of the PLS-DAs for the fatty acid data set, the metabolites increased in the mutant nematodes were α-linolenic acid, arachidonic acid, nonadecanoic acid, di-homo-γ-linolenic acid, heptadecenoic acid, γ-linolenic acid and docosanoic acid and there was a decrease in a branched chain form of palmitate.
Metabolomic analysis of liver from the 5 month PPAR-α null mouse revealed a number of changes similar to those observed in the nhr-49 mutant C. elegans strain. 1H NMR spectroscopy of the aqueous soluble metabolites in conjunction with PLS-DA showed the PPAR-α null samples could be differentiated from age matched controls through an increase in lactate (δ 1.30-1.34) and a decrease in glucose concentration (δ 3.38-3.42), (δ 3.72-3.76), (δ 3.90-3.94) and (δ 5.20-5.24) (R2= 0.88, Q2= 0.47) [Figure 2c]. Similarly, PLS-DA of the fatty acid composition, following analysis by GC-MS, was able to distinguish PPAR-α null samples from controls by an increase in concentration of arachidonic acid, linoleic acid, eicosanoic acid, lauric acid, and di-homo γ-linolenic acid, and a decrease in concentration of palmitic acid, and a branched chain 16:0 fatty acid (R2= 0.81, Q2= 0.25; Figure 2d).
To compare directly the mutants, PLS-DA models were constructed of the aqueous and lipid phase datasets.
Discussion
In this study we have demonstrated the versatility of a combined high resolution 1H NMR spectroscopy and GC-MS based profiling approach to study metabolic changes in C. elegans. The approach detected and quantified ~100 metabolites of which 86 were identified. Both approaches were rapid and cheap on a per sample basis, indicating the approach could be used as a rapid screening tool of C. elegans mutants. In this study the metabolic changes in C .elegans nhr-49 mutants have been compared with the PPAR-α null mouse, demonstrating the functional similarity between these genes.
Previous studies looking at metabolism in the PPAR-α null mouse during fasting have described a significant reduction in fatty acid metabolism via β oxidation which has been attributed to a loss of regulation of genes that encode enzymes critical to this pathway (15), while we have previously detected a milder phenotype in fed animals (16). In this study we observed an increase in linoleic and di-homo-γ-linolenic acid in PPAR-α null livers indicative of reduced β oxidation. This was observed despite the fact these animals were fed ad libitum signifying an underlying metabolic deficit in these animals. Similar changes were observed in the nhr-49 mutant nematodes with an increase in the β oxidation precursors di-homo-γ-linolenic acid, α-linolenic and γ-linolenic acid suggesting a similar loss of fatty acid oxidation. This demonstrates that the mechanisms of fatty acid metabolism are largely conserved between mammals and C. elegans making the nematode worm a highly versatile tool for future metabolic studies.
One notable difference between our present study and that of Van Gilst and colleagues (2005a) examining the nhr-49 mutant was that we detected no change in stearate, despite a marked increase being detected in the previous study. Similar increases in stearate have been detected in fat-7 (F10D2.9; homologous with mammalian stearoyl-CoA desaturase and induced by nhr-49) and nhr-80 mutants (7, 17). However, stearate also increases during fasting and the expression of fat-7 varies markedly during development and fasting, suggesting that the concentration of stearate may be a sensitive marker for the physiological and developmental state of the nematodes.
In the nhr-49 nematodes the perturbation in fat metabolism was accompanied by a decrease in glucose and an increase in lactate and alanine, indicative of an increase in the relative ratio of glycolysis to gluconeogenesis. Similar changes were also detected in the liver tissue of PPAR-α knock out mice in this study and in younger animals (16). In mice, PPAR-α is known to control pyruvate dehydrogenase kinase 4 (PDK4) activity, and this can limit flux through the Krebs cycle by inhibiting pyruvate dehydrogenase (18). Loss of PPAR-α prevents PDK4 expression, and this may lead to an active form of pyruvate dehydrogenase in the liver causing increased glucose utilisation. A similar interaction may exist between nhr-49 and PDK in nematodes. Another plausible hypothesis is that there may be reduced flux through the glycolylate pathway arising from a decrease in precursor fatty acid β oxidation products, in particular acetyl Co-A, decreasing glucose synthesis. Decreased levels of acetyl Co-A would also limit the de novo synthesis of fatty acids in turn accounting for the observed decrease in 16:0 fatty acids.
In conclusion, our combined metabolomic approach has demonstrated that nhr-49 has a role in regulating lipid synthesis, β-oxidation of fatty acids, glycolysis and gluconeogenesis in a similar manner to the role of PPAR-α in the mouse.
Supplementary Material
Figure 3.
Gas-chromotogram showing 36 standard FAMEs used in compound identification. Key: (1) 6:0, (2) 8:0, (3) 10:0, (4) 11:0, (5) 12:0, (6) 13:0, (7) 14:0, (8) 9c-14:1, (9) 15:0, (10) 10c-15:1, (11) 16:0, (12) 9c-18:1, (13) 17:0, (14) 10c-17:1, (15) 18:0, (16) 9c-18:1, (17) 9t-18:1, (18) 9t, 12t-18:2, (19) 9c, 12c-18:2, (20) 6c, 9c, 12c-18:3, (21) 9c, 12c, 15c-18:3, (22) 20:0, (23) 11c-20:1, (24) 21:0, (25) 11c, 14c-20:2, (26) 8c, 11c, 14c-20:3, (27) 5c, 8c, 11c, 14c-20:4, (28) 11c, 14c, 17c-20:3, (29) 22:0, (30) 13c-22:1, (31) 5c, 8c, 11c, 14c, 17c-20:5, (32) 23:0, (33) 13c, 16c-22:2, (34) 24:0, (35) 15c-24:1, and (36) 4c, 7c, 10c, 13c, 16c, 19c-22:6.
Acknowledgements
This study was supported by grants from the Biotechnology and Biological Sciences Research Council and Selcia Ltd (HJA and JLG), and the European Union (European Commission, FP6 Contract No. 003956 (OAHJ). JLG is a Royal Scoiety (UK) University Research Fellow. EAM was supported by a Programme Grant from Cancer Research UK. We would like to thank Rob Shaw for help with C. elegans husbandry.
References
- 1.Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 2.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 3.Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, Westerhoff HV, et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 4.Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–6. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- 5.Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 6.Jones GL, Sang E, Goddard C, Mortishire-Smith RJ, Sweatman BC, Haselden JN, Davies K, Grace AA, Clarke K, Griffin JL. A functional analysis of mouse models of cardiac disease through metabolic profiling. J Biol Chem. 2005;280:7530–7539. doi: 10.1074/jbc.M410200200. [DOI] [PubMed] [Google Scholar]
- 7.Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood WB. The Nematode Caenorhabditis elegans. Cold Spring Harbor Press; New York: 1988. [Google Scholar]
- 10.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgley M, D'Souza A, Moulder G, McKay S, Shen B, al e. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res. 2002;30:e52. doi: 10.1093/nar/gnf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun S, Kalinowski H-O, Berger S. 100 and more basic NMR experiments : a practical course. VCH; Weinheim: 1996. [Google Scholar]
- 13.Bundy JG, Osborn D, Weeks JM, Lindon JC, Nicholson JK. An NMR-based metabonomic approach to the investigation of coelomic fluid biochemistry in earthworms under toxic stress. FEBS Lett. 2001;500:31–35. doi: 10.1016/s0014-5793(01)02582-0. [DOI] [PubMed] [Google Scholar]
- 14.Bundy JG, Spurgeon DJ, Svendsen C, Hankard PK, Osborn D, Lindon JC, Nicholson JK. Earthworm species of the genus Eisenia can be phenotypically differentiated by metabolic profiling. FEBS Lett. 2002;521:115–120. doi: 10.1016/s0014-5793(02)02854-5. [DOI] [PubMed] [Google Scholar]
- 15.Reddy JK, Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- 16.Atherton HJ, Bailey NJ, Zhang W, Taylor J, Major H, Shockcor J, Clarke K, Griffin JL. A combined 1H-NMR spectroscopy- and mass spectrometry-based metabolomic study of the PPAR-alpha null mutant mouse defines profound systemic changes in metabolism linked to the metabolic syndrome. Physiol Genomics. 2006;27:178–186. doi: 10.1152/physiolgenomics.00060.2006. [DOI] [PubMed] [Google Scholar]
- 17.Brock TJ, Browse J, Watts JL. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006;2:e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu P, Peters JM, Harris RA. Adaptive increase in pyruvate dehydrogenase kinase 4 during starvation is mediated by peroxisome proliferator-activated receptor alpha. Biochem Biophys Res Commun. 2001;287:391–396. doi: 10.1006/bbrc.2001.5608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.