Abstract
Ploidy represents the number of chromosome sets in a cell. Although gametes have a haploid genome (n), most mammalian cells have diploid genomes (2n). The diploid status of most cells correlates with the number of probable alleles for each autosomal gene and makes it difficult to target these genes via mutagenesis techniques. Here we describe a 7-week protocol for the derivation of mouse haploid embryonic stem cells (hESCs) from female gametes that also outlines how to maintain the cells once derived. We detail additional procedures that can be used with cell lines obtained from the mouse Haplobank, a biobank of >100,000 individual mouse hESC lines with targeted mutations in 16,970 genes. hESCs can spontaneously diploidize and can be maintained both in haploid and diploid states. Mouse hESCs are genomically and karyotypically stable, are innately immortal and isogenic, and can be derived in an array of differentiated cell types; they are thus highly amenable to genetic screens and to defining molecular connectivity pathways.
Keywords: haploid ESC, genetic screen, Haplobank, haploid stem cell
Introduction
The packing and organization of genomes into chromosomes is central to the maintenance and transmission of genes during evolution1. In eukaryotes, ploidy (defined as the number of chromosome sets, n, of each cell) can range from a haploid genome n=1 chromosome in the male of the Jack jumper ant (Myrmecia pilosula)2 to a staggering ~1900 x n= ~15,600 chromosomes (29,64 x 106 chromosomes) in the ciliated protozoan Oxytricha trifallax3. However, within mammals, the large majority of species maintain a diploid (2n) organization for most somatic cells and a haploid (n) status for gametes; an exception being made by the tetraploid vizcacha rat Tympanoctomys barrerae4. The somatic/diploid and germline/haploid configuration is beneficial and enables genetic variation to be increased through sexual reproduction5, whilst conferring protection from deleterious mutations in the long-lived somatic tissues by allowing the presence of back-up alleles as well as providing a template for DNA damage repair6. Thus, to maintain normal physiology, mammalian cells have developed mechanisms that act throughout the cell cycle to prevent the occurrence of cells with abnormal chromosome numbers (aneuploid or polyploid)7 as these are a hallmark of genomic instability, and oncogenic transformation8.
Most somatic cells maintain their diploid chromosome number through mitotic division, although non-oncogenic mosaic polyploidy can happen in selected tissues during development (e.g. placenta) or in adult life (e.g. liver, muscle or brain)9. By contrast, germ cells (oocytes or sperm) are terminally differentiated cells incapable of mitotic self-renewal in vivo. Although methods of derivation and maintenance in cell culture for somatic cells are very well established, the derivation and culture of oocyte-derived hESCs has only recently been achieved10–13 (also see preview by Sagy and Benvenisti14). Here, we detail an experimental protocol for the derivation, maintenance and use of mouse hESCs.
Development and applications of the protocol
The haploid budding yeast (Saccharomyces cerevisiae) has been used for a long time as a model organism for genetic experiments, as its genome can be easily manipulated for targeted or random mutagenesis15.
In vivo, mouse N-Ethyl-N-Nitrosourea mutagenesis was until recently the only method used to perform random mutagenesis screens in mammalian haploid cells16. Although very informative, these screens require extensive resources, space and time for phenotypic mutants of interest to be identified. More recently, the discovery of KBM7, a near-haploid leukemic cancer cell line17,18, and its further engineering to remove one extra copy of chromosome 8 (ref. 19) and parts of chromosome 15 (ref. 20) created the HAP1 cell line, representing the first human cell line with a haploid genome to be propagated in culture. Limitations of the HAP1 cells relate to their transformed nature and the presence of various genomic alterations, including the BCR-ABL1 translocation, confounding some studies that aim to understand physiologic genetic pathways in normal cells.
In vitro activation of oocytes for parthenogenetic embryogenesis and later generation of hESCs was initially done in the 1920s by Pincus, who observed that some unfertilized rabbit oocytes could spontaneously undergo variable degrees of development that were morphologically indistinguishable from the development of fertilized oocytes21. Subsequent work expanded on these methods in mouse models and showed that exposing a mouse oocyte to electric- or hyaluronidase-mediumted activation mimics fertilization and promotes cell division towards the formation of parthenogenetic blastocysts containing a mixture of haploid and diploid cells22,23. Further elaboration on the methods of oocyte activation in mouse showed that this can be achieved in many ways, from spontaneous activation upon mechanical handling to thermal, electric or chemical activation24. Ultimately, investigators showed strontium chloride (SrCl2) to be the only known parthenogenetic-activating agent that induces repetitive intracellular calcium releases25–27 in a manner similar to those following normal fertilization by spermatozoa28.
Moreover, it was observed that parthenogenesis may occur spontaneously in mice29 and women30, leading to the generation of benign ovarian teratomas composed of a mixture of diploid and haploid cells, thus highlighting how haploid cells can persist after parthenogenesis. Collectively, these observations led to the development of methods for oocyte activation, growth and expansion of parthenogenetic blastocysts and subsequent isolation and maintenance of haploid mammalian ESCs from the mouse10,11,31–33, the monkey34, the rat35 and the human12,13. Analogously, by transferring sperm into an enucleated oocyte, haploid androgenetic ESCs have also been derived36,37. Parthenogenetic haploid ESCs are fully competent for functional contribution to the germline38.
The generation of mouse and human hESCs has opened the door to functional random mutagenesis screens39 that have shown that these cells can be successfully used, for example, for the identification of loss-of-function and separation-of-function mutants via approaches such as ethyl methanesulfonate mutagenesis40,41, clustered regularly interspaced short palindromic repeats (CRISPR) screens33,42,43 or transposon-induced mutagenesis42. The use of haploid cells in point mutagenesis screens open an important avenue for identification of separation-of-function mutants40. Using hESCs in CRISPR screens allows higher efficiency, as one confounding factor in such genome-wide approaches in diploid cells is the presence of heterozygous deletions. Moreover, the hESCs are amenable to differentiation in different cell types, which in the future will allow screening in a multitude of cell lineages43. Furthermore, the generation of such cells allowed the creation of a cell bank (Haplobank: www.haplobank.at) of >100,000 publicly available individual mouse hESC lines carrying conditional and reversible disruption events in 16,970 mouse genes42.
Limitations of the protocol
Mouse strain background can play a defining role in the success of hESC derivation. We have derived hESCs from C57BL/6J, NOD/ShiLtJ and 129S1/SvImJ genetic backgrounds, and have observed that the success rate was higher for C57BL/6J and NOD/ShiLtJ than for 129S1/SvImJ ones. Another important limitation of the method is the fact that mouse and human hESCs spontaneously diploidize upon serial passage. Thus, to maintain them in a haploid format, cells need to be periodically flow-sorted from time to time (Boxes 1–3; Figs. 1–3). Notably, as shown by others53, we have also observed that the success of hESC derivation can be influenced by the knockout background; for example, Tp53 knockout hESCs are more stable in culture than wild-type (WT) cells. Another important note is that, with adaptation, hESCs become more stable and it takes a longer time for them to diploidize (~3 weeks, depending on background and number of cells sorted). Upon diploidization, ESCs remain stable and no longer duplicate their genome (i.e. they do not readily become polyploid). In all the clones that we have karyotyped, we observed low rates of chromosomal abnormalities involving mouse chromosome 8, mouse chromosome 11 and loss of chromosome X, consistent with observations of ESCs derived from fertilized embryos44,45. In addition, the experimenter needs to keep in mind that the parthenogenetic hESCs derived from oocytes will always contain an X chromosome and never a Y chromosome.
Box 1. Quantification of cell cycle profile of newly derived ESCs and flow cytometric analysis.
Timing 1.5 h for 10 samples
Additional reagents
Propidium iodide solution (PI; Sigma-Aldrich; cat. no. P4864).
CAUTION
PI is a health hazard if swallowed and is a mutagen. Consult safety and hazard sheets provided by the seller.
Sphero Rainbow fluorescent particles (Spherotech, cat. no. RCP-20-5).
Equipment
Falcon round-bottom tubes (polypropylene, 5 ml, snap-cap; BD, cat. no. 352063).
Filters (30 μm, green; CellTrics; Sysmex, cat. no. 25004-0042-2316)
Flow cytometer (CytoFLEX S; Beckman Coulter; model no. B75442)
Reagent setup
FACS buffer. FACS buffer is 0.5% BSA in 1x PBS. It can be stored for up to 1 month at 4 ºC.
PI solution. PI solution is 10 μg/ml PI and 250 μg/ml RNase in FACS buffer. Make fresh.
Procedure: quantification of cell cycle profile
-
1 |
(Optional) Collect medium from each well into a separate 15-ml conical tube and place it on ice.
-
2 |
(Optional) Wash the plate with 2 ml of 1x PBS per well and collect it in the same respective tube.
CRITICAL STEP
Steps 1 and 2 are important if floating cells (i.e. dead or mitotic) are to be taken in account; otherwise, the experimenter can just remove the medium, wash with 1x PBS and proceed to step 3.
-
3 |
Add 0.5 ml of trypsin-EDTA (for a well of a 6-cm plate) and incubate the plate for a few min at 37 °C.
-
4 |
Transfer trypsinized cells from the well to the tube from step 2, if available.
-
5 |
Filter the cells through a 30-μm filter to remove any cell clumps and count the cells.
CRITICAL STEP
Proceed with 1 million-5 million cells, depending on how many cells are available. For consistency of comparison with diploid control across the gates, it is important to keep all the samples at the same cell concentration (so that voltage settings on the flow cytometer do not need to be changed).
-
6 |
Centrifuge for 5 min at 450g at RT and discard the supernatant.
-
7 |
Resuspend the cells in 5 ml of ice-cold 1x PBS containing 1 mg/ml BSA.
-
8 |
Centrifuge for 5 min at 450g at RT, discard the supernatant and resuspend the cells in the residual liquid.
-
9 |
Add 4 ml of ice-cold 70% ethanol dropwise, using the vortex.
-
10 |
Incubate the samples at -20 °C for at least 30 min.
Pause point: Cells can be kept at -20 °C for weeks.
-
11 |
Centrifuge for 5 min at 450g at RT and discard the supernatant.
-
12 |
Resuspend the cells in 0.5 ml of PI and transfer the cells to a FACS tube.
Pause point: Cells can be kept at 4 °C overnight.
Procedure: Flow cytometric analysis
Timing: 30 min for 10 samples
-
13 |
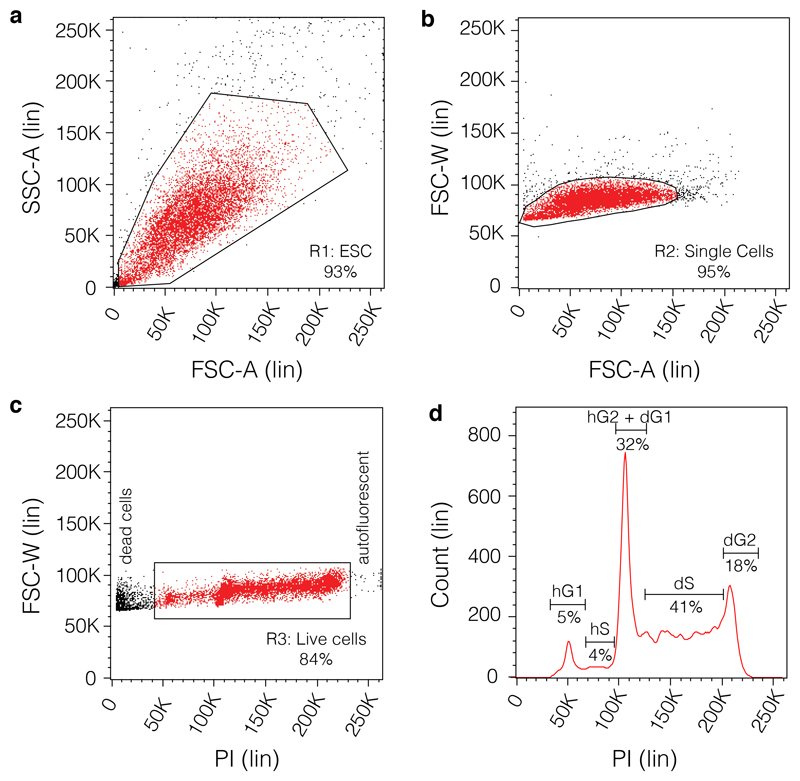
Using the diploid control, acquire events with side scatter area on a linear scale (Lin) (SSC-A)-Lin versus forward scatter area (FSC-A)-Lin axes. The SSC-A-Lin versus FSC-A-Lin plot should reveal one population. Use the voltage and gain settings to centre the main population and create the first gate (R1: ESCs) (Fig 1a).
-
14 |
Isolate doublets by plotting forward scatter width (FSC-W)-Lin versus FSC-A-Lin and set gate R2 around single cells (Figure 1b).
CRITICAL STEP
In the case of haploid/diploid cells, the doublet cell population will reveal two overlapping populations, haploid (n; below) and diploid (2n; above); when the first experimental sample is run, double-check that the haploid population is not excluded).
-
15 |
Isolate the live cells by plotting FSC-W-Lin versus PI stain to identify dead cells (left side) and events caused by autofluorescence (right side) and remove them by setting gate R3 on the PI population (Figure 1c).
CRITICAL STEP
In the case of haploid/diploid cells, the doublet cell population will reveal two populations overlapping at the 100k mark: haploid (n; at 50k-100k) and diploid (2n; 100k-225k); when the first experimental sample is run, double-check that the haploid population is not excluded.
-
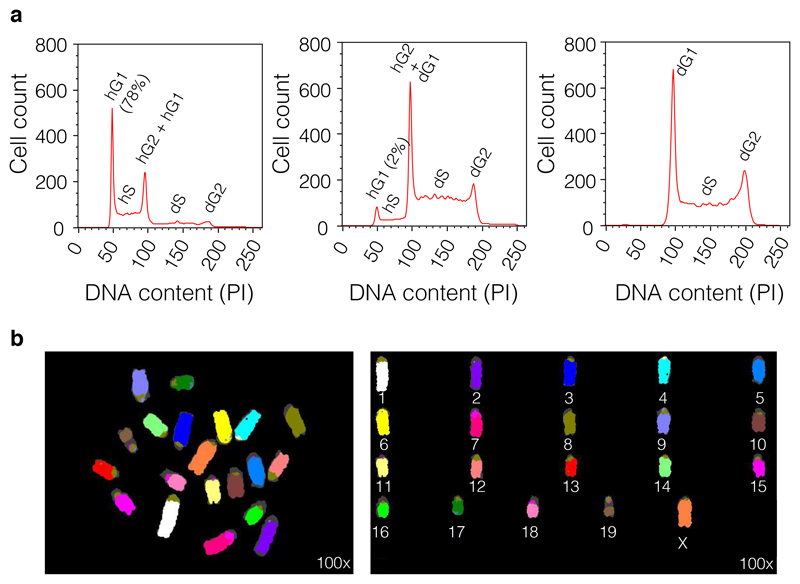
16 |On Count-Lin versus PI-Lin axes, resolve the cell cycle profile. Acquire at least 10,000 events. On a cell line that contains a mix of haploid/diploid cells the following will be evident (Figure 1d):
- Haploid G1 phase of the cell cycle (hG1) at the 50K mark;
- Haploid S phase of the cell cycle (hS1) between the 50K and 100K marks;
- Overlapping haploid G2 phase of the cell cycle (hG2) and diploid G1 phase of the cell cycle (dG1) populations at the 100K mark;
- Diploid S phase of the cell cycle (dS) between 100K and 200K marks;
- Diploid G2 phase of the cell cycle (dG2) at the 200K mark.
Figure 2a shows the cell cycle profiles of three random WT clones.
CRITICAL
It is critical to first run the diploid-only control to recognize the diploid cell cycle peaks. Some experimental samples might be pure haploid populations, and without a diploid control, it will be impossible to tell them apart.
-
17 |
Save the experiment settings and experimental analysis on the flow cytometer.
Box 3. Sorting of haploid ESC.
Timing 1 h
Before preparing for the sort, bear in mind that you should always have a fully diploid control ESC cell line available to stain side by side to be able to calibrate the cell cycle profiles on the sorting machine. Before starting the cell harvest procedure, fill two Falcon round-bottom tubes to the edge with warm DMEM culture medium (from Step 52) and leave them at RT in a hood for at least 30 min (the serum from the medium will prevent the sorted cells from sticking to the walls and not being collected in the collection medium). Gelatinize T75 flasks using 5 ml of 0.02% gelation solution each for 5 min. Prepare a bucket of ice to store and transport the cells to and from the sorter.
Additional reagents
70% Ethanol (Fisher Bioreagents, cat. no. BP8201-1).
CAUTION
Ethanol is flammable and an irritant if swallowed. Consult afety and Hazard sheets that are provided by the seller.
Hoechst 33342, trihydrochloride, trihydrate (10 mg/ml solution in water (Invitrogen; Thermo Fischer Scientific cat. no. H3570).
CAUTION
Hoechst 33342 is a health hazard if swallowed and a mutagen. Consult safety and hazard sheets provided by the seller.
Verapamil hydrochloride (working solution is 100 μmol/l, Sigma-Aldrich; cat. no. V4629)
CAUTION
Verapamil hydrochloride is acutely toxic if swallowed and an environmental hazard. Consult safety and hazard sheets provided by the seller.
Sphero Rainbow Fluorescent particles, (Spherotech, cat. no. RCP-20-5).
Equipment
Falcon round-bottom tubes (polypropylene, 5 ml, snap-cap; BD, cat. no. 352063).
Filters (CellTrics 30 μm, green; Sysmex, cat. no. 25004-0042-2316).
Flow cytometer (CytoFLEX S; Beckman Coulter; model no. B75442).
Cell sorter (BD Influx, BD Biosciences; cat. no. 646500).
Reagent setup
FACS buffer. FACS buffer is 0.5% BSA in 1x PBS. The solution can be stored for up to 1 month at 4 ºC.
Hoechst working solution. Hoechst working solution is 1 μg/ml Hoechst in DMEM culture medium (e.g., add 1 μl of 10 mg/ml Hoechst 33342 stock for each 100 μl of DMEM culture medium). Make fresh.
Procedure
-
1 |
Wash the cells with 1x PBS, trypsinize them using 1 ml of trypsin-EDTA and take up the cells in DMEM culture medium (10 ml for a T75 flask) by pipetting up and down a couple of times.
-
2 |
USe 30-μm filters to remove cell clumps; filter into a new 15-ml conical tube.
-
3 |
Spin the tube down at 450g for 5 min at RT; discard the supernatant and resuspend in appropriate volume of NDiff N2B27 culture medium (0.5-ml minimum for a T25 flask or 1-ml minimum for a T75 flask).
-
4 |
Prepare Hoechst working solution in DMEM culture medium (e.g. 3 μl in 300 μl of DMEM culture medium).
CRITICAL STEP
For some applications, such as point mutagenesis screens, it is advised to avoid the use of Hoechst dye for setting up the gates and instead to sort by size only.
-
5 |
For each millileter of cell suspension, add 100 μl of 1 μg/ml Hoechst working solution to a final Hoechst concentration of 10 μg/ml. Incubate the mixture in the tissue culture incubator for up to 30 min (at 37 °C, 5% CO2) with gentle resuspension every 10 min.
-
6 |
(Optional) If you wish, add verapamil (an inhibitor of calcium pumps and hence any G1 to S transition) to the staining buffer to promote the identification of hG1 phase cells.
-
7 |
Filter into a new flow cytometry tube using 30-μm filters to remove cell clumps. Aspirate the medium from the collection tubes, allowing 0.5 ml of residual medium in each tube and transport the collection tubes, the diploid control tube and the sample tubes to the sorting machine for sorting. Sorting can take 2 – 3 h (depending on how many cells are sorted).
CRITICAL STEP
As with any cell sorting protocol, make sure the sorting machine is clean before sorting so that contamination with other cells or pathogens is avoided.
-
8 |
Align the optical light path of the flow cytometer before cell analysis using 3-μm beads (Sphero Rainbow Fluorescent particles) with minimum peak coefficient of variance for all fluorescence channels.
CRITICAL STEP
In parallel, stain as above the same (isogenic) cell line that has been allowed to diploidize previously. Using small aliquots (e.g. 100 μl), prepare a 1:1 mixture from the haploid line for sorting and the isogenic diploid line. This step will allow the clear differentiation between haploid/diploid populations.
-
9 |
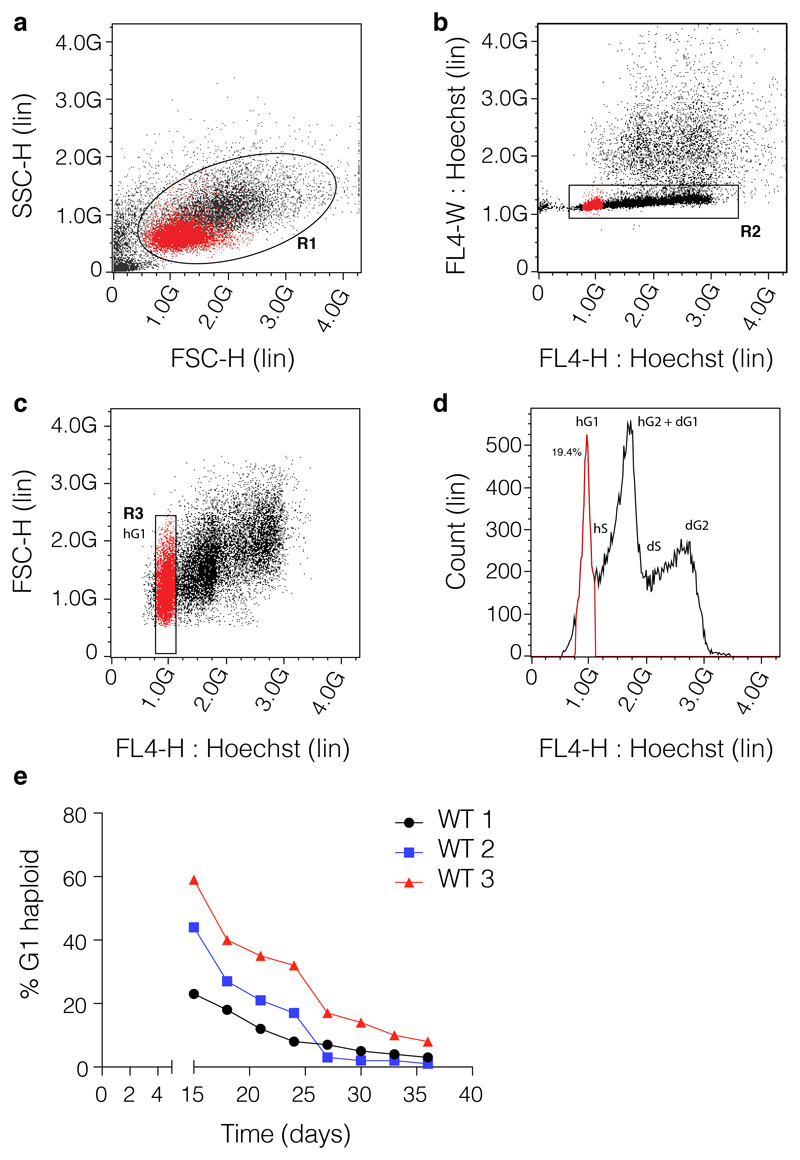
Sort the G1 haploid population, following the gating strategy depicted in Fig. 3 and collect as many cells as possible. Haploid cells can be sorted as single cells in individual wells in 96-well plates. The clones selected this way have different haploid latency times (Fig. 3d). If, for downstream experimental purposes (e.g. point mutagenesis), the use of Hoechst dye is not desired, establish the gates with a small subpopulation of Hoechst-stained cells, and then sort the bulk of the cells by size (mouse hESCs are approximatively half the size of diploid isogenic ESCs). Precautions must be taken to avoid mistakenly gating out the diploids as doublets.
CRITICAL STEP
Sort the bulk of the hG1 population and avoid the sub-hG1 population, which might contain cells that have lost chromosome X (mESCs are reported to randomly loose the X chromosome45).
-
10 |
Spin the sorted cells down (450g, 5 min at RT) and resuspend in hESC medium.
-
11 |
Plate the cells in appropriate flasks or dishes containing hESC medium and allow for recovery.
CRITICAL STEP
The hESCs can be sorted as a bulk population or as single-cell clones. Although the sorting of a bulk population reliably obtains almost pure haploid populations, sorting of individual cells has different rates of success due to the long time these cells have to be in culture before they can be expanded for banking. An example of the haploid status of three individual clones from sorting three individual WT cells is presented in Fig. 3e.
Figure 1. Representative flow cytometric analysis of the cell cycle profile.
(a) The samples harvested and fixed as described in the protocol from Box 2 are acquired on SSC-A-Lin versus FSC-A-Lin plot to reveal the ESCs population. The gate R1 is centred around the main population representing the bulk of the ESCs. (b) Doublets are isolated by setting up gate R2 around the single cells by on an FSC-W-Lin versus FSC-A-Lin plot. In the case of the haploid/diploid cells the doublet cell population will reveal two overlapping populations, haploid (n; below; note that as explained in the text using the right controls this population can be sorted by size only without the use of a dye) and diploid (2n; above). (c) The live cells are identified by plotting gate R3 on FSC-W-Lin versus PI to identify and isolate dead cells (left side) and events caused by auto-fluorescence (right side). (d) Representative cell cycle profile of a clone that contains a mix of haploid and diploid cells. On Count-Lin versus PI-Lin axes the following cell cycle phases can be identified: the haploid G1 phase of the cell cycle (hG1), the haploid S phase of the cell cycle (hS1), the overlapping haploid G2 phase of the cell cycle (hG2) and diploid G1 phase of the cell cycle (dG1), the diploid S phase of the cell cycle (dS) and the diploid G2 phase of the cell cycle (dG2).
Figure 3. Sorting of haploid cells.
The sorting of the haploid ESCs follows similar set-up as for the cell cycle analysis on fixed cells. (a) The haploid/diploid cell line is stained with Hoechst 33342 and gate R1 is centred around the main population representing the bulk of the ESCs (SSC-H-Lin vs. FSC-H-Lin). (b) Doublets or clumps are removed by setting gate R2 around the main cell population (FL4-W: Hoechst-Lin vs. FL4-H: Hoechst-Lin); care must be taken so that diploids are not mistaken as doublets. A 1:1 haploid: diploid isogenic mix is recommended to be used always to set up the gates. (c) The cells gated on R2 are subsequently resolved on an FSC-H-Lin vs. FL4-H: Hoechst-Lin plot and haploid G1 (hG1) population is identified and sorted using gate R3. Gate R3 is built tightly around the hG1 peak to avoid sorting of cells with lower DNA content (that might be aneuploidy). (d) The profile of the hG1 (gate R3) on the Count-Lin vs. FL4: Hoechst-Lin representing the cell cycle profile. (e). Graph presenting the overtime %hG1 status of 3 individual WT clones derived by individually sorting from the same WT line. Data was obtained by flow-cytometry profiling using PI. 10,000 cells (events) were acquired for each cell line at each time point.
Experimental design considerations
A workflow of the procedure is outlined in Fig. 4. Female mice should be matched by genetic background, and ideally the experimenter should start with no less than five individual mice. The use of mice should be compliant with local animal research regulations as well as the ARRIVE guidelines46. If the derivation is done from specific knockout lines, to obtain both WT control and mutants in the same derivation, heterozygous mice should be used. Although an elaborate technique, by following the procedure described herein, the derivation of hESCs is highly reproducible.
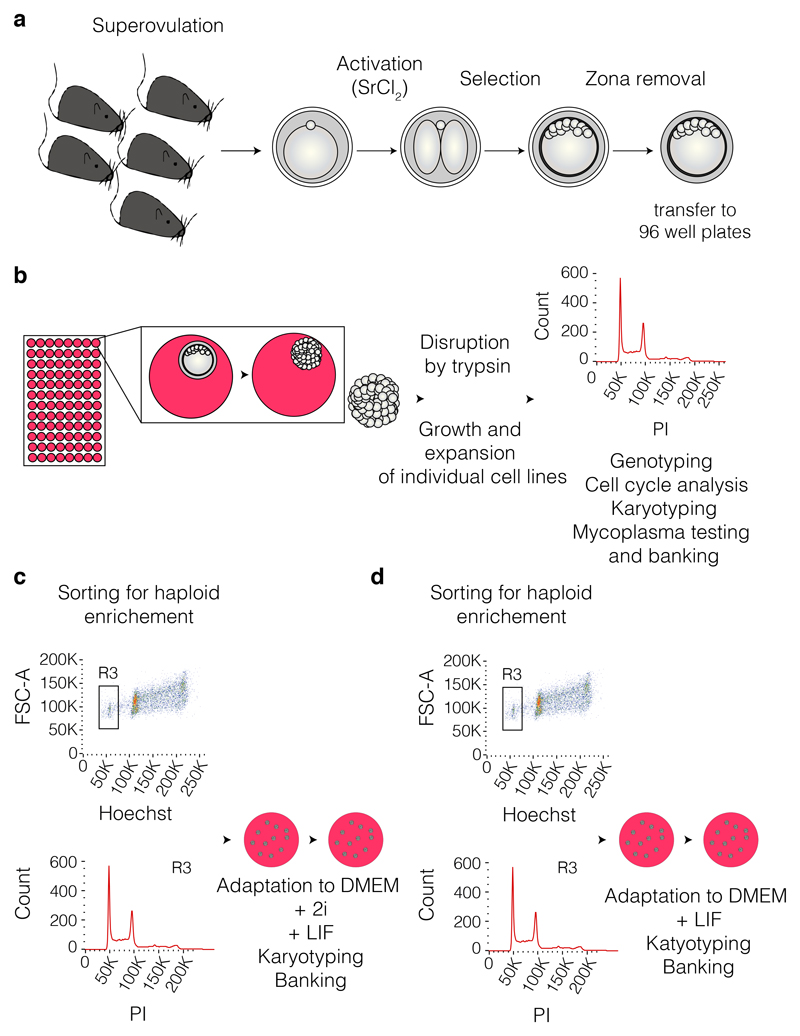
Figure 4. Workflow diagram of hESC derivation.
(a) Female mice of selected genotypes are selected, superovulated and used for the harvest of oocytes. The oocytes are then activated by the use of strontium chloride (SrCl2) and passed through rounds of selection for identification of blastocysts that can be used for zona pellucida removal and later transferred individually in NDiff-N2B27/2i/LIF culture medium in 96 well plates (steps 1 to 52). (b) The individual clones are allowed to expand to form defined colonies that are disrupted by trypsinisation and grown and expanded into individual cell lines (steps 53 to 62). The established cell lines are then quality checked including: haploid status (for cell cycle analysis see Box 1), karyotyped (see Box 2), mycoplasma tested and banked. (c and d) The haploid cell lines can be adapted to DMEM/2i/LIF culture or only DMEM/LIF culture while enriching for the haploid content by sorting (see Box 3).
Materials
Biological materials
Female mice, ideally between 3 and 6 weeks of age (e.g., C57BL/6J mice; The Jackson Laboratory, stock no. 000664)
CAUTION
All procedures performed on mice must be carried out following national and institutional guidelines. In the UK, Home Office regulations must be followed with appropriate establishment, project and personal licensing in place. Experiments using mouse models should be performed in accordance with the ARRIVE guidelines46. Approval for our experiments was granted by the UK Home Office.
Reagents
Strontium chloride hexahydrate (Cl2Sr·6H2O; Sigma-Aldrich, cat. no. 204463-10G).
CAUTION
Strontium chloride hexahydrate is harmful if swallowed and an irritant. Consult safety and hazard sheets provided by the seller.
Ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid tetrasodium salt (C14H20N2O10Na4; EGTA tetrasodium; Sigma-Aldrich, cat. no. E8145).
KSOM Mouse Embryo Culture Medium with phenol red (KSOM medium; GlobalStem, cat. no. GSM 5140).
M2 medium with HEPES, without penicillin and streptomycin, liquid, sterile-filtered (M2; Sigma-Aldrich, cat. no. M7167).
Pregnant mare's serum gonadotropin (PMSG; Prospec Protein Specialists, cat. no. HOR 272).
Human chorionic gonadotropin (HGC; 1,500 IU; under the brand name Chorulon, MSD Animal Health, cat. No. VM10708/4301)
Hyaluronidase from bovine testes Type I-S (lyophilized powder, 400-1000 U/mg solid; Sigma-Aldrich, cat. no. H3506-100MG).
Mettler buffer sachets (pH4.01 Pk30 and pH 7.01 Pk30; Scientific Laboratory Supplies, cat. nos. PHM0144 and PHM0142).
Dulbecco's PBS (DPBS; Thermo Fisher, cat. no. 14190094).
Acidic Tyrodes solution (Millipore, cat. no. MR-004-D).
Hydrochloric acid solution (HCl; 1.0 N, bioreagent, suitable for cell culture; Sigma-Aldrich, cat. no. H9892).
CAUTION
Hydrochloric acid solution is corrosive and harmful if swallowed, as well as an irritant. Consult safety and hazard sheets provided by the seller.
NDiff N2B27 medium (StemCells, cat. no. SCS-SF-NB-02).
GSK3 inhibitor: CHIR99021 (CHIR; Abcam, cat. no. ab120890).
CAUTION
CHIR is acutely toxic if swallowed and an irritant. Consult safety and hazard sheets provided by the seller.
MEK inhibitor: PD0325901 (PD; Selleckchem, cat. no. S1036).
CAUTION
PD is acutely toxic if swallowed. Consult safety and hazard sheets provided by the seller.
ESGRO leukemia inhibitory factor supplement for mouse ES cell culture (LIF; EMD Millipore-Merk, cat. no. ESC1107).
DMEM, high glucose with UltraGlutamine (Lonza; cat. no. BE12-604F/U1).
Gelatin solution (Sigma-Aldrich, cat. no. G1393).
PBS (1x; Lonza; cat. no. BE17-516F).
2-Mercaptoethanol (Sigma-Aldrich, cat. no. M6250).
CAUTION
2-Mercaptoethanol is classified as an acute toxin, health hazard, environmental hazard, and a DNA mutagen. Consult safety and hazard sheets provided by the seller; use only under a chemical cabinet.
Dimethyl sulfoxide (DMSO; Sigma-Aldrich; cat. no. D2650).
FBS (Gibco; Thermo Fischer Scientific, cat. no. A31608-02).
Bovine Albumin Fraction V (BSA; 7.5% (wt/vol) solution; Gibco; Thermo Fischer Scientific, cat. no. 15260037).
MEM non-essential amino acids solution (NEAA, 100x; Gibco; Thermo Fischer Scientific; cat. no. 11140-050).
CAUTION
NEAA is an irritant if swallowed. Consult safety and hazard sheets provided by the seller.
Penicillin-streptomycin-glutamine (100x; Gibco; Thermo Fischer Scientific; cat. no. 10378-016).
CAUTION
Penicillin-streptomycin-glutamine is a health hazard if swallowed. It should be disposed by appropriate routes. Consult safety and hazard sheets provided by the seller.
Penicillin-streptomycin (10,000 U/ml; Gibco; Thermo Fischer Scientific; cat. no. 15140122).
CAUTION
Penicillin-streptomycin is a health hazard if swallowed. It should be disposed by appropriate routes. Consult safety and hazard sheets provided by the seller.
Sodium Pyruvate (100 mM; Gibco; Thermo Fischer Scientific; cat. no. 11360-039).
Trypsin-EDTA (0.05% (wt/vol), phenol red; Gibco; Thermo Fischer Scientific; cat. no. 25300-054).
Trypan blue solution (0.4% (wt/vol) in PBS, pH 7.5 ± 0.5; Corning, cat. no. 25-900-CI).
CAUTION
Trypan blue solution is acutely toxic if swallowed and is a health hazard. Consult safety and hazard sheets provided by the seller.
Equipment
Stereomicroscope (Leica, cat. no. M125).
CO2 incubator (Eppendorf/New Brunswick Galaxy 48R incubator, cat. no. CO48R230 or Panasonic cat. no. KM-CC17RU2).
Tissue culture biological safety cabinet (NuAire LabGard ES cat. no. NU-437).
pH meter (Five Easy FE20; Mettler Toledo, cat. no. 10385343).
Fine forceps (Student Dumont no. 5 forceps; Interfocus, cat. no. 91150-20).
Fine scissors (straight/large loop; Interfocus, cat. no. 14040-10).
Pasteur pipettes (Scientific Laboratory Supplies Ltd, cat. no. PIP4172).
Tissue culture-grade flasks or plates (Corning, cat. no. 430641U or 430167)
Falcon center-well culture dish for in vitro fertilization (60mm; Scientific Laboratory Supplies, cat. no. 353653 or similar).
Aspirator tube assemblies for calibrated microcapillary pipettes (Sigma-Aldrich, cat. no. A5177-5EA).
Millex GS 0.22-μm filter (EMD Millipore, cat no. SLGS033SB)
T25 flasks (Corning, cat no. 431464U)
Reagent setup
Strontium chloride hexahydrate solution
Prepare 100mM Strontium chloride hexahydrate solution by using an analytical balance with precision of 0.1mg to weigh 26 mg of Cl2Sr·6H2O and add it to 1 ml of KSOM medium. Make up fresh and use within 24 h. Store at 4 °C.
CAUTION
Cl2Sr·6H2O may cause serious eye damage or respiratory irritation.
EGTA
Prepare 0.5 M EGTA using an analytical balance with precision of 0.1 mg to weigh 234 mg of EGTA tetrasodium and add it to 1 ml of KSOM medium. pH adjustment of the EGTA solution is important; the pH will initially be ~10. Add 1 M HCl to the 1 ml of EGTA solution in increments of 200 μl, adding a total of 600 μl and using the pH meter to ensure the pH = 8. Make up fresh and use within 24hrs. Store at 4 °C.
Oocyte Activation Medium
Add 150 μl of 100 mM strontium chloride hexahydrate solution and 12 μl of 0.5 M EGTA to 3 ml of KSOM medium and filter-sterilize. Store at 4 °C for use the following day or put 2 ml in a 60-mm centre-well culture dish, surrounded by PBS in outer well, and pre-equilibrate to 37 °C for use on the same day.
Hyaluronidase solutions
Make a stock solution of hyaluronidase by adding 3.53 ml of M2 to 100 mg lyophilised powder. Gently agitate the solution, ensuring that the powder is fully dissolved. Make working concentrations by adding 100 μl of 30 mg/ml stock hyaluronidase solution to 900 μl of M2 to create a 3 mg/ml solution. Filter-sterilize, divide into 200-μl aliquots and store at -20 °C for up to 1 year. For use in removal of oocyte cumulus cells, add one 200-μl aliquot of the solution to 1.8 ml M2.
Pregnant mare's serum gonadotropin
Prepare PMSG in a clean class 2 safety cabinet. Reconstitute the 5,000 international units (IU) of lyophilised powder by mixing it with 80 ml of PBS, ensuring that the powder is completely dissolved. Divide the solution into 1.5-ml aliquots and store at -20 °C to use for up to 3 months.
Human chorionic gonadotropin
Prepare HCG in a clean class 2 safety cabinet. Reconstitute the 1,500 IU of lyophilised powder by mixing it with 30ml of PBS, ensuring that the powder is completely dissolved. Divide the solution into 1.5-ml aliquots and store at -20 °C to use for up to 3 months.
NDiff N2B27 culture medium
To 94 ml NDiff N2B27 medium, add 5 ml of 7.5% BSA, 1 ml of penicillin-streptomycin (100X), 10 μl of LIF, 1 μl of 2-mercaptoethanol, 2i (10 μl of PD (10 mM stock; stored at 4°C) and 30 μl of CHIR (10 mM stock; stored at -20°C)). NDiff N2B27 culture medium can be stored at 4 °C for up to 1 month.
CAUTION
2-Mercaptoethanol is classified as a severely toxic chemical and a DNA mutagen. Consult safety and hazard sheets provided by the seller; use only under a chemical cabinet.
DMEM culture medium
To 500 ml of DMEM high-glucose medium (1 bottle), add 75 ml of FBS, 6 ml of NEAA (100X), 6 ml of sodium pyruvate (100 mM), 6 ml of penicillin-streptomycin-glutamine (100X), 60 μl of LIF, and 6 μl of 2-mercaptoethanol. (Optional) If 2i addition is required (see Procedure) add 60 μl of PD (10 mM stock; stored at 4 °C) and 180 μl of CHIR (10 mM stock; stored at -20°C). DMEM culture medium can be stored at 4°C for up to 1 month.
CAUTION
2-Mercaptoethanol is classified as a severely toxic chemical and a DNA mutagen. Consult safety and hazard sheets provided by the seller; use only under a chemical cabinet.
0.02% Gelatin solution
Allow 2% (vol/vol) gelatin solution to completely liquefy at 37 °C and make 0.02% gelatin working solution with 1x PBS (e.g. 5 ml of 2% gelatin in 500 ml of 1x PBS). The solution can be stored for up to 1 year at room temperature (RT; 20-25 °C).
Equipment setup
Stereo microscope with heated stage
Manipulation of embryos is done using a stereomicroscope with heated stage and a zoom capacity of up to 100x magnification.
Humidified incubator
Incubation of oocytes in oocyte activation medium and all subsequent culture is done in a humidified incubator set to 37 °C, 5% CO2.
Procedure
Superovulation
Timing: 30-40 min
-
1 |
Identify female mice to use as donors for oocyte production and set aside in advance. The mice should be superovulated between the ages of 3 and 4 weeks.
CAUTION
All procedures on mice must be carried out in accord with relevant local and national regulations. In the UK, this means in accordance with Home Office regulations and with appropriate establishment, project and personal licensing in place.
CRITICAL STEP
Note that the best age for superovulation varies from strain to strain but usually lies between 3 and 6 weeks of age, during the prepubescent stage of development47.
-
2 |
Inject the mice at 3 pm, removing the PMSG hormone (6.25 IU, 0.1 ml for each female mouse) from the freezer 15-20 min before the injections are scheduled, in order to allow the hormone to defrost and warm to RT.
-
3 |
Once completely defrosted, draw up the hormone into a 1-ml syringe, making sure there are no air bubbles in the syringe. Attach a 27-gauge needle to the syringe, hold it with the needle facing up and depress the syringe plunger to expel air from the dead space created.
-
4 |
Scruff the mouse and turn it so its stomach is facing upwards and its head is slightly tilted back.
-
5 |
With your free hand, hold the syringe so the needle is bevel side up. Carefully insert the needle into the peritoneal cavity of the mouse, to the left or right of the midline of the mouse, making sure not to hit the bladder.
-
6 |
Following your approved technique for mouse i.p injection, administer 6.25 IU of PMSG (0.1 ml). After preparing the PMSG solution, we draw it up in the syringe, keeping in mind the dead volume in the needle. We gently remove the animal from the cage and restrain appropriately in the head-down position. We disinfect the lower right quadrant of the abdomen with 70% ethanol and inject by inserting the needle with the bevel facing up at ~30° angle to horizontal, toward the head in order to avoid damage to the urinary bladder, cecum and other abdominal organs. We wait briefly before withdrawing the needle, to make sure the liquid does not seep out.
-
7 |
Place the mouse into an empty cage and then repeat Steps 5 and 6 for each mouse until finished. Once all mice have been injected, they can then be returned to their original cages.
-
8 |
Dispose of the needle and syringe in a sharps bin.
CAUTION
Extreme caution is required at all times when handling used sharps. Dispose sharps directly without capping.
-
9 |
46-48 h after PMSG administration, repeat Steps 2-8, but inject with 5 IU of HCG (0.1 ml), rather than PMSG.
Dissection and oocyte harvest
Timing: 50 – 60 min
-
10 |
Set up four single Falcon center-well culture dishes per line; one containing oocyte activation medium and three with KSOM medium for washing. There should be 1 ml of respective culture medium in the centre well, with 2-3 ml of DPBS in the outer well.
-
11 |
Filter 10 ml of M2 into a Falcon tube, using a 0.22 μm filter, and place the tube in an incubator to warm to 37 °C.
-
12 |
Remove one vial of hyaluronidase (working stock solution) from the -20 °C freezer and thaw it in an incubator, warming to 37 °C.
-
13 |
Place a 50-mm dish containing 2-3 ml of M2 or 0.9% DPBS on a heat blanket or warming stage set to 37 °C.
-
14 |
Cull a female by cervical dislocation and lay the mouse on its back, exposing the abdomen.
-
15 |
Make a midline incision up and though the abdominal wall, followed by the peritoneal sac, to expose the internal organs, pushing the gut up and out toward the chest revealing the reproductive organs.
-
16 |
Grasp the upper end of one of the uterine horns with fine forceps and gently pull the uterus, oviduct, ovary, and fat pad taut and away from the body cavity. This will reveal a fine membrane, the mesometrium, which connects the reproductive tract to the body wall and carries a prominent blood vessel. Make a hole in the membrane close to the oviduct with the closed tips of a pair of fine forceps or scissors.
-
17 |
Reposition the forceps and cut the uterus near the oviduct.
-
18 |
Transfer the oviduct and attached segment of uterus to the 50-mm dish containing DPBS at 37 °C.
CRITICAL
It is important to keep oocytes at 37 °C at all stages to maintain oocyte viability and chances of successful parthenogenesis.
-
19 |
Repeat Steps 14-18 for all females; oviducts from several mice can be collected in the same dish.
-
20 |
Prepare a 35-mm dish containing 2 ml of filtered M2 for washing oocytes after exposure to hyaluronidase. Keep the dish at 37 °C.
-
21 |
Pipette 1.8 ml of the pre-warmed M2 into a 35-mm culture dish.
-
22 |
Move 1x oviduct into the dish with the 1.8 ml of filtered M2 and ‘pop’ the swollen ampulla to release the cumulus mass, then discard the oviduct. Repeat until all oviducts have been processed. (Supplementary Video 1).
-
23 |
Add all the hyaluronidase working stock solution from the vial and gently swirl/agitate the dish to help disperse the cumulus mass or pipette up and down gently several times with a 1-ml tip, then wait for the cumulus cells to start falling away leaving individual oocytes.
CRITICAL STEP
Hyaluronidase can have a detrimental effect on the oocyte, so the oocytes should be collected and washed quickly. Reducing the amount of hyaluronidase added to the M2 will increase the time it takes embryos to become free from cumulus cells but will allow more time for users less familiar with the protocol to collect the oocytes.
-
24 |
Pick up the individual oocytes with a transfer pipette and place into the 2 ml of warmed M2, repeating until all oocytes are collected.
-
25 |
Wash oocytes through 6x drops of pre-warmed, filtered M2. Once washed, the oocytes can be kept together in a drop of M2 and assessed for viability.
Activation
Timing: 2-3 h
-
26 |
Remove oocytes that have lysed or fragmented or that have a large perivitelline space (shrunken cell). A lysed cell of an oocyte will look darker in comparison to a healthy oocyte. Fragmentation of a single cell can occur and can vary from a section to the entire cell. A larger perivitelline space can be obvious when the oocyte is placed next to healthy examples. Sometimes the increased perivitelline space is all around the circumference of the cell; other times it is more obvious on one side of the oocyte. If in doubt, the oocytes can be retained and put in activation medium to be removed later, if necessary. Aspirate the oocytes around a drop of M2 with a mouth pipette, rolling the oocytes to allow viewing from a variety of angles under a bench-top stereomicroscope to make the assessment thorough.
Troubleshooting advice can be found in Table 1.
Table 1. Troubleshooting guidance.
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 26 | There are too many embryos to easily process in 50-60 min | More mice superovulated than is necessary | Only superovulate 10-15 mice per session |
| 27 | Embryos are being lost when manipulated using a mouth pipette | Pipette tip is flicked against cell culture dish | Use a cell culture dish with low edges or an unpturned dish with drops of medium |
| Embryos are sucked up beyond the tip of the pipette | Aspirate a small amount of M2 into the pipette tip and include a small air bubble to sit behind the embryos when manipulating | ||
| 39 | Embryos are not lysed but cell division looks abnormal | Embryo patterning differs in the absence of sperm entry | Allow the embryos to culture for longer |
| 42 | Blastocyst is close to breaching it's zona and hatching | The blastocysts were at an optimal stage for acid treatment over night | Perform the zona removal, being prepared to take them out of the acid sooner |
| 46 | The zona does not dissolve rapidly | The acid Tyrodes is not sufficiently warm | Warm the acid in an incubator at 37 °C and use a heat plate on the microscope |
| 49 | It is hard to visualise the dissolving of the zona | Magnification of the microscope is insufficient | Use x100 magnification capable objective |
| 50 | Zona disappears but then reappears when in M2 | Variation in osmotic gradient between mediums may give the impression the zona is shrinking away | Ensure the acid is suitably warmed |
| 52 | Embryos are lost when retrieving them from the vial | Too much medium in vial | Use no more than 50 μl in the vial. |
| Flush the inner sides and lid of the vial with M2 to dislodge and collect embryos that may have got attached to these surfaces |
-
27 |
Move any oocytes identified as subviable to a separate drop of M2 for final assessment. Discard sub-viable oocytes (Supplementary Video 2).
Troubleshooting advice can be found in Table 1.
-
28 |
Transfer all viable oocytes to the pre-incubated oocyte activation medium and place in a tissue culture incubator for 90 min at 5% CO2, 37 °C.
CRITICAL
Viable oocytes must be placed in to the oocyte activation medium for the stated time and without delay to optimise chances of successful parthenogenic activation.
-
29 |
Remove the oocytes from the oocyte activation medium by gently mouth pipetting and wash by successive placement in three dishes of pre-equilibrated KSOM medium.
-
30 |
Observe the oocytes for polar body extrusion, which indicates successful activation. Once the oocytes are in the oocyte activation medium it takes up to 90 min for all the oocytes to become activated (polar body extrusion).
-
31 |
Culture the oocytes for 72 hours.
Selection of blastocysts
Timing: 2-3 d
CRITICAL
This section of the protocol is outlined in Fig 5.
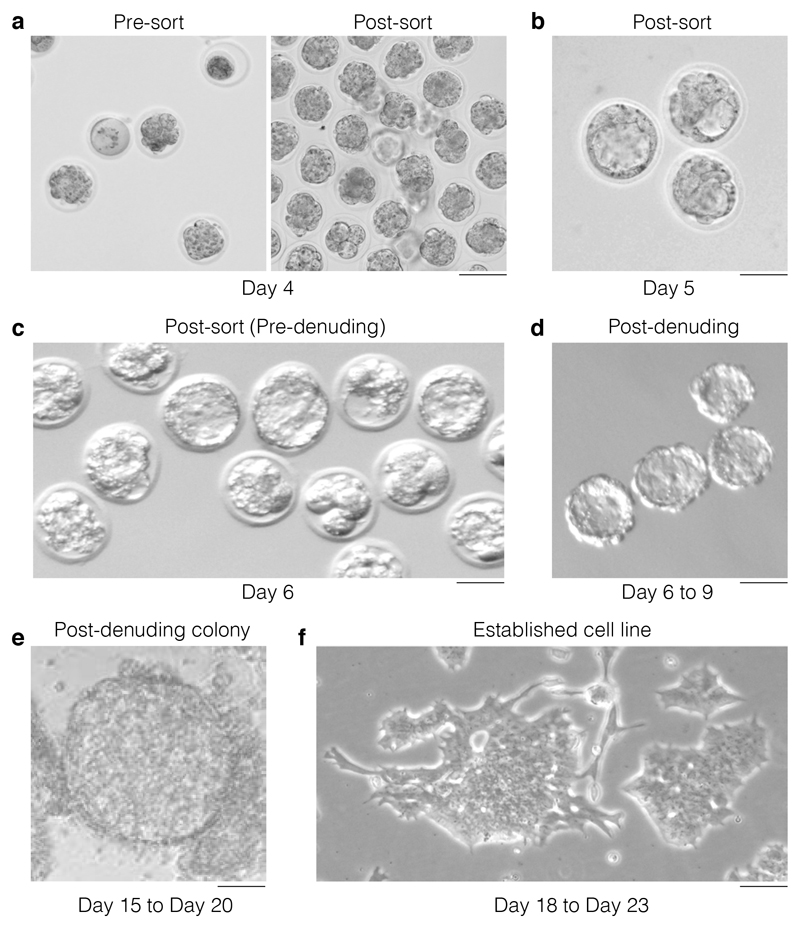
Figure 5. Morphological selection during the hESC derivation.
(a) On day 4 post harvesting and activation, embryos will be at a variety of developmental stages with some embryos looking sub-viable and some lysed (left panel: pre-sort). Upon removal of sub-viable or lysed embryos only the embryos that show viable characteristics are retained (right panel: post-sort). Scale bar 250 μm. (b) On day 5, the embryos that have developed to blastocyst and are suitable for zona pellucida removal are identified. Only select embryos that have a well expanded blastocoel for zona removal. Scale bar 250 μm. (c) The check and selection of blastocysts for zona removal is repeated on day 6 (representative images). Scale bar 250 μm. (d) Representative image depicting the morphologic aspect of blastocyst post-zona removal (days 6 to 9). These individual embryos are transferred to 96 well plates at this point. Scale bar 250 μm. (e) Representative image of a colony that has formed from a post-denuding blastocyst and that is ready for dissociation using trypsin. Scale bar 250 μm. (f) Representative image of an established hESC clone/cell line that is ready for quality control. Scale bar 250 μm.
-
32 |
On day 3 post activation, embryos will be at a variety of developmental stages: some embryos will appear subviable and some may have lysed (Fig 5a). Remove lysed embryos, but retain those in which the cells have not divided cleanly, as they may still progress to blastocysts.
-
33 |
Move 8-cell embryos to KSOM medium supplemented with 2i (CHIR (3 μM) and PD (1 μM)), pre-equilibrated to 5% CO2, 37 °C and continue to culture.
-
34 |
On day 4, there will be some embryos that have developed to blastocysts that are suitable for zona pellucida removal (Figure 5b). Select only embryos that have a well-expanded blastocoel for zona removal.
-
35 |
Repeat steps 32-34 (the check and selection of blastocysts for zona removal) on days 5 and 6 (Figure 5c).
Zona pellucida removal
Timing: 30 min
CRITICAL
When handling embryos, process one line at a time to make sure that strains do not become mixed, and label all dishes and vials containing embryos with strain information or a unique identifier.
-
36 |
Pre-warm acidic Tyrode’s solution to 37 °C.
-
37 |
Put two ~200-μl drops of acidic Tyrode’s solution and two ~200-μl drops of M2 on a dish suitable for working under a stereomicroscope.
-
38 |
Select the blastocysts for zona removal and move to one of the M2 drops.
CRITICAL STEP
Most blastocysts should be available for zona pellucida removal on this day. Move the remaining embryos into fresh, pre-equilibrated KSOM medium for further culture.
-
39 |
Transfer the blastocysts to the first drop of acidic Tyrode’s solution and observe the dissolving of the zona. The zona should take 30-70 s to dissolve.
Troubleshooting advice can be found in Table 1.
-
40 |
Briefly pipette the blastocysts up and down with a mouth pipette to ensure the zona is completely removed and transfer the blastocysts to the second drop of M2 (Supplementary Video 3 and Supplementary Video 4).
-
41 |
Observe the blastocysts in M2 and manipulate them with a new mouth pipette to check that the zona has been completely removed (Figure 5d; Supplementary Video 4). If the zona remains intact after inspection, repeat steps 38-41 using a second drop of acid.
-
42 |
When the zonae have been removed, immediumtely transfer the blastocysts to a fresh dish of M2.
Troubleshooting advice can be found in Table 1.
-
43 |
Wash the blastocysts through an additional 2 dishes of M2.
-
44 |
If transporting the blastocysts, pipette 50 μl of M2 into a 1.5-ml microcentrifuge tube and label it with strain and embryo number. Using a mouth pipette and M2, load an air bubble to be positioned behind the blastocyst and transfer embryos to the vial. The bubble behind the blastocysts should come from the pipette, indicating the blastocysts are deposited into the vial.
ESC cell line establishment
Timing: 10 - 20 d
-
45 |
Coat plates with 0.02% gelatin for 5 min (100 μl per well). It is not necessary to wash the plates after removal of gelatin solution.
-
46 |
Transfer denuded embryos to individual wells of 96-well plates containing 250 μl of NDiff N2B27 culture medium (see ‘Reagent setup’ section).
Troubleshooting advice can be found in Table 1.
-
47 |
Culture the embryos until they grow into visible cellular colonies (~1 mm in size; this takes up to 10 d in culture) (Figure 5e).
CRITICAL STEP
Pay particular attention to retaining the colony if medium change is required (if the medium starts to become yellow in colour), as the colony will not attach strongly to the bottom of the dish.
-
48 |
Transfer a unique colony to 15 μl of trypsin-EDTA and, after 2-3 min, pipette up and down, using 15 μl of DMEM culture medium to inactivate the trypsin-EDTA, and transfer to another well of a 96-well gelatin-coated plate containing 250 μl of NDiff N2B27 culture medium.
CAUTION
hESCs diploidize faster in DMEM culture medium than in NDiff N2B27 culture medium. Do not try to spin down the cells after trypsinization, as it is very easy to lose the lines when doing this.
CRITICAL STEP
Most blastocysts will have expanded to cellular colonies by this point, but some might need additional days of culture.
-
49 |
In 3 d, or upon confluency, trypsinise the cells as described in Step 48 and re-plate in a gelatin-coated 24-well plate; at this point the hESCs will form colonies (Figure 5e).
Troubleshooting advice can be found in Table 1.
-
50 |
In 3 d, or upon confluency, again trypsinise as described in Step 48 (increasing the trypsin-EDTA volume accordingly to cover the bottom of the well; e.g., 50 μl of trypsin-EDTA) and block trypsinization with an equal volume of DMEM culture medium. Transfer the cells to a gelatin-coated 6-well plate; at this point, flow cytometry cell cycle analysis can be performed to assess the haploid status of each clone (Box 1)
Troubleshooting advice can be found in Table 1.
-
51 |
In 3 d or upon confluency, trypsinise the cells yet again as described in Step 48 and transfer them to a gelatin-coated 10-cm dish or T25 flask. The first sorting for haploid enrichment can also be performed (Box 3). Plate sorted cells in a gelatin-coated T25 flask.
-
52 |
Upon regrowth, trypsinise the cells (as in Step 48) and split them among several gelatin-coated flasks or 10-cm dishes (1 million cells/flask). In 2 d, freeze down some cells in 1 ml of 1:9 DMSO/FBS freezing medium, and transfer them to a liquid nitrogen storing unit. From this point onwards, one of the clones can be adapted to DMEM with 2i and LIF. If desired, the cells can be adapted to DMEM with LIF alone.
Troubleshooting advice can be found in Table 1.
-
53 |
(Optional) Repeat Steps 51 and 52 to increase the number of sorted cells.
-
54 |
Depending on how many cells have been sorted, perform expansion of haploid populations and freeze down for banking of cell lines.
Troubleshooting
Troubleshooting advice can be found in Table 1.
Timing
It is recommended that the superovulation be scheduled for Tuesday and Thursday, with injections at 3 PM, as this allows mouse dissection, oocyte harvest and activation on a Friday, followed by embryo culture over the weekend and assessment for blastocysts during the weekdays of the following week.
Steps 1 to 8, first injection of female donors and incubation: 2d; 30-40 min hands-on
Step 9, second injection of female donors and incubation: 2d; 30-40 min hands-on
Steps 10-31, dissection of female donors; removal and activation of oocytes: ~3-4 h
Steps 32 and 33, removal of lysed embryos, transfer of 8-cell embryos to KSOM medium supplemented with 2i, and continued culture: x d; x min hands-on
Step 34 and 35, selection of embryos for zona removal: 2 d; 50-60 min hands-on
Step 36-44, zona pellucida removal: 30 min
Steps 45-47, embryo transfer and culture: 10 d
Step 48, passage to 96-well plates: ~1 d, ~ 15 min per cell line
Step 49, passage to 24-well plates: ~3 d; ~15 min per cell line hands-on
Step 50, passage to six-well plates ~3 d; ~15 min per cell line hands-on
Step 51 and 52, passage to T25 flask or 10 cm dish, cell cycle profile, genotyping and mycoplasma testing: ~5 d; ~15 min per cell line hands-on
Step 53, (optional) repetition of steps 51 and 52: ~5 d; ~15 min per cell line hands-on
Step 54, expansion of haploid populations and freezing for banking: ~3 d; ~15 min per cell line hands-on
Box 1, quantification of cell cycle profile of newly derived ESCs and flow cytometric analysis: 1.5 h for 10 samples
Box 2, chromosomal analysis of hESCs: 1 d + 3 h
Box 2. Chromosomal analysis of hESCs.
Timing 1 d plus 3 h
Additional reagents
KaryoMAX Colcemid Solution in HBSS (10 μg/ml stock; Thermo Fisher cat. no. 15210040).
CAUTION
This solution is acutely toxic if swallowed as well as a health hazard. Consult safety and hazard sheets provided by the seller.
KCl (Sigma-Aldrich; cat. no. P9333).
Acetic acid (>99.7%; Sigma-Aldrich; cat. no. 695092-500ML-D).
CAUTION
Acetic acid is acutely toxic if swallowed; it is flammable and corrosive. Consult safety and hazard sheets provided by the seller.
Methanol (>99.8%, methanol/acetic acid (3:1); Sigma-Aldrich; cat. no. 179337-2.5L-D).
CAUTION
Methanol is acutely toxic if swallowed; it is flammable. Consult safety and hazard sheets provided by the seller.
Antifade Mounting Medium with DAPI (Vectashield laboratories; cat. no. H-1200).
CAUTION
The mounting medium is a mutagen and a health hazard if swallowed. Consult safety and hazard sheets provided by the seller.
Equipment
Glass histology slides (75x25 mm) (ThermoFisher; cat. no. 4951PLUS4).
Glass coverslips (60x24 mm) (ThermoFisher; cat. no. Q10143263NR).
Water bath (JB Nova unstirred water bath, Grant, cat. no. SAP12)
Reagent setup
Hypotonic solution. Add 5.6 g of KCl to 100 ml of H2O to make a 0.75M stock; dilute the stock 10x to make a working solution. The working solution should be made fresh; the stock can be stored at RT for up to 1 year.
Methanol/acetic acid fixative. Mix methanol and acetic acid in a 3:1 ratio (i.e. mix 30 ml of methanol with 10 ml of acetic acid). Always make fresh.
CAUTION
Work under the chemical hood to prepare the fixative. The fixative is acutely toxic if swallowed and is flammable.
Sample preparation procedure
-
1 |
1 day before karyotyping, prepare a gelatinised plate by adding 5 ml of 0.02% gelatin to a 10-cm plate and leaving it for 5 min before plating. Then passage 1 million hESCs onto the gelatinised plate.
-
2 |
Add microscope slides to a jar containing 70% ethanol (this will degrease the slides and allow optimal adherence).
-
3 |
24 h later, add 200 μl of Colcemid Solution to the culture to achieve a final concentration of 0.2 μg/ml. Return the flask to the incubator and continue incubating for 45 min.
-
4 |
Prepare a working hypothonic solution (10X dilution of the stock), 4 ml for each sample, and place it in a 37 °C water bath.
CRITICAL STEP
The hypotonic solution needs to be 37 °C for optimal hypotonization.
-
5 |
Prepare the methanol/glacial acetic acid (3:1) fixative solution and place it on ice.
-
6 |
Wash the plate vigorously two to three times with the medium covering the cells and save it in an individually labeled 15-ml conical tube (one for each cell line).
CRITICAL STEP
Most metaphases will be loosely attached to the plate and will be obtained at this step.
-
7 |
Wash the plate gently with 1x PBS and trypsinise the hESCs, using 1.5 ml of trypsin-EDTA for 1 - 2 min in the incubator.
-
8 |
Wash the cells with the medium from the 15-ml conical tube (step 6) and place them in the same conical tube; this medium will also stop trypsin action.
-
9 |
Centrifuge the cells at 450g for 5 min at RT and discard the supernatant by inverting the conical tube.
-
10 |
Add 6 ml of working hypotonic solution and mix well by pipetting up and down.
-
11 |
Incubate the cells for 6 min at 37 °C.
CRITICAL STEP
Optimal hypotonisation is highly dependent on this incubation step. Under-hypotonisation will not break the cell wall. Over-hypotonisation will lead to chromosomes dissociating from the metaphase plate. If working with more than four samples, stagger them at this step so that only four samples are processed at a time.
-
12 |
Stop the hypotonization by adding 1 ml of cold methanol/acetic acid fixative to each sample and mix by gentle inversion two to three times.
-
13 |
Centrifuge the cells at 450g for 5 min and discard the supernatant by inverting the conical tube.
-
14 |
Resuspend the cell pellet in the residual liquid by flicking the tube.
CRITICAL STEP
It is important to resuspend the cells well at this step; otherwise, clumps will form and it will be hard to resolve single metaphase spreads.
-
15 |
Using a Pasteur pipette, carefully add 2 ml of methanol/acetic acid fixative solution dropwise, with gentle mixing to avoid clumping. Add an additional 6 ml of fixative and mix by gentle inversion of the tube.
CRITICAL STEP
It is important to gently add the fixative at the first fixation, as this will have a big impact on the metaphases (chromosomes can dissociate from the metaphase plate). The fixative must be made fresh.
-
16 |
Incubate the metaphase preparations on ice for 30 min.
-
17 |
Centrifuge the cells (450g, 5 min, 4 °C) and discard the supernatant.
Pause Point
At this step, the metaphase preparations can be stored at -20 °C for years.
-
18 |
Repeat steps 13-16 three times to clear the cytoplasm.
-
19 |
Before spotting on slides, resuspend the pellet in a small amount (0.5 ml) of methanol/acetic acid fixative (this volume may need to be adjusted slightly according to cell number).
-
20 |
Spot two to three drops in a row on each slide. Allow the slides to dry. Humidity will affect spreading of metaphases (higher humidity will increase spreading). If fluorescence in situ hybridization (FISH) analysis is desired, follow the protocol by Rens W. et al. 48.
-
21 |
Mount the slides using Antifade mounting medium with DAPI, cover with a coverslip and seal. A representative karyotype of an hESC clone is presented in Fig. 2b.
Box 3, sorting of hESCs: 1 h
Box 4, gene trap flipping: 3-4 d
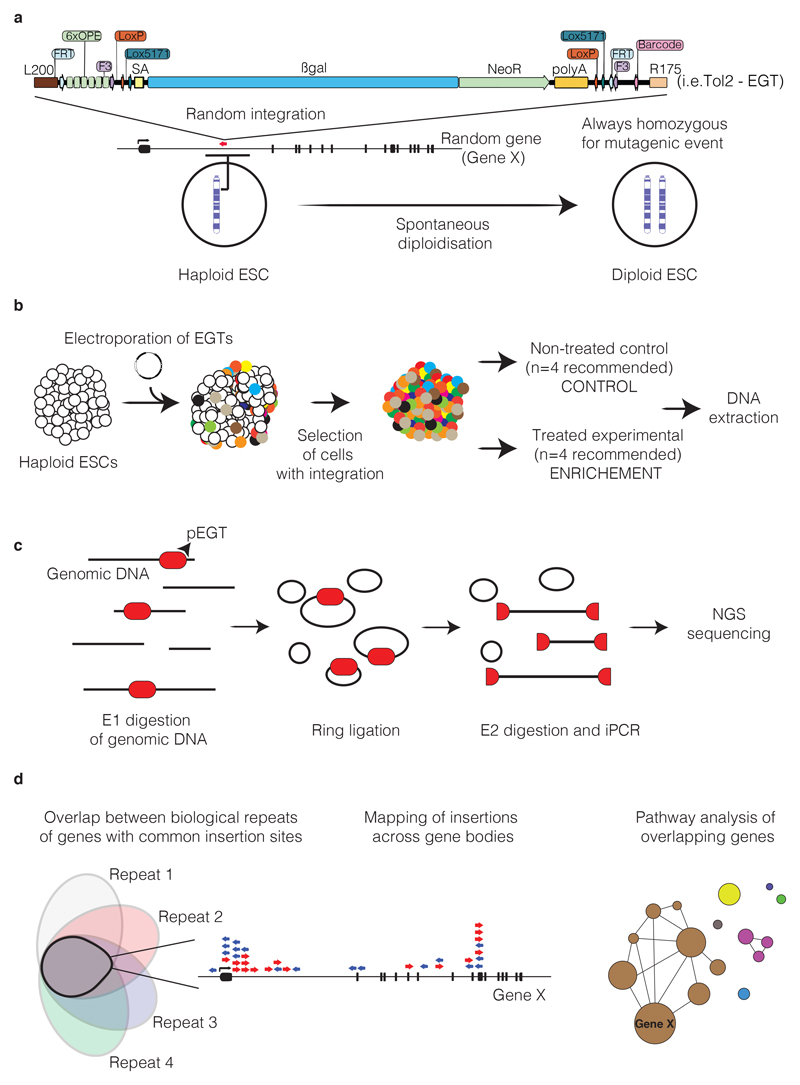
Box 4. The Haplobank and gene trap flipping.
The Haplobank (www.haplobank.at) contains >100,000 cell lines covering ~17,000 genes in the mouse genome and thus represents the largest homozygous ESC library available to the scientific community to date42. These cell lines are on/off systems that allow for conditional induction of gene disruption and subsequent reversal, permitting fast complementation to be performed for validation. Moreover, these mutants were engineered in single cell-derived clones and are thus isogenic, therefore allowing improved reproducibility across different comparisons, not only in one laboratory, but across the scientific community. Such a resource will permit the broad and robust interrogation of the functional genome.
Below we highlight information to help with the decision making when selecting clones from the Haplobank website and provide a protocol for gene trap flipping. The constructs used for disrupting are presented in Supplementary Fig. 1a and their sequences are available as Supplementary Data.
Decision points: Selection of clones from the Haplobank website
Sense: disruptive (knockout; WT conditional ready).
Antisense: non-disruptive (WT; knockout conditional ready).
Clones highlighted in green: homozygous integration sites are confirmed.
Clones highlighted in blue: only one gene trap insertion (should be first choice).
Clones highlighted in grey: more than one gene trap insertion (should be second choice).
1 intron, intron, 5'UTR (untranslated region), CDS (coding region), and noncoding exons: disruptive mutations. If possible, we recommend going for an insertion in the first intron (1intron). Keep in mind that a mutation in the coding sequence is phenotypically irreversible.
Upstream, InterDown and InterUp carry the gene trap in an intergenic region and cannot be regarded as knockouts.
The choice of the mutagen is then the last thing: Tol2GT/Retro/Lenti are enhanced gene traps, whereas Tol2 is a polyA-enhanced gene trap that carries EGFP.
The insertion site is predicted with high certainty to be disruptive for all RefSeq transcripts of this gene. The splice acceptor is integrated in a genomic locus common to all RefSeq transcripts (intronic and exonic) and before 50% of the longest open reading frame.
Integration sites were identified using three different approaches: two inverse PCRs using the two different restriction enzymes, E1 (Supplementary Fig. 1b, Tables 2 and 3) and a barcode PCR, all followed by NGS. If the mapping strategy was successful, it has a green light.
Gene trap flipping: Infection of murine ESCs
Timing: 3 - 4 d
Additional reagent
Hexadimethrine bromide (Polybrene; Sigma-Aldrich, cat. No. H9268)
CAUTION
Hexadimethrine bromide is harmful if swallowed.
Procedure
CRITICAL
Gene trap cassettes harbouring disruptive splice acceptor sites are integrated either in sense or antisense relative to corresponding gene transcripts. This will lead to disruptive or non-disruptive integrations, respectively. Inverted LoxP and FRT sites flank the gene trap cassette, enabling the orientation to be reversed twice, by transiently infecting cells with Cre or Flp recombinase-expressing plasmids (double-flex system). A thereby-generated isogenic sister clone can be used as a perfect internal control. For the production of retrovirus containing the Cre- or Flp-expressing vector, the Cre/Flp-expressing plasmid should be linked with a selection marker (e. g. mCherry, Cre-puro) to evaluate infection efficiency and/or select for infected cells.
CRITICAL
Before infection the ESCs should be propagated by standard procedures and should be passaged several times before infection and/or sorted for haploid content if desired. The maps of these constructs are presented in Supplementary Fig. 1 and respective sequences in Supplementary Data.
-
1 |
3 - 4h before infection, seed ~250,000 cells per 6-well plate or 50,000 cells per 24-well plate for infection in triplicates.
-
2 |
Distribute the viral supernatant on the ESCs. Infection efficiency can be increased by adding 2 μg/ml polybrene to the culture medium. Alternatively, if you use a frozen virus stock, thaw an aliquot (on ice) and recalculate infection efficiency using the chosen selection marker (e.g., mCherry, Cre-puro).
-
3 |
8-12 h post infection, exchange the medium. Check infection efficiency by flow cytometry 48 h post infection or change to selection media and select until selection control wells are empty (e.g. with puromycin it takes 2-3 d).
-
4 |
Confirm the successful inversion of the gene trap cassette by PCR or western blot for the target gene.
Box 5, transposon mutagenesis in hESCs: 10-15 d
Box 5. Transposon mutagenesis in hESCs.
Timing: 10-15 d
Various vectors can be used to stably integrate gene trap cassettes into genomic DNA of haploid cells. Among them, virus is most commonly used. However, integration sites for viral integrations cluster strongly and thus result in poor genome saturation42. Moreover, integration sites relative to gene bodies vary for different vector systems. Transposon mutagenesis is an attractive alternative method for integrating gene trap vectors into genomes, as it not only achieves better genome saturation, it also circumvents virus work and associated risks.
Single transposon integrations are generated only by strongly compromising on integration efficiency. When electroporating for complex library preparation (e.g. to generate 50 million independently mutated cells), the experimenter needs to electroporate at least 50 independent reactions. Using Tol2 transposons achieves good single-integration events at 0.5 μg of transposon plasmid and 10 μg transposase plasmid; however, this may vary depending on transposon system and transposase variant, electroporator, cell line, and medium conditions.
The challenge for transposon mutagenesis is to achieve single integration events for each cell. Electroporation of DNA is best suited to single integrations; nevertheless, careful titration of transposon containing plasmids must be undertaken. The following protocol describes one possible approach, namely use of an AMAXA 2D electroporator (Equipment) and a Mouse ES cell Nucleofection Kit (Reagents). The workflow is outlined diagrammatically in Fig. 6.
Additional reagents
Mouse Embryonic Stem Cell Nucleofector™ Kit (Lonza, cat. no. VPH-1001) or human version (cat. no. VAPH-5022).
DNeasy Blood & Tissue Kits (50 DNeasy Mini Spin Columns, cat. no. 69504; 4 DNeasy 96 Plates, cat. no. 69581).
CAUTION
Buffer AL and AL/E (provided in the kit) are irritants if swallowed. Buffer AW1 (provided in the kit) is acutely toxic if swallowed and an irritant. Proteinase K (provided in the kit) is acutely toxic if swallowed and a health hazard.
Isopropanol (AppliChem, cat. no. A3928.0500PE).
CAUTION
Isopropanol is flammable and is an irritant if swallowed. Consult safety and hazard sheets provided by the seller.
Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, cat. no. P7589)
T4 DNA Ligase (Roche, cat. no. 10716359001)
Phusion High-Fidelity DNA Polymerase (New England Biolabs, cat. no. 174M0530S)
QIAquick PCR Purification Kit (Qiagen, cat. no. 28104).
CAUTION
Kit reagents are flammable and irritants if swallowed. Consult safety and hazard sheets provided by the seller.
QIAquick Gel Extraction Kit (Qiagen cat. no. 28706).
CAUTION
Kit reagents are flammable and irritants if swallowed. Consult safety and hazard sheets provided by the seller.
NlaIII, MseI, PacI and Sbf1 (restriction enzymes; New England Biolabs, cat. nos. R0125S, R0525S, R0547S and R0642S, respectively).
CutSmart Buffer (New England Biolabs, cat no. B7204S).
TaqI (methyltransferase enzyme; New England Biolabs, cat. no. M0219S).
NextSeq 550 v2.5 Kits (Illumina, cat. nos. 20024906 (75 cycles), 20024907 (150 cycles), 20024908 (300 cycles))
Tris-HCl (10 mM, pH 8.0, Teknova, cat. no. T1173)
EDTA disodium salt, (0.005 M, Astral Scientific, cat. no. E-155-500ML)
NaCl (100mM, Jena Bioscience, cat. no. BU-114-100)
SDS (10%, Fisher Bioreagents, cat. no. BP2436-1)
CAUTION
SDS is an irritant if swallowed. It causes serious eye damage. Consult safety and hazard sheets provided by the seller.
-
Proteinase K from Tritirachium album (Sigma, cat. no. P2308-25MG).
CAUTION
Proteinase K is an irritant if swallowed. Consult safety and hazard sheets provided by the seller.
Agencourt AMPure XP beads (Beckman Coulter; cat. no. #A63882).
Agarose gel (Sigma-Aldrich, cat. no. A9539-100G)
RNase A (Qiagen, cat. no. 19101)
SpeedBeads (GE Healthcare, cat. no. 1915210401030)
dNTP mix (Sigma-Aldrich, cat. no. D7295)
Equipment
Electroporator (Nucleofector™ 2b Device, Lonza, cat. no. AAB-1001).
Covaris Sonicator; (1 × 96 microtube plate, adaptive focused acoustics fiber; Covaris, cat. no. 520069)
Next generation sequencer (Illumina, model no. NextSeq 550); for further details see http://www.illumina.com. NGS requires extensive expertise. We recommend the use of an NGS facility.
Reagent setup
Genomic DNA lysis buffer. Genomic DNA lysis buffer is 10 mM Tris-HCl, pH 8.0; 5 mM EDTA;100 mM NaCl; 1.0% SDS. The buffer can be stored at RT for up to 6 months. 1 mg/ml proteinase K should be added fresh.
Procedure for electroporation of gene traps and screening
-
1 |
Expand haploid ESCs using standard conditions and sort for haploid cells (Box 3) a few days before electroporation.
-
2 |
Harvest the cells by trypsinization, stop trypsinization by adding 10 ml of culture medium and centrifuge the cells using a 15- or 50-ml centrifuge tube, depending on cell volume, for 5 min at 20g at RT.
-
3 |
In the meantime, prepare electroporation cuvettes and set the electroporator to program A-013. (Alternative programs suggested by the supplier can also be tested.)
-
4 |
Prepare and label cell culture dishes, then fill them with ES cell medium.
-
5 |
Aspirate the supernatant, take up the cell pellet in 1x PBS, and count the cells.
-
6 |
Add 7 million cells to independent 15-ml centrifuge tubes and fill up with 1 x PBS to 15 ml. Use one tube per planned electroporation plus one tube for control.
-
7 |
Centrifuge cells for 5 min at 20g at RT. In the meantime, prepare DNA solutions (step 8).
-
8 |
In separate 1.5-ml tubes, mix 90 μl of electroporation mix (also see manufacturer’s notes), 10 μg of transposase containing plasmid, and various amounts of transposon containing plasmid. We recommend testing 0, 0.25, 0.5, 1, 2.5, and 5 μg. Ensure that the total added volume does not exceed 10 μl. If the volume is between 10 and 20 μl, add 10x PBS at 1/9 of the volume of DNA. The total volume should not exceed 20 μl.
-
9 |
Aspirate 1 x PBS from the ES cell-containing tubes. Briefly centrifuge the tubes at 100 g for 10-20 s at RT and fully aspirate the 1x PBS.
-
10 |
Take up DNA solution and use it to carefully resuspend the cell pellet and immediately place the cells in an electroporation cuvette.
-
11 |
Electroporate the cells.
-
12 |
Carefully aspirate the cells from the cuvette, using the provided plastic Pasteur pipette, and pipette directly into the cell culture medium of a prepared dish.
-
13 |
24 h post electroporation, begin antibiotic selection for integration events. Upon completion of selection, the cells can be used for cryopreservation and usage at a later time point.
Library preparation and sequencing
For library preparation enzyme 1 (E1) is used to fragment the genome (Table 2). Because the recognition sequence for E1 is also present in the terminal repeat of the gene trap vector, it is possible to retrieve the exact integration site of the gene trap cassette within the genome by circularizing E1-digested genomic DNA (ring ligation) and subsequently amplifying the genomic region by inverse PCR (iPCR) using primers “US” and “DS” (see Tables 2 and 3 and Supplementary Fig. 1b). To improve iPCR efficiency, a linearization step using E2 should be used, which re-opens the rings generated previously. Moreover, each integration site can be mapped by using two different E1 enzymes. We recommend splitting the samples and use both E1 enzymes on each sample in parallel. The directionality of the mapping strategies for different gene traps is important, as this affects the assignment of a particular insertion to the sense and antisense strand in the genome, respectively. The DS primer is common for all PCR reactions, and binds in the gene trap. The US primer contains a sequence that will bind to the oligo present on the NGS flow cell surface, an index of 4-8 bases, and then a sequence that binds in the gene trap.
The mutagenesis systems and enzymes used are shown in Table 2, and iPCR primer sequences used for library amplification and sequencing are shown in Table 3.
The INDEX is a custom barcode (4-8 base pairs) that allows the unique identification of all the PCR reactions from one complex sample. For each sample, two different US primers with two different indices need to be used: one for the PCR reaction of the genomic DNA digested by E1-1 and one for the PCR reaction of the genomic DNA digested by E1-2. For NGS, all samples can be mixed and loaded into one NGS flow cell. The index sequence will allow the identification of the original samples. The workflow for library preparation is shown below.
Library preparation and sequencing procedure
-
1 |
Lyse cell pellet according to pellet size in 2-10 ml genomic DNA lysis buffer (see REAGENT SET UP).
-
2 |
Incubate at 55°C overnight.
-
3 |
Add 1:1,000 RNAse A for 1h at 37°C.
-
4 |
Precipitate the genomic DNA (gDNA) using isopropanol, and spool the DNA, using heat-sealed Pasteur pipettes.
-
5 |
Wash with 70% ethanol twice and resuspend in 0.1-0.2 ml Tris/EDTA (TE Buffer).
-
6 |Digest the samples with E1enzymes (Table 2) in parallel (two separate reactions) using the following master mix:
Component Volume Final concentration gDNA 100ng/μl 40 μl 50ng/μl 10x CutSmart 8 μl 1x Enzyme1 3 μl 150U/μl dH2O 29 μl -
7 |
Incubate the digestion mixtures at 37°C (65°C for TaqI) overnight.
-
8 |
Purify the restriction digests (Qiagen PCR kit or Agencourt AMPure XP beads).
-
9 |
Elute in 100 μl in total.
-
10 |Perform ring ligation (RL) using the following master mix:
Component Volume Final concentration E1-gDNA 100 μl 8.3% 10x Ligation Buffer 120 μl 1x T4 DNA Ligase 4 μl 1.6 U/μl dH2O 976 μl -
11 |
Incubate over night at 16°C.
-
12 |
Heat-inactivate T4 DNA Ligase at 65 °C for 15 min.
-
13 |Linearize the samples with enzyme E2 at 37 °C for 2 h using the following master mix:
Component Volume Final concentration RL-E1-gDNA 1200 μl Enzyme 2 2 μl ~0.016 U/μl -
14 |
Purify restriction digest (Qiagen PCR kit or magnetic Speedbeads) and elute in 100 μl H2O in total.
-
15 |Perform the iPCR reaction. Use 5-10 reactions per sample and process half or all of the digest according to the expected DNA amount. See the table below for setting up the master mix.
Component Volume Final concentration E2-RL-E1-gDNA from step 7 10 μl 20% primer US 100 μM 0.1 μl 0.2 μM primer DS 100 μM 0.1 μl 0.2 μM 10 mM dNTP mix 1 μl 0.2 mM 10x pol. buffer 5 μl 1x 20x polymerase 3 μl 1.2x dH2O 30.8 μl Use the PCR cycler parameters given below:Cycles Denature Anneal Extend 1 95 ºC, 3 min 2-38 95 ºC, 15 sec 61 ºC, 25 sec 72 ºC, 75 s 39 72 ºC, 5 min CRITICAL
After amplification, there will be large molar amounts of amplicons, therefore, there is a very high danger of contaminating other experiments with post-PCR samples. We recommend using different pipette sets, rooms and equipment for post-PCR samples.
-
16 |
Pool all the PCR products from one sample (5-10 reactions) and analyse 20 μl on an agarose gel. A smear band beginning at around 400 base pairs is expected.
-
17 |
Quantify the DNA amount and mix all PCR samples (both separated digests for all 5 samples) in such a way that you use the same DNA amount from each.
-
18 |
Load that mixture on another agarose gel and cut the part of the smear between 400 and 800 bp.
-
19 |
Extract the DNA from the gel using Qiagen’s Gel Extraction Kit.
-
20 |
Perform NGS.
Statistical analysis
To increase reliability of the analysis for each biological replicate two sequencing libraries should be constructed: one for sequencing into the adjacent genome 5’ of the insertion site, and one into the 3’adjacent genome. Pearson correlation should be used to compare the read counts obtained by sequencing the two libraries and should allow reliable identification for each insertion. The statistical analysis can be done using the R framework49 to map the reads for each insertion site, and Gaussian kernel convolution50 can be used to identify common insertion sites. By using co-occurrence analysis in R and using a Fisher's exact test the overlap of common insertion sites across the biological repeats can be performed to identify strong gene candidates. For each gene the insertion sites can be mapped to identify position and direction of insertions across the gene body (e.g. gene X). These candidates can be subsequently used for DAVID analysis51 to generate a network of genes nodes, with the node sizes being representative of the total number of common insertion sites (Fig 6d).
Anticipated results
Typically, cells will undergo cell division before expression of transposase. Therefore, individual colonies appearing in selection will very often be mixed colonies, with individual cells harboring independent integration events. Thus, to test for multiplicity of infection (MOI), pick colonies, trypsinize cells (of each condition) subsequent to completion of selection (days 5-7) and reseed them at clonal density (100 cells/10 cm dish, 500 cells/15 cm dish). Pick 20-50 colonies per condition at days 10-12 after reseeding and expand them in 96-well dishes. Genomic DNA can be prepared in bulk or in 96 wells, depending on screen setup using either genomic DNA lysis buffer or Qiagen DNeasy Blood & Tissue Kits. Optionally, acoustic shearing of DNA can be carried out using the Covaris system. It is most convenient to perform a PCR across a random barcode integrated within the transposon and then subject it to Sanger sequencing. Clean sequencing tracks are indicative of single integrations, whereas overlaying sequence tracks indicate multiple integration events. Please note that in the presence of transposase, transposons may be remobilized and integrated in a new genomic site. Even though these DNA transposons work via a cut-paste mechanism, mobilization in G2 phase of cell cycle can in some cases result in two independent transposon integration sites within a cell that carry the same random genetic barcode. Alternatively, perform iPCR to map integration sites. Please note, that efficiency of iPCR is only ~80%, so some double integrations may be missed.
For generation of complex pools of cells, proceed with screening for phenotypes of interest. Note that loss of protein may take up to 7- 10 d for stable proteins in cases of late integration. The number and location of integrated transposons per cell can be identified by library sequencing and analysis. Library preparation can be performed in various ways (see review by Elling and Penninger39). We recommend the iPCR method described here, which we have successfully used42. Alternatively, the transposon insertion site sequencing can be done also by QiSeq method as described by Friedrich and collaborators52.
Before executing NGS, please consider the expected complexity. You will receive 100 - 200 million reads from one single lane. As this is usually multiple times more than the complexity of your input, you might be oversampling the experiment. This can – by misalignment – cause artefacts. For that reason, it can be advantageous to combine several experiments in 1 lane by indexing. Subdividing single experiments is not advisable, as the data are not simply additive. To optimally use NGS, consider the scale of input and experiment. If you wish to quantitatively use the NGS data, the 200 million reads must stem from 200 million independent genomes. If the complexity of input is significantly lower, the experiment can be scaled accordingly.
Anticipated results
The protocol presented here will allow derivation and maintenance of mouse hESCs. Although the protocol for derivation of hESCs is lengthy (about 7 weeks), it allows reliable and reproducible derivation of hESCs. In our experience, mouse hESCs can be successfully established in gelatin-coated plates, with only minor improvements when deriving them on plates containing irradiated feeders. The bottleneck for successful cell line production is the quality of the embryos selected. This can vary depending on mouse strain, mouse age, handling of oocytes, choice of which oocytes to select, successful activation, subsequent decision of which blastocysts to select and successful zona pellucida removal. In our experience, from 1,534 oocytes, we had overall success rates of 1 blastocyst for each 10.5 oocytes (1 blastocyst for every 3.95 oocytes, if we do not include oocytes that do not progress within 24 h and that were unlikely to be viable in the first place). Our activation success rate is 1 per 2.65 oocytes and our average success rates for cell line production is 1 cell line for every 1.4 blastocysts used. Although there might be a minor advantage to using feeders, for users that do not have much experience with handling ESCs on feeders, culture without feeders is the most straightforward to undertake, as it allows easier identification of the hESC colonies. We recommend that before freezing down the cellular clones, a mycoplasma test be performed. We have not had any mycoplasma contamination so far.
The associated procedures allow the screening of clones that contain haploid populations (Box 1), karyotyping of the isolated hESC clones (Box 2) and sorting for enrichment of hESCs (Box 3). We also describe considerations for use of hESCs imported from the Haplobank (Box 4; Tables 2 and 3). Haploid cells can be subsequently successfully used for genetic screens (see Box 5 and Fig. 6 for one such example). Similar principles can be used for the maintenance and propagation of human haploid ESCs.
Table 2. Mutagenesis systems and enzymes.
| Name | Enzymes 1 (E1-1; E1-2) | Enzyme 2 (E2) | Mapping strategy |
|---|---|---|---|
| Lenti-EGT | NlaIII; TaqI | PacI | 5’ |
| Retro-EGT | NlaIII; MseI | SbfI | 5’ |
| Tol2-EGT | NlaIII; TaqI | PacI | 3’ |
| Tol2-polyA-EGT | NlaIII; TaqI | PacI | 3’ |
EGT, enhanced gene trap
Table 3. iPCR primer sequences.
| Name | Sequence |
|---|---|
| Primer DS | AATGATACGGCGACCACCGAGATCTACACGAGCCAGAACCAGAAGGAACTTGA |
| Primer US | CAAGCAGAAGACGGCATACGAGATINDEXGTGACTGGAGTTCAGACGTGTGCTCTTC |
| Lenti-EGT | CAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCA |
| Retro-EGT (1:1 mix) | GAGTGATTGACTACCCGTCAGCGGGGGTCTTTCA and TGAGTGATTGACTACCCACGACGGGGGTCTTTCA |
| Tol2-EGT Tol2-polyA-EGT | CACTTGAGTAAAATTTTTGAGTACTTTTTACACCTCTG |
Figure 6. Representative diagram of a transposon screen in hESCs.
(a) Diagram showing a map of one gene trap construct. The red arrow depicts a random integration event into a random gene (Gene X) and points to the direction of integration. As the integration is achieved in haploid ESC, upon diploidization the insertion will also be duplicated thus resulting always in homozygous representation. In screening strategies using transposons but also CRISPR-Cas9 this represents an important advantage. (b) Screening using transposon approaches rely on the electroporation of the transposon into the haploid ESC and selection of cells that have successfully integrated the constructs. After around 10 days in culture to allow the respective proteins to be depleted, these cells can be subsequently used for different screening strategies where a comparison between control untreated samples and treated sample is made. We recommend an n=4 technical replicates over 2 biological replicates for each group. (c) Library preparation for inverse PCR (iPCR) requires digestion of the genomic DNA using enzymes E1. Upon digestion the DNA fragments containing part of the enhanced gene trap containing DNA fragments (pEGT; red) are ring ligated followed by digestion of the ring DNA constructs containing EGT’s followed by next generation sequencing (NGS). See also Supplementary Figure 1b. (d) Schematic representation of a possible screening outcome. The overlap between the common insertion sites across the 4 biological replicates can be represented as a Venn diagram. The enriched transposons (presumably one transposon on each cell) can be mapped to the respective genome locations for each gene. Based on this enrichment and based upon statistical analysis across the technical repeats by comparing the treatment groups to untreated groups, a gene ranking statistical score can be generated. The top candidate genes can subsequently be used for DAVID pathway analysis for example, where each gene circle size can be representative of the number of insertions. Moreover, mapping the insertions over the gene bodies can inform on specific clustering around the promoter or maybe around a relevant domain region. For the statistical analysis see also the protocol by Friedrick et al. 201752.
Supplementary Material
Editorial Summary.
This protocol describes how to derive and maintain mouse haploid embryonic stem cells from female gametes. Additional procedures that can be carried out with cell lines obtained from the mouse Haplobank are also described.
Figure 2. Qualitative control of the haploid ESCs.
(a) Representative examples of the cell cycle profiles of three individual ESC cell lines at the first cell cycle analysis post derivation. While some clones might be more stable and thus highly enriched for haploid ESCs (left side profile), most will be a mix of haploid/diploid cells (middle profile) while some will be more unstable and be fully diploid (right side profile). (b) Fluorescent in situ hybridisation analysis of a representative haploid clone shows a stable n=20 karyotype (100X magnification).
Acknowledgments
MW, BD, BF, BLN and DJA are supported by the Wellcome Trust through core funding to the Wellcome Trust Sanger Institute (WT098051). Research in the SPJ laboratory is funded by Cancer Research UK (Programme grant C6/A18796), Wellcome Trust Investigator Award (206388/Z/17/Z). Core funding is provided by CRUK (C6946/A14492) and the Wellcome Trust (WT092096). Research in JMP laboratory is funded by Advanced ERC and Era of Hope/DoD grants. Research in the GB laboratory is funded by a UK Dementia Research Institute fellowship (MC_PC_17111).
Footnotes
Competing financial interests
JMP is a founder of JLP Health.
Data availability
All data presented in the manuscript are available from the corresponding authors upon reasonable request.
Author contributions
EU, MW and GB performed experimental analysis and procedures through-out and wrote the manuscript. BD and DJA helped MW with set-up of blastocyst work. BLN assisted with flow-cytometry with help from JVF and SPJ. BF and FY performed the FISH and karyotyped the cell lines. JRV wrote the transposon induced mutagenesis protocol with help from EU. GB and JMP conceived the idea of this manuscript. All authors commented on the manuscript.
References
- 1.Maderspacher F. Theodor Boveri and the natural experiment. Current Biology. 2008;18:R279–R286. doi: 10.1016/j.cub.2008.02.061. [DOI] [PubMed] [Google Scholar]
- 2.Crosland MWJ, Crozier RH. Myrmecia pilosula, an Ant with Only One Pair of Chromosomes. Science. 1986;231 doi: 10.1126/science.231.4743.1278. 1278–1278. [DOI] [PubMed] [Google Scholar]
- 3.Swart EC, et al. The Oxytricha trifallax Macronuclear Genome: A Complex Eukaryotic Genome with 16,000 Tiny Chromosomes. PLOS Biology. 2013;11:e1001473. doi: 10.1371/journal.pbio.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallardo MH, Bickham JW, Honeycutt RL, Ojeda RA, Köhler N. Discovery of tetraploidy in a mammal. Nature. 1999;401 doi: 10.1038/43815. 341–341. [DOI] [PubMed] [Google Scholar]
- 5.Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 6.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nature Cell Biology. 2017;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Davoli T, de Lange T. The Causes and Consequences of Polyploidy in Normal Development and Cancer. Annual Review of Cell and Developmental Biology. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 10.Elling U, et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sagi I, et al. Derivation and differentiation of haploid human embryonic stem cells. Nature. 2016;532:1–19. doi: 10.1038/nature17408. [DOI] [PubMed] [Google Scholar]
- 13.Zhong C, et al. Generation of human haploid embryonic stem cells from parthenogenetic embryos obtained by microsurgical removal of male pronucleus. Cell Research. 2016;26:743–746. doi: 10.1038/cr.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sagi I, Benvenisty N. Haploidy in Humans: An Evolutionary and Developmental Perspective. Dev Cell. 2017;41:581–589. doi: 10.1016/j.devcel.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 15.Botstein D, Fink GR. Yeast: An Experimental Organism for 21st Century Biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordes SP. N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express. Microbiol Mol Biol Rev. 2005;69:426–439. doi: 10.1128/MMBR.69.3.426-439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotecki M. Isolation and Characterization of a Near-Haploid Human Cell Line. Experimental Cell Research. 1999;252:273–280. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- 18.Carette JE, et al. Haploid Genetic Screens in Human Cells Identify Host Factors Used by Pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 19.Carette JE, et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Essletzbichler P, et al. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Research. 2014;24:2059–2065. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pincus G. Observations on the living eggs of the rabbit. Proc R Soc Lond B. 1930;107:132–167. [Google Scholar]
- 22.Tarkowski AK, Witkowska A, Nowicka J. Experimental Parthenogenesis in the Mouse. Nature. 1970;226:162–165. doi: 10.1038/226162a0. [DOI] [PubMed] [Google Scholar]
- 23.Graham CF. Parthenogenetic Mouse Blastocysts. Nature. 1970;226:165–167. doi: 10.1038/226165a0. [DOI] [PubMed] [Google Scholar]
- 24.Kaufman MH. Early Mammalian Development: Parthenogenetic studies. 1983;14 [Google Scholar]
- 25.Fraser LR. Strontium supports capacitation and the acrosome reaction in mouse sperm and rapidly activates mouse eggs. Gamete Research. 1987;18:363–374. doi: 10.1002/mrd.1120180410. [DOI] [PubMed] [Google Scholar]
- 26.Kline D, Kline JT. Repetitive calcium transients and the role of calcium in exocytosis and cell cycle activation in the mouse egg. Developmental Biology. 1992;149:80–89. doi: 10.1016/0012-1606(92)90265-i. [DOI] [PubMed] [Google Scholar]
- 27.Mikich AB, et al. Calcium oscillations and protein synthesis inhibition synergistically activate mouse oocytes. Mol Reprod Dev. 1995;41:84–90. doi: 10.1002/mrd.1080410113. [DOI] [PubMed] [Google Scholar]
- 28.Cuthbertson KSR, Whittingham DG, Cobbold PH. Free Ca2+ increases in exponential phases during mouse oocyte activation. Nature. 1981;294:754–757. doi: 10.1038/294754a0. [DOI] [PubMed] [Google Scholar]
- 29.Stevens LC, Varnum DS. The development of teratomas from parthenogenetically activated ovarian mouse eggs. Developmental Biology. 1974;37:369–380. doi: 10.1016/0012-1606(74)90155-9. [DOI] [PubMed] [Google Scholar]
- 30.Linder D, McCaw BK, Hecht F. Parthenogenic Origin of Benign Ovarian Teratomas. N Engl J Med. 1975;292:63–66. doi: 10.1056/NEJM197501092920202. [DOI] [PubMed] [Google Scholar]
- 31.Leeb M, Perry ACF, Wutz A. Establishment and Use of Mouse Haploid ES Cells. Curr Protoc Mouse Biol. 2017;5:155–185. doi: 10.1002/9780470942390.mo140214. [DOI] [PubMed] [Google Scholar]
- 32.Shuai L, et al. Generation of Mammalian offspring by haploid embryonic stem cells microinjection. Curr Protoc Stem Cell Biol. 2014;31:1A.6.1–15. doi: 10.1002/9780470151808.sc01a06s31. [DOI] [PubMed] [Google Scholar]
- 33.Balmus G, et al. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat Commun. 2019;10 doi: 10.1038/s41467-018-07729-2. 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H, et al. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Research. 2013;23:1187–1200. doi: 10.1038/cr.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, et al. Generation and Application of Mouse-Rat Allodiploid Embryonic Stem Cells. Cell. 2016;164:279–292. doi: 10.1016/j.cell.2015.11.035. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, et al. Generation of Genetically Modified Mice by Oocyte Injection of Androgenetic Haploid Embryonic Stem Cells. Cell. 2012;149:605–617. doi: 10.1016/j.cell.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Li W, et al. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature. 2012;490:407–411. doi: 10.1038/nature11435. [DOI] [PubMed] [Google Scholar]
- 38.Leeb M, Wutz A. Germline potential of parthenogenetic haploid mouse embryonic stem cells. Development. 2012;139:3301–3305. doi: 10.1242/dev.083675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elling U, Penninger JM. Genome wide functional genetics in haploid cells. FEBS Lett. 2014;588:2415–2421. doi: 10.1016/j.febslet.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 40.Forment JV, et al. Genome-wide genetic screening with chemically mutagenized haploid embryonic stem cells. Nat Chem Biol. 2017;13:12–14. doi: 10.1038/nchembio.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzog M, et al. Detection of functional protein domains by unbiased genome-wide forward genetic screening. Sci Rep. 2018;8 doi: 10.1038/s41598-018-24400-4. 6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elling U, et al. A reversible haploid mouse embryonic stem cell biobank resource for functional genomics. Nature. 2017;550:114. doi: 10.1038/nature24027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Q, et al. Derivation of Haploid Neural Stem Cell Lines by Selection for a Pax6-GFPReporter. Stem Cells and Development. 2018;27:479–487. doi: 10.1089/scd.2017.0193. [DOI] [PubMed] [Google Scholar]
- 44.Kim YM, Lee J-Y, Xia L, Mulvihill JJ, Li S. Trisomy 8: a common finding in mouse embryonic stem (ES) cell lines. Mol Cytogenet. 2013;6:3. doi: 10.1186/1755-8166-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morey R, Laurent LC. Getting off the ground state: X chromosome inactivation knocks down barriers to differentiation. Cell Stem Cell. 2014;14:131–132. doi: 10.1016/j.stem.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS biology. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brownstein DG. Manipulating the Mouse Embryo: A Laboratory Manual. In: Nagy Andras, Gertsenstein Marina, Vintersten Kristina, Behringer Richard., editors. The Quarterly Review of Biology. Third Edition. Vol. 78. Cold Spring Harbor Laboratory Press; 2003. 365–365. [Google Scholar]
- 48.Rens W, Fu B, O'Brien PCM, Ferguson-Smith M. Cross-species chromosome painting. Nat Protoc. 2006;1:783–790. doi: 10.1038/nprot.2006.91. [DOI] [PubMed] [Google Scholar]
- 49.Team RC. R: A language and environment for statistical computing. 2013. [Google Scholar]
- 50.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting Statistically Significant Common Insertion Sites in Retroviral Insertional Mutagenesis Screens. PLoS Comput Biol. 2006;2:e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Da Wei, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 52.Friedrich MJ, et al. Genome-wide transposon screening and quantitative insertion site sequencing for cancer gene discovery in mice. Nat Protoc. 2017;12:289–309. doi: 10.1038/nprot.2016.164. [DOI] [PubMed] [Google Scholar]
- 53.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.